Abstract
Cholesterol accumulation and removal are regulated by two different transcription factors. SREBP-2 (sterol-regulatory-element-binding protein-2) is best known to up-regulate genes involved in cholesterol biosynthesis and uptake, whereas LXR (liver X receptor) is best known for up-regulating cholesterol efflux genes. An important cholesterol efflux gene that is regulated by LXR is the ATP-binding cassette transporter, ABCA1 (ATP-binding cassette transporter-A1). We have previously shown that statin treatment down-regulated ABCA1 expression in human macrophages, probably by inhibiting synthesis of the LXR ligand 24(S),25-epoxycholesterol. However, it was subsequently reported that ABCA1 expression is down-regulated by SREBP-2 through binding of SREBP-2 to an E-box element in ABCA1's proximal promoter. As statin treatment induces SREBP-2 activation, this may provide an alternative explanation for the statin-mediated down-regulation of ABCA1. In the present study, we employed a set of CHO (Chinese-hamster ovary) mutant cell lines to investigate the role of SREBP-2 in the regulation of ABCA1. We observed increased ABCA1 mRNA levels in SREBP-2-overexpressing cells and decreased levels in cells lacking a functional SREBP-2 pathway, which were restored when the SREBP-2 pathway was reinstated. Moreover, ABCA1 gene expression was positively associated with synthesis of 24(S),25-epoxycholesterol in these cell lines. In studies using a human ABCA1 promoter reporter assay, mutation of the E-box motif had a similar response as the wild-type construct to either statin treatment or addition of 24(S),25-epoxycholesterol. By contrast, these responses were completely ablated when the DR4 element to which LXR binds was mutated. These results support the idea that 24(S),25-epoxycholesterol and statin treatment influence ABCA1 transcription via supply of an LXR ligand and not through an SREBP-2/E-box-related mechanism. In addition, our results indicate a critical role of SREBP-2 as a positive regulator of ABCA1 gene expression by enabling the generation of oxysterol ligands for LXR.
Keywords: ATP-binding-cassette transporter-A1 (ABCA1), E-box, 24(S), 25-epoxycholesterol, liver X receptor (LXR), statin, sterol-regulatory-element-binding protein 2 (SREBP-2)
Abbreviations: ABCA1, ATP-binding cassette transporter-A1; CHO cell, Chinese-hamster ovary cell; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; LDL, low-density lipoprotein; LDL-R, LDL receptor; LPDS, lipoprotein-deficient serum; LXR, liver X receptor; NBS, newborn calf serum; PBGD, porphobilinogen deaminase; RT, reverse transcriptase; QRT–PCR, quantitative (‘real-time’) RT–PCR; SREBP, sterol-regulatory-element-binding protein; SCAP, SREBP cleavage activating protein; SRE, sterol-regulatory element
INTRODUCTION
There are three main levels of control in the regulation of cholesterol homoeostasis: cholesterol synthesis, uptake and efflux. The transcription factor SREBP-2 (sterol-regulatory-element-binding protein 2) regulates the expression of many genes involved in cholesterol synthesis [e.g. HMG-CoA (3-hydroxy-3-methylglutaryl-CoA) reductase] and uptake [LDL-R (low-density-lipoprotein receptor)]. Activation of SREBP-2 is dependent on the cholesterol status of the cell [1]. When SCAP [SREBP cleavage activating protein] senses low cholesterol levels in the endoplasmic reticulum, it escorts full-length SREBP-2 to the Golgi apparatus, where two proteases act sequentially to liberate the active N-terminal transcription factor. This is then delivered to the nucleus for binding to SREs (sterol-regulatory elements) in the promoter of target genes [1]. The classical SRE sequence was identified as ATCACCCCAC [2]. Unique to the SREBPs is their ability to bind to and activate classic palindromic E-box (CANNTG)-containing promoters as well as nonpalindromic SREs [3].
An important facet of regulation of cholesterol efflux is via the ligand-activated transcription factor LXR (liver X receptor). LXR forms a heterodimer with the retinoid X receptor, which together bind to a variation of a consensus sequence (DR4) TGACCGNNNNTAACCT in the promoter of ABCA1 (ATP-binding-cassette transporter-A1) and other genes, causing up-regulated expression [4,5]. ABCA1 is an ATP-binding cassette protein that is critical in the initial stages of cholesterol efflux from peripheral cells back to the liver for eventual excretion. Patients lacking functional ABCA1 have virtually absent high-density lipoprotein levels and a greater propensity to develop premature atherosclerosis [6,7].
A common link between SREBP-2- and LXR-mediated processes is their regulatory response to certain oxidized cholesterol derivatives (oxysterols). Certain oxysterols inhibit SREBP-2 activation while serving as ligands for LXR [8–13]. The common physiological oxysterols are derived either from internalized lipoproteins or by de novo synthesis. Specific enzymes required for the synthesis of 20(S)-, 22(R)-, 24(S)- and 27-hydroxycholesterol are cell-type-specific. Both 20(S)-hydroxycholesterol and 22(R)-hydroxycholesterol are synthesized by the adrenals, 24(S)-hydroxycholesterol is synthesized exclusively in the brain, whereas 27-hydroxycholesterol is abundant in liver cells and macrophages [14,15]. 24(S),25-Epoxycholesterol is a potent LXR ligand that is synthesized uniquely via a shunt in the mevalonate pathway parallelling de novo cholesterol synthesis [16]. We have previously shown in human macrophages [17] that statins (HMG-CoA reductase inhibitors) down-regulate ABCA1 gene expression and ABCA1-mediated cholesterol efflux. We attributed the down-regulatory effect of statins on ABCA1 expression to the inhibition of 24(S),25-epoxycholesterol production. In a subsequent study in human vascular endothelial cells, Zeng et al. [18] reported a novel link between SREBP-2 and ABCA1. The authors found that binding of SREBP-2 to an E-box element in the ABCA1 promoter down-regulated its expression, an effect that was enhanced during serum deprivation. Considering that statins increase, while oxysterols inhibit, SREBP-2 activation [12], their effects on ABCA1 expression could potentially be mediated via direct effects on SREBP-2 activation rather than on ligand activation of LXR.
In the present study, we investigated the role of SREBP-2 in the regulation of the LXR-target gene ABCA1. We present compelling data that show that SREBP-2 is in fact required for ABCA1 transcription by maintaining a supply of endogenous oxysterol ligands for LXR and that this effect is independent of the E-box motif.
MATERIALS AND METHODS
Reagents
Chemicals and reagents used are listed below with the supplier. From GE Healthcare (formerly Amersham Biosciences): [1-14C]-acetic acid sodium salt (specific radioactivity: 56 mCi/mmol). From GlaxoSmithKline: GW534511X. From Invitrogen: DH5α competent cells, Dulbecco's modified Eagle's medium/Ham's F-12 medium (50:50 mixture), OptiMEM, fetal calf serum, L-glutamine, Lipofectamine™ 2000, NBS (newborn calf serum), oligo(dT) and penicillin/streptomycin. From New England Biolabs: KpnI and XhoI. From Promega: pGL3-basic vector and phRL-TK Renilla plasmid. From Sigma: compactin (also called mevastatin), PMA and TRI Reagent; oligonucleotides were synthesized by Sigma-Genosys. From Steraloids: cholesterol, 24(S), 25-epoxycholesterol and 25-hydroxycholesterol. Other reagents used were: iQ SyBr Green Supermix (Bio-Rad), TO901317 (Cayman Chemicals), analytical- or HPLC-grade solvents (EM Science) and T4 DNA ligase (Fermentas). Cholesterol complexed with methyl-β-cyclodextrin was prepared as described in [19]. LPDS (lipoprotein-deficient serum) was prepared from NBS [20]. CHO cells (Chinese-hamster ovary cells) (wild-type and mutants) and the mammalian expression plasmid, pTK3-SCAP [21], were generously provided by Dr Michael S. Brown and Dr Joseph L. Goldstein (UT Southwestern, Dallas, TX, U.S.A.). A reporter construct (pGL3-hABCA1) containing a fragment of the human ABCA1 promoter (−928 bp to +101 bp) was a gift from Dr Alan Tall (Columbia University, New York, NY, U.S.A.). A reporter construct (pGL3-TK) was a gift from Dr Malcolm Lyons (Western Australian Institute of Medical Research, Perth, WA, Australia).
Cell culture
All cell types used were grown at 37 °C in a 5% CO2 atmosphere. CHO cells (wild-type and mutants) were cultured in Dulbecco's modified Eagle's medium/Ham's F-12 medium (1:1 mixture) with various additions as previously described [22,23]. For production of the stably transfected cell line, 13A/pSCAP, SRD-13A cells (lacking SCAP) were transfected with pTK3-SCAP [21], which restores the ability of SRD-13A cells to grow in the absence of cholesterol. Surviving colonies were cloned by limiting dilution [19]. CHO cells were plated at 2.5×105 cells/ml and grown to approx. 80% confluence prior to treatment. Treatments for all cell types were conducted for 24 h in media containing 5% (v/v) LPDS for CHO cell lines. SRD-1 cells are routinely cultured in the presence of 25-hydroxycholesterol [22] and were pre-incubated for 1 h prior to treatment with media containing 5% LPDS to remove this oxysterol, which can weakly activate LXR [24]. The human monocytic cell line, THP-1, was plated at 1.2×106 cells/ml in the presence of PMA (50 ng/ml; 3 days) to promote differentiation into macrophages. All treatments were added to cells in 100% ethanol, DMSO or water and compared with vehicle-only controls. No cell toxicity was observed for any treatment at the concentrations employed.
RNA isolation and gene expression analysis by quantitative RT (reverse transcriptase)–PCR
Cells were harvested for total RNA using TRI Reagent according to the manufacturer's instructions. Concentrations of total RNA were measured by spectrophotometry (Nanodrop ND-100 spectrophotometer; Biolab). RT–PCR was performed according to the manufacturer's protocol for SuperScript III First Strand cDNA Synthesis kit (Invitrogen). QRT–PCR [quantitative (‘real-time’) RT–PCR] was performed using iQSupermix (Bio-Rad) on an ABI 7700 sequence detector and analysed using ABI Prism sequence detector software v1.6.3 (PE Biosystems). Primer pairs used for the amplification reaction of various genes from cDNAs are listed in Table 1. PCR products were verified by sequencing. The change in gene expression levels was determined by normalizing mRNA levels of the gene of interest to the mRNA level of the house-keeping gene, PBGD (porphobilinogen deaminase). Melting curve analysis was performed to confirm production of a single product in each reaction.
Table 1. Primer sequences for gene expression analysis.
ABCA1 and PBGD are mouse (Mus musculus) genes, and LDLR is a hamster (Cricetulus griseus) gene.
| Gene | GenBank® accession no. | Direction | Primer sequence | Size (bp) | Reference |
|---|---|---|---|---|---|
| ABCA1 | NM_013454 | Forward | 5′-ATAGCAGGCTCCAACCCTGAC-3′ | 103 | [33] |
| Reverse | 5′-GGTACTGAAGCATGTTTCGATGTT-3′ | ||||
| LDLR | M94387.1 | Forward | 5′-AAGGAGAAGGACACTGTTCC-3′ | 246 | Present study |
| Reverse | 5′-ATGCTGGAGATAGAGTGGAG-3′ | ||||
| PBGD | NM_013551 | Forward | 5′-AGATTCTTGATACTGCACTC-3′ | 192 | Present study |
Cholesterol and 24(S),25-epoxycholesterol synthesis assay
Cells were metabolically labelled with [1-14C]acetic acid for 24 h during treatments as previously described [17]. Cells were harvested, saponified, and the neutral lipid extracts were separated by TLC. Bands corresponding to authentic cholesterol and 24(S),25-epoxycholesterol were visualized by a phosphoimager (18–72 h exposure). Positive identification of the band corresponding to 24(S),25-epoxycholesterol has previously been performed chemically and by MS [17].
Plasmid construction
The insert for 6× SRE-luc was constructed by annealing complementary sequences of a 2× SRE oligonucleotide [2] with KpnI and XhoI restriction sites synthesized at the 5′- and 3′-ends respectively (Sigma-Genosys). Annealed insert and pGL3-TK vector were then sequentially digested with KpnI and XhoI. Digested insert and vector were then ligated with T4 DNA ligase and transformed into the DH5α Escherichia coli strain. Positive clones were verified by sequencing.
Human ABCA1 promoter activity assay
Promoter analysis was performed using a fragment of the hABCA1 promoter that was linked to the firefly luciferase reporter gene [4]. Mutations in pGL3-hABCA1 were generated using the QuikChange® site-directed mutagenesis kit (Stratagene). Mutagenic primers were: E-box-mut [18]: 5′-GGGCCCCGGCTCCACGgaCTTTCTGCTGAG-3′; DR4-mut1 [4]: 5′-GAGGGAGAGCACAGGCTTTGtgtGATAGTAACCTCTGCGCTCG-3′; DR4-mut2 [4]: 5′-CACAGGCTTTGtgtGATAGTAACtaCTGCGCTCGGTGCAGC-CGAATC-3′.
Bases in lower-case letters are the designated mutations in the underlined regions corresponding to the E-box and DR4 element respectively. Mutagenesis was performed according to the manufacturer's protocol, and sequences were verified. Reporter plasmids (250 ng/well) were transfected for 24 h into cells using Lipofectamine™ 2000 (1 μl/well). The phRL-TK Renilla internal control plasmid (25 ng/well) was co-transfected for normalization of transfection efficiency. After treatment (24 h), cells were washed and resuspended in 100 μl of 1× passive lysis buffer (Promega). Luciferase assays were performed using the Dual Luciferase Assay Reporter system according to the manufacturer's instructions in a TD20/20 or Veritas luminometer (Turner Designs). Results were expressed as changes in luciferase activity relative to vehicle-treated controls.
Data presentation and statistics
Data are presented as means±S.E.M. or half-range. All results are representative of two to five separate experiments. Statistical analyses were performed to find correlations (Pearson's) between two sets of continuous variables or when differences were not immediately obvious (paired t test). A P value less than 0.05 was considered statistically significant.
RESULTS
Studies in CHO cells indicate that a functional SREBP pathway is required for ABCA1 gene expression
To investigate the importance of SREBP-2 for ABCA1 gene expression, we utilized CHO cells overexpressing or lacking a functional SREBP-2 pathway. For mRNA expression analyses, the LDL-R, a classical SREBP-2 target gene [2], served as a positive control for changes associated with SREBP-2. The archetypal statin, compactin, was employed as a tool to increase SREBP processing and to inhibit endogenous sterol synthesis [17].
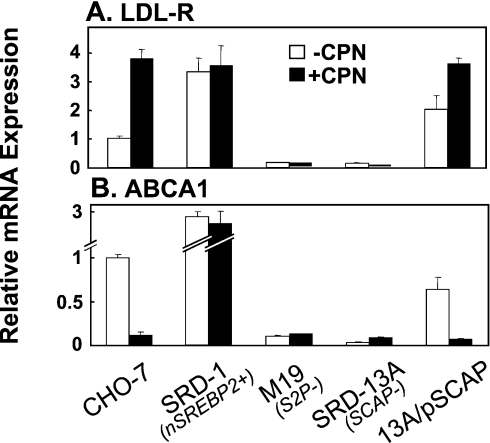
In agreement with our findings in human macrophages [17], incubation of CHO-7 wild-type cells with compactin stimulated LDL-R mRNA expression (Figure 1A) and decreased ABCA1 mRNA expression (Figure 1B). If ABCA1 gene expression is down-regulated by SREBP-2, as proposed by Zeng et al. [18], we would expect overexpression of SREBP-2 to down-regulate ABCA1. Conversely, the opposite would be true for mutant cells lacking a functional SREBP-2 pathway. SRD-1 cells, a mutant cell line overexpressing the N-terminus of SREBP-2 (constitutively active) [22,25], showed increased mRNA expression for both LDL-R and ABCA1. The cell lines, M19 and SRD-13A, are effectively SREBP-null. M19 cells [26] have a deletion in the gene encoding the Site-2 Protease which releases the active N-terminus of SREBPs from the Golgi apparatus [27]. SRD-13A cells have a deletion in the gene encoding SCAP, a protein that escorts SREBP from the endoplasmic reticulum to the Golgi apparatus for proteolytic processing [23]. mRNA expression of LDL-R and ABCA1 were virtually absent from these two SREBP-defective cell lines. Compactin treatment had no effect on LDL-R and ABCA1 mRNA expression in any of the mutant cell lines. Reinstatement of SCAP by stable transfection into SRD-13A cells (13A/pSCAP) restored gene expression of both the LDL-R and ABCA1. In addition, reinstatement of SCAP also restored responses to compactin treatment, as LDL-R and ABCA1 mRNA expressions were increased and decreased respectively. The trends observed for LDL-R gene expression were expected for that of an SREBP-2 target gene. However, in contrast with Zeng et al. [18], our results suggest that a functional SREBP-2 processing pathway may be required for transcription of ABCA1.
Figure 1. A functional SREBP pathway is required for ABCA1 gene expression.
Various CHO cell lines were incubated in the absence or presence of compactin (CPN; 5 μM) for 24 h. Cell lines were CHO-7 (wild-type), SRD-1 (overexpressing the nuclear form of SREBP-2), M19 (lacking Site-2 Protease), SRD-13A (lacking SCAP) and 13A/pSCAP (an SRD-13A cell line stably expressing pTK3-SCAP). mRNA levels for (A) LDL-R and (B) ABCA1 were measured using QRT–PCR. Data are presented relative to vehicle-treated controls and are means+S.E.M. (three replicate cultures representative of two separate experiments).
SREBP-2 is required for the synthesis of 24(S),25-epoxycholesterol
ABCA1 mRNA expression in CHO-7 wild-type cells was down-regulated by statin treatment (Figure 1B) consistent with our previous findings, which we attributed to the inhibition of LXR ligand synthesis. The level of ABCA1 down-regulation by compactin treatment in CHO-7 paralleled the levels of ABCA1 mRNA expression observed in the SREBP-defective cell lines M19 and SRD-13A. This suggests that the SREBP-defective cells may be lacking LXR ligands. LXR ligands can be derived from two main sources: de novo synthesis from acetate [e.g. 24(S),25-epoxycholesterol] or cholesterol (either endogenously or exogenously sourced). We metabolically labelled various cell lines with [1-14C]acetate, separated extracted sterols by TLC, and visualized synthesized cholesterol and 24(S),25-epoxycholesterol by phosphoimaging. Positive identification of the band corresponding to 24(S),25-epoxycholesterol has previously been performed chemically and by MS (see supplementary data of [17] at http://atvb.ahajournals.org/cgi/content/full/01.ATV.0000148707.93054.7d/DC1).
Synthesis of both sterols was inhibited by compactin treatment (results not shown). The intensity of bands corresponding to cholesterol and 24(S),25-epoxycholesterol synthesis in CHO-7 cells were comparable with THP-1 macrophages, which we previously reported to synthesize 24(S),25-epoxycholesterol [17].
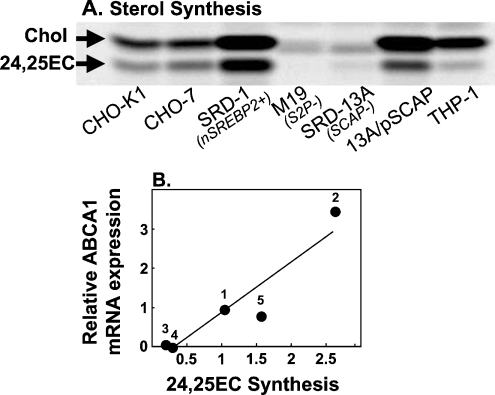
Comparing the various CHO cell lines, synthesis of cholesterol and 24(S),25-epoxycholesterol reflected the differences observed for ABCA1 mRNA expression between the various CHO mutant cell lines (Figure 2A; cf. Figure 1B). In particular, SRD-1 cells produced appreciably more cholesterol and 24(S),25-epoxycholesterol than any other cell type examined, whereas synthesis in the SREBP-defective cells, M19 and SRD-13A, was negligible. 13A/pSCAP cells with a reinstated SREBP pathway had cholesterol and 24(S),25-epoxycholesterol synthesis restored. Figure 2(B) illustrates that 24(S),25-epoxycholesterol synthesis in the CHO cell lines was positively correlated with relative expression of ABCA1. These results support the idea that SREBP pathway activity is positively associated with LXR ligand production, notably 24(S),25-epoxycholesterol, and ABCA1 gene expression.
Figure 2. SREBP-2 is required for the synthesis of 24(S),25-epoxycholesterol (24,25EC).
Various CHO cell lines and PMA-differentiated THP-1 macrophages were metabolically labelled by incubation with [1-14C]acetate. Neutral lipid extracts were separated by TLC and bands corresponding to authentic cholesterol and 24(S),25-epoxycholesterol were visualized by a phosphoimager. (A) This phosphoimage is representative of three separate experiments. (B) Values are means from the three separate experiments represented by Figures 1(B), 3(F) and 2A. Key: 1, CHO-7; 2, SRD-1; 3, M19; 4, SRD-13A; 5, 13A/pSCAP. Equation of the line for (B): y=1.32x−0.36; R2=0.88; P<0.02.
Addition of oxysterol, cholesterol and synthetic LXR ligands restores ABCA1 gene expression in cells lacking a functional SREBP-2 pathway
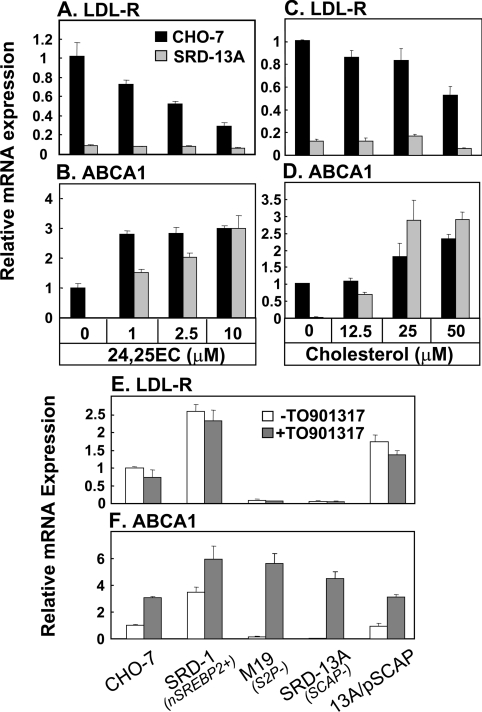
We then determined if added 24(S),25-epoxycholesterol could restore ABCA1 gene expression in cells lacking a functional SREBP-2 pathway. CHO-7 (wild-type) and SRD-13A cells (SCAP-null) were treated with increasing concentrations of 24(S),25-epoxycholesterol. LDL-R mRNA expression in CHO-7 cells was decreased in a dose-dependent manner (Figure 3A), whereas expression remained low for SRD-13A cells. Addition of 24(S),25-epoxycholesterol increased ABCA1 mRNA expression for both CHO-7 and SRD-13A cells (Figure 3B).
Figure 3. Addition of oxysterols and a synthetic LXR ligand restores ABCA1 gene expression in cells lacking SREBP-2.
CHO-7 cells (wild-type) or SRD-13A cells (lacking SCAP) were incubated with increasing concentrations of (A, B) 24(S),25-epoxycholesterol or (C, D) cholesterol complexed with methyl-β-cyclodextrin. mRNA levels for (A, C) LDL-R and (B, D) ABCA1 were measured using QRT–PCR. Data are presented relative to vehicle-treated controls and are means+S.E.M. (three replicate cultures representative of two separate experiments). Various CHO cell lines were incubated in the absence or presence of TO901317 (1 μM) for 24 h. Cell lines were CHO-7 (wild-type), SRD-1 (overexpressing the nuclear form of SREBP-2), M19 (lacking Site-2 Protease), SRD-13A (lacking SCAP), and 13A/pSCAP (an SRD-13A cell line stably expressing pTK3-SCAP). mRNA levels for (E) LDL-R and (F) ABCA1 were measured using QRT–PCR. Data are presented relative to vehicle-treated controls and are means+S.E.M. (three replicate cultures representative of two separate experiments).
Since many cell types are capable of generating oxysterols from cholesterol, we examined if cholesterol loading also corrects ABCA1 mRNA expression in SREBP-defective cells. CHO-7 and SRD-13A cells were incubated with increasing concentrations of cholesterol made soluble by complexing with methyl-β-cyclodextrin. CHO-7 LDL-R mRNA expression decreased in a concentration-dependent manner, whereas again expression remained low for SRD-13A cells (Figure 3C). ABCA1 mRNA expression was increased for both CHO-7 and SRD-13A cells (Figure 3D), suggesting that LXR ligands may be derived from cholesterol in these cells. These results suggest that synthesis of 24(S),25-epoxycholesterol in the mevalonate pathway is important for ABCA1 transcription when an alternative source of LXR ligand is limiting. When cellular cholesterol status is increased, LXR ligands can probably be sourced from cholesterol in certain cell types, including CHO-7 cells.
We next employed a non-sterol synthetic LXR ligand TO901317, which has no known effects on SREBP-2 [12]. Hence, addition of TO901317 had no significant effect on LDL-R mRNA expression (Figure 3E), but up-regulated ABCA1 expression in all the CHO cell lines overexpressing or lacking SREBP-2 (Figure 3F). In particular, TO901317 restored ABCA1 expression in the SREBP-2-defective cell lines, M19 and SRD-13A (Figure 3F), consistent with the idea that an active SREBP-2 pathway is important for ABCA1 transcription by enabling LXR ligand synthesis.
SREBP-2 is required for ABCA1 promoter activation, and the effects are mediated at the LXR response element
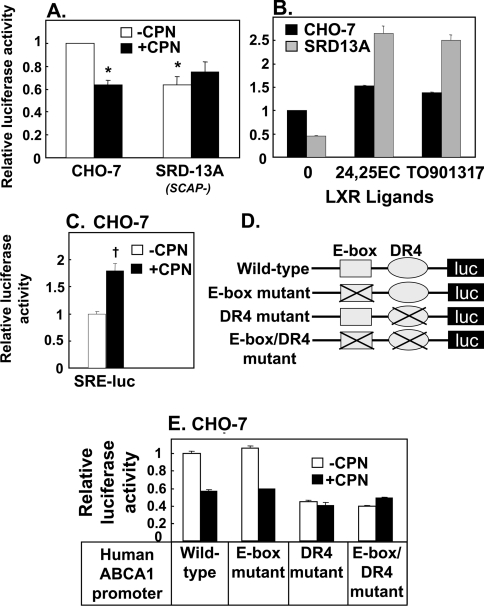
To assess ABCA1 transcription directly, transient transfection assays were performed using a human ABCA1 promoter-driven luciferase reporter [4] together with the normalizing vector phRL-TK into CHO-7 or SRD-13A cells. In agreement with ABCA1 mRNA expression data (Figures 1B and 3F), ABCA1 promoter activity was lower in SRD-13A cells relative to the wild-type CHO-7 cells (Figure 4A). Compactin treatment decreased ABCA1 promoter activity in CHO-7 cells, close to that observed in SRD-13A cells. As we observed for ABCA1 mRNA expression (Figures 3F and 3B), treatment with LXR ligands, 24(S),25-epoxycholesterol and TO901317, increased ABCA1 promoter activity in the SREBP-defective cells, SRD-13A cells (Figure 4B).
Figure 4. SREBP-2 is required for ABCA1 promoter activation and the effects are mediated by the LXR response element.
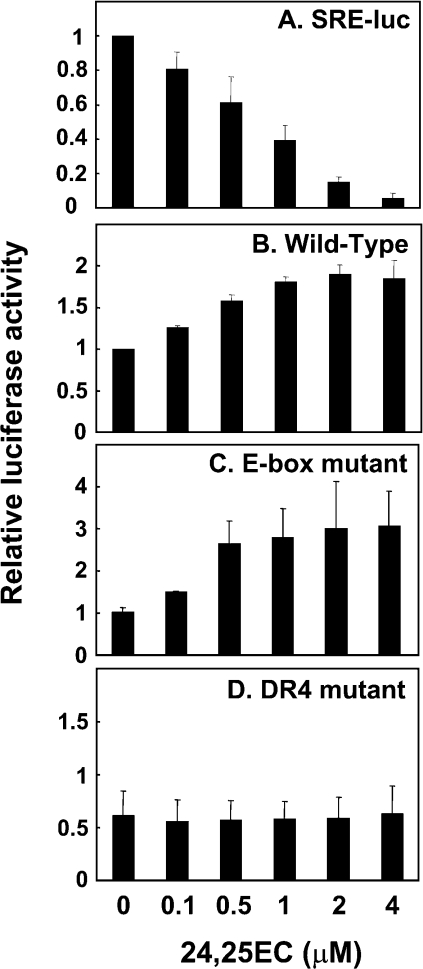
CHO-7 cells and SRD-13A cells were transiently transfected for 24 h with phRL-TK Renilla internal control plasmid together with either (A, B) pGL3-hABCA1-wild-type or (C) SRE-luc (CHO-7 cells only). (D) Schematic illustration of the E-box and DR4 elements within the proximal −200 bp promoter region of a human ABCA1 promoter-driven luciferase (luc) reporter. (E) CHO-7 cells were transiently transfected with phRL-TK Renilla internal control plasmid together with pGL3-hABCA1: wild-type, E-box mutant, DR4 mutant, E-box/DR4 mutant. Following transfection, cells were incubated for 24 h in the absence or presence of (A, C, E) compactin (5 μM) or (B) 24(S),25-epoxycholesterol (10 μM) or TO901317 (1 μM). Data are presented relative to the wild-type construct, vehicle-treated control condition, and for (A) are means+S.E.M. (n=5 separate experiments; *P≤0.05 compared with CHO-7 untreated cells); for (C), data are means+S.E.M. (n=3 separate experiments; †P=0.005); for (B, E), data are means+S.E.M. (three replicate cultures representative of two or three separate experiments).
Compactin treatment may decrease ABCA1 promoter activity by inhibiting production of an exogenous LXR ligand [17] or by increasing SREBP-2 activation and repression via the E-box [18]. Firstly, we used six tandem repeats of the SRE driving a luciferase reporter (SRE-luc) to confirm that compactin treatment activates SREBP-mediated transcription in our system (Figure 4C). To determine if the down-regulation of ABCA1 transcription by compactin is due to decreased LXR ligand production or increased SREBP activation, we mutated the E-box and/or DR4 element (to which LXR binds) in the human ABCA1 promoter (Figure 4D). When mutations were introduced into the E-box element, compactin treatment still had a down-regulatory effect on ABCA1 promoter activity (Figure 4E). However, promoter activity of the ABCA1 construct with mutations in the DR4 element to which LXR binds was lower compared with wild-type and mutated E-box promoter constructs and was unaffected by compactin treatment. This confirms our previous hypothesis [17] that statins exert their down-regulatory effect on ABCA1 expression by interfering with LXR-mediated transcription, most likely by inhibiting the LXR ligand, 24(S),25-epoxycholesterol.
Compactin treatment substantially inhibited ABCA1 gene expression (∼90%) (Figure 1A), and yet in Figure 4(A) the same treatment only lowered ABCA1 promoter activity by approx. 40%. A likely explanation for this apparent discrepancy is the relatively high levels of basal promoter activity in the luciferase assay. Thus promoter activities were comparable when cells expressing the wild-type construct were treated with compactin or when the critical DR4 motif was mutated (Figure 4E).
Incubation of CHO-7 cells with incremental concentrations of 24(S),25-epoxycholesterol showed dose-dependent changes in promoter activity. SRE-luc reporter activity was decreased (Figure 5A), while ABCA1 promoter activity was increased, for both the wild-type and mutated E-box constructs (Figures 5B and 5C). By contrast, mutation of the DR4 element completely ablated the response of the human ABCA1 promoter to 24(S),25-epoxycholesterol (Figure 5D).
Figure 5. Addition of 24(S),25-epoxycholesterol increases ABCA1 promoter activation via the LXR response element independent of the E-box.
CHO-7 cells were transiently transfected for 24 h with phRL-TK Renilla internal control plasmid together with either (A) SRE-luc or (B–D) pGL3-hABCA1: (B) wild-type, (C) E-box mutant, or (D) DR4 mutant. Following transfection, cells were incubated for 24 h in the absence or presence of indicated concentrations of 24(S),25-epoxycholesterol. Data are presented relative to the wild-type construct, vehicle-treated control condition, and for (A, B) are means+S.E.M. (n=3 separate experiments); for (C, D) data are means+half range (n=2 separate experiments).
Taken together, these results confirm current concepts that 24(S),25-epoxycholesterol and statin treatments influence ABCA1 transcription via supply of oxysterol ligands for LXR and not through an SREBP-2/E-box-related mechanism. Therefore SREBP-2 does not negatively regulate ABCA1 expression through the E-box element in this system. These results further reinforce the assertion that SREBP-2 is important for the generation of LXR ligands for ABCA1 transcription.
DISCUSSION
In the present study, we explored the role of SREBP-2 in the regulation of the LXR-target gene ABCA1. Contrary to the findings of Zeng et al. [18] in vascular endothelial cells, we present compelling data that show that SREBP-2 is in fact required for ABCA1 transcription by maintaining a supply of endogenous oxysterol ligands for LXR. Below we present several lines of evidence in support of this contention.
Our studies in CHO mutant cell lines suggest that SREBP-2 does not influence ABCA1 gene expression as a negative regulator. We observed increased ABCA1 mRNA levels in SREBP-2-overexpressing cells (Figures 1B and 3F), reduced ABCA1 expression and promoter activity in an SREBP-2-lacking cell line (Figures 1B, 3F, 4A and 4B), and also compactin decreased ABCA1 promoter activity even when the E-box element was mutated (Figure 4E). These results indicate a critical role of SREBP-2 as a positive regulator of ABCA1 gene expression. Through induction of the mevalonate pathway and subsequent production of an endogenous ligand for LXR and/or perhaps through up-regulation of the LDL-R for lipoprotein cholesterol uptake, SREBP-2 indirectly up-regulates ABCA1. We showed that CHO cells can synthesize the potent LXR ligand, 24(S),25-epoxycholesterol, in an SREBP-2-dependent manner (Figure 2A) and that reinstatement of a functional SREBP-2 pathway in SREBP-defective SRD-13A cells (by stably transfecting SCAP to create 13A/pSCAP cells) restores 24(S),25-epoxycholesterol synthesis (Figure 2A) and ABCA1 transcription (Figure 1B). In cells lacking the SREBP-2 pathway, ABCA1 gene expression and/or promoter activity were restored by addition of 24(S),25-epoxycholesterol or a synthetic LXR ligand (Figure 4B). The importance of LXR was further emphasized by the fact that an intact DR4 element was essential for ABCA1 promoter activity, even when the E-box element was mutated (Figure 4E). Therefore we propose that SREBP-2 does not work as a negative regulator, but acts as an indirect positive regulator by permitting the generation of oxysterol ligands for LXR. SREBP-2 is required to induce the mevalonate pathway to generate LXR ligands, notably 24(S),25-epoxycholesterol. Although statins induce the SREBP-2 pathway, they inhibit 24(S),25-epoxycholesterol production by blocking HMG-CoA reductase. Hence, statins lower the activity of ABCA1 gene expression by inhibiting the mevalonate pathway and subsequent formation of this LXR ligand.
Two studies in the mouse macrophage cell line RAW 264.7 [28,29] have shown that the E-box element is a repressor of ABCA1 promoter activity. Mutagenesis of the E-box element increases constitutive ABCA1 activity, consistent with trends we observed in CHO cells (e.g. Figure 5C versus Figure 5B) and THP-1 human macrophages (see Supplementary Figure S1 at http://www.BiochemJ.org/bj/400/bj4000485add.htm), yet neither of the previous studies [28,29] have detected any binding of SREBP-2 to the E-box element in the ABCA1 promoter. DNA–protein interactions at the E-box element were examined through gel shift assays. While supershift experiments with antibodies specific for transcription factors that are known to bind E-boxes did not detect any SREBP, Yang et al. [29] independently demonstrated that repression at the ABCA1 promoter was caused by a triple complex involving USF-1 (upstream stimulatory factor-1), USF-2 and Fra2 (fos-related antigen 2). Thus any down-regulatory effect associated with the E-box element in the ABCA1 promoter is likely to be independent of SREBP-2.
A role for SREBP-2 in the positive regulation of LXR-target genes has been implied in two previous studies [12,30]. Forman et al. [30] showed that products of the mevalonate pathway can serve as ligands for LXR. Inhibition of the mevalonate pathway using a statin decreased LXR reporter activity, and this could be overcome by addition of mevalonate or oxysterols [20(S)-hydroxycholesterol and 22(R)-hydroxycholesterol]. Similarly, DeBose-Boyd et al. [12] used statins to explore the regulation of an LXR-target gene, SREBP-1c, in rat hepatoma cells. SREBP-1c expression was decreased by compactin, whereas addition of mevalonate and LXR ligands [including 22(R)-hydroxycholesterol] was able to reverse the down-regulatory effect. It was concluded that transcription of SREBP-1c in hepatocytes required activation of LXR by an oxysterol intermediate in the mevalonate pathway, and 24(S),25-epoxycholesterol was proposed as a candidate LXR ligand [12]. However, our current study provides the first direct evidence for a link between 24(S),25-epoxycholesterol and LXR-target gene transcription, notably in relation to the important cholesterol efflux gene, ABCA1.
In terms of LXR ligands, we have focused on 24(S),25-epoxycholesterol, but it is known that cholesterol can also give rise to oxysterol LXR ligands [31,32]. Thus the addition of cholesterol, like addition of 24(S),25-epoxycholesterol, restored ABCA1 gene expression in SREBP-defective cells (Figure 3D). In the present study, we did not attempt to characterize the oxysterols derived from cholesterol by CHO cells.
In conclusion, SREBP-2 is central to the co-ordinated regulation of cholesterol homoeostasis, well known for its regulatory role in cholesterol uptake and cholesterol synthesis. Our current work provides new evidence suggesting an additional role for SREBP-2 in positively regulating ABCA1, a key gene involved in cholesterol efflux. We have demonstrated that SREBP-2 is required for the generation of LXR ligands within the cell, notably 24(S),25-epoxycholesterol, and that in the absence of SREBP-2, ABCA1 gene expression is abolished. Co-ordinated control of a cholesterol efflux gene together with genes involved in cholesterol synthesis and uptake may represent a ‘safety valve’ to prevent cholesterol overload in the cell.
Online data
Acknowledgments
We thank Dr Michael S. Brown and Dr Joseph L. Goldstein for generously providing cell lines and reagents, and Dr Alan Tall for his gift of the human ABCA1 promoter-driven luciferase reporter. We are grateful to Robin (Ximing) Du for generating the 13A/pSCAP stable cell line in-house, and to Dr Ingrid Gelissen for critically reading this paper. This work was supported by a grant from the NHMRC (National Health and Medical Research Council; no. 350828). C. M. Q. was supported by the NHMRC Atherosclerosis Program (222722).
References
- 1.Goldstein J. L., DeBose-Boyd R. A., Brown M. S. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 2.Briggs M. R., Yokoyama C., Wang X., Brown M. S., Goldstein J. L. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J. Biol. Chem. 1993;268:14490–14496. [PubMed] [Google Scholar]
- 3.Kim J. B., Spotts G. D., Halvorsen Y. D., Shih H. M., Ellenberger T., Towle H. C., Spiegelman B. M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix–loop–helix domain. Mol. Cell. Biol. 1995;15:2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costet P., Luo Y., Wang N., Tall A. R. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 2000;275:28240–28245. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- 5.Edwards P. A., Kennedy M. A., Mak P. A. LXRs; oxysterol-activated nuclear receptors that regulate genes controlling lipid homeostasis. Vasc. Pharmacol. 2002;38:249–256. doi: 10.1016/s1537-1891(02)00175-1. [DOI] [PubMed] [Google Scholar]
- 6.Tall A. R., Wang N. Tangier disease as a test of the reverse cholesterol transport hypothesis. J. Clin. Invest. 2000;106:1205–1207. doi: 10.1172/JCI11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soumian S., Albrecht C., Davies A. H., Gibbs R. G. ABCA1 and atherosclerosis. Vasc. Med. 2005;10:109–119. doi: 10.1191/1358863x05vm593ra. [DOI] [PubMed] [Google Scholar]
- 8.Willy P. J., Umesono K., Ong E. S., Evans R. M., Heyman R. A., Mangelsdorf D. J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 9.Peet D. J., Janowski B. A., Mangelsdorf D. J. The LXRs: a new class of oxysterol receptors. Curr. Opin. Genet. Dev. 1998;8:571–575. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- 10.Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXRα. Proc. Natl. Acad. Sci. U.S.A. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teboul M., Enmark E., Li Q., Wikstrom A. C., Pelto-Huikko M., Gustafsson J. A. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc. Natl. Acad. Sci. U.S.A. 1995;92:2096–2100. doi: 10.1073/pnas.92.6.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeBose-Boyd R. A., Ou J., Goldstein J. L., Brown M. S. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1477–1482. doi: 10.1073/pnas.98.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janowski B. A., Shan B., Russell D. W. The hypocholesterolemic agent LY295427 reverses suppression of sterol regulatory element-binding protein processing mediated by oxysterols. J. Biol. Chem. 2001;276:45408–45416. doi: 10.1074/jbc.M108348200. [DOI] [PubMed] [Google Scholar]
- 14.Bjorkhem I. Do oxysterols control cholesterol homeostasis? J. Clin. Invest. 2002;110:725–730. doi: 10.1172/JCI16388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schroepfer G. J., Jr Oxysterols: modulators of cholesterol metabolism and other processes. Physiol. Rev. 2000;80:361–554. doi: 10.1152/physrev.2000.80.1.361. [DOI] [PubMed] [Google Scholar]
- 16.Spencer T. A., Gayen A. K., Phirwa S., Nelson J. A., Taylor F. R., Kandutsch A. A., Erickson S. K. 24(S),25-Epoxycholesterol. Evidence consistent with a role in the regulation of hepatic cholesterogenesis. J. Biol. Chem. 1985;260:13391–13394. [PubMed] [Google Scholar]
- 17.Wong J., Quinn C. M., Brown A. J. Statins inhibit synthesis of an oxysterol ligand for the liver X receptor in human macrophages with consequences for cholesterol flux. Arterioscler. Thromb. Vasc. Biol. 2004;24:2365–2371. doi: 10.1161/01.ATV.0000148707.93054.7d. [DOI] [PubMed] [Google Scholar]
- 18.Zeng L., Liao H., Liu Y., Lee T. S., Zhu M., Wang X., Stemerman M. B., Zhu Y., Shyy J. Y. Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J. Biol. Chem. 2004;279:48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 19.Brown A. J., Sun L., Feramisco J. D., Brown M. S., Goldstein J. L. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell. 2002;10:237–245. doi: 10.1016/s1097-2765(02)00591-9. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein J. L., Basu S. K., Brown M. S. Receptor-mediated endocytosis of low-density lipoprotein in cultured cells. Methods Enzymol. 1983;98:241–260. doi: 10.1016/0076-6879(83)98152-1. [DOI] [PubMed] [Google Scholar]
- 21.Nohturfft A., Brown M. S., Goldstein J. L. Sterols regulate processing of carbohydrate chains of wild-type SREBP cleavage-activating protein (SCAP), but not sterol-resistant mutants Y298C or D443N. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12848–12853. doi: 10.1073/pnas.95.22.12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metherall J. E., Goldstein J. L., Luskey K. L., Brown M. S. Loss of transcriptional repression of three sterol-regulated genes in mutant hamster cells. J. Biol. Chem. 1989;264:15634–15641. [PubMed] [Google Scholar]
- 23.Rawson R. B., DeBose-Boyd R., Goldstein J. L., Brown M. S. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J. Biol. Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 24.Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. Structural requirements of ligands for the oxysterol liver X receptors LXRα and LXRβ. Proc. Natl. Acad. Sci. U.S.A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J., Sato R., Goldstein J. L., Brown M. S. Sterol-resistant transcription in CHO cells caused by gene rearrangement that truncates SREBP-2. Genes Dev. 1994;8:1910–1919. doi: 10.1101/gad.8.16.1910. [DOI] [PubMed] [Google Scholar]
- 26.Hasan M. T., Chang C. C., Chang T. Y. Somatic cell genetic and biochemical characterization of cell lines resulting from human genomic DNA transfections of Chinese hamster ovary cell mutants defective in sterol-dependent activation of sterol synthesis and LDL receptor expression. Somat. Cell Mol. Genet. 1994;20:183–194. doi: 10.1007/BF02254759. [DOI] [PubMed] [Google Scholar]
- 27.Rawson R. B., Zelenski N. G., Nijhawan D., Ye J., Sakai J., Hasan M. T., Chang T. Y., Brown M. S., Goldstein J. L. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 28.Langmann T., Porsch-Ozcurumez M., Heimerl S., Probst M., Moehle C., Taher M., Borsukova H., Kielar D., Kaminski W. E., Dittrich-Wengenroth E., et al. Identification of sterol-independent regulatory elements in the human ATP-binding cassette transporter A1 promoter: role of Sp1/3, E-box binding factors, and an oncostatin M-responsive element. J. Biol. Chem. 2002;277:14443–14450. doi: 10.1074/jbc.M110270200. [DOI] [PubMed] [Google Scholar]
- 29.Yang X. P., Freeman L. A., Knapper C. L., Amar M. J., Remaley A., Brewer H. B., Jr, Santamarina-Fojo S. The E-box motif in the proximal ABCA1 promoter mediates transcriptional repression of the ABCA1 gene. J. Lipid Res. 2002;43:297–306. [PubMed] [Google Scholar]
- 30.Forman B. M., Ruan B., Chen J., Schroepfer G. J., Jr, Evans R. M. The orphan nuclear receptor LXRα is positively and negatively regulated by distinct products of mevalonate metabolism. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu X., Menke J. G., Chen Y., Zhou G., MacNaul K. L., Wright S. D., Sparrow C. P., Lund E. G. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J. Biol. Chem. 2001;276:38378–38387. doi: 10.1074/jbc.M105805200. [DOI] [PubMed] [Google Scholar]
- 32.Lund E. G., Menke J. G., Sparrow C. P. Liver X receptor agonists as potential therapeutic agents for dyslipidemia and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2003;23:1169–1177. doi: 10.1161/01.ATV.0000056743.42348.59. [DOI] [PubMed] [Google Scholar]
- 33.Field F. J., Born E., Mathur S. N. Stanol esters decrease plasma cholesterol independently of intestinal ABC sterol transporters and Niemann–Pick C1-like 1 protein gene expression. J. Lipid Res. 2004;45:2252–2259. doi: 10.1194/jlr.M400208-JLR200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.