Abstract
Lipid rafts and caveolae are biochemically similar, specialized domains of the PM (plasma membrane) that cluster specific proteins. However, they are morphologically distinct, implying different, possibly complementary functions. Two-dimensional gel electrophoresis preceding identification of proteins by MS was used to compare the relative abundance of proteins in DRMs (detergent-resistant membranes) isolated from HUVEC (human umbilical-vein endothelial cells), and caveolae immunopurified from DRM fractions. Various signalling and transport proteins were identified and additional cell-surface biotinylation revealed the majority to be exposed, demonstrating their presence at the PM. In resting endothelial cells, the scaffold of immunoisolated caveolae consists of only few resident proteins, related to structure [CAV1 (caveolin-1), vimentin] and transport (V-ATPase), as well as the GPI (glycosylphosphatidylinositol)-linked, surface-exposed protein CD59. Further quantitative characterization by immunoblotting and confocal microscopy of well-known [eNOS (endothelial nitric oxide synthase) and CAV1], less known [SNAP-23 (23 kDa synaptosome-associated protein) and BASP1 (brain acid soluble protein 1)] and novel [C8ORF2 (chromosome 8 open reading frame 2)] proteins showed different subcellular distributions with none of these proteins being exclusive to either caveolae or DRM. However, the DRM-associated fraction of the novel protein C8ORF2 (∼5% of total protein) associated with immunoseparated caveolae, in contrast with the raft protein SNAP-23. The segregation of caveolae from lipid rafts was visually confirmed in proliferating cells, where CAV1 was spatially separated from eNOS, SNAP-23 and BASP1. These results provide direct evidence for the previously suggested segregation of transport and signalling functions between specialized domains of the endothelial plasma membrane.
Keywords: caveolae, detergent-resistant membrane (DRM), human umbilical-vein endothelial cells (HUVEC), lipid raft, sub-proteomics, vesicular transport
Abbreviations: BASP1, brain acid soluble protein 1; C8ORF2, chromosome 8 open reading frame 2; CAV1, caveolin-1; DIC, differential interference contrast; DRM, detergent-resistant membrane; eNOS, endothelial nitric oxide synthase; ER, endoplasmic reticulum; GFP, green fluorescent protein; GNB, guanine nucleotide-binding protein; GPI, glycosylphosphatidylinositol; GRP78, 78 kDa glucose-regulated protein; HRP, horseradish peroxidase; HUVEC, human umbilical-vein endothelial cells; IB, immunoisolation buffer; IPG, immobilized pH gradient; mAb, monoclonal antibody; MES, 2-(N-morpholino)ethanesulfonic acid; MBS, MES-buffered saline; NAP-22, neuronal axonal myristoylated membrane protein of 22 kDa; PDI, protein disulfide-isomerase; PM, plasma membrane; PTRF, polymerase I and transcript-release factor; RT, room temperature; SNAP-23, 23 kDa synaptosome-associated protein; SPFH, stomatin/prohibitin/flotillin/HflK; vWF, von Willebrand factor
INTRODUCTION
Research in the last decade has changed our view of the mammalian PM (plasma membrane), which is no longer considered as a uniform lipid bilayer containing randomly distributed membrane and associated proteins. Instead, the PM is considered to contain organized structures such as cholesterol- and glycosphingolipid-enriched microdomains, termed lipid rafts [1]. By sequestering specific proteins, while excluding others, these specialized domains provide a dynamic scaffold for organizing cellular processes such as signal transduction, vesicular transport, cholesterol homoeostasis and even internalization of pathogens like viruses and bacteria [2–4]. Whereas definitive proof for the existence of lipid rafts remains to be obtained [5], caveolae represent a specialized subset of rafts with a morphologically distinct flask-shaped structure, 50–100 nm in diameter [3]. Caveolin, which oligomerizes and coats the cytoplasmic surface of caveolae, acts as a scaffolding protein and appears to negatively regulate signalling [6]. In contrast with lipid rafts, caveolae are not present in all cell types. Caveolae with the CAV1 (caveolin-1) isoform are particularly abundant in endothelial cells, where they regulate permeability (transcytosis), angiogenesis and vascular tone [7], which includes the well-studied regulation of nitric oxide signalling by modulating eNOS (endothelial nitric oxide synthase) activity. Surprisingly, in various studies, CAV1-knockout mice are viable while displaying abnormalities which clearly demonstrate the involvement of caveolae in physiology and pathological conditions, such as cancer and cardiovascular disease [8–10].
A dynamic and balanced interaction between rafts and caveolae has been suggested to regulate important processes, especially in human endothelial cells [10]. Separately studying their protein composition could therefore provide further functional insight. However, due to their similar biochemical properties, protein compositions of caveolae and rafts are generally not studied separately [11–13], even though caveolae and lipid rafts are morphologically distinct, implying different or even complementary functions.
At present no methodological consensus exists for the isolation of microdomains such as caveolae and lipid rafts. Resistance to detergent solubility at low temperature is the most widely used method to obtain pure fractions uncontaminated by other components, such as the ER (endoplasmic reticulum) [10,14,15]. Although the resulting DRMs (detergent-resistant membranes) are not thought to be identical with lipid rafts as they exist in the intact cell [16], these fractions most accurately reflect raft protein composition. The use of Triton X-100 appears the most stringent compared with other detergents [17] and yields the highest enrichment, 2000-fold for CAV1, being far superior to other, non-detergent-based methods [12,18,19]. Our optimized proteomics protocols provide enhanced representation of membrane proteins [18], thereby enabling the definition of the detergent-resistant protein scaffold of these domains.
We used one- and two-dimensional analyses to visualize and compare the relative abundance and surface exposure of proteins within DRM fractions and immunoseparated caveolae isolated from resting HUVEC (human umbilical-vein endothelial cells). Additionally, we characterized the cellular distribution of selected proteins within different classes in both confluent and subconfluent cells. These include SNAP-23 (23 kDa synaptosome-associated protein), C8ORF2 (chromosome 8 open reading frame 2) and BASP1 (brain acid soluble protein 1), which are relatively unknown proteins in or even new to human endothelial caveolae and rafts. Together, the presented data suggest a spatial and temporal segregation of transport and signalling functions between caveolae and raft proteomes of human endothelial cells, further supporting the proposed dynamic and interactive regulatory behaviour of these specialized membrane domains.
EXPERIMENTAL
Materials
Antibodies were purchased from the following sources: anti-CAV1 polyclonal antibody, mAb (monoclonal antibody) clone 2234, anti-transferrin receptor mAb and anti-eNOS mAb (BD Transduction Laboratories, Lexington, KY, U.S.A.); anti-SNAP-23, anti-PDI (protein disulfide-isomerase) and anti-GFP (green fluorescent protein) (Abcam, Cambridge, U.K.); antisera against two separate, synthetic peptides of human C8ORF2 were raised in rabbits by the Eurogentec Double-X program (Eurogentec, Seraing, Belgium). Reagents MES [2-(N-morpholino)ethanesulfonic acid] and Triton X-100 were from Sigma (St. Louis, MO, U.S.A.).
Cell culture
HUVEC were isolated and cultured as described in [18,20], using culture medium 199 (Gibco-BRL, Paisley, Scotland, U.K.), supplemented with 20% (v/v) foetal bovine serum, 50 μg/ml heparin (Sigma), 6.5–25 μg/ml Endothelial Cell Growth Supplement (Sigma) and 100 units/ml penicillin/streptomycin (Gibco-BRL).
Preparation of caveolae-enriched DRM fractions
DRM fractions were prepared as described previously [18,21]. Briefly, cells were homogenized at 4 °C with 1% (v/v) Triton X-100 in MBS (MES-buffered saline; 25 mM MES, pH 6.5, and 150 mM NaCl), supplemented with mammalian protease inhibitor cocktail (Sigma). The lysate was adjusted to 80% sucrose in MBS, overlaid with a 30–5% sucrose step gradient and centrifuged for 18 h at 36000 rev./min in a Kontron TST41.14 rotor at 4 °C.
Biotinylation of cell-surface-exposed proteins
Cultured cells were washed twice with ice-cold MBS+ (MBS supplemented with 2 mM CaCl2 and 1 mM MgCl2), followed by incubation for 30 min at 4 °C with 500 μg/ml sulfosuccinimidyl-6-(biotinamido) hexanoate [sulfo-NHS (N-hydroxysuccinimido)-LC (long chain)-biotin; Pierce Biotechnology, Rockford, IL, U.S.A.] in MBS+. The reaction was stopped by washing for 5 min with 100 mM glycine in MBS+. After washing twice with MBS, DRM fractions were prepared as described above.
Immunopurification of caveolae from total DRM fractions
Protein G-coupled Dynabeads (Dynal Biotech, Oslo, Norway) were washed twice with wash buffer (50 mM Tris/HCl, pH 8.1, and 150 mM NaCl) and incubated for 10 min at RT (room temperature) with 2.5 μg of antibody (anti-caveolin mAb clone 2234) and washed twice with wash buffer. After washing twice with cross-link buffer (200 mM triethanolamine, pH 8.2), the antibodies were cross-linked to the Protein G beads with 20 mM dimethyl pimelimidate dihydrochloride (Pierce) in cross-link buffer for 30 min at RT. The reaction was stopped by incubating for 15 min at RT with 50 mM Tris/HCl (pH 7.5) and 150 mM NaCl and the beads were subsequently washed three times with IB (immunoisolation buffer; 50 mM Tris/HCl, pH 8.1, 150 mM NaCl and 0.5% Triton X-100). Prepared beads were pre-eluted for 2 min with 30 μl of 100 mM citrate (pH 2.4) and washed three times with IB. Cross-linked beads were incubated with 25 μg of isolated DRM proteins, resuspended in IB overnight at 4 °C. Unbound material was collected and the beads were washed three times with ‘high-salt’ wash buffer (50 mM Tris/HCl, pH 8.1, 500 mM NaCl and 0.5% Triton X-100). Bound material was eluted for 5 min at RT with 0.5% SDS, 40 mM HCl and afterwards neutralized with 1.5 M Tris (pH 8.8). Immunoisolated material was processed for one- or two-dimensional gel electrophoresis as described in [18].
Electrophoresis and immunoblotting
Two-dimensional gel electrophoresis, staining and imaging was performed with our optimized protocols as described in [18] using first dimension (pH 4–7) linear IPG (immobilized pH gradient) strips and second dimension SDS/10% polyacrylamide and SDS/12% polyacrylamide gels. For one-dimensional sample analysis, either equal volumes from gradient fractions were used, or protein concentrations were first quantified to determine the volume required for 10 μg of protein, followed by SDS/PAGE and transfer to nitrocellulose for immunoblot analysis. Wash buffer was composed of 10 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.1% Tween 20 (TBS-T) supplemented with 5% non-fat dry milk for the blocking solution and antibody diluents. Primary antibodies were used at 1:1000–1:5000 dilutions. HRP (horseradish peroxidase)-conjugated secondary antibodies (1:10000 dilution; Bio-Rad, Hercules, CA, U.S.A.) were used in combination with a chemiluminescent substrate [ECL® (enhanced chemiluminescence), Amersham Biosciences, Uppsala, Sweden]. For the detection of biotinylated proteins with ECL®, membranes were washed with PBS and 0.1% Tween 20 (PBS-T), while blocking and incubation with avidin–HRP (Amersham Biosciences) were performed in PBS-T, supplemented with 1% casein. Specific signals were detected using a Lumi-Imager (Roche Molecular Biochemicals, Mannheim, Germany) and the density of visualized bands was quantified using ImageQuant TL software (Amersham Biosciences).
BASP1 GFP-construct generation and transfection
The complete open reading frame for BASP1 (NM_006317) was amplified from a HUVEC cDNA library, using specific primers containing a mutated STOP codon and restriction sites (EcoRI and BamHI), and was TA-cloned into the pGEM-T vector. The vector containing the BASP1 cDNA was sequenced and the released encoding fragment, using EcoRI and BamHI, was fused in-frame at the N-terminal end of GFP using the pEGFP-N1 vector (Clontech, Palo Alto, CA, U.S.A.). HUVEC were transfected using optimized protocols of the Nucleofector® system (Amaxa Biosystems).
Transmission and freeze fracture electron microscopy
HUVEC were cultured as described above on fibronectin-coated thermanox slides and fixed in a solution of 4% (w/v) paraformaldehyde and 1% (v/v) glutaraldehyde in 100 mM phosphate buffer (pH 7.4). For transmission electron microscopy, post-fixation was performed in 1% osmium tetroxide followed by dehydration and embedding in LX-112 epoxy resin according to standard procedures. Ultrathin sections were counterstained with uranyl acetate and observed with a Philips EM-420 transmission electron microscope (Philips, Eindhoven, The Netherlands). For freeze fracture replicas, fixed cells were cryoprotected with 2.3 M sucrose in 100 mM phosphate buffer (pH 7.4) and frozen in liquid ethane at 90 K. Freeze fracturing was performed in a BAF300 (BAL-TEC, Balzers, Liechtenstein) at a vacuum of <10−5 Pa and at a temperature of 150 K. Cells were replicated with 2 nm platinum at 45° and 20 nm carbon was deposited at 90°. After cleaning with household bleach, the replicas were examined in an EM-420 transmission electron microscope (Philips).
Immunofluorescence microscopy
HUVEC were grown on fibronectin-coated coverslips. The slides were coated with 1% (w/v) gelatin and cross-linked with 0.5% (v/v) glutardialdehyde before coating with fibronectin. Cells were washed twice with HBSS (Hanks balanced salt solution), fixed for 20 min in HBSS containing 2% paraformaldehyde and washed three times with PBS. The cells were then treated with permeabilization buffer (PBS, 0.2% BSA and 0.1% Triton X-100) for 10 min, washed with PBS and incubated with specific antibodies. The bound primary antibodies were visualized with tagged secondary antibodies (FITC- and Cy5-conjugated goat anti-mouse or anti-rabbit IgG; Jackson Immunochemicals). Cells were then washed with PBS (three times) and slides were mounted with Mowiol (Calbiochem, La Jolla, CA, U.S.A.). Confocal laser scanning microscopy was performed using Leica (Leica Microsystems, Heidelberg, Germany) and/or Bio-Rad instruments.
Identification of proteins by MS
Protein-containing gel slices were S-alkylated, digested with sequencing grade trypsin (Roche Molecular Biochemicals, Indianapolis, IN, U.S.A.) and extracted by the method of Shevchenko et al. [22] with modifications [18]. Extracted peptides were concentrated using a SpeedVac and the pellet was taken up in 5 μl of 1% formic acid and 60% acetonitrile. The peptide solution was mixed with an equal volume of 10 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma Chemical Co.) solution in acetonitrile/ethanol (1:1, v/v) with 1% trifluoroacetic acid and 1 mM of ammonium acetate. One microlitre was spotted on target and allowed to dry at RT. MALDI-TOF-MS (matrix-assisted laser-desorption ionization–time-of-flight MS) spectra were acquired on a Micromass M@LDI (Micromass, Wythenshawe, U.K.). Peptide sequencing [MS/MS (tandem MS)] was performed on a QSTAR-XL equipped with an oMALDI interface (Applied Biosystems/MDS Sciex, Toronto, ON, Canada). The resulting peptide spectra were used to search a non-redundant protein sequence database (Swiss-Prot/TREMBL) using the Proteinprobe program (Micromass) or using the MASCOT search engine (http://www.matrixscience.com).
RESULTS
Caveolae in cultured human endothelial cells
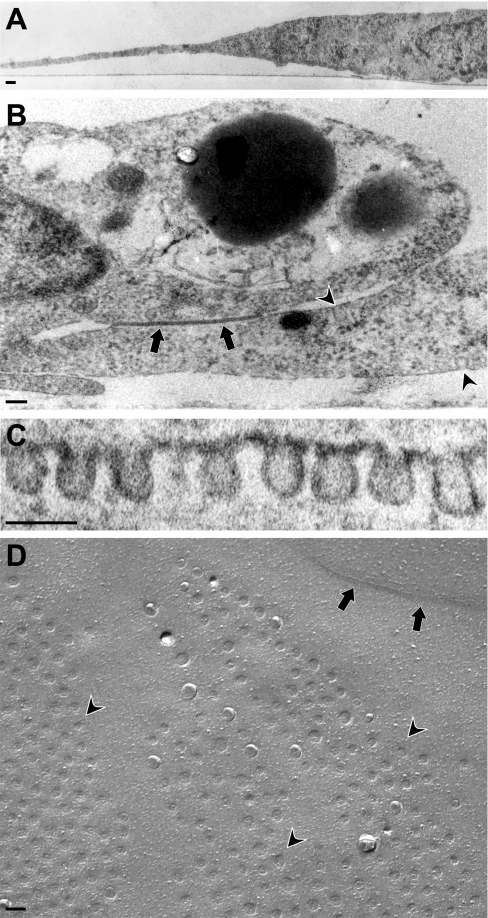
Whereas lipid rafts are assumed to be universally present in many different cell types, the presence and abundance of caveolae at the surface of endothelial cells seem to vary in order to modulate transcytosis and regulate signalling. Several papers have suggested a lack of caveolae in cultured human endothelial cells including HUVEC [23,24]. Ultrastructurally, caveolae can be distinguished from rafts by their characteristic flask-like shape. Using the growth conditions as described in the Experimental section, cultured endothelial cells grow as flat, stretched cells (Figure 1A), in continuous closed confluent layers displaying junctional structures (indicated by arrows in Figure 1B), highly similar to arteries in vivo [25]. In our hands, confluent endothelial cells, which have adopted a characteristic ‘cobble-stone’ appearance, contain abundant caveolae, which are uniform in size, approx. 75 nm (Figure 1C) and localize close to these junctional cell contact sites (indicated by arrowheads in Figure 1B). The abundance of caveolae at the surface was visualized using freeze fracture electron microscopy (Figure 1D). Extended patches of caveolae (arrowheads) were found, often in proximity to junctional structures (indicated by arrows).
Figure 1. Morphological examination of caveolae in cultured confluent human primary endothelial cells.
Transmission electron microscopy reveals endothelial cells as (A) flat and stretched cells (B) forming closed confluent layers displaying junctional structures (arrow), and caveolae (arrowheads). (C) Caveolae are abundant at the PM, optimally visualized by (D) freeze fracture EM revealing patches of caveolae (arrowheads) in proximity of juntional structures (arrows). Scale bar indicates 100 nm.
Sub-proteomics and distribution analysis of known and novel proteins in DRM fractions
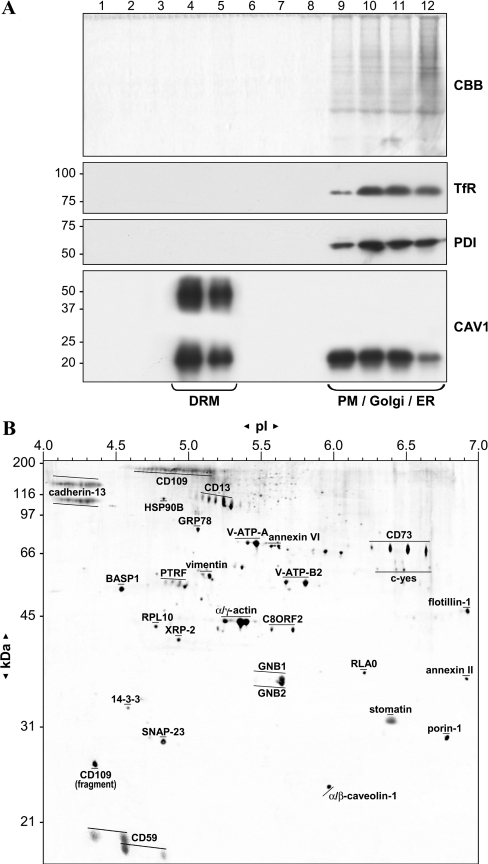
CAV1-enriched DRM fractions were prepared using an established technique based on resistance to Triton X-100 at 4 °C, and subsequent flotation in sucrose density gradients [21]. Equal aliquots of the sucrose-gradient fractions were subjected to SDS/PAGE and analysed by colloidal Coomassie staining and anti-CAV1 Western blotting. Staining for total protein and CAV1 (Figure 2A) revealed that approx. 99% of cellular protein was retained in the input fractions 9–12, as compared with floating fractions 4–5, which were over 2000-fold enriched in CAV1. Additional CAV1 immunoreactive bands, possibly representing CAV1 dimers were only detected in the floating fractions. In contrast, markers from other compartments, such as the PM, Golgi apparatus, lysosomes and ER, are excluded [26] as illustrated by immunoblotting for PDI (ER marker) and transferrin receptor (PM marker).
Figure 2. Sub-proteomics of caveolin-enriched, DRM fractions.
HUVEC were fractionated by sucrose-gradient ultracentrifugation as described in the Experimental section and equal amounts of each fraction were analysed by SDS/PAGE, stained with (A) Coomassie Brilliant Blue (CBB) and immunoblotted for transferrin receptor (TfR) for PM, PDI for ER and CAV1 for caveolae. Molecular masses (in kDa) are indicated. DRM fractions 4 and 5 were further analysed by two-dimensional gel electrophoresis, followed by excision of protein spots and identification by MS as described. (B) A composite two-dimensional DRM map is depicted with protein names labelling (stretches of) identified proteins. Molecular masses and pI markers are indicated.
The floating DRM fractions were processed for an extended sub-proteomics two-dimensional analysis using first dimension (pH 4–7) linear IPG strips and second dimension SDS/10% polyacryalmide and SDS/12% polyacryalmide gels utilizing our optimized protocols [18]. Representative two-dimensional images obtained using 10 and 12% gels were combined in Adobe Photoshop for maximum molecular mass coverage and horizontal resolution of observed proteins in a single image. Compared with our initial technical analysis, we have re-analysed each protein spot for unambiguous identification by peptide sequencing (MS/MS) using current protein databases. This culminated in the confirmation of 22 proteins, additional identification of six proteins [CD109, CD13, HSP90B (heat-shock protein 90B), PTRF (polymerase I and transcript-release factor), c-Yes and RLA0], removal of one protein [VAT2 (vesicular amine transporter 2)] and corrected spot annotation for three proteins (V-ATP-B2, annexin II and flotillin-1). The combined results of DRM two-dimensional analysis and subsequent identification of proteins by MS are depicted in Figure 2(B). Several functional groups of identified proteins can be distinguished, such as structural proteins (CAV1, flotillin-1 and stomatin), GPI (glycosylphosphatidylinositol)-linked proteins (CD13, CD59, CD73, CD109 and cadherin-13), signal transduction proteins [GNB1 (guanine nucleotide-binding protein 1), GNB2, 14-3-3ϵ and c-Yes] and proteins involved in vesicular transport (SNAP-23, annexin VI and V-ATPase). Proteins that were previously uncharacterized with respect to their membrane localization or even novel to human endothelial DRM fractions include 14-3-3ϵ, cadherin-13, XRP-2, PTRF, SNAP-23, C8ORF2 and BASP1. We also identified a specific CD109 fragment, containing only the extreme C-terminal (GPI-anchored) part of the protein. Two less characterized proteins (SNAP-23 and BASP1) and a novel protein (C8ORF2) were selected for further study with two endothelial caveolae and raft proteins (CAV1 and eNOS) serving as well-characterized controls.
Subcellular distribution of proteins encountered in DRM fractions
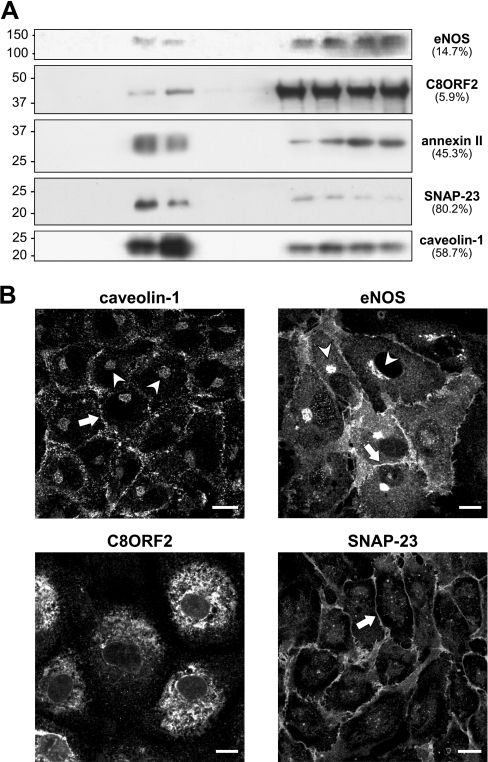
Several of the proteins identified in the DRM fractions have also been described to be present at other subcellular sites. Immunoblotting with antibodies against various identified proteins, of which a small selection is depicted in Figure 3(A), confirmed that none of these proteins are exclusive to the floating DRM fractions. In addition, the distribution of these proteins between floating and non-floating fractions is highly variable as indicated in Figure 3(A). The enrichment of SNAP-23 (80.2%) for example is much higher than for protein C8ORF2 (5.9%). Immunostaining for CAV1 and eNOS in the non-floating fractions could represent the Golgi apparatus. Visualization by confocal microscopy (Figure 3B) reveals perinuclear Golgi stain for both eNOS and CAV1 (arrowheads) in addition to the expected staining of the PM (arrows), which is also the primary location of SNAP-23 (arrow). Notably, CAV1 was predominantly detected as a punctate stain at the PM representing caveolae, in contrast with a more uniform membrane signal for eNOS and SNAP-23, indicating predominant lipid raft/PM localization. In marked contrast, the novel C8ORF2 protein seems predominantly present in the ER, with only a minor subfraction present at the cell surface (Figure 3B).
Figure 3. Distribution analysis of known and novel proteins in DRM.
(A) Equal aliquots of each sucrose-gradient fraction were separated by SDS/PAGE and immunoblotted for several known and novel identified proteins in DRM, illustrating the varying degree of relative enrichment (indicated as a percentage of total signal). (B) Confluent HUVEC were fixed and analysed by confocal microscopy to reveal the cellular distribution of each protein using specific antibodies. Indicated are the PM (arrows) and the Golgi apparatus (arrowheads). Scale bars represent 10 μm.
Cell-surface biotinylation identifies surface-exposed DRM proteins
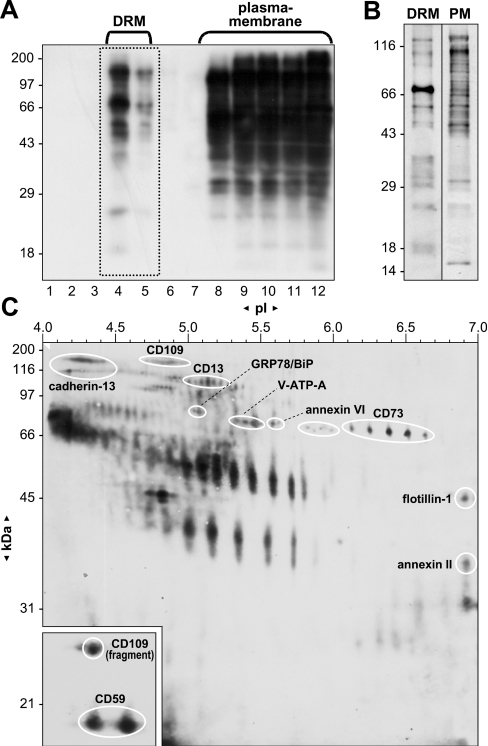
To further study subcellular localization and to characterize potential raft- and caveolae-associated proteins, our initial sub-proteome analysis was extended with the identification of surface-exposed proteins. For surface labelling of cultured cells, a non-permeant biotin derivative was used, containing a negatively charged SO4 group which prevents the reagent from passing through the cell membrane, keeping biotinylation localized at the cell surface. After surface labelling and subsequent isolation of DRM proteins, density-gradient fractions were run on SDS/PAGE, transferred to nitrocellulose followed by detection of biotin-labelled proteins using an avidin–HRP conjugate and a chemiluminescent substrate (Figure 4A). Floating fractions 4–5 potentially represent labelled and therefore surface-exposed caveolae and/or raft proteins, whereas bottom fractions 8–12 represent the remainder of labelled PM proteins. Figure 4(B) clearly shows in enhanced detail the remarkably different pattern of labelled proteins between pooled floating and non-floating DRM fractions, corrected for equal protein loading. This indicates clearly that indeed a subset of proteins is represented in the floating DRM fraction compared with the remainder of surface-exposed PM proteins. Next, two-dimensional protein maps of pooled surface-labelled DRM proteins were generated, visualizing proteins both by biotin detection and silver staining. By digitally overlaying the surface biotinylated DRM two-dimensional map (Figure 4C) with silver-stained duplicates (see Figure 2B for reference), protein patterns could be compared, leading to the identification of several proteins, with the marked exception of many highly biotinylated proteins present in small quantities. Identified proteins known to be exposed at the cell surface include the GPI-linked proteins cadherin-13, CD13, CD59, CD73 and CD109, but also the proteins annexin II and VI, GRP78/BiP (78 kDa glucose-regulated protein/immunoglobulin heavy-chain-binding protein) and V-ATPase subunit A. The identified C-terminal fragment of CD109 was also labelled. Surface labelling of flotillin-1 is unexpected but nonetheless considered genuine since other (abundant) proteins such as actin, CAV1, GNB1 and GNB2 are clearly not labelled. Additionally, a number of proteins and spot clusters visualized after biotin detection were either undetectable by silver staining or their overlapping position could not warrant unambiguous identification. From these gels, we estimate that approx. 65% of proteins contain surface-exposed elements, indicating a strong enrichment for extracellularly exposed proteins in these cholesterol- and glycosphingolipid-rich domains.
Figure 4. Identification of surface-exposed proteins in DRM by surface biotinylation.
(A) Confluent HUVEC were surface biotinylated, fractionated by sucrose-density ultracentrifugation and equal amounts of each fraction were separated by SDS/PAGE. Labelled proteins were detected using an avidin–HRP conjugate and a chemiluminescent substrate after transfer to nitrocellulose. (B) Equal protein loading and detection of fractions 8–12, representing labelled PM proteins and fractions 4–5, representing labelled DRM proteins, more clearly illustrate the discriminating labelling pattern. (C) Surface-labelled proteins from DRM fractions 4–5 (boxed in A) were further resolved by two-dimensional gel electrophoresis followed by blotting and biotin detection. Inset represents a shorter exposure time to equalize the signals due to more efficient blotting of smaller proteins. Molecular masses and pI markers are indicated. Proteins were identified by digital overlaying of a series of biotin- and silver-stained duplicate gels.
Differential localization of SNAP-23 and C8ORF2 after immunoseparation of caveolae from DRM
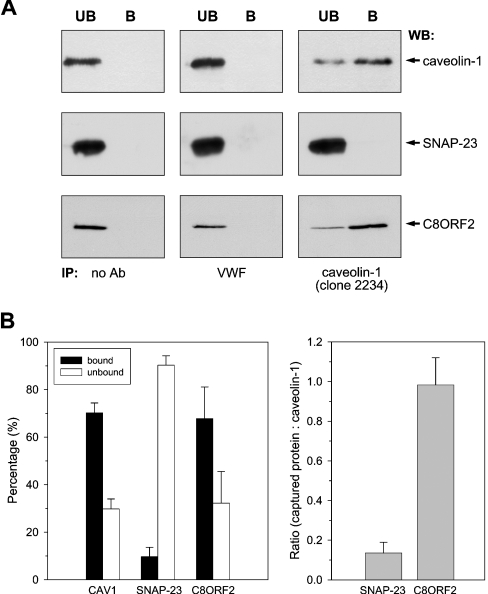
Previously it was shown that non-detergent-based methods for specific caveolae isolation, including cationic silica, yield poor separation from other membrane compartments [12,18]. Cold-detergent methods do yield highly enriched fractions, but proteomics studies so far have not discriminated between DRM and caveolae proteomes. Therefore an immunoisolation procedure was deployed, separating caveolae from DRM, to study the distribution of SNAP-23, C8ORF2 and other proteins. The CAV1 antibody clone 2234, which recognizes the cage-like structure of caveolae [27], was cross-linked to magnetic beads and used to separate caveolae from caveolin-enriched floating DRM fractions. By using this approach, approx. 75% of all CAV1 could routinely be detected in the bound (B) fraction representing caveolae (Figure 5). When using beads alone or beads with an irrelevant but endothelial-specific antibody [directed against vWF (von Willebrand factor)], all CAV1 signal remained in the unbound (UB) fractions (Figure 5A). Next, the compartmentalization of two of the identified proteins was studied, being SNAP-23, a target SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor), mediating the targeting and subsequent fusion of vesicles to the PM [28], and C8ORF2, a novel protein of unknown function, belonging to the SPFH (stomatin/prohibitin/flotillin/HflK) domain-containing family [29]. Figure 5(A) shows representative results of triplicate CAV1 immunoseparation experiments, including controls, followed by CAV1, SNAP-23 and C8ORF2 immunodetection. More than 90% of SNAP-23 remained in the unbound fractions, whereas C8ORF2 was co-immunoisolated together with CAV1. Thus, combined with the fact that approx. 80% of total SNAP-23 protein is present in enriched fractions, this potentially indicates SNAP-23 to be a raft resident protein in confluent endothelial cells. In contrast, the reproducible co-immunoisolation of C8ORF2 in similar ratios with CAV1 (Figure 5B) suggests a distinct caveolar localization for this fraction of the novel protein (6% of total cellular level).
Figure 5. Immunoseparation of caveolae from DRM and differential localization of SNAP-23 and C8ORF2.
Caveolae were immunoseparated from isolated DRM (∼25 μg) in triplicate using CAV1 mAb (clone 2234) cross-linked to magnetic beads. Bound (B) and unbound (UB) fractions were resolved by SDS/PAGE and immunoblotted for CAV1, SNAP-23 and C8ORF2. (A) Using anti-CAV1 clone 2234, approx. 75% of total CAV1 can be captured, while CAV1 remains in the unbound fraction when using either an irrelevant endothelial-specific antibody (vWF) or no antibody. Most of the SNAP-23 remains in the unbound fraction, while C8ORF2 is co-immunoisolated with CAV1. (B) Quantification of the reproducible CAV1 immunoisolation efficiency and divergent capture ratios of SNAP-23 and C8ORF2, compared with CAV1.
Caveolar proteome consists mainly of transport and structural proteins
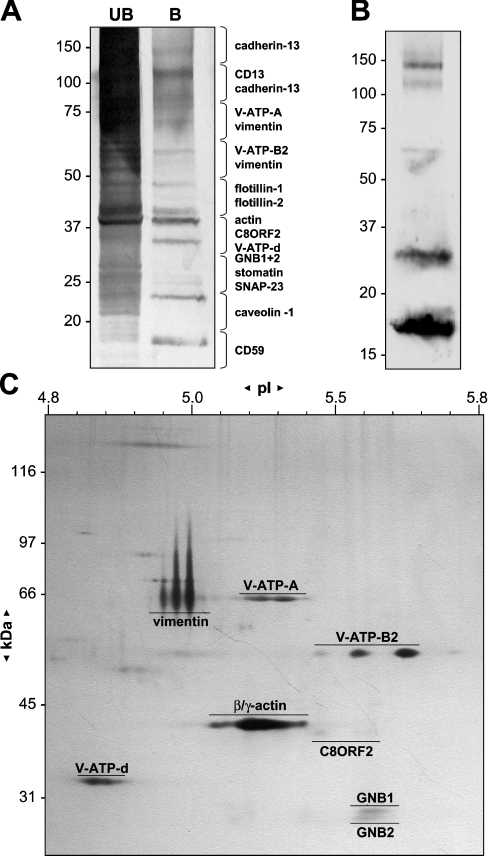
The specific caveolae immunoseparation method was further utilized to analyse the principal components of endothelial caveolae. After immunopurification of caveolae from pooled floating detergent-insoluble fractions, the bound and unbound fractions were separated by SDS/PAGE and silver-stained (Figure 6A). The gel lanes were divided into equally sized gel pieces, treated by in-gel digestion with trypsin and extracted peptides were identified by peptide mass fingerprinting and peptide sequencing using MS/MS. Figure 6(A) depicts the protein bands present in the unbound (UB) and bound (B) fractions visualized by silver-staining, together with a list of proteins identified from each gel piece. Among the most abundant proteins found roughly two categories can be distinguished. The first group constitutes structural proteins, such as CAV1, vimentin and actin. The second contains proteins involved in vesicular transport, such as components of the V-ATPase (subunits A, B2 and d) with the identified subunit B2 representing the ‘brain-specific’ isoform, as opposed to the more ubiquitous B1 isoform (not detected). Minor contaminating quantities of (potential) raft proteins, stomatin, flotillin-1 and -2, cadherin-13, CD13, GNB1, GNB2 and SNAP-23, were also found. In addition to caveolar C8ORF2, a significant amount of CD59 was found in caveolae (Figure 6A), being one of the few surface-exposed proteins in caveolae as shown by surface biotinylation preceding immunoisolation (Figure 6B). Analysis of the same bound fraction by two-dimensional gel electrophoresis, of which an enlarged fragment is shown in Figure 6(C), yields a more realistic view of average abundance of the different proteins and also reveals differently shaped and sized vimentin spots. Apparently, only transport and structural proteins are relatively abundant resident constituents of caveolae, whereas other proteins are minor or possibly transient constituents.
Figure 6. Analysis and identification of caveolae proteins by surface biotinylation, one- and two-dimensional gel electrophoresis.
(A) Caveolae were immunoseparated from DRM and bound (B) and unbound (UB) fractions were resolved by SDS/PAGE and silver-stained. The gel lanes were sliced into equally sized pieces and analysed by MS/MS. Proteins identified on the basis of MS/MS peptide sequences from the bound fraction are listed alongside the corresponding gel piece. (B) Labelled caveolae proteins as visualized by biotin detection after immunoblotting of immunoseparated proteins (bound fraction) from DRM fractions, isolated from surface-biotinylated HUVEC, clearly showing the abundant presence of CD59 (band at 18 kDa). (C) Part of a silver-stained two-dimensional gel representing captured caveolae proteins, revealing differently shaped and sized vimentin spots. Identified proteins, molecular masses and pI markers are indicated.
Spatial segregation of SNAP-23 and eNOS from caveolin in (sub)confluent cells
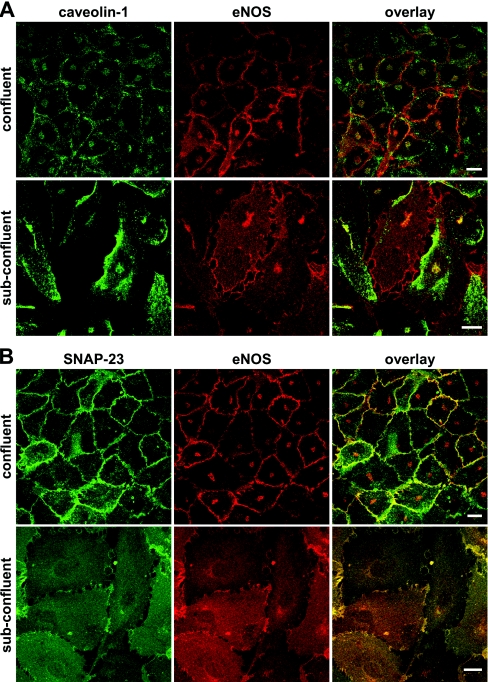
To confirm the biochemical findings, we further analysed the dynamic cellular distribution of several of the identified proteins. As caveolae number and cellular distribution seem to be strongly influenced by growth conditions and external stimuli [30], the localizations of CAV1, eNOS and SNAP-23 were compared between contact-inhibited (confluent) and proliferating (subconfluent) cells using confocal microscopy. Localization of fluorescent signals was quite comparable between CAV1 and eNOS in confluent cells, apart from the more punctate stain for CAV1 at the PM as compared with eNOS (Figure 7A). When subconfluent, however, eNOS is localized at the PM on filopodia-like extensions providing cell–cell contact, whereas an intracellular punctate CAV1 staining is detected directly beneath the PM without any significant co-localization. Co-localization of CAV1 and eNOS could only be detected at the Golgi apparatus. Whereas eNOS staining follows the contour of the cell, staining with CAV1 makes cells appear single. Co-immunostaining of eNOS and SNAP-23 (Figure 7B) however shows an almost identical staining pattern in both confluent and subconfluent cells. Although fluorescent signals for SNAP-23 show minimal Golgi staining, both eNOS and SNAP-23 are detected at the PM as a continuous stain in confluent cells and on filopodia-like extensions of the PM at cell–cell contact sites in subconfluent cells. The lack of co-localization of CAV1 with SNAP-23 and eNOS in subconfluent cells clearly indicates spatio-functional differences. Although CAV1 seems to overlap at the PM with SNAP-23 and eNOS due to poor resolution of immune fluorescence, our biochemical analysis demonstrated that this segregation is retained in confluent cells.
Figure 7. Spatial segregation of SNAP-23 and eNOS from CAV1 in (sub)confluent cells.
Confluent and subconfluent HUVEC, grown on coverslips, were fixed and subjected to confocal fluorescent microscopy using specific antibodies to (A) CAV1 and (B) SNAP-23 in comparison with eNOS. Note the clear spatial separation of CAV1 in subconfluent cells from the similar localization patterns of SNAP-23 and eNOS at the PM. Scale bars represent 10 μm.
Membrane localization and induction of filopodia by raft protein BASP1
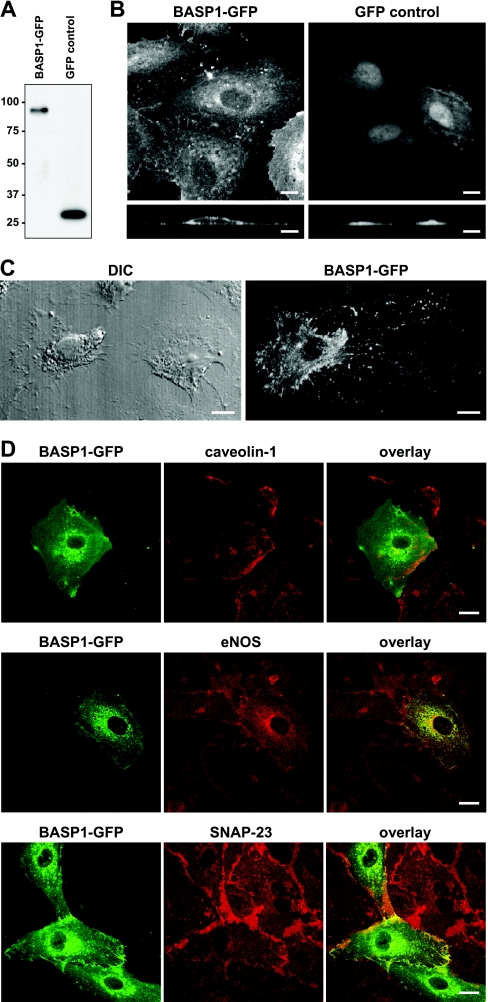
Next, we studied BASP1, being one of the less characterized proteins, identified in our endothelial DRM, but not in caveolar fractions. BASP1 is a myristoylated membrane protein originally found in neuronal axon termini forming the growth tips of elongating axons, but has also been detected in non-neural tissues [31]. Studies with the bovine and rat homologue NAP-22 (neuronal axonal myristoylated membrane protein of 22 kDa) showed the ability of this protein to initiate lipid raft formation by sequestering cholesterol and to regulate neurite outgrowth [32]. We therefore used a BASP1–GFP fusion product to study the localization and possible role of BASP1 in proliferating endothelial cells. A vector encoding BASP1–GFP was constructed and expression was visualized 24 h after transfection by confocal microscopy or anti-GFP Western blotting. Transfection with GFP alone was used as a control. Immunoblotting of BASP1–GFP cell lysates showed protein expression with the known abnormally low mobility of the protein in SDS/PAGE (Figure 8A). Although the calculated mass of the protein is 23 kDa, the native protein migrates at 50 kDa (see Figure 2A) and at 80 kDa in its GFP-hybrid form (Figure 8A). Visualization of BASP1–GFP by immunofluorescence in subconfluent cells (Figure 8B) shows predominant membrane localization, with BASP1–GFP also being present on cell–cell contact sites, but not in the nucleus, in contrast with the GFP control. Figure 8(C) shows in greater detail the fine network of numerous filopodia-like protrusions of the PM between two adjacent cells with patchy BASP1–GFP expression, combined with DIC (differential interference contrast). Next, we compared BASP1 localization relative to CAV1, eNOS and SNAP-23 using confocal microscopy. Figure 8(D) shows that staining patterns for BASP1 are virtually identical with those for SNAP-23 and eNOS, both at the PM and intracellularly, whereas again, there is a clear spatial segregation of BASP1 from CAV1 (Figure 8D), as was previously observed for both SNAP-23 and eNOS (Figure 7).
Figure 8. Co-localization of BASP1–GFP with raft proteins eNOS and SNAP-23 in endothelial cells.
HUVEC were transiently transfected with vectors encoding GFP-tagged BASP1 or GFP alone. Protein expression was examined by (A) immunoblotting of total cell lysates followed by detection with anti-GFP antibody or (B) analyses of fluorescent GFP signal by confocal microscopy. (C) Visualization of cell morphology by DIC and detection of GFP signal in sparsely seeded cells transiently expressing the BASP1–GFP construct. (D) Cellular distribution of BASP1–GFP examined by confocal laser scanning microscopy compared with eNOS, SNAP-23 and CAV1, using specific antibodies. Scale bars represent 10 μm.
DISCUSSION
Here, we describe the identification and subcellular distribution of the high to medium abundant DRM proteins in HUVEC. The specific identification of the caveolar residents by immunoisolation and immunofluorescence showed that lipid raft and caveolae proteomes are biochemically and physically separated in these confluent human endothelial cells. We show that none of these proteins appear to be exclusive to these membrane compartments, as their significant enrichment in PM DRM varies from over 80% to as little as 5% as shown by cellular distribution analysis using confocal microscopy and immunoblotting. Combining these observations, SNAP-23 could be considered a raft resident while the ER protein GRP78, whose presence in cell-surface rafts was confirmed by surface labelling, represents a minor, possibly transient component. When comparing our endothelial DRM proteome to similar published analyses from other cell types an interesting, unifying picture emerges. The major components of these compartments are the same in different cell types, with only ill-characterized differences in the minor constituents. Still, various important functions are attributed to lipid rafts and caveolae for specific cell types [23,33,34]. Whereas a substantial number of cell-specific surface-exposed PM proteins exist, as recently shown for endothelial cells [35], specialized microdomains seem to provide a general, universal scaffold to support cell-specific functions. Nevertheless, due to endothelial heterogeneity that provides specific endothelial function to different organs and vascular beds, it is likely that the detailed composition of various endothelial microdomains will display specific differences compared with our observations in HUVEC.
All of the proteins identified in our sub-proteomic analysis can be categorized as being raft or raft-associated, based on the classification by Mann and co-workers in HeLa cells [12]. We have now separated caveolae from DRM using a CAV1 immunoisolation approach that excluded over 90% of raft proteins and resulted in an 18000-fold enrichment for CAV1. Subsequent analyses of associated proteins reveals caveolae to be rather empty structures with surprisingly little protein diversity, with V-ATPase identified as the major caveolar protein in addition to CAV1, actin, an alternative molecular form of vimentin, and specific minor amounts of C8ORF2. Interestingly, in the quantitative study of Foster et al. [12], CAV1 was not classified as a true raft protein, but was unexpectedly classified as being raft-associated, based on its poor response to >95% cholesterol depletion. As the same holds true for V-ATPase and C8ORF2, this specific set of ‘raft-associated’ proteins could therefore very well represent the true residents of caveolae. The same holds true for the specific, possibly glycosylated form of vimentin we exclusively observed in caveolae (Figure 6C), but never in whole cell lysate proteomes or raft fractions (Figure 2B).
The different members of the V-ATPase complex appear to be one of the most clear and somewhat surprising residents of caveolae. V-ATPase is a multi-subunit proton pump providing vacuolar acidification, required for many cellular processes including prohormone processing, protein degradation and membrane transport [36]. Possibly, V-ATPase itself is capable of interacting with the involved signalling pathways, and certain domains of V-ATPase may directly interact with the machinery required for vesicle trafficking [39]. Dynamin was shown to mediate the fission of caveolae from the PM [37], but blocking of V-ATPase is reported to inhibit caveolae-mediated potocytosis [38], implicating this protein in the regulation of caveolar transport. Various V-ATPase subunit isoforms exist [39] of which we identified the V-ATPase B-subunit exclusively as the B2 or ‘brain’ isoform. Potentially, the existence of caveolae or endothelial subunit isoforms would allow specific modulation of V-ATPase activity, as has been demonstrated for other cell types [39,40].
We are the first to show that the PM-localized fraction (∼6%) of C8ORF2 is specifically located in caveolae, although most of this novel protein can be found in the ER. The function of C8ORF2 is unknown at present, but it contains an evolutionarily conserved SPFH domain [29], which is possibly involved in controlling signalling events, proliferation and cellular senescence. Potentially, this would therefore represent the caveolae-specific member of this family, as opposed to its well-known raft-associated family members stomatin and flotillin, and indicate a putative direct ER–caveolae route.
CD59 was the only major resident GPI-linked protein that we detected in caveolae. GPI-linked proteins are generally recognized as resident raft proteins, only entering caveolae upon activation [41,42], although it was shown that GPI-linked proteins require CAV1 and the transcytosis mechanism to reach the cell surface after leaving the Golgi [43,44]. Functional characterization of CD59 has focused on its inhibitory role in the complement activation system, its role in T-cell receptor activation [45] and downstream tyrosine kinase activity. Recently, CD59 was implicated in EGF (epidermal growth factor) receptor signalling, possibly involving endocytosis [46]. Further functional studies will be necessary to uncover the role of CD59 in caveolae-related processes such as transport and signalling.
Confocal microscopy revealed a complete spatial segregation between CAV1 and the potential raft residents SNAP-23, eNOS and BASP1, especially in subconfluent cells. Overexpressed BASP1 not only co-localized with SNAP-23 and eNOS, and segregated from CAV1, but also seemed to be involved in the formation of cellular contact. Besides the potential raft organizational properties of BASP1 and its involvement in signalling as inferred from the bovine and rat homologue NAP-22 [32], the observed spatial segregation in proliferating cells most clearly reflects the functional separation between caveolae and lipid rafts as first proposed by Schnitzer et al. [42]. Caveolae and CAV1 are important regulators of eNOS signalling in endothelial cells where NO is essential for VEGF (vascular endothelial growth factor)-driven angiogenesis, vascular permeability and tumour growth [23,47], which seems to require spatial uncoupling of rafts and caveolae in proliferating cells.
In unstimulated HUVEC, caveolae appear relatively empty transcytosis vesicles, containing primarily structural proteins and vesicular transport components. Furthermore, the primary scaffold of endothelial raft resident proteins of medium to high abundance seems to lack cell specificity. Still, many reports have reported the presence of various receptors and other proteins in these compartments using highly sensitive detection by Western blotting. Recent reports have elaborated the notion that caveolae are not signalling centres but rather function to promote rapid receptor turnover and attenuation of signalling by combining scavenging of activated components with specific endocytotic events [48–50]. The concept of caveolae being transport vesicles capable of fine-tuning signalling is in line with results obtained from CAV1-knockout mouse studies, which lack a substantial phenotype until challenged [10]. This supports the emerging view of signalling requiring precise spatial and temporal regulation to evoke a unique biological response. The evidence we present for the spatial segregation of transport and signalling functions between caveolae and raft proteomes in HUVEC underlines the importance of a balanced raft–caveola interaction in signal transduction, membrane trafficking and growth regulation.
Acknowledgments
We thank H. van Veen (Department of Cell Biology, University of Amsterdam) for technical assistance with electron microscopy, H. Dekker (Mass Spectrometry Group, University of Amsterdam) for valuable help with peptide sequencing and D. Speijer (Department of Medical Biochemistry, University of Amsterdam) for technical support and corrections of this paper. This study was supported by Research Support grant 902-26-201 to A. J. G. H. from ZonMW (The Hague, The Netherlands) and Molecular Cardiology Program grant M93.007 from the Netherlands Heart Foundation (The Hague, The Netherlands).
References
- 1.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Simons K., Toomre D. Lipid rafts and signal transduction. Nat. Rev. Mol. Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 3.Parton R. G. Caveolae-from ultrastructure to molecular mechanisms. Nat. Rev. Mol. Cell Biol. 2003;4:162–167. doi: 10.1038/nrm1017. [DOI] [PubMed] [Google Scholar]
- 4.Mañes S., del Real G., Martínez-A C. Pathogens: raft hijackers. Nat. Rev. Immunol. 2003;3:557–568. doi: 10.1038/nri1129. [DOI] [PubMed] [Google Scholar]
- 5.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 6.Razani B., Woodman S. E., Lisanti M. P. Caveolae: from cell biology to animal physiology. Pharmacol. Rev. 2002;54:431–467. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 7.Frank P. G., Woodman S. E., Park D. S., Lisanti M. P. Caveolin, caveolae, and endothelial cell function. Arterioscler. Thromb. Vasc. Biol. 2003;23:1161–1168. doi: 10.1161/01.ATV.0000070546.16946.3A. [DOI] [PubMed] [Google Scholar]
- 8.Drab M., Verkade P., Elger M., Kasper M., Lohn M., Lauterbach B., Menne J., Lindschau C., Mende F., Luft F. C., et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452. doi: 10.1126/science.1062688. [DOI] [PubMed] [Google Scholar]
- 9.Razani B., Engelman J. A., Wang X. B., Schubert W., Zhang X. L., Marks C. B., Macaluso F., Russell R. G., Li M., Pestell R. G., et al. Caveolin-1 null mice are viable but show evidence of hyperproliferative and vascular abnormalities. J. Biol. Chem. 2001;276:38121–38138. doi: 10.1074/jbc.M105408200. [DOI] [PubMed] [Google Scholar]
- 10.Cohen A. W., Hnasko R., Schubert W., Lisanti M. P. Role of caveolae and caveolins in health and disease. Physiol. Rev. 2004;84:1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 11.Blonder J., Hale M. L., Lucas D. A., Schaefer C. F., Yu L. R., Conrads T. P., Issaq H. J., Stiles B. G., Veenstra T. D. Proteomic analysis of detergent-resistant membrane rafts. Electrophoresis. 2004;25:1307–1318. doi: 10.1002/elps.200405891. [DOI] [PubMed] [Google Scholar]
- 12.Foster L. J., De Hoog C. L., Mann M. Unbiased quantitative proteomics of lipid rafts reveals high specificity for signaling factors. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li N., Shaw A. R., Zhang N., Mak A., Li L. Lipid raft proteomics: analysis of in-solution digest of sodium dodecyl sulfate-solubilized lipid raft proteins by liquid chromatography-matrix-assisted laser desorption/ionization tandem mass spectrometry. Proteomics. 2004;4:3156–3166. doi: 10.1002/pmic.200400832. [DOI] [PubMed] [Google Scholar]
- 14.Shogomori H., Brown D. A. Use of detergents to study membrane rafts: the good, the bad, and the ugly. Biol. Chem. 2003;384:1259–1263. doi: 10.1515/BC.2003.139. [DOI] [PubMed] [Google Scholar]
- 15.Raimondo F., Ceppi P., Guidi K., Masserini M., Foletti C., Pitto M. Proteomics of plasma membrane microdomains. Expert Rev. Proteomics. 2005;2:793–807. doi: 10.1586/14789450.2.5.793. [DOI] [PubMed] [Google Scholar]
- 16.Lichtenberg D., Goni F. M., Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sprenger R. R., Speijer D., Back J. W., De Koster C. G., Pannekoek H., Horrevoets A. J. Comparative proteomics of human endothelial cell caveolae and rafts using two-dimensional gel electrophoresis and mass spectrometry. Electrophoresis. 2004;25:156–172. doi: 10.1002/elps.200305675. [DOI] [PubMed] [Google Scholar]
- 19.McMahon K. A., Zhu M., Kwon S. W., Liu P., Zhao Y., Anderson R. G. Detergent-free caveolae proteome suggests an interaction with ER and mitochondria. Proteomics. 2005;6:143–152. doi: 10.1002/pmic.200500208. [DOI] [PubMed] [Google Scholar]
- 20.Jaffe E. A., Nachman R. L., Becker C. G., Minick C. R. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 1973;52:2745–2756. doi: 10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown D. A., Rose J. K. Sorting of GPI-anchored proteins to glyolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 23.Gratton J. P., Bernatchez P., Sessa W. C. Caveolae and caveolins in the cardiovascular system. Circ. Res. 2004;94:1408–1417. doi: 10.1161/01.RES.0000129178.56294.17. [DOI] [PubMed] [Google Scholar]
- 24.D'Alessio A., Al-Lamki R. S., Bradley J. R., Pober J. S. Caveolae participate in tumor necrosis factor receptor 1 signaling and internalization in a human endothelial cell line. Am. J. Pathol. 2005;166:1273–1282. doi: 10.1016/S0002-9440(10)62346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee W. C., Chao W. T., Yang V. C. Effects of high-cholesterol diet on the interendothelial clefts and the associated junctional complexes in rat aorta. Atherosclerosis. 2001;155:307–312. doi: 10.1016/s0021-9150(00)00578-5. [DOI] [PubMed] [Google Scholar]
- 26.Sargiocomo M., Sudol M., Tang Z., Lisanti M. P. Signal transduction molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J. Cell Biol. 1993;122:789–808. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh P., Schnitzer J. E. Immunoisolation of caveolae with high affinity antibody binding to the oligomeric caveolin cage. Toward understanding the basis of purification. J. Biol. Chem. 1999;274:23144–23154. doi: 10.1074/jbc.274.33.23144. [DOI] [PubMed] [Google Scholar]
- 28.Salaün C., James D. J., Greaves J., Chamberlain L. H. Plasma membrane targeting of exocytic SNARE proteins. Biochim. Biophys. Acta. 2004;1693:81–99. doi: 10.1016/j.bbamcr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 29.Tavernarakis N., Driscoll M., Kyrpides N. C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 30.Navarro A., Anand-Apte B., Parat M. O. A role for caveolae in cell migration. FASEB J. 2004;18:1801–1811. doi: 10.1096/fj.04-2516rev. [DOI] [PubMed] [Google Scholar]
- 31.Mosevitsky M. I., Capony J. P., Skladchikova G. Yu., Novitskaya V. A., Plekhanov A. Yu., Zakharov V. V. The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physico-chemical properties. Biochimie. 1997;79:373–384. doi: 10.1016/s0300-9084(97)80032-6. [DOI] [PubMed] [Google Scholar]
- 32.Epand R. M., Maekawa S., Yip C. M., Epand R. F. Protein-induced formation of cholesterol-rich domains. Biochemistry. 2001;40:10514–10521. doi: 10.1021/bi010897s. [DOI] [PubMed] [Google Scholar]
- 33.Razzaq T. M., Ozegbe P., Jury E. C., Sembi P., Blackwell N. M., Kabouridis P. S. Regulation of T-cell receptor signalling by membrane microdomains. Immunology. 2004;113:413–426. doi: 10.1111/j.1365-2567.2004.01998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maekawa S., Iino S., Miyata S. Molecular characterization of the detergent-insoluble cholesterol-rich membrane microdomain (raft) of the central nervous system. Biochim. Biophys. Acta. 2003;1610:261–270. doi: 10.1016/s0005-2736(03)00023-3. [DOI] [PubMed] [Google Scholar]
- 35.Durr E., Yu J., Krasinska K. M., Carver L. A., Yates J. R., Testa J. E., Oh P., Schnitzer J. E. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat. Biotechnol. 2004;22:985–992. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
- 36.Sun-Wada G. H., Wada Y., Futai M. Diverse and essential roles of mammalian vacuolar-type proton pump ATPase: toward the physiological understanding of inside acidic compartments. Biochim. Biophys. Acta. 2004;1658:106–114. doi: 10.1016/j.bbabio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Henley J. R., Krueger E. W., Oswald B. J., McNiven M. A. Dynamin-mediated internalization of caveolae. J. Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mineo C., Anderson R. G. A vacuolar-type proton ATPase mediates acidification of plasmalemmal vesicles during potocytosis. Exp. Cell Res. 1996;224:237–242. doi: 10.1006/excr.1996.0133. [DOI] [PubMed] [Google Scholar]
- 39.Sun-Wada G. H., Wada Y., Futai M. Lysosome and lysosome-related organelles responsible for specialized functions in higher organisms, with special emphasis on vacuolar-type proton ATPase. Cell Struct. Funct. 2003;28:455–463. doi: 10.1247/csf.28.455. [DOI] [PubMed] [Google Scholar]
- 40.Visentin L., Dodds R. A., Valente M., Misiano P., Bradbeer J. N., Oneta S., Liang X., Gowen M., Farina C. A selective inhibitor of the osteoclastic V-H(+)-ATPase prevents bone loss in both thyroparathyroidectomized and ovariectomized rats. J. Clin. Invest. 2000;106:309–318. doi: 10.1172/JCI6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayor S., Rothberg K. G., Maxfield F. R. Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking. Science. 1994;264:1948–1951. doi: 10.1126/science.7516582. [DOI] [PubMed] [Google Scholar]
- 42.Schnitzer J. E., McIntosh D. P., Dvorak A. M., Liu J., Oh P. Separation of caveolae from associated microdomains of GPI-anchored proteins. Science. 1995;269:1435–1439. doi: 10.1126/science.7660128. [DOI] [PubMed] [Google Scholar]
- 43.Sotgia F., Razani B., Bonuccelli G., Schubert W., Battista M., Lee H., Capozza F., Schubert A. L., Minetti C., Buckley J. T., et al. Intracellular retention of glycosylphosphatidyl inositol-linked proteins in caveolin-deficient cells. Mol. Cell. Biol. 2002;22:3905–3926. doi: 10.1128/MCB.22.11.3905-3926.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polishchuk R., Di Pentima A., Lippincott-Schwartz J. Delivery of raft-associated, GPI-anchored proteins to the apical surface of polarized MDCK cells by a transcytotic pathway. Nat. Cell Biol. 2004;6:297–307. doi: 10.1038/ncb1109. [DOI] [PubMed] [Google Scholar]
- 45.Korty P. E., Brando C., Shevach E. M. CD59 functions as a signal-transducing molecule for human T cell activation. J. Immunol. 1991;146:4092–4098. [PubMed] [Google Scholar]
- 46.Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. A proteomics strategy to elucidate functional protein–protein interactions applied to EGF signaling. Nat. Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 47.Bauer P. M., Yu J., Chen Y., Hickey R., Bernatchez P. N., Looft-Wilson R., Huang Y., Giordano F., Stan R. V., Sessa W. C. Endothelial-specific expression of caveolin-1 impairs microvascular permeability and angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 2005;102:204–209. doi: 10.1073/pnas.0406092102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matveev S. V., Smart E. J. Heterologous desensitization of EGF receptors and PDGF receptors by sequestration in caveolae. Am. J. Physiol. Cell Physiol. 2002;282:C935–C946. doi: 10.1152/ajpcell.00349.2001. [DOI] [PubMed] [Google Scholar]
- 49.Legler D. F., Micheau O., Doucey M. A., Tschopp J., Bron C. Recruitment of TNF receptor 1 to lipid rafts is essential for TNFα-mediated NF-kappaB activation. Immunity. 2003;18:655–664. doi: 10.1016/s1074-7613(03)00092-x. [DOI] [PubMed] [Google Scholar]
- 50.Di Guglielmo G. M., Le Roy C., Goodfellow A. F., Wrana J. L. Distinct endocytic pathways regulate TGF-beta receptor signalling and turnover. Nat. Cell Biol. 2003;5:410–421. doi: 10.1038/ncb975. [DOI] [PubMed] [Google Scholar]