Abstract
Subcellular localization of PKA (cAMP-dependent protein kinase or protein kinase A) is determined by protein–protein interactions between its R (regulatory) subunits and AKAPs (A-kinase-anchoring proteins). In the present paper, we report the development of the Amplified Luminescent Proximity Homogeneous Assay (AlphaScreen™) as a means to characterize AKAP-based peptide competitors of PKA anchoring. In this assay, the prototypic anchoring disruptor Ht31 efficiently competed in RIIα isoform binding with RII-specific and dual-specificity AKAPs (IC50 values of 1.4±0.2 nM and 6±1 nM respectively). In contrast, RIα isoform binding to a dual-specific AKAP was less efficiently competed (IC50 of 156±10 nM). Characterization of two RI-selective anchoring disruptors, RIAD (RI-anchoring disruptor) and PV-38 revealed that RIAD (IC50 of 13±1 nM) was 20-fold more potent than PV-38 (IC50 of 304±17 nM) and did not compete in the RIIα–AKAP interaction. We also observed that the kinetics of RII displacement from pre-formed PKA–AKAP complexes and competition of RII–AKAP complex formation by Ht31 differed by an order of magnitude when the component parts were mixed in vitro. No such difference in potency was seen for RIα–AKAP complexes. Thus the AlphaScreen assay may prove to be a valuable tool for detailed characterization of a variety of PKA–AKAP complexes.
Keywords: A-kinase-anchoring protein (AKAP), Amplified Luminescent Proximity Homogeneous Assay (AlphaScreen™), anchoring disruptor, cAMP-dependent protein kinase (PKA)
Abbreviations: AKAP, A-kinase-anchoring protein; C, catalytic subunit; D/D domain, dimerization/docking domain; GST, glutathione S-transferase; PKA, cAMP-dependent protein kinase or protein kinase A; R, regulatory subunit; hR, recombinant human R subunit; RIAD, RI-anchoring disruptor; S/B ratio, signal to background ratio; SPR, surface plasmon resonance
INTRODUCTION
PKA (cAMP-dependent protein kinase or protein kinase A) phosphorylates a variety of substrate proteins and is involved in the regulation of many different intracellular events. The PKA holoenzyme consists of an R (regulatory) subunit dimer which associates with two C (catalytic) subunits to form an inactive holoenzyme (R2C2) (for reviews, see [1–3]). Four different isoforms (RIα, RIβ, RIIα and RIIβ) of the R subunit have been identified, of which RIα and RIIα are the most ubiquitously expressed in cells and tissues. cAMP binds to the R subunit in a positively co-operative fashion, triggering dissociation of the C subunit. The active enzyme is then free to phosphorylate target substrates within its vicinity. Compartmentalization of the PKA holoenzyme favours the localized action of this broad-specificity kinase by placing the enzyme in close proximity to a subset of its target substrates. This is achieved through protein–protein interactions of the R subunit with AKAPs (A-kinase-anchoring proteins), a diverse and growing family of scaffolding proteins that target PKA to distinct subcellular compartments and towards specific substrates (for reviews, see [2,4]).
Dimerization of the R subunit is mediated through the formation of an antiparallel X-type four-helix bundle in the N-terminal region [5,6]. Once formed, this domain also serves as the docking site for different AKAPs and is therefore referred to as the D/D (dimerization/docking) domain. The D/D domain has a hydrophobic character and interacts with an amphipathic helix constituting the A-kinase-binding site found in all typical AKAPs. Different AKAPs were initially isolated as RII-binding proteins, whereas RI was not thought to interact with AKAPs. Subsequently, dual-specific anchoring proteins that can bind both RI and RII with nanomolar affinity have been identified [7–9], in addition to a few RI-selective AKAPs [10–12]. This suggests that both PKA isoforms can be targeted via interactions with AKAPs. However, there appears to be structural differences between the anchoring surfaces of RIα and RIIα [13]. These structural differences are reflected in the different kinetics and specificities of their AKAP binding [14].
The inhibitor Ht31 [15], constituting the amphipathic helix of AKAP-Lbc [16], is considered to be the prototypic anchoring disruptor peptide. However, this reagent delocalizes both PKA holoenzyme subtypes by displacing RI and RII from AKAPs [14,17]. Consequently, there has been interest in designing high-affinity isoform-specific anchoring disruptor peptides that exclusively target the type I or type II PKA holoenzymes [18–21]. These studies have emphasized a need for a methodology that characterizes the kinetics of such peptides on a high-throughput platform.
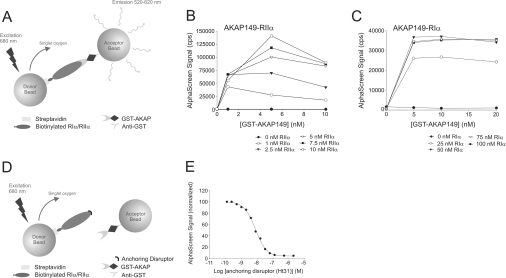
We became interested in developing a medium-to-high-throughput screening assay for characterizing different AKAP-binding sites. The Amplified Luminescent Proximity Homogeneous Assay (AlphaScreen™; PerkinElmer) [22,23] is a bead-based technology for screening biomolecular interactions in a microplate format [24]. The assay utilizes two different bead types, described as donor beads and acceptor beads, and the surfaces of these beads are coated with reactive aldehyde groups to which biomolecules can be covalently conjugated. A specific biomolecular interaction brings donor and acceptor beads in close proximity, and a photosensitizer in the donor beads then excites the acceptor beads that deliver output photons. Upon excitation at 680 nm, the photosensitizer converts ambient oxygen into a hyperexcited singlet oxygen state that can diffuse up to 200 nm in solution and interact with chemiluminescent groups on a proximate acceptor bead. This transfer of energy ultimately emits light at 520–620 nm, which constitutes the AlphaScreen signal (Figure 1A).
Figure 1. Principle of the PKA–AKAP anchoring competition/disruption assay using AlphaScreen technology.
(A) Streptavidin-coated donor beads and anti-GST-coated acceptor beads are brought into close proximity when the biotinylated R–GST–AKAP complex conjugates to the respective beads. Upon laser light excitation, singlet oxygen is produced by the photosensitizer in the donor bead and can diffuse to the acceptor bead, giving rise to a chemiluminescent/fluorescent signal, i.e. the AlphaScreen signal. The concentration-dependent interactions of RIIα–AKAP149 (B) and RIα–AKAP149 (C) assessed by AlphaScreen are shown. The different graphs show an increase in signal with increasing concentrations of biotinylated R subunit dimer. cps, counts per second. (D) A specific anchoring disruptor, competing with AKAP for binding to the R subunit dimer is capable of displacing the binding between the R subunit and the AKAP. Thus the acceptor and donor beads are no longer in close proximity and the AlphaScreen signal decreases. (E) A representative concentration-dependent inhibition curve of an anchoring disruptor (Ht31) that competes with AKAP149 for binding to RIIα with an IC50 of 7.6 nM (average IC50 for all experiments was 6±1 nM). The interaction partners were used at a 1 nM concentration in the assay.
In the present paper, we demonstrate the use of this technology as a tool to screen and characterize selective peptide antagonists of AKAPs that bind to the D/D domain in RIα and RIIα. Furthermore, this method also serves as a well-suited tool to measure the isoform specificity of these peptides and can be used to characterize new AKAP-binding sites. We have used the peptide Ht31 as a prototype to establish the method, and have expanded our analysis to include two recently developed RIα-selective anchoring disruptors [19,20].
MATERIALS AND METHODS
Expression and purification of recombinant proteins
The recombinant human R subunits of PKA (hRIα and hRIIα) were overexpressed in Escherichia coli BL21(DE3)-RIL CodonPlus cells (Stratagene) and purified via absorption to 8-AHA-cAMP–agarose [8-(6-aminohexyl)aminoadenosine-3′-,5′-cyclic monophosphate immobilized on agarose] for hRIIα and Rp-8-AHA-cAMPS–agarose [8-(6-aminohexyl)aminoadenosine-3′-,5′-cyclic monophosphorothioate, Rp-isomer, immobilized on agarose] (both from Biolog) for hRIα. AKAP149 (amino acids 285–387) and AKAP95 (amino acids 387–692), both containing the amphipathic helix constituting the A-kinase-binding domain, were expressed as GST (glutathione S-transferase) fusion proteins in E. coli BL21(DE3) and were purified using reduced glutathione–agarose beads (Sigma–Aldrich) as described previously [14]. The purified recombinant proteins were dialysed extensively against 20 mM Mops and 150 mM NaCl, pH 7, and the concentration was determined using the Bradford protein assay and SDS/PAGE (10% gels) using BSA as a standard.
Synthesis of peptides
The peptides used were Ht31 (DLIEEAASRIVDAVIEQVKAAGAY), RIAD (RI-anchoring disruptor) (LEQYANQLADQIIKEATE) and PV-38 (FEELAWKIAKMIWSDVFQQC). Peptides were synthesized on an Intavis Model MultiPep (Intavis Bioanalytical Instruments AG) and purified by HPLC. The purity and mass were analysed further by MS. The peptides were dissolved in PBS, except for PV-38 which was dissolved in PBS with 37.5% DMSO. The concentration of peptides was determined by amino acid analysis using an amino acid analyser from Applied Biosystems.
Biotinylation of the R subunit of PKA
RIα and RIIα were biotinylated according to the manufacturer's protocol. A 20-fold molar excess of EZ-Link® NHS-LC [succinimidyl-6-(biotinamido)-6-hexanamidohexanoate] biotin (Pierce) was added to the R subunit and was incubated on ice for 2 h before passing the reaction mixture through a NAP-5 column (Sephadex™ G-25 medium) (Amersham Biosciences) equilibrated with 20 mM Mops and 150 mM NaCl, pH 7. The concentration of biotinylated protein was determined using the Bradford protein assay, and the protein was divided into aliquots and stored at −80 °C for further use.
AlphaScreen assay
All assays were carried out in 384-well white opaque plates (PerkinElmer), and the assay reagents were added and mixed using a Matrix Impact2™ Electronic Pipettor (Matrix Technologies). Unless otherwise stated, 20 nM biotinylated RIα and 20 nM GST–AKAP149, or 1 nM RIIα and 1 nM GST–AKAP149/GST–AKAP95, were diluted in assay buffer (25 mM Hepes, 100 mM NaCl, pH 7.4, and 0.1% BSA), mixed to a final volume of 15 μl and incubated on ice for 15 min.
For the competition assays, peptide antagonist was added together with the selected AKAP. A time course was carried out in order to determine the time required to reach reaction equilibrium. Incubation for 15 min (on ice) of binding partners together with the selected peptide proved to be sufficient for the competition assay (results not shown). Importantly, increasing the incubation time did not decrease the AlphaScreen signal. With respect to the disruption of pre-formed PKA–AKAP complexes, it was necessary to establish the time required to reach equilibrium for each peptide separately. This was carried out by using the established 15 min incubation time for complex formation before peptide was added and the mixture was incubated (on ice) for different time periods, ranging from 15 min to 4 h (results not shown). From this it was evident that equilibrium of both the RIα and RIIα assays were reached within the first 15 min for all the peptides tested, and an incubation step of 30 min was therefore sufficient and was included in all set-ups. A concentration–response curve was obtained with 1:2 serial dilution of peptide (always in duplicate), starting at 2000 nM, unless otherwise stated.
After the selected incubation times, 5 μl of anti-GST-coated acceptor beads (PerkinElmer), diluted in assay buffer (20 μg/ml final concentration of beads in assay), were added and incubated on ice for 30 min before 5 μl of streptavidin-coated donor beads (PerkinElmer), diluted in assay buffer (20 μg/ml final concentration of beads in assay), were added to a final volume of 25 μl. After the addition of donor beads, plates were incubated at room temperature (20 °C) for 60 min (RII) or 90 min (RI) before being read on an EnVision™ multiplate reader (PerkinElmer). An identical set-up where all the incubation steps were performed at room temperature was run in parallel to control for temperature effects; however, no difference could be observed. In consideration of the limited stability of the R subunits, the first incubation steps were carried out on ice.
Owing to the light-sensitivity of the beads, all assay steps were performed under subdued lighting, and the incubation steps were carried out in the dark.
The IC50 values, defined as the concentration of antagonist needed to inhibit R–AKAP binding by 50%, were estimated by non-linear regression analyses using SigmaPlot (SPSS).
RESULTS
The AlphaScreen assay system is designed so that the streptavidin-coated donor beads and the anti-GST-coated acceptor beads have to be at a distance of less than 200 nm in solution in order to produce an optimal signal (for details on the technology, see the Introduction and Figure 1). In the present study, we established the assay using biotinylated RIα/RIIα and GST-fused truncated AKAP95/AKAP149 immobilized on these beads via streptavidin and anti-GST respectively. When the beads are brought into close proximity by an R–AKAP complex, they will produce an AlphaScreen signal (Figure 1A). A peptide antagonist that competes with the AKAP for binding to the D/D domain in the R subunit of PKA will reduce the AlphaScreen signal in a concentration-dependent manner (Figures 1D and 1E).
Assay design
Initial experiments determined the time required for the R–AKAP complex formation to reach equilibrium. This was shown to be a rapid process and was consistent with previous evidence from SPR (surface plasmon resonance) studies demonstrating that there is a very high on rate for these biomolecular interactions [14].
It was also important to define the optimal concentration of binding partners that produces a robust AlphaScreen signal with a high S/B (signal to background) ratio in the competition/disruption assays. Consequently, five different concentrations of RIα (0–100 nM) and six different concentrations of RIIα (0–10 nM) were incubated with increasing concentrations of the dual-specific AKAP149 (0–20 nM for RIα, Figure 1C; 0–10 nM for RIIα, Figure 1B) or the RII-specific AKAP95 (results not shown). Concentration-dependent increases in signal were observed for both AKAP149 and AKAP95 interactions with RIIα. However, a signal decrease was observed at all concentrations of R when the AKAP149 concentration exceeded 5 nM (Figure 1B). Similar discrepancies in the linearity of the assay were observed when high concentrations of AKAP95 were used as the acceptor (results not shown). This phenomenon, known as a ‘hooking effect’, is most likely to be due to the formation of non-signal producing complexes because the beads are saturated with acceptor protein. A maximum signal was achieved at 10 nM RIIα and 5 nM AKAP95/AKAP149. Nevertheless, a strong signal and S/B ratios of 142 for AKAP149 and 41 for AKAP95 were obtained even at the lowest concentration tested, i.e. 1 nM of both AKAP and RIIα. Therefore we decided to establish the binding assay using this low concentration in order to minimize the influence of artefactual effects that could arise from higher protein concentrations.
The interaction of RIα with AKAP149 is of lower affinity, and higher concentrations of interacting partners were therefore required. AlphaScreen signal was observed at concentrations of 25–100 nM RIα and 5–20 nM AKAP149 (Figure 1C). The signal was in general less dependent on increasing concentrations of either interaction partner. Furthermore, for this interaction, no significant decrease in signal was observed at the highest concentrations used (Figure 1C). Since no further signal increase was observed for concentrations higher than 25 nM of RIα (Figure 1C), 20 nM RIα and 20 nM of AKAP149 were used in the subsequent assays, producing an S/B ratio of approx. 30. In the absence of either RIα or RIIα subunit, no signal was detected from the AKAP-bound acceptor bead (Figures 1B and 1C).
Peptide antagonists as competitors of the RIα/RIIα–AKAP interaction
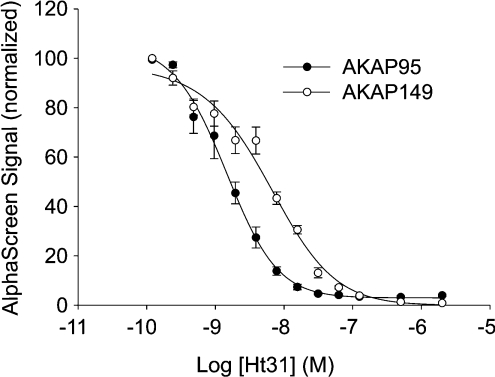
As seen in Figure 2, the dual-specific peptide Ht31 competed with both AKAP149 and AKAP95 in their interaction with RIIα in a concentration-dependent manner, with IC50 values of 6±1 nM and 1.4±0.2 nM respectively. Moreover, concentrations of 30–100 nM Ht31 were required to fully abolish the AlphaScreen signal (Figure 2).
Figure 2. Concentration-dependent inhibition by Ht31 of AlphaScreen signal from AKAP95 (●) and AKAP149 (○) binding to RIIα.
In the assay, 1 nM biotinylated RIIα and 1 nM GST–AKAP were used with Ht31 peptide concentrations ranging from 2000 to 0.12 nM. For experimental conditions, see the Materials and methods section. The IC50 values were calculated for each AKAP by non-linear regression analysis using SigmaPlot. Results are means±S.E.M. for three independent experiments performed in duplicate.
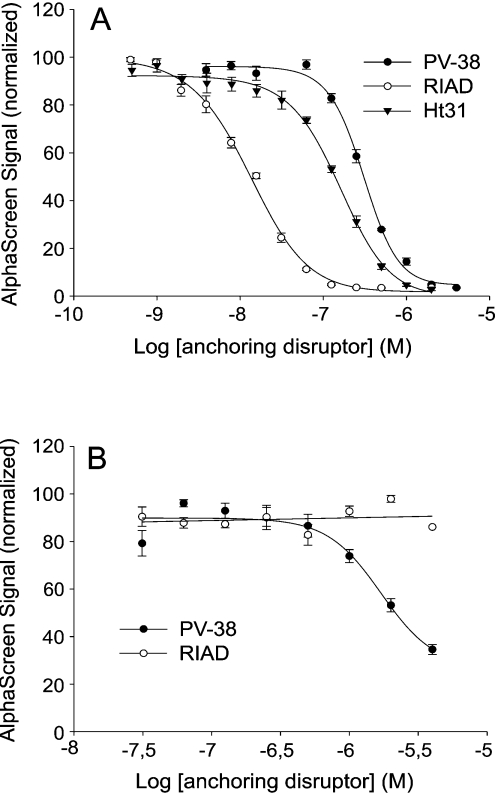
Figure 3(A) depicts similar data for RIα, with an IC50 value for inhibition of 156±10 nM. In this case, the kinetics of two other RI-specific peptides, RIAD and PV-38 [19,20], were also examined. RIAD, with an IC50 value of 13±1 nM, was approximately one order of magnitude more potent than Ht31 in competing with AKAP149 in the interaction with RIα, whereas PV-38 was less potent than Ht31 with an IC50 value of 304±17 nM (Figure 3A). Concentrations of 100 nM (RIAD) to 1000 nM (PV-38) were required to bring the AlphaScreen signal down to baseline levels (Figure 3A). In both Figures 2 and 3, slope differences in the inhibition curves were seen. The reason for this is not known, but may reflect differences in co-operativity, possibly at the level of the assay system. Furthermore, the RI selectivity of RIAD and PV-38 was examined by attempts to compete with AKAP149 for interaction with RIIα using increasing concentrations of peptides. No significant decrease in AlphaScreen signal was observed even at 4000 nM RIAD peptide (Figure 3B). In contrast, approx. 60% of the AlphaScreen signal was disrupted at this concentration of PV-38 (Figure 3B). The working concentration of DMSO in the experiments with PV-38 was 0.5% (at 2000 nM), and to test whether such concentrations of DMSO influenced the assay, 0.5% DMSO was also used in the competition assay with Ht31. Importantly, this did not interfere with the AlphaScreen signal or affect the IC50 value.
Figure 3. Concentration-dependent inhibition of AlphaScreen signal due to competition by selective peptide antagonists of the RIα–AKAP149 (A) and RIIα–AKAP149 (B) interactions.
Increasing concentrations of the peptides Ht31 (▼), RIAD (○) or PV38 (●) were incubated with 20 nM biotinylated RIα and 20 nM GST–AKAP149 (A) or 1 nM RIIα and 1 nM GST–AKAP149 (B). Peptide concentrations ranged from 2000 to 0.48 nM (A) and from 4000 to 30 nM (B). IC50 values were calculated for each peptide antagonist by non-linear regression analysis using SigmaPlot. Results are means±S.E.M. for three independent experiments performed in duplicate.
Peptide antagonists as disruptors of the RIα/RIIα–AKAP complex
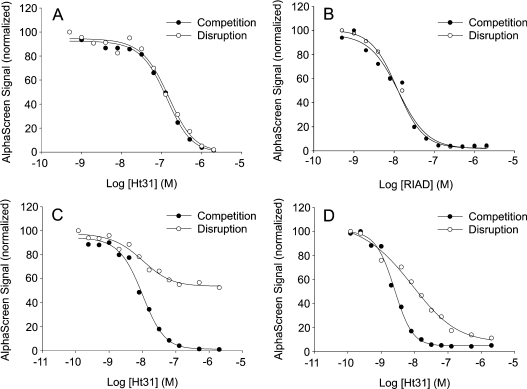
An efficient anchoring antagonist peptide that works in situ not only competes with the ongoing complex formation, but also disrupts pre-formed R–AKAP complexes. Kinetic studies by SPR have revealed Kd values of 0.5 and 5.9 nM for RIIα interactions with AKAP149 and AKAP95 respectively [14]. In contrast, RIα was demonstrated to have a much lower affinity for AKAP149 with a Kd of 185 nM, primarily due to a higher off rate [14]. The rate by which a peptide antagonist is capable of displacing R–AKAP complexes may then be determined by its intrinsic affinity for the R subunit and the exchange rate of the R–AKAP complex. The ability of Ht31 and RIAD to displace R–AKAP interactions is shown in Figure 4. Figures 4(A) and 4(B) show the effect of increasing concentrations of Ht31 and RIAD respectively on RIα–AKAP149 competition and disruption. As seen from the Figures, the curves are completely overlapping, indicating that the exchange rate is sufficiently high to reach complete equilibrium even with pre-formed complexes.
Figure 4. Concentration-dependent inhibition of the AlphaScreen signal due to competition (●) or anchoring disruption (○) by Ht31 of AKAP149 interaction with RIα (A), RIAD of AKAP149 interaction with RIα (B), Ht31 of AKAP149 interaction with RIIα (C) and Ht31 of AKAP95 interaction with RIIα (D).
For experimental conditions, see Figure 2 and the Materials and methods section. Results are means of duplicates from one single experiment performed in parallel and are representative of three independent experiments.
Figures 4(C) and 4(D) show similar experiments with RIIα–AKAP149 and RIIα–AKAP95 respectively. As expected, during competition, Ht31 brought the AlphaScreen signal all the way down to baseline level. When Ht31 was used to disrupt pre-formed complexes, however, it only displaced 50% of the RIIα-bound AKAP149 (Figure 4C), indicating that the RIIα–AKAP149 complexes had so low an exchange rate that displacement only occurred for a fraction of the complexes during the duration of the assay.
Figure 4(D) shows the effect of Ht31 on RIIα–AKAP95 complexes. In this case, displacement of pre-formed complexes was almost 100% at high concentrations of peptide. However, the slope differences of the competition and displacement curves are probably related to a somewhat lower exchange rate of the AKAP–R complex under the experimental conditions used, compared with that seen with RIα in Figure 4(A).
DISCUSSION
We have successfully developed an in vitro screening tool for efficient characterization of peptide disruptors that block the binding of PKA to different AKAPs. Both RIα and RIIα were set up to interact with selective AKAP reporters in the AlphaScreen assay. The AlphaScreen technology is principally used by the pharmaceutical industry in high-throughput screening of low-molecular-mass drug candidates. Here we report use of this technology for measurements of protein–protein interactions, notably R–AKAP interactions, and for characterization of selective anchoring disruptors of these complexes by competition/disruption. With this reporter assay established, it will be possible to screen a large number of putative anchoring disruptors for their effect on a particular R–AKAP interaction. The different AKAP reporters display different affinities for the R subunit [14], giving rise to differences in AlphaScreen signal and distinct IC50 values.
The anchoring disruptors used in the present study were the peptides Ht31, representing the amphipathic helix of AKAP-Lbc, and two RI-specific amphipathic helix sequences optimized for selectivity and specificity, all competing with AKAPs for binding to the D/D domain of PKA. Ht31 efficiently competed the dual-specific AKAP149 and the RII-specific AKAP95 interaction with RIIα, as well as the AKAP149 interaction with RIα (Figures 2 and 3). The affinities of Ht31 were greater for the RIIα–AKAP interaction than for the RIα–AKAP interaction, as shown previously using other methods [14]. Although competition is easily assessed, displacement of pre-formed complexes may be more relevant for the use of anchoring disruptor peptides in situ. Although Ht31 efficiently displaced RIα from the interaction with AKAP149, the more stable complexes with RIIα could not be as efficiently disrupted (Figure 4). A potential explanation for this observation is that RIIα is released from the complex with a lower off rate. In contrast, RIIα–AKAP95 complexes could be completely displaced, albeit at higher concentrations.
Another aspect of our study was to assess applicability of this technology for screening different anchoring disruptors by using RIα–AKAP149 in competition with different RI-interacting peptides and measuring kinetic properties. RIAD was demonstrated to be ∼10- and ∼20-fold more efficient than Ht31 and PV-38 respectively. This finding illustrates the utility of this screening protocol as a means to search for anchoring disruptors with predefined properties. Consequently, it could become a standard method for screening peptides and compound libraries to identify new antagonists of these protein–protein interactions.
One clear advantage of AlphaScreen over other screening platforms available today is the sensitivity of this type of assay [22]. In the AlphaScreen assay, each donor bead can generate approx. 60000 singlet oxygen molecules per second, resulting in a high degree of signal amplification and very high sensitivity [22]. Furthermore, the system is designed in such a manner that it keeps the background signal very low, mainly because the excitation wavelength of the laser is above the detection wavelength for the luminescence signal. Hence, the high signal together with the low background results in a favourable S/B ratio, making this assay technology preferable to other technologies, particularly because low physiologically relevant concentrations of the interacting partners can be used.
Mapping of affinities in the AlphaScreen assay system involves screening for relative IC50 values between the compounds or antagonists in question. Such affinities were shown to be very reproducible within the assay itself. Importantly, the experimentally obtained IC50 values reflect both (i) the affinity of a given peptide for the D/D domain in the R subunit, i.e. high affinity will result in a lower IC50 value, and (ii) the affinity of the given AKAP for the D/D domain, i.e. more anchoring disruptor peptide will be required to compete a tightly bound AKAP than a loosely bound AKAP. Thus the selection of R–AKAP pairs for the assay affects sensitivity, and a combination of a high-affinity peptide together with a less stringent R–AKAP interaction will give the most efficient competition with the lowest IC50 value. The different disruption efficiency for RIα– and RIIα–AKAP complexes is probably related to the mode of binding and hence produces differences in kinetics. For example, the dissociation rate of AKAP149 from R is approx. 100-fold higher for the RIα subunit than for the RIIα subunit [14]. The more dynamic state of the RIα–AKAP complex probably accounts for the ease by which RIα is displaced from AKAP149, using either Ht31 or the high-affinity RI-specific peptide RIAD (Figure 4). Furthermore, the differences in RI and RII binding to the amphipathic helix of the AKAPs are also clear from structural data. Although both subunits fold into an X-type four-helix bundle constituting the D/D domain, RII has a pre-formed localized binding surface of primarily hydrophobic character, whereas the docking surface of RI involves charged residues [13].
From these findings, it is apparent that the disruption assay is a more efficient means of reporting on the general efficiency of anchoring disruptor peptides than the competition assay. This information is particularly important when these peptides are used in situ as reagents to assess the role of anchored PKA complexes in the control of cAMP-responsive events. In line with this, recently developed high-affinity RII-selective peptides which are more potent competitors than Ht31, such as AKAP-IS (AKAP-in silico) and AKAP7-L314E, have been designed [18,21] that should be characterized in future studies for their ability to both compete in ongoing complex formation and disrupt pre-formed complexes.
Although most of the AKAPs found in Nature are RII-specific, some AKAPs display dual binding to both RI and RII [7–9] and a few AKAPs appear to be selective for RI [10–12], although the binding domains of the latter class of AKAPs have not been defined. Therefore it would be advantageous to design an anchoring disruptor peptide with high affinity for RI that at the same time discriminates towards RII. PV-38, derived from D-AKAP2, was made in an attempt to resolve this task [19]. This peptide binds RI with ∼100-fold higher affinity than RII as measured using a fluorescence anisotropy binding assay [19]. However, when comparing the IC50 values of PV-38 and Ht31, the former was shown to be 2-fold less effective in competing in RIα complex formation (Figure 3A). In contrast, the RIα-specific peptide RIAD [20] competes AKAP149 10-fold more efficiently than Ht31, making this a very interesting starting point for development of peptidomimetics and drug design. Furthermore, RIAD was shown in the present work to be strongly RI-selective with no effect on competing in RIIα binding to AKAP149 (Figure 3B). This is in contrast with PV-38 which also, to some extent, competed in RII binding to AKAP149, although an accurate IC50 value could not be obtained due to the very high concentrations of peptide required and the lack of complete displacement of RII by PV-38 (Figure 3B).
In conclusion, the AlphaScreen technology can be used to follow disruption of protein–protein interaction by anchoring disruptors, with a focus on PKA–AKAP interactions in the present paper. Importantly, it can be used for peptidomimetics and, in the future, possibly also aid in the characterization of small-molecular substances (<500 Da) designed to replace the peptides as anchoring disruptors using chemical biology approaches.
Acknowledgments
We are grateful to Dr Vidar Hansson for helpful discussions and critical comments on the manuscript. This work was supported by grants from the Functional Genomics Programme, The Research Council of Norway, The Norwegian Cancer Society, Novo Nordisk Foundation Committee, the European Union (RTD grant number QLK3-CT-2002-02149) and the Bundesministerium für Bildung und Forschung (grant number 01Gr0441, NGFN) to F. W. H.
References
- 1.Francis S. H., Corbin J. D. Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 1994;56:237–272. doi: 10.1146/annurev.ph.56.030194.001321. [DOI] [PubMed] [Google Scholar]
- 2.Taskén K., Aandahl E. M. Localized effects of cAMP mediated by distinct routes of protein kinase A. Physiol Rev. 2004;84:137–167. doi: 10.1152/physrev.00021.2003. [DOI] [PubMed] [Google Scholar]
- 3.Taylor S. S., Buechler J. A., Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu. Rev. Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 4.Wong W., Scott J. D. AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 5.Banky P., Newlon M. G., Roy M., Garrod S., Taylor S. S., Jennings P. A. Isoform-specific differences between the type Iα and IIα cyclic AMP-dependent protein kinase anchoring domains revealed by solution NMR. J. Biol. Chem. 2000;275:35146–35152. doi: 10.1074/jbc.M003961200. [DOI] [PubMed] [Google Scholar]
- 6.Newlon M. G., Roy M., Morikis D., Hausken Z. E., Coghlan V., Scott J. D., Jennings P. A. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 1999;6:222–227. doi: 10.1038/6663. [DOI] [PubMed] [Google Scholar]
- 7.Huang L. J., Durick K., Weiner J. A., Chun J., Taylor S. S. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L. J., Durick K., Weiner J. A., Chun J., Taylor S. S. Identification of a novel protein kinase A anchoring protein that binds both type I and type II regulatory subunits. J. Biol. Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 9.Reinton N., Collas P., Haugen T. B., Skålhegg B. S., Hansson V., Jahnsen T., Taskén K. Localization of a novel human A-kinase-anchoring protein, hAKAP220, during spermatogenesis. Dev. Biol. 2000;223:194–204. doi: 10.1006/dbio.2000.9725. [DOI] [PubMed] [Google Scholar]
- 10.Angelo R., Rubin C. S. Molecular characterization of an anchor protein (AKAPCE) that binds the RI subunit (RCE) of type I protein kinase A from Caenorhabditis elegans. J. Biol. Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- 11.Küssel-Andermann P., El-Amraoui A., Safieddine S., Hardelin J. P., Nouaille S., Camonis J., Petit C. Unconventional myosin VIIA is a novel A-kinase-anchoring protein. J. Biol. Chem. 2000;275:29654–29659. doi: 10.1074/jbc.M004393200. [DOI] [PubMed] [Google Scholar]
- 12.Li H., Degenhardt B., Tobin D., Yao Z. X., Taskén K., Papadopoulos V. Identification, localization, and function in steroidogenesis of PAP7: a peripheral-type benzodiazepine receptor- and PKA (RIα)-associated protein. Mol. Endocrinol. 2001;15:2211–2228. doi: 10.1210/mend.15.12.0736. [DOI] [PubMed] [Google Scholar]
- 13.Banky P., Roy M., Newlon M. G., Morikis D., Haste N. M., Taylor S. S., Jennings P. A. Related protein–protein interaction modules present drastically different surface topographies despite a conserved helical platform. J. Mol. Biol. 2003;330:1117–1129. doi: 10.1016/s0022-2836(03)00552-7. [DOI] [PubMed] [Google Scholar]
- 14.Herberg F. W., Maleszka A., Eide T., Vossebein L., Taskén K. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 15.Carr D. W., Hausken Z. E., Fraser I. D., Stofko-Hahn R. E., Scott J. D. Association of the type II cAMP-dependent protein kinase with a human thyroid RII-anchoring protein: cloning and characterization of the RII-binding domain. J. Biol. Chem. 1992;267:13376–13382. [PubMed] [Google Scholar]
- 16.Diviani D., Soderling J., Scott J. D. AKAP-Lbc anchors protein kinase A and nucleates Gα12-selective Rho-mediated stress fiber formation. J. Biol. Chem. 2001;276:44247–44257. doi: 10.1074/jbc.M106629200. [DOI] [PubMed] [Google Scholar]
- 17.Burton K. A., Johnson B. D., Hausken Z. E., Westenbroek R. E., Idzerda R. L., Scheuer T., Scott J. D., Catterall W. A., McKnight G. S. Type II regulatory subunits are not required for the anchoring-dependent modulation of Ca2+ channel activity by cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11067–11072. doi: 10.1073/pnas.94.20.11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alto N. M., Soderling S. H., Hoshi N., Langeberg L. K., Fayos R., Jennings P. A., Scott J. D. Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4445–4450. doi: 10.1073/pnas.0330734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burns-Hamuro L. L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G. L., Cantor C. R., Braun A., Taylor S. S. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. U.S.A. 2003;100:4072–4077. doi: 10.1073/pnas.2628038100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson C. R., Lygren B., Berge T., Hoshi N., Wong W., Taskén, Scott J. D. Delineation of type I protein kinase a selective signaling events using an RI anchoring disruptor (RIAD) J. Biol. Chem. 2006;281:21535–21545. doi: 10.1074/jbc.M603223200. [DOI] [PubMed] [Google Scholar]
- 21.Hundsrucker C., Krause G., Beyermann M., Prinz A., Zimmermann B., Diekmann O., Lorenz D., Stefan E., Nedvetsky P., Dathe M., et al. High-affinity AKAP7δ-protein kinase A interaction yields novel protein kinase A anchoring disruptor peptides. Biochem. J. 2006;396:297–306. doi: 10.1042/BJ20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ullman E. F., Kirakossian H., Singh S., Wu Z. P., Irvin B. R., Pease J. S., Switchenko A. C., Irvine J. D., Dafforn A., Skold C. N., Wagner D. B. Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc. Natl. Acad. Sci. U.S.A. 1994;91:5426–5430. doi: 10.1073/pnas.91.12.5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ullman E. F., Kirakossian H., Switchenko A. C., Ishkanian J., Ericson M., Wartchow C. A., Pirio M., Pease J., Irvin B. R., Singh S., et al. Luminescent oxygen channeling assay (LOCI™): sensitive, broadly applicable homogeneous immunoassay method. Clin. Chem. 1996;42:1518–1526. [PubMed] [Google Scholar]
- 24.Gesellchen F., Prinz A., Zimmermann B., Herberg F. W. Quantification of cAMP antagonist action in vitro and in living cells. Eur. J. Cell Biol. 2006;85:663–672. doi: 10.1016/j.ejcb.2006.01.009. [DOI] [PubMed] [Google Scholar]