Abstract
We have purified to homogeneity two forms of a new serine protease inhibitor specific for elastase/chymotrypsin from the ovary gland of the desert locust Schistocerca gregaria. This protein, greglin, has 83 amino acid residues and bears putative phosphorylation sites. Amino acid sequence alignments revealed no homology with pacifastin insect inhibitors and only a distant relationship with Kazal-type inhibitors. This was confirmed by computer-based structural studies. The most closely related homologue is a putative gene product from Ciona intestinalis with which it shares 38% sequence homology. Greglin is a fast-acting and tight binding inhibitor of human neutrophil elastase (kass=1.2×107 M−1·s−1, Ki=3.6 nM) and subtilisin. It also binds neutrophil cathepsin G, pancreatic elastase and chymotrypsin with a lower affinity (26 nM≤Ki≤153 nM), but does not inhibit neutrophil protease 3 or pancreatic trypsin. The capacity of greglin to inhibit neutrophil elastase was not significantly affected by exposure to acetonitrile, high temperature (90 °C), low or high pH (2.5–11.0), N-chlorosuccinimide-mediated oxidation or the proteolytic enzymes trypsin, papain and pseudolysin from Pseudomonas aeruginosa. Greglin efficiently inhibits the neutrophil elastase activity of sputum supernatants from cystic fibrosis patients. Its biological function in the locust ovary gland is currently unknown, but its physicochemical properties suggest that it can be used as a template to design a new generation of highly resistant elastase inhibitors for treating inflammatory diseases.
Keywords: greglin, Kazal inhibitor, neutrophil elastase, pacifastin, Schistocerca gregaria, serine protease
Abbreviations: Abz-APEEIMRRQ-EDDnp, O-aminobenzoyl-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-ethylenediamine 2,4-dinitrophenyl; HNE, human neutrophil elastase; OMTKY-III, turkey ovomucoid trypsin inhibitor third domain; α1-Pi, human α1-protease inhibitor; MeOsuc-(Ala)2-Pro-Ala-TBE, methoxysuccinyl-Ala-Ala-Pro-Ala-thiobenzyl ester; PPE, porcine pancreatic elastase; RP-HPLC, reverse-phase HPLC; SLPI, human secretory leucocyte protease inhibitor; Suc-(Ala)3-p-NA, succinyl-Ala-Ala-Ala-p-nitroanilide
INTRODUCTION
Neutrophil elastase is thought to be the major protease involved in tissue destruction during inflammation. This is especially true for lung diseases such as cystic fibrosis, COPD (chronic obstructive pulmonary disease) and ARDS (acute respiratory distress syndrome), which are accompanied by considerable recruitment of neutrophils. Anti-inflammatory therapy using protease inhibitors may be an efficient means of reducing the unopposed proteolytic activity [e.g. HNE (human neutrophil elastase) activity] that occurs when neutrophils invade inflammatory sites (reviewed in [1]). A number of studies using natural and recombinant inhibitors have had mixed results due to ineffective administration, or the inactivation of the inhibitor by mechanical or physicochemical constraints in the local environment [1]. There is thus a need for molecular structures that possess the physicochemical and mechanical properties required for use in vivo.
At least 2500 sequences homologous with those of known protease inhibitors can be retrieved from data banks, and assigned to one of 48 families [2]. The classical inhibitors of HNE and other neutrophil proteases are members of families I1 (Kazal), I2, I3, (Kunitz), I4 (serpin) and I17 (elafin). The most prominent physiological inhibitor of HNE is α1-Pi (human α1-protease inhibitor; I4 family), the recombinant form of which has been given by inhalation or intravenously to treat cystic fibrosis and α1-Pi deficiency. Several other inhibitors, including MNEI (monocyte and neutrophil elastase inhibitor), SLPI (human secretory leucocyte protease inhibitor) and elafin/pre-elafin, have also been given as aerosols or systemically to inhibit neutrophil elastase. Most of these natural inhibitors are sensitive to oxidation and/or proteolytic degradation by other proteases that are present at inflammatory sites [1]. Recombinant protease inhibitors based on the sequences of natural endogenous inhibitors have been developed to overcome some of these drawbacks and a few have proved to have beneficial anti-inflammatory properties [3].
Synthetic low Mr elastase inhibitors have also been developed, but their side effects and limited efficacy are barriers to further work on them [1].
Insects have many more serine proteases than do humans, but they have relatively few serine protease inhibitors [4,5]. A new family of low Mr protease inhibitors, the pacifastins, was found previously in arthropods [6]. They all have six cysteine residues arranged in a conserved disulfide pattern [6]. Most of them preferentially inhibit chymotrypsin and trypsin. Five pacifastin inhibitors have been purified from the ovary of the desert locust Schistocerca gregaria [7]. We have now isolated and purified to homogeneity two forms of a novel inhibitor of higher Mr, greglin, from the same material. The protein is a powerful neutrophil elastase inhibitor that also inhibits pancreatic elastase and chymotrypsin-like proteases.
We used computational tools to look for sequence homologies with other proteinaceous inhibitors and to predict the secondary and tertiary structures of greglin.
EXPERIMENTAL
Materials
S. gregaria ovaries were obtained as previously described [7,8]. PPE (porcine pancreatic elastase; EC 3.4.21.36), HNE (EC 3.4.21.37), proteinase 3 (EC 3.4.21.76), cathepsin G (EC 3.4.21.20) and human α1-Pi were purified [9,10] or obtained from Athens Research and Technology (Athens, GA, U.S.A.). Porcine pancreatic trypsin (EC 3.4.21.4) and subtilisin Carlsberg (EC 3.4.21.62) were obtained from Sigma (St Quentin Fallavier, France). Bovine pancreatic chymotrypsin (EC 3.4.21.1) was purchased from Worthington (Lakewood, NJ, U.S.A.) and papain (EC 3.4.22.2) from Roche Diagnostics (Meylan, France). The active sites of all the above proteases were titrated using published methods [11,12–14]. The concentrations given here refer to active protein concentrations. Pseudolysin (EC 3.4.24.26) and endoprotease Glu-C (EC 3.4.21.19) were from Calbiochem (VWR, Strasbourg, France) and Sigma respectively. Suc-(Ala)3-p-NA (succinyl-Ala-Ala-Ala-p-nitroanilide), MeOsuc-(Ala)2-Pro-Ala-TBE (methoxysuccinyl-Ala-Ala-Pro-Ala-thiobenzyl ester), BAPNA (benzoyl-Arg-p-nitroanilide), Suc-(Ala)2-Pro-Phe-p-NA and MeOsuc-Lys-(pico)-Ala-Pro-Val-p-NA [methoxysuccinyl-Lys(2-picolinoyl)-Ala-Pro-Val-p-nitroanilide], the chromogenic substrates of the various proteases, were purchased from Bachem (Bubendorf, Switzerland). The HNE fluorogenic substrate Abz-APEEIMRRQ-EDDnp (O-aminobenzoyl-Ala-Pro-Glu-Glu-Ile-Met-Arg-Arg-Gln-ethylenediamine 2,4-dinitrophenyl) was used as described previously [15].
Sputum samples were collected from adult cystic fibrosis patients after informed consent. Approval was obtained from the Ethics Committee of our Institution.
Purification of greglin
The detailed purification procedure can be found in the Supplementary material section at http://www.BiochemJ.org/bj/400/bj4000467add.htm. Briefly, a homogenate of 150 S. gregaria ovaries was precipitated at between 50 and 80% solid ammonium sulfate and the resulting pellet suspended and fractionated by gel filtration. The collected fractions were checked for their anti-elastase activity using Suc-(Ala)3-p-NA as a substrate. The elastase-inhibiting fractions were pooled and chromatographed on an anion-exchange column. The bound material was eluted with a 0–300 mM gradient of NaCl and the fractions containing inhibitor detected as above and further chromatographed on a Mono Q HR 5/5 column. The two major peaks were further purified by chromatography on the same column and their purity checked by SDS gel (12%) electrophoresis and RP-HPLC (reverse-phase HPLC) using a C4 Brownlee cartridge.
Kinetic measurements
Experimental details related to the measurements of enzymes activities on chromogenic and fluorogenic substrates are given in the Supplementary material section.
Determination of active inhibitor concentrations
The active concentration of each inhibitor was determined with active-site-titrated HNE. Increasing quantities of inhibitor were incubated at 25 °C with 0.38 μM enzyme in 990 μl of reaction mixture for 10 min. The remaining enzyme activities were measured by adding 10 μl of 50 mM Suc-(Ala)3-p-NA. Under these conditions the decrease in HNE activity as a function of both inhibitor concentrations was linear up to approx. 90% inhibition. We used the linear portion of the inhibition curve to deduce the active concentration of both inhibitory species, assuming a 1:1 binding stoichiometry.
Determination of Ki, the equilibrium dissociation constant, for the complexes formed between greglin and various proteases
Constant amounts of a given protease were reacted with increasing amounts of inhibitor in 990 μl of buffered reaction mixture for 20 min. The remaining enzymic activities were measured by adding 10 μl of appropriate substrate solution [16]. Further details are given in the Supplementary material section.
Determination of kass and kdiss, the association and dissociation rate constants for the interaction of HNE with greglin
The association and dissociation kinetics for the reaction of HNE with greglin 1 and 2 were investigated by the progress curve method [16] using a stopped-flow apparatus (SFM3, Bio-Logic, Claix, France) [16,17]. Data acquisition and processing were done with the manufacturer's BioKine software.
Assessment of the residual greglin inhibitory activity after incubation with non-target proteolytic enzymes
HNE (2.8 nM) was incubated with 14 nM native or protease-treated inhibitor for 10 min at 37 °C in 212 μl of buffer [50 mM Hepes, pH 7.4, 150 mM NaCl and 0.05% (v/v) Igepal CA-630]. The residual enzyme activity was measured by adding 2 μl of 0.2 mM Abz-APEEIMRRQ-EDDnp.
Inhibition by greglin of the HNE activity in cystic fibrosis sputum supernatant
The sputum samples collected from adult cystic fibrosis patients were homogenized in 10 mM PBS (pH 7.4) (3 ml/g of sputum) and centrifuged at 10000 g for 10 min at 4 °C. The concentrations of active HNE in these samples were deduced by comparing their enzyme activities with that of pure, active-site-titrated HNE using Abz-APEEIMRRQ-EDDnp (1.33 μM final concentration). Buffered reaction mixtures containing greglin (3–36 nM final concentration) were incubated with samples of sputum supernatant containing HNE (30 nM final concentration) for 10 min at 37 °C. Residual HNE activity was measured spectrofluorimetrically using Abz-APEEIMRRQ-EDDnp as a substrate.
Determination of the primary structure of greglin
The complete amino acid sequences of the two reduced and alkylated greglin isoforms were determined by automated N-terminal sequencing of the purified inhibitors and the enzymatically and chemically cleaved forms using an Applied Biosystems Procise pulsed liquid sequencer with the chemicals and program recommended by the manufacturer. Inhibitor (1 nmol) was reduced with dithiothreitol and alkylated with 2 μl of 4-vinyl pyridine (see the Supplementary material section). Samples were desalted by RP-HPLC and incubated (4.6 nM final concentration) with trypsin (2.0 nM final) or chymotrypsin (1.5 nM final concentration) in appropriate buffers. The products were separated by RP-HPLC, freeze-dried and sequenced.
The reduced, alkylated inhibitors (1.5 nM final concentration) were also incubated in the dark at 20 °C for 18 h with 75 mg/ml cyanogen bromide in 70% (v/v) formic acid. The products were separated by RP-HPLC, freeze-dried and sequenced.
C-terminal sequences were determined by MS using a Bruker BIFLEX IIIÔ mass spectrometer (Bremen, Germany) in linear positive-ion mode. Samples were prepared by the sandwich method [18]. Purified peptides were incubated at 25 °C for 3 h with 10 ng/μl carboxypeptidase P (Sigma) in 10 μl of 50 mM sodium citrate buffer (pH 4.0).
Prediction of the secondary and tertiary structures of greglin
Sequence analysis tools
The BLAST (http://www.ebi.ac.uk/blastall/) and MEROPS (a protease database; http://merops.sanger.ac.uk/) suites of programs were used to look for homologies in the sequence databases. We used T-coffee (http://igs-server.cnrs-mrs.fr/Tcoffee/tcoffee_cgi/index.cgi) to generate multiple sequence alignments, and NetPhos (http://www.cbs.dtu.dk/services/NetPhos/) to predict phosphorylation sites.
Secondary structure prediction
Seconday structural elements were predicted using sspro [19], nnpredict [20], psipred, sam and jufo through the Robetta server [21] and sable and profsec through the GeneSilico metaserver (http://genesilico.pl/meta).
These programs were selected to cover the whole range of methods (neural network, hidden Markov chain, position specific and profile matrices, etc.) and parameters (solvent representation, amino acid properties, etc.) that are currently available.
Tertiary structure prediction
To predict the tertiary structure of greglin, we used the automated GeneSilico metaserver (http://genesilico.pl/meta) that uses results from the inbgu, 3dpssm, ffas, mgenthreader, sam, sparks, fugues and 3dpssm servers. The Robetta server was used to build the structure of greglin starting from its primary sequence alone [21].
SwissPdbViewer (http://au.expasy.org/spdbv/) was used to generate multiple structural alignments and to superimpose the models obtained.
Greglin physicochemical properties
Oxidation by N-chlorosuccinimide
Greglin (13 μM final concentration) was incubated at room temperature with N-chlorosuccinimide (125 μM final concentration) in 50 mM Hepes buffer (pH 7.4), 150 mM NaCl and 0.05% Igepal CA-630 [(octylphenoxy)polyethoxyethanol] for 20 min. Excess oxidant was removed by RP-HPLC on a C4 column and oxidation was checked by MS using a M@LDI L/R Waters micromass apparatus. The anti-PPE activity of the desalted inhibitor was assayed as above.
Effect of pH and temperature on inhibitor activity
Greglin was incubated for 20 h at 37 °C in 0.5 M glycine/HCl buffer (pH 2.5); 50 mM Hepes buffer (pH 7.4); and in 0.5 M Tris-base (pH 11.0), and its residual inhibitory activity assayed in the Hepes buffer. The effect of temperature was investigated by incubating greglin (3.85 μM) in Hepes buffer (pH 7.4) for 1 h at temperatures from 37 to 90 °C and then measuring its residual inhibitory activity at 37 °C.
Reaction of greglin with PPE and HNE under acidic conditions
The greglin reactive bond (P1–P1′) was tentatively identified by incubating the inhibitor with target enzymes at low pH and looking for putative cleavage sites by N-terminal sequencing, as described for soya-bean trypsin inhibitor [22]. Experimental details are provided in the Supplementary material section.
Susceptibility of greglin activity to proteolytic enzymes
The susceptibility of greglin anti-elastase activity was investigated by incubating the inhibitor with serine (trypsin, endoprotease Glu-C), cysteine (papain) and metallo (pseudolysin) proteases. The mixtures were then separated by RP-HPLC and the inhibitory properties of the eluted peaks analysed (see the Supplementary material section).
RESULTS
Purification of two greglin isoforms from a crude homogenate of S. gregaria ovaries
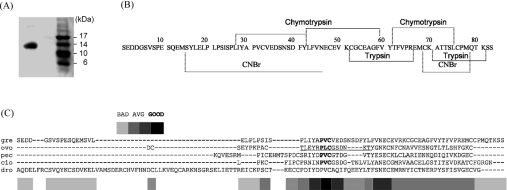
We isolated two forms of an anti-PPE protein referred to as ‘greglin’ from the S. gregaria ovaries using a combination of salt precipitation, size-exclusion and anion-exchange chromatographies. The inhibitory activity of greglin was used to monitor its presence along the purification procedure. The protein was eluted from the Mono Q column as two close peaks that were further purified by RP-HPLC on a Brownlee C4 column for sequence analysis, and gave a single band when run on an SDS gel (Figure 1A). A homogenate of 150 ovaries gave approx. 4 mg of purified greglin.
Figure 1. Purification of greglin.
(A) SDS gel electrophoresis of purified greglin after ion-exchange chromatography. (B) Complete amino acid sequence of greglin obtained from N-terminal sequencing of overlapping peptides produced by chemical and proteolytic cleavages followed by RP-HPLC separation of each greglin isoform after reduction and alkylation. Proteolytically cleaved and chemically cleaved fragments are underlined. (C) Alignment of the sequences of greglin (gre) and Kazal-related homologues calculated using T-coffee [45] [OMTKY-III (ovo), porcine PEC-60 (pec), C. intestinalis (cio) and D. melanogaster (dro)]. The inhibitory site of OMTKY-III (P4 to P3′) is underlined. The scores resulting from the multiple sequence analysis are identified as bad, average (AVG) and good.
Primary structure
The sequence of each greglin isoform was obtained by sequencing overlapping peptides produced by cleaving the reduced alkylated RP-HPLC-purified proteins with cyanogen bromide (Met14 and Met68), trypsin (Lys52, Lys70 and Lys81) and chymotrypsin (Leu27, Tyr42 and Tyr6) (Figure 1B). Both sequences contained 83 identical amino acid residues except that one sequence had no identifiable residues at positions 8, 11 and 15, whereas the other had serine residues at these positions. This strongly suggests that the serine residues in the first sequence are glycosylated or phosphorylated. The Mr calculated from the greglin sequence was 9229, while MS gave a minimal Mr of 9600 and a maximal Mr of 9844, consistent with a triphosphorylation. Intermediate masses consistent with mono- and di-phosphorylation were also recorded. The Net Phos program of the Expasy server predicted the phosphorylation of the serine residues at positions 6, 8, 11 and 15 and 37 in agreement with sequence data that found no serine residues at positions 8, 11 and 15 in one isoform, i.e. those with the best predictive scores. But there may well be additional post-translational modifications that could explain the differences between the mass deduced from amino acid analysis and those obtained experimentally. We identified six cysteine residues unambiguously by N-terminal sequencing after 4-vinyl-pyridine alkylation. The C-terminal sequence was confirmed by carboxypeptidase P digestion plus MS analyses of the products. The data indicated a decrease of 303 in the Mr, consistent with the KSS C-terminal sequence. In addition, cleavage of the native inhibitor with trypsin gave a molecule of lower Mr (−175), in agreement with the release of the two C-terminal serine residues. Incubation of the trypsin-reacted products with carboxypeptidase P reduced the Mr by a further 128, confirming the presence of a lysine residue at position 81.
We found no significant relationship between the whole sequence of greglin and any of the protease inhibitors searching the MEROPS database [23]. However, using only part of the sequence (residues Pro26 to Cys69), the program retrieved two putative inhibitors from Ciona intestinalis (O97362) and from Drosophila melanogaster (Q9VE57) which share 38 and 36% homology respectively with this part of the greglin sequence.
We tentatively aligned the greglin sequence with these two peptides and with OMTKY-III (turkey ovomucoid trypsin inhibitor third domain), a typical Kazal inhibitor [24], and with the Kazal-related peptide PEC-60 [25], identified by fold recognition methods as the closest structural homologue to greglin (see the next section). The best scores were obtained with the C. intestinalis and D. melanogaster peptides; these were the only two that gave significant E values (Table 1). However, only three of six cysteine residues in greglin aligned with those in the Kazal-related molecules (Figure 1C). This suggests a different disulfide bridge pattern, but we failed to identify this pattern due to the resistance of greglin to proteolysis. We then used computer-based structural studies to tentatively assign greglin to a family of peptidase inhibitors.
Table 1. Sequence alignments of greglin residues Pro26 to Cys69 with Kazal-related inhibitors from C. intestinalis, D. melanogaster, pig PEC-60 and OMTKY-III using the LALIGN program from Fasta.
Homologous proteins were taken to be those for which the aligned amino acid sequences were related with an E value of 0.001 or less. Seqences were from Swiss-Prot, with ID numbers as follows: O97362, C. intestinalis; Q9VE57, D. melanogaster; P37109, pig PEC-60; and P68390, OMTKY-III.
| Sequence comparison | Alignment (%) | E value |
|---|---|---|
| Greglin–O97362 | 36 | 0.0005 |
| Greglin–Q9VE57 | 33 | 0.00091 |
| O97362–Q9VE57 | 27 | 0.014 |
| Greglin–P37109 | 34 | 1.7 |
| Greglin–P68390 | 27 | 1.400 |
Secondary and tertiary structure predictions
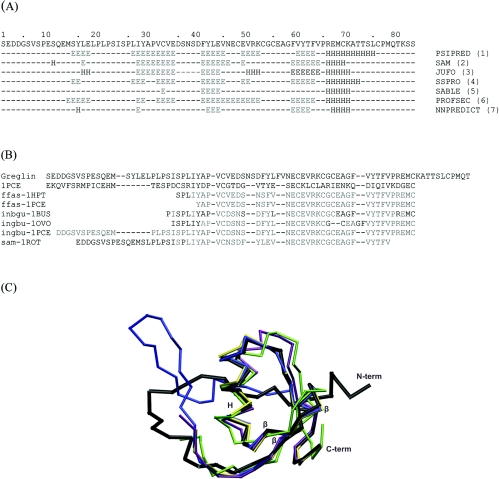
The secondary structure elements of the full-length greglin sequence (83 amino acids) were predicted using seven different programs (Figure 2A). All the programs except SABLE agreed on the presence of three main β-strands in greglin at positions 27–35, 41–45 and 59–64. They also predicted an α-helix starting at Arg66 up to Leu76. The characteristic scaffold of the Kazal family of inhibitors consists of a central α-helix, an adjacent three-stranded β-sheet and a proteinase-binding loop cross-connected through disulfide bridges 1–5, 2–4 and 3–6, whereas inhibitors of the pacifastin family have β-strands only as secondary structure elements [26]. The predicted secondary structure of greglin is thus more closely related to that of Kazal inhibitors than to that of pacifastin inhibitors.
Figure 2. Prediction of the greglin secondary and tertiary structures.
(A) The full amino acid sequence of greglin is displayed on the first line. Each of the other seven lines reports the secondary structure predicted by the different programs used. The residues predicted to be involved in β-strands are annotated with the letter ‘E’. The residues that are predicted to be in helical regions are annotated with the letter ‘H’. (B) Fold recognition sequences and fold recognition structural alignments. Sequence alignments are based on the structural alignment between 1PCE and a subset of ‘intermediate’ models generated by the Genesilico server from the best hits of the fold recognition search. The first line is the greglin sequence, which was fitted manually to the template structure (1PCE). The identifier at the beginning of each line refers to the program (ffas, inbgu and sam) and the structure (1HPT, 1PCE, 1BUS, 1OVO and 1ROT) used to build each of the ‘intermediate’ models. SwissPdbViewer (Domain Fit) was used to calculate the structural superimposition and generate the corresponding alignment. Residues in light grey are the residues of the intermediate models that were aligned with those of the template 1PCE by the fitting operations. (C) Superimposition of PEC-60 (1PCE, black) and the six best intermediate models for the greglin sequence. Only the carbon-α (traces) of the polypeptides are displayed. The colours used are as follows: ffas-1HPT, yellow; inbgu-1BUS, green; ingbu-1OVO, purple; sam-1ROT, blue. The structures corresponding to ffas-1PCE and ingbu-1PCE can be superimposed on those represented, and are not visible. The secondary structures of the template 1PCE are identified by the letter ‘H’ for the α-helix and ‘β’ for the β-strands.
Because few sequence homologies were found by searching sequence databases (max. 38%), we used protein fold recognition methods via the ‘GeneSilico’ metaserver. The folding of greglin is predicted to contain α-helices and β-sheets (‘a+b’ group of the SCOP classification; http://scop.mrc-lmb.cam.ac.uk/scop/). Most of the fold-recognition programs identified PEC-60 (PDB ID: 1PCE) as the closest structural homologue of greglin. Intermediate models (those not containing the variable loops) that correspond to the best hits of the fold recognition searches were aligned structurally to PEC-60 using the tool ‘Domain fit’ of ‘SwissPdbViewer’ (Figure 2B). The cores are readily superimposed, but there are deviations in the loops, and the N-terminal region of 1PCE could only be aligned with one model (Figure 2C). We manually improved the structural alignment of greglin using insertions and deletions (Figure 2B). However, only one disulfide bridge was generated by this approach, while there are three disulfide bridges in the folds of the Kazal-type inhibitors. Therefore greglin appears to be only distantly related to the Kazal-type inhibitors and it could well have a novel folding pattern.
Inhibition of serine proteases by greglin
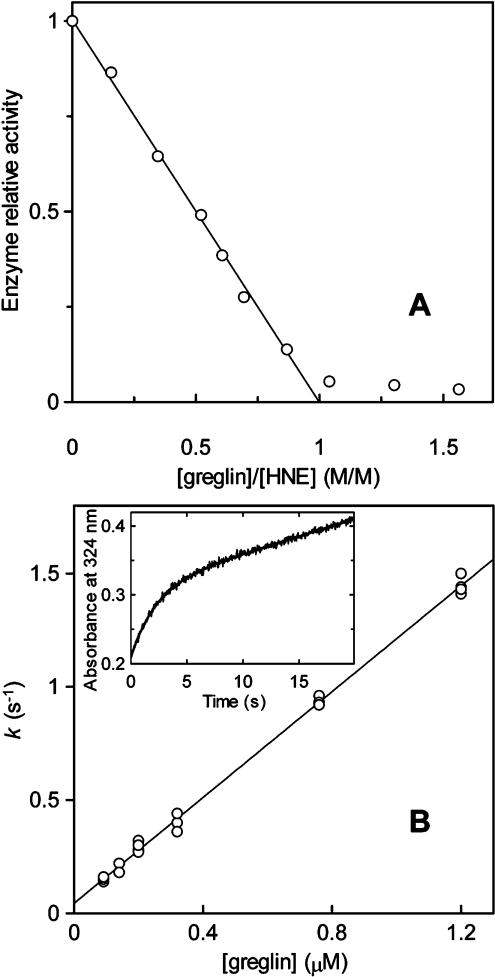
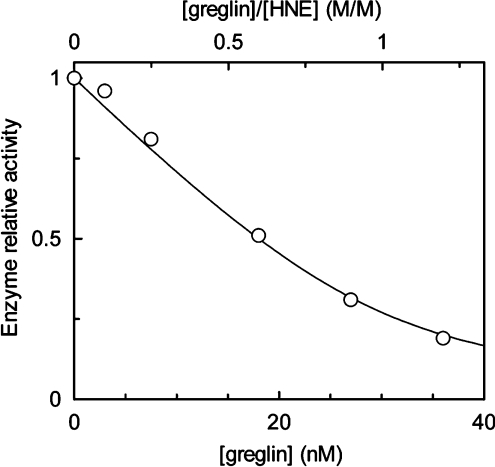
The inhibitory activity of greglin was assayed towards a series of serine proteases to delineate its specificity. In addition to PPE used to monitor its purification, those used included the related human neutrophil proteases cathepsin G and proteinase 3, porcine trypin, bovine pancreatic chymotrypsin and subtilisin Carlsberg. Preliminary experiments showed that both greglin isoforms reacted similarly with the enzymes tested, so that both were equally used in the following kinetic experiments. Preliminary experiments also showed that HNE forms a much tighter complex with greglin than does PPE. We used this property to titrate the inhibitor. Increasing quantities of the inhibitor were reacted with 0.38 μM HNE as describe in the Experimental section. The residual enzyme activity decreased linearly with the greglin concentration up to approx. 90% inhibition, indicating that the inhibitor binds the enzyme tightly under the experimental conditions used (Figure 3), and suggesting a value of Ki at least two orders of magnitude lower than the HNE concentration used, in accordance with theory [20]. This experiment could not be used to determine Ki but allowed accurate titration of the inhibitor by fitting the data of the linear portion of the curve to the best theoretical straight line, whose intercept with the x-axis was used to calculate the concentration of the active greglin (Figure 3A). Since the present titration data could not be used to measure Ki, the equilibrium dissociation constant for the greglin–HNE complex, this parameter was obtained from kass and kdiss, the association and dissociation rate constants. These parameters were measured using the progress curve method. Enzyme and inhibitor were reacted under pseudo-first order conditions ([I]0≥10×[E]0) in the presence of 0.2 mM MeOsuc-(Ala)2-Pro-Ala-TBE with stopped- flow mixing and detection. The rate of HNE inhibition was investigated at various inhibitor concentrations (Figure 3B).
Figure 3. Kinetic properties of greglin on HNE.
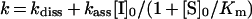 |
(1) |
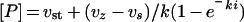 |
(2) |
The inset of Figure 3(B) shows a typical progress curve used to follow the time course of HNE inhibition. In all cases a pre-steady-state of product accumulation preceded its release at a constant rate, indicating a reversible interaction between enzyme and inhibitor. Non-linear regression analyses of the progress curves (Figure 3B) yielded k, the apparent pseudo-first order rate constant for the approach to equilibrium [20].
k increased linearly as a function of the inhibitor concentration (Figure 3B), suggesting that no reaction intermediate accumulates over the range of inhibitor concentrations used. Inhibition was therefore analysed assuming that HNE and greglin associate via a simple bimolecular mechanism: the second order rate constant kass was calculated from the slope of the straight line on Figure 3(B), its intercept with the ordinate yielding kdiss. We found kass=1.23±0.16×107 M−1·s−1 and kdiss=0.043±0.010 s−1. Ki=3.49±1.25×10−9 M was deduced from Ki=kdiss/kass.
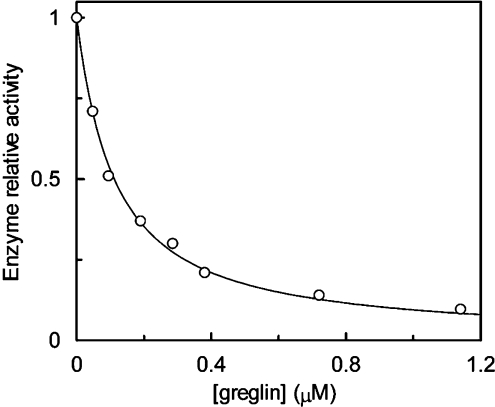
The Ki for the complexes between greglin and the other enzymes were obtained from equilibrium titration experiments with experimental conditions compatible with concave inhibition curves. The effects of increasing greglin concentrations on PPE activity (18 nM final concentration) are illustrated in Figure 4. The residual enzyme activity was assessed by adding the substrate Suc-(Ala)3-p-NA to the equilibrium reaction mixtures. The rate of product release accelerated for approx. 30 s and reached a steady state. This time course of substrate breakdown indicates three things: first, that the enzyme partially dissociates from a reversible complex with inhibitor as a result of disturbance by substrate of the initial equilibrium; secondly, that substrate and inhibitor compete for binding to PPE; and thirdly, that a new equilibrium between PPE, greglin, substrate and their complexes is reached. The plot of the steady-state rate of substrate hydrolysis versus the inhibitor concentration was analysed [16] to obtain Ki(app), the substrate-dependant equilibrium dissociation constant for the greglin–PPE complex (Figure 4). The true Ki value was obtained from Ki=Ki(app)/(1+[S]0/Km), where [S]0 is the initial substrate concentration and Km the Michaelis constant obtained by classical means.
Figure 4. Effect of increasing quantities of greglin on the activity of 18 nM PPE.
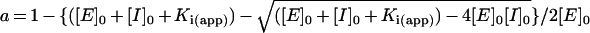 |
(3) |
This procedure was used to study the interaction of greglin with bovine chymotrypsin, subtilisin Carlsberg and the human cathepsin G. The Ki values for all enzyme–inhibitor pairs are shown in Table 2. Porcine trypsin and proteinase 3 were not inhibited under these conditions.
Table 2. Equilibrium dissociation constant (Ki) for the interaction of greglin with the various proteases.
| Enzyme | Ki (nM) |
|---|---|
| PPE | 58.3±12 |
| HNE | 3.6±2.1* |
| Protease 3 | NI† |
| Cathepsin G | 153.5±10 |
| Chymotrypsin | 26.7±10.8 |
| Subtilisin | 0.68±0.10 |
| Trypsin | NI† |
*Calculated from kass and kdiss (see text).
†NI, no inhibition.
Greglin physicochemical properties
Resistance to proteolysis
Greglin was incubated with HNE and PPE at low pH to locate the enzyme-binding site through a peptide bond cleavage [22]. Sequence analysis of the RP-HPLC-fractionated mixtures revealed that greglin had been N-terminally truncated at Glu10, Gln12 and Leu27 by PPE and at Met14 by HNE. The N-terminally cleaved inhibitor (minus 26 residues) retained full inhibitory activity towards PPE or HNE at pH 7.4. The observation that the P1–P26 N-terminal segment is not involved in inhibition agrees with the total lack of homology between this segment and the sequences of other protease inhibitors.
Native greglin was also incubated with other proteases, including serine (trypsin and Glu-C), cysteine (papain) and metallo (pseudolysin) proteases, at various molar ratios. The resulting mixtures were fractionated by RP-HPLC and the main peaks sequenced and analysed for their HNE-inhibitory properties. Again the cleavage sites were located within the N-terminal segment (P1–P26), except that trypsin cleaved at K81 before the penultimate C-terminal residue of the protein. Greglin remained a potent HNE inhibitor even after prolonged incubation and fractionation by RP-HPLC, whatever the protease used. We conclude that N-terminal (minus 26) and C-terminal (minus 3) truncations of greglin do not alter its inhibitory properties.
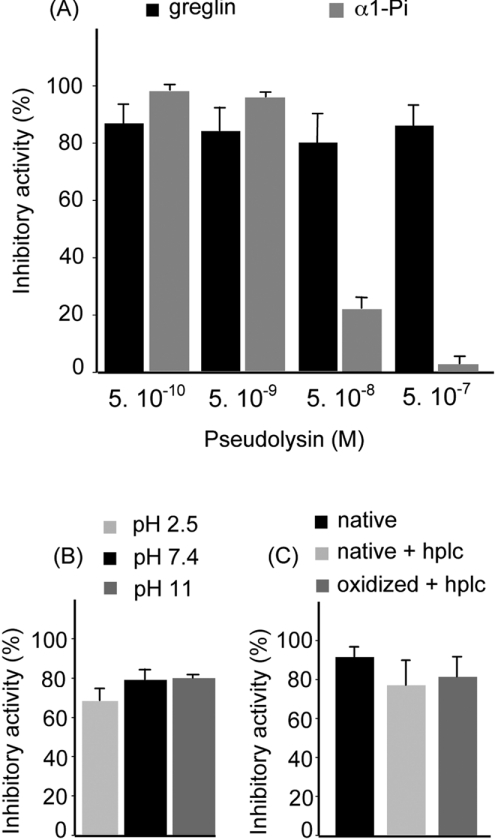
The resistance of greglin anti-HNE activity to pseudolysin was also compared with that of α1-Pi, the major physiological inhibitor of HNE. Figure 5(A) shows the effect of incubating greglin and α1-PI with various amounts of pseudolysin for 15 min on their anti-HNE activity. Greglin retained most of its inhibitory capacity even at the highest enzyme concentration used, whereas α1-Pi activity was reduced by 80% at [E]=50 nM or completely lost at [E]=500 nM (Figure 5A).
Figure 5. Physiochemical properties of greglin.
(A) Effect of pseudolysin on the anti-HNE activities of greglin and α1-Pi. Inhibitors (4 μM final concentration) were incubated with pseudolysin (0.5 nM–0.5 μM) as indicated in the text. Results show the median and maximal values (n≥2). (B) Resistance of greglin activity to oxidation and acetonitrile. Greglin 2 (77 μM final concentration) was incubated with 125 μM N-chlorosuccinimide. Residual greglin inhibitory activity was then measured and compared with those of the untreated inhibitor and inhibitor after RP-HPLC. The HNE concentration was adjusted so that inhibition by native greglin was incomplete (90%). Results show the median and maximal values (n≥2). (C) Resistance of greglin activity to acid and base conditions. Solutions of purified greglin were incubated at pH 2.5, pH 7.4 and pH 11.0 at 37 °C overnight and their capacity to inhibit neutrophil elastase measured. Results show the median and maximal values (n≥2).
pH and temperature stability
The recovery of inhibitory activity after RP-HPLC demonstrated that the two forms of greglin resist denaturation by acid and acetonitrile. We investigated the resistance of the inhibitor to low and high pH by incubating it overnight at pH 2.5 and at pH 11.0. The inhibitory properties were not significantly altered (Figure 5B). Similarly, we found no significant loss of activity when the inhibitor was incubated in Hepes buffer for 1 h at temperatures from 37 to 90 °C (results not shown).
Sensitivity to oxidation and reduction
Chemical and physiological oxidizing systems can alter the biological activity of many proteins by converting their exposed methionine residues into methionine sulfoxide. This can dramatically lower the activity of protein protease inhibitors, especially those having a methionine residue at position P1 or P1′ of their reactive site [27–29]. Since greglin contains three methionine residues, its activity might be altered by oxidants. The inhibitor was incubated with N-chlorosuccinimide and its oxidation checked by MS. The observed Mr was consistent with the oxidation of the three methionine residues, but the HNE-inhibitory capacity of oxidized greglin was unaltered (Figure 5C). This absence of significant change in the greglin anti-HNE property suggests that no methionine residue occupies a critical position in the inhibitor reactive site. However, greglin lost all its inhibitory capacity once its disulfide bridges had been reduced by 10 mM dithiothreitol and alkylated with iodoacetamide. The greglin tertiary structure therefore is essential for inhibition, which suggests that the sequence from the first to the last cysteine residues (Cys33 to Cys76) is required for inhibitory activity.
Inhibition of HNE in cystic fibrosis sputum
Purulent sputum is a complex biological medium containing elements (oxidants and proteolytic enzymes) able to decrease the activity of local physiological inhibitors [30]. We reacted increasing amounts of greglin with fixed amounts of cystic fibrosis sputum supernatant to delineate the anti-HNE efficiency of the inhibitor in sputum. The remaining HNE activity was measured by adding the fluorescent substrate Abz-APEEIMRRQ-EDDnp to the equilibrium mixtures. The relative enzyme activity decreased curvilinearly with the quantity of inhibitor (Figure 6), as anticipated from the Ki value in Hepes buffer (Table 2), the greglin concentration, and the protease concentration (30 nM) that gave a [HNE]/Ki≈8.5 [16]. Since inhibition of HNE by greglin in sputum is best described by the Ki measured in this environment, we analysed the inhibition data as indicated in the legend of Figure 4 to determine this parameter. The concave inhibition curve shown in Figure 6 was generated using the best estimate of Ki(app). After correction for the substrate (Km=15 μM, determined by classical methods), we found Ki=3.1±0.43 nM, a value close to that determined by stopped-flow kinetics with the purified enzyme–inhibitor system. Thus the sputum components did not inactivate the anti-HNE function of greglin during the time of the experiment and did not dissociate the enzyme–inhibitor complex.
Figure 6. Inhibition of sputum elastase activity by greglin.
Aliquots of homogenized sputum containing a constant concentration of HNE were reacted with increasing quantities of greglin. The final enzyme concentration (30 nM) was estimated as described in the text and Abz-APEEIMRRQ-EDDnp was used as the substrate. Under the experimental conditions used, the decrease in HNE activity versus the inhibitor concentration is concave, therefore the data could be analysed as described in the legend of Figure 4 to obtain Ki(app). The curve was generated using eqn (3) and Ki(app)=3.28 nM.
DISCUSSION
Anti-inflammatory therapy using protease inhibitors has long been suggested as an efficient means of controlling the unopposed proteolytic activity that occurs at inflammatory sites. Neutrophil elastase is one of the main proteases released from activated and dying PMNs (polymorphonuclear cells) and it is involved in tissue destruction and inflammation. A variety of natural, recombinant and synthetic inhibitors have been used as anti-inflammatory drugs to target HNE, but the results to date have not been entirely satisfactory [1]. New elastase inhibitors that have different structural and/or physicochemical properties from those described so far could lead to new, therapeutically useful elastase inhibitors.
Arthropods and other invertebrates are a rich source of protease inhibitors. Several belonging to the Kunitz and serpin families have been isolated, especially from haemolymph [31]. Two new families of low-molecular-mass serine protease inhibitors were discovered previously: one in silkworms [32] and the other in locusts [6,7,17,33] and crayfish [34]. The function of these serine protease inhibitors and their distribution in invertebrates, especially arthropods, remains a matter of debate; but this could be due to the far larger number of serine proteases in insects than in humans and other vertebrates [4]. Five serine protease inhibitors, designated SGPI 1–5, have been isolated from the ovaries of the locust (S. gregaria) [7]; they are members of the pacifastin family of peptidase inhibitors. These low Mr (∼35 residues) inhibitors are found only in invertebrates and they all have a similar cysteine array [5]. Aqueous homogenates of mature locust ovaries also contain potent trypsin, chymotrypsin and elastase inhibitory activities. The trypsin and chymotrypsin inhibitor is a 14 kDa, heat-stable peptide that has been purified and characterized [8], but the protein(s) responsible for the anti-elastase activity is still unidentified.
The protease inhibitors we have purified from the locust ovary are two forms of a single protein, greglin. They have similar inhibitory behaviours towards all the tested proteases. Greglin reversibly inhibits enzymes of the elastase and chymotrypsin families. It forms tight complexes with subtilisin and HNE, but has no effect on the activities of trypsin or proteinase 3. To our knowledge, no other potent HNE inhibitor has been purified from locusts. Most of the known locust inhibitors belong to the pacifastin family and preferentially inhibit trypsin and chymotrypsin [8,17]. The sequence of greglin is only distantly related to that of any other protease inhibitor. The putative Kazal-type inhibitor of C. intestinalis is the most closely related, as the C-terminal region of the sequence is 38% homologous. The function of the greglin N-terminal extension that takes no part in inhibition remains unexplained, but its sensitivity to proteolysis contrasts with that of the inhibitory C-terminal domain. Although this domain has some physicochemical properties in common with other low Mr inhibitors, it also has features that can be exploited in drug development.
The stopped-flow kinetics of HNE inhibition by greglin show that the latter is a fast acting inhibitor that binds the enzyme via a simple bimolecular mechanism under the experimental conditions used. The HNE–greglin complex formation is governed by the second-order rate constant kass=1.23×107 M−1·s−1, a value that is similar to or higher than that reported for the main physiological inhibitors of this enzyme: α1-PI (1.3×107 M−1·s−1 [35], which irreversibly inhibits its target proteases, and the reversible inhibitors SLPI (6.4×106 M−1·s−1 [36]) and elafin (3.6×106 M−1·s−1 [37]). The rate of HNE inhibition by greglin is also similar to that measured for eglin c (kass=1.3×107 M−1·s−1 [38]), another elastase inhibitor from the leech. The Ki for the HNE–greglin complex is significantly higher than those for the interactions between HNE and the reversible inhibitors SLPI (0.3 nM [36]) and elafin (0.2 nM [37]). Despite this relatively low affinity, greglin concentrations of approx. 10−8 M efficiently inhibit the elastase activity in purulent sputum.
Greglin also resists oxidation, while α1-PI, SLPI and elafin, the major protease inhibitors of the lung, do not. The HNE-inhibitory activity of these three human proteins is reduced by their interaction with the oxidant species produced by the myeloperoxidase–H2O2 system of triggered neutrophils [39], or with N-chlorosuccinimide [27,28]. Oxidative inactivation of the inhibitors is due to the conversion of a methionine residue involved in protease binding into methionine sulfoxide. As greglin is not inactivated by incubation with N-chlorosuccinimide, it probably has no methionine residue in such a critical position (P1 or P1′) in its reactive site. This also suggests that greglin would retain its antielastase activity in a chronic inflammation environment with high concentrations of oxidant species and lysosomal proteinases produced by continuously recruited neutrophils.
Pseudolysin, a metalloenzyme that contributes to the virulence of Pseudomonas aeruginosa, can break down many proteins, including α1-PI and secretory leucocyte protease inhibitor. Both inhibitors are inactivated by limited cleavage in their reactive site regions [40,41]. This mechanism, in which inhibitors behave as substrates, may cause a significant loss of active inhibitors and promote the breakdown of lung connective tissues by neutrophil proteases. We find that greglin resists inactivation by several proteolytic enzymes, including high concentrations of trypsin, papain and pseudolysin. This is important for the development of a therapeutic system, since Ps. aeruginosa infection accompanies the inflammation of airways in patients with cystic fibrosis [42].
The predicted secondary and tertiary structures suggest that greglin is more closely related to the Kazal inhibitors than to pacifastin inhibitors. However, the distribution of cysteine residues within the greglin sequence does not fit the disulfide pattern found in classical Kazal inhibitors, which all contain three disulfide bridges. Although the inhibitory site of Kazal inhibitors lies within the Cys2–Cys4 disulfide loop, computer-based analysis predicts that the greglin inhibitory site lies in the N-terminal extended strand, part of which is not necessary for inhibition. We therefore believe that greglin is folded in a manner different from that of classical Kazal-type inhibitors. However, non-classical Kazal-type inhibitors with different folding [43] or different positioning of their cysteine residues [44] have been described. Our results and observations suggest that it is difficult to assign a fold to the greglin sequence using computer-based methods alone. For example, none of the ab initio models constructed by the Robetta server (http://robetta.bakerlab.org/) was folded like any known inhibitor (results not shown).
The resolution of the greglin three-dimensional structure or the production of recombinant forms with single mutation points at the putative inhibitory site will be needed before we can assign this inhibitor to a family and understand the molecular basis of its physicochemical properties.
Online data
Acknowledgments
We thank ‘Vaincre la mucoviscidose’ for financial support, Luiz Juliano (Universidade Federal, São Paulo, Brazil) for synthesizing the Abz-APEEIMRRQ-EDDnp substrate, Valérie Labas (Department of Proteomics, Institut National de la Recherche Agronomique, Tours-Nouzilly, France) for performing mass spectrum analyses, Sylvie Attucci for her help in preparing this paper, Jose W. Saldanha and Janusz Bujnicki for fruitful discussions and Owen Parkes for editing the English text prior to submission. Funding for N. R. and E. H. was provided by the National Program for Research in Functional Genomics in Norway (FUGE) and the Research Council of Norway.
References
- 1.Tremblay G. M., Janelle M. F., Bourbonnais Y. Anti-inflammatory activity of neutrophil elastase inhibitors. Curr. Opin. Investig. Drugs. 2003;4:556–565. [PubMed] [Google Scholar]
- 2.Rawlings N. D., Tolle D. P., Barrett A. J. Evolutionary families of peptidase inhibitors. Biochem. J. 2004;378:705–716. doi: 10.1042/BJ20031825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Honore S., Attalah H. L., Azoulay E., Soussy C. J., Saudubray F., Harf A., Brochard L., Delclaux C. Beneficial effect of an inhibitor of leukocyte elastase (EPI-hNE-4) in presence of repeated lung injuries. Shock. 2004;22:131–136. doi: 10.1097/01.shk.0000126861.77543.d0. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C., Overall C. M. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;3:509–519. doi: 10.1038/nrm858. [DOI] [PubMed] [Google Scholar]
- 5.Simonet G., Claeys I., Franssens V., De Loof A., Broeck J. V. Genomics, evolution and biological functions of the pacifastin peptide family: a conserved serine protease inhibitor family in arthropods. Peptides. 2003;24:1633–1644. doi: 10.1016/j.peptides.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Simonet G., Claeys I., Broeck J. V. Structural and functional properties of a novel serine protease inhibiting peptide family in arthropods. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002;132:247–255. doi: 10.1016/s1096-4959(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 7.Hamdaoui A., Wataleb S., Devreese B., Chiou S. J., Vanden Broeck J., Van Beeumen J., De Loof A., Schoofs L. Purification and characterization of a group of five novel peptide serine protease inhibitors from ovaries of the desert locust, Schistocerca gregaria. FEBS Lett. 1998;422:74–78. doi: 10.1016/s0014-5793(97)01585-8. [DOI] [PubMed] [Google Scholar]
- 8.Hamdaoui A., Schoofs L., Wateleb S., Bosch L. V., Verhaert P., Waelkens E., De Loof A. Purification of a novel, heat-stable serine protease inhibitor protein from ovaries of the desert locust, Schistocerca gregaria. Biochem. Biophys. Res. Commun. 1997;238:357–360. doi: 10.1006/bbrc.1997.7283. [DOI] [PubMed] [Google Scholar]
- 9.Martodam R. R., Baugh R. J., Twumasi D. Y., Liener I. E. A rapid procedure for the large scale purification of elastase and cathepsin G from human sputum. Prep. Biochem. 1979;9:15–31. doi: 10.1080/00327487908061669. [DOI] [PubMed] [Google Scholar]
- 10.Shotton Elastase. Methods Enzymol. 1970;19:113–140. [Google Scholar]
- 11.Boudier C., Bieth J. G. The proteinase: mucus proteinase inhibitor binding stoichiometry. J. Biol. Chem. 1992;267:4370–4375. [PubMed] [Google Scholar]
- 12.Komiyama T., Gron H., Pemberton P. A., Salvesen G. S. Interaction of subtilisins with serpins. Protein Sci. 1996;5:874–882. doi: 10.1002/pro.5560050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rao N. V., Wehner N. G., Marshall B. C., Gray W. R., Gray B. H., Hoidal J. R. Characterization of proteinase-3 (PR-3), a neutrophil serine proteinase. Structural and functional properties. J. Biol. Chem. 1991;266:9540–9548. [PubMed] [Google Scholar]
- 14.Barrett A. J., Kembhavi A. A., Brown M. A., Kirschke H., Knight C. G., Tamai M., Hanada K. L-trans-Epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korkmaz B., Attucci S., Hazouard E., Ferrandiere M., Jourdan M. L., Brillard-Bourdet M., Juliano L., Gauthier F. Discriminating between the activities of human neutrophil elastase and proteinase 3 using serpin-derived fluorogenic substrates. J. Biol. Chem. 2002;277:39074–39081. doi: 10.1074/jbc.M202918200. [DOI] [PubMed] [Google Scholar]
- 16.Bieth J. G. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol. 1995;248:59–84. doi: 10.1016/0076-6879(95)48007-2. [DOI] [PubMed] [Google Scholar]
- 17.Kellenberger C., Boudier C., Bermudez I., Bieth J. G., Luu B., Hietter H. Serine protease inhibition by insect peptides containing a cysteine knot and a triple-stranded beta-sheet. J. Biol. Chem. 1995;270:25514–25519. doi: 10.1074/jbc.270.43.25514. [DOI] [PubMed] [Google Scholar]
- 18.Kussmann M., Lassing U., Sturmer C. A., Przybylski M., Roepstorff P. Matrix-assisted laser desorption/ionization mass spectrometric peptide mapping of the neural cell adhesion protein neurolin purified by sodium dodecyl sulfate polyacrylamide gel electrophoresis or acidic precipitation. Eur. J. Mass Spectrom. 1997;32:483–493. doi: 10.1002/(SICI)1096-9888(199705)32:5<483::AID-JMS502>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 19.Baldi P., Brunak S., Frasconi P., Soda G., Pollastri G. Exploiting the past and the future in protein secondary structure prediction. Bioinformatics. 1999;15:937–946. doi: 10.1093/bioinformatics/15.11.937. [DOI] [PubMed] [Google Scholar]
- 20.Kneller D. G., Cohen F. E., Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J. Mol. Biol. 1990;214:171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 21.Kim D. E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sealock R. W., Laskowski M., Jr Enzymatic replacement of the arginyl by a lysyl residue in the reactive site of soybean trypsin inhibitor. Biochemistry. 1969;8:3703–3710. doi: 10.1021/bi00837a032. [DOI] [PubMed] [Google Scholar]
- 23.Rawlings N. D., Tolle D. P., Barrett A. J. MEROPS: the peptidase database. Nucleic Acids Res. 2004;32:D160–D164. doi: 10.1093/nar/gkh071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laskowski M., Jr, Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 25.Liepinsh E., Berndt K. D., Sillard R., Mutt V., Otting G. Solution structure and dynamics of PEC-60, a protein of the Kazal type Inhibitor family, determined by nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1994;239:137–153. doi: 10.1006/jmbi.1994.1356. [DOI] [PubMed] [Google Scholar]
- 26.Gaspari Z., Ortutay C., Perczel A. A simple fold with variations: the pacifastin inhibitor family. Bioinformatics. 2004;20:448–451. doi: 10.1093/bioinformatics/btg451. [DOI] [PubMed] [Google Scholar]
- 27.Johnson D., Travis J. The oxidative inactivation of human α1-proteinase inhibitor. Further evidence for methionine at the reactive center. J. Biol. Chem. 1979;254:4022–4026. [PubMed] [Google Scholar]
- 28.Nobar S. M., Zani M. L., Boudier C., Moreau T., Bieth J. G. Oxidized elafin and trappin poorly inhibit the elastolytic activity of neutrophil elastase and proteinase 3. FEBS J. 2005;272:5883–5893. doi: 10.1111/j.1742-4658.2005.04988.x. [DOI] [PubMed] [Google Scholar]
- 29.Vogelmeier C., Hubbard R. C., Fells G. A., Schnebli H. P., Thompson R. C., Fritz H., Crystal R. G. Anti-neutrophil elastase defense of the normal human respiratory epithelial surface provided by the secretory leukoprotease inhibitor. J. Clin. Invest. 1991;87:482–488. doi: 10.1172/JCI115021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matheson N. R., Wong P. S., Travis J. Enzymatic inactivation of human α1-proteinase inhibitor by neutrophil myeloperoxidase. Biochem. Biophys. Res. Commun. 1979;88:402–409. doi: 10.1016/0006-291x(79)92062-x. [DOI] [PubMed] [Google Scholar]
- 31.Polanowski A., Wilusz T. Serine proteinase inhibitors from insect hemolymph. Acta Biochim. Pol. 1996;43:445–453. [PubMed] [Google Scholar]
- 32.Pham T. N., Hayashi K., Takano R., Itoh M., Eguchi M., Shibata H., Tanaka T., Hara S. A new family of serine protease inhibitors (Bombyx family) as established from the unique topological relation between the positions of disulphide bridges and reactive site. J. Biochem. (Tokyo) 1996;119:428–434. doi: 10.1093/oxfordjournals.jbchem.a021259. [DOI] [PubMed] [Google Scholar]
- 33.Boigegrain R. A., Pugniere M., Paroutaud P., Castro B., Brehelin M. Low molecular weight serine protease inhibitors from insects are proteins with highly conserved sequences. Insect Biochem. Mol. Biol. 2000;30:145–152. doi: 10.1016/s0965-1748(99)00109-5. [DOI] [PubMed] [Google Scholar]
- 34.Liang Z., Sottrup-Jensen L., Aspan A., Hall M., Soderhall K. Pacifastin, a novel 155-kDa heterodimeric proteinase inhibitor containing a unique transferrin chain. Proc. Natl. Acad. Sci. U.S.A. 1997;94:6682–6687. doi: 10.1073/pnas.94.13.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frommherz K. J., Faller B., Bieth J. G. Heparin strongly decreases the rate of inhibition of neutrophil elastase by α1-proteinase inhibitor. J. Biol. Chem. 1991;266:15356–15362. [PubMed] [Google Scholar]
- 36.Boudier C., Bieth J. G. Mucus proteinase inhibitor: a fast-acting inhibitor of leucocyte elastase. Biochim. Biophys. Acta. 1989;995:36–41. doi: 10.1016/0167-4838(89)90230-6. [DOI] [PubMed] [Google Scholar]
- 37.Ying Q. L., Simon S. R. Kinetics of the inhibition of human leukocyte elastase by elafin, a 6-kilodalton elastase-specific inhibitor from human skin. Biochemistry. 1993;32:1866–1874. doi: 10.1021/bi00058a021. [DOI] [PubMed] [Google Scholar]
- 38.Braun N. J., Bodmer J. L., Virca G. D., Metz-Virca G., Maschler R., Bieth J. G., Schnebli H. P. Kinetic studies on the interaction of eglin c with human leukocyte elastase and cathepsin G. Biol. Chem. Hoppe Seyler. 1987;368:299–308. doi: 10.1515/bchm3.1987.368.1.299. [DOI] [PubMed] [Google Scholar]
- 39.Carp H., Janoff A. Potential mediator of inflammation. Phagocyte-derived oxidants suppress the elastase-inhibitory capacity of α1-proteinase inhibitor in vitro. J. Clin. Invest. 1980;66:987–995. doi: 10.1172/JCI109968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson D. A., Carter-Hamm B., Dralle W. M. Inactivation of human bronchial mucosal proteinase inhibitor by Pseudomonas aeruginosa elastase. Am. Rev. Respir. Dis. 1982;126:1070–1073. doi: 10.1164/arrd.1982.126.6.1070. [DOI] [PubMed] [Google Scholar]
- 41.Morihara K., Tsuzuki H., Harada M., Iwata T. Purification of human plasma α1-proteinase inhibitor and its inactivation by Pseudomonas aeruginosa elastase. J. Biochem. (Tokyo) 1984;95:795–804. doi: 10.1093/oxfordjournals.jbchem.a134671. [DOI] [PubMed] [Google Scholar]
- 42.Wretlind B., Pavlovskis O. R. Pseudomonas aeruginosa elastase and its role in pseudomonas infections. Rev. Infect. Dis. 1983;5(Suppl. 5):S998–S1004. doi: 10.1093/clinids/5.supplement_5.s998. [DOI] [PubMed] [Google Scholar]
- 43.Lauber T., Schulz A., Schweimer K., Adermann K., Marx U. C. Homologous proteins with different folds: the three-dimensional structures of domains 1 and 6 of the multiple Kazal-type inhibitor LEKTI. J. Mol. Biol. 2003;328:205–219. doi: 10.1016/s0022-2836(03)00245-6. [DOI] [PubMed] [Google Scholar]
- 44.Hemmi H., Kumazaki T., Yoshizawa-Kumagaye K., Nishiuchi Y., Yoshida T., Ohkubo T., Kobayashi Y. Structural and functional study of an Anemonia elastase inhibitor, a ‘nonclassical’ Kazal-type inhibitor from Anemonia sulcata. Biochemistry. 2005;44:9626–9636. doi: 10.1021/bi0472806. [DOI] [PubMed] [Google Scholar]
- 45.Notredame C., Higgins D. G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.