Abstract
Hyperphosphorylated tau proteins accumulate in the paired helical filaments of neurofibrillary tangles seen in such tauopathies as Alzheimer's disease. In the present paper we show that tau turnover is dependent on degradation by the proteasome (inhibited by MG132) in HT22 neuronal cells. Recombinant human tau was rapidly degraded by the 20 S proteasome in vitro, but tau phosphorylation by GSK3β (glycogen synthase kinase 3β) significantly inhibited proteolysis. Tau phosphorylation was increased in HT22 cells by OA [okadaic acid; which inhibits PP (protein phosphatase) 1 and PP2A] or CsA [cyclosporin A; which inhibits PP2B (calcineurin)], and in PC12 cells by induction of a tet-off dependent RCAN1 transgene (which also inhibits PP2B). Inhibition of PP1/PP2A by OA was the most effective of these treatments, and tau hyperphosphorylation induced by OA almost completely blocked tau degradation in HT22 cells (and in cell lysates to which purified proteasome was added) even though proteasome activity actually increased. Many tauopathies involve both tau hyperphosphorylation and the oxidative stress of chronic inflammation. We tested the effects of both cellular oxidative stress, and direct tau oxidative modification in vitro, on tau proteolysis. In HT22 cells, oxidative stress alone caused no increase in tau phosphorylation, but did subtly change the pattern of tau phosphorylation. Tau was actually less susceptible to direct oxidative modification than most cell proteins, and oxidized tau was degraded no better than untreated tau. The combination of oxidative stress plus OA treatment caused extensive tau phosphorylation and significant inhibition of tau degradation. HT22 cells transfected with tau–CFP (cyan fluorescent protein)/tau–GFP (green fluorescent protein) constructs exhibited significant toxicity following tau hyperphosphorylation and oxidative stress, with loss of fibrillar tau structure throughout the cytoplasm. We suggest that the combination of tau phosphorylation and tau oxidation, which also occurs in tauopathies, may be directly responsible for the accumulation of tau aggregates.
Keywords: oxidative stress, phosphorylation, proteasome, RCAN1, tau protein
Abbreviations: Cdk, cyclin-dependent kinase; CFP, cyan fluorescent protein; CLSM, confocal laser-scanning microscopy; CsA, cyclosporin A; DMEM, Dulbecco's modified Eagle's medium; DTT, dithiothreitol; GFP, green fluorescent protein; GSK3β, glycogen synthase kinase 3β; HNE, 4-hydroxy-2-nonenal; I-2, PP (protein phosphatase) 1 inhibitor 2; OA, okadaic acid; PHF, paired helical filament; PP, protein phosphatase
INTRODUCTION
The brain of an Alzheimer's disease patient exhibits two main types of lesions: extracellular neuritic plaques and intracellular neurofibrillary tangles. Plaques represent extracellular deposits of amyloid peptides. Neurofibrillary tangles are bundles of abnormally phosphorylated tau proteins containing PHFs (paired helical filaments). The six isoforms of the tau protein [1] play important roles in stabilizing microtubules; the ‘railway’ of axonal transport. How and why plaques and neurofibrillary tangles develop is not fully understood. The actual function of tau depends on its phosphorylation status. Tau can be phosphorylated by several kinases, and it is highly phosphorylated during brain development [2]. Phosphorylation of tau occurs in different regions of the protein. The mechanism by which tau binds (and rebinds) to microtubules seems to be regulated via phosphorylation and dephosphorylation at specific sites [3–6]. GSK3β (glycogen synthase kinase 3β), a proline-directed serine/threonine kinase, ubiquitously expressed in mammalian tissues, has been implicated as a major tau kinase in both normal and Alzheimer's disease brain [7]. Epitopes phosphorylated by GSK3β are among the phoshorylation sites of PHF-tau seen in Alzheimer's disease, and GSK3β has been strongly implicated in tau hyperphosphorylation [8].
Dephosphorylation of tau is also crucial for the normal functional status of the protein. PP (protein phosphatase) 1, PP2A and PP2B (calcineurin) are responsible for this process [9]. PP2B is a calcium/calmodulin-activated serine/threonine phosphatase, regulated by RCAN1 (previously also known as Adapt78, DSCR1 or calcipressin1). RCAN1 overexpression leads to inhibition of PP2B and increased tau phosphorylation [10]. PP2A appears to be a major phosphatase regulating tau phosphorylation in the brain [11,12], but downregulation of PP2A alone, in animal brains, does not produce PHFs [12,13]. Thus other phosphatases, or additional regulatory mechanisms, may be responsible for the development of PHFs. Cdk (cyclin-dependent kinase) 5 is also involved in tau dephosphorylation through its interaction with I-2 (PP1 inhibitor-2). Cdk5 phosphorylates I-2, causing the inhibitor to dissociate from PP1. Thus by dissociating I-2 from PP1, Cdk5 actually de-represses PP1 activity and promotes tau dephosphorylation [14].
Increasing evidence suggests that an inhibition of the proteasome might be involved in Alzheimer's disease. Inhibition or dysfunction of the proteasome system, which degrades damaged proteins, can cause protein aggregation, and this has been suggested as a mechanism for neurodegeneration in Alzheimer's disease [15]. The proteasome does appear to play a major role in the turnover of the tau protein [16,17], although PHF-tau appears to actually inhibit the proteasome. It has been suggested that proteasome dysfunction in Alzheimer's disease brain may result from the inhibitory binding of PHF-tau [15]. Whether phosphorylation of the proteasome itself (or its regulatory subunits) might affect tau degradation has not been carefully studied.
Oxidative stress is a major pathological component of neuro-degenerative conditions such as Parkinson's [18] and Alzheimer's [19] diseases. Alzheimer's disease involves the formation of significant markers of oxidative stress, such as 8OHDG (8-hydroxy-2′-deoxyguanosine), HNE (4-hydroxy-2-nonenal) [20,21] and intraneuronal accumulations of oxidatively damaged proteins [22]. Oxidation markers are also found in the accumulated tau protein aggregates seen in Alzheimer's disease [23]. The effects of oxidative stress on phosphorylation of the tau protein are complex, and appear to be somewhat contradictory [24–26]. Under some conditions, H2O2 (hydrogen peroxide) can decrease tau phosphorylation via activation of Cdk5, leading to increased phosphorylation of I-2 which results in PP1 de-repression/activation and decreased tau phosphorylation [21]. On the other hand, H2O2 can also induce the upregulation of the RCAN1 protein (product of the RCAN1 gene), leading to inhibition of PP2B and an increased phosphorylation of tau [10].
In addition to these complex effects of H2O2 on phosphorylation, we have also found that whereas mild oxidative stress typically increases the proteolytic susceptibility of intracellular proteins, severe oxidation promotes protein aggregation and cross-linking, and results in decreased proteolysis [27–30]. It therefore seemed important to determine the overall effects of oxidative stress on phosphorylation of the tau protein, how tau phosphorylation influences its degradation by the proteasome, and how tau degradation is actually catalysed under oxidative conditions. In the present study, we have examined these questions using two different neuronal cell lines (PC12 and HT22 cells), and we have also tested the toxicity of tau protein aggregates in these cells under normal, oxidizing and hyperphosphorylating conditions.
EXPERIMENTAL
Materials
All medium was purchased from Invitrogen or Gibco BRL; serum was from Biochrom KG; and medium supplements were from Seromed, Gibco BRL or PAA. The different inhibitors were obtained from Affiniti (proteasome inhibitor MG132), Sigma [CsA (cyclosporin A)] and Calbiochem [OA (okadaic acid)]. Other materials were purchased from Perkin Elmer, Sigma and Amersham. The various antibodies used were obtained from BD Pharmingen (Tau-5), Autogen Bioclear UK (Alzheimer's disease specific antibody AT100), Biosource (phospho-tau pSer199/Ser202 and pThr231), and secondary antibodies were from Amersham. PHF-1 was a gift from Dr P. Davies (Albert Einstein College, Bronx, NY, U.S.A.) [31].
Cell culture
HT22 cells were maintained in T75 (75 cm2) flasks using DMEM (Dulbecco's modified Eagle's medium) supplemented with 10% foetal calf serum, 1% glutamine and 0.35% glucose under an atmosphere of 5% CO2 at 37 °C. PC12 cells were maintained in T75 flasks using RPMI medium supplemented with 10% horse serum, 5% foetal calf serum and 1% glutamine. Cells were subcultured before reaching confluence and the medium was changed three times per week. Cells were dissociated and seeded into T25 (25 cm2) or T75 flasks 24 h before experiments were started.
Isolation of recombinant tau from bacterial cells
Recombinant tau, pEThtau40 [provided by Professor E. Mandelkow (DESY, Hamburg, Germany) and Professor E. M. Mandelkow (Max Planck Unit for Structural Molecular Biology)], was expressed in Escherichia coli BLN21(DE3) pLysS. The bacteria were grown overnight at 37 °C in 300–350 ml Luria–Bertani medium and 0.4 mM IPTG (isopropyl β-D-thiogalactoside) was added to start translation. Bacteria were pelleted at 4 °C, supernatants were removed, and the pellets were resuspended in 1–3 ml of PBS containing a protease inhibitor cocktail. To lyse the cells, 2 mg/ml of lysozyme was added to each pellet and incubated for 30 min on ice. The mixture was then boiled for 30 min followed by a 30 min centrifugation. Recombinant tau was then concentrated from the supernatants in centricon tubes [Amicon, 10000 MWCO (molecular mass cut-off)].
Regulated RCAN1 expression in PC12 cells
The RCAN1 gene was overexpressed in PC12 tet-off cells, using the RCAN1 transgene system we have described previously [10,32]. This system provides regulated, high-level expression of the RCAN1 transgene in response to the withdrawal of doxycycline from the medium.
Transfection of tau–GFP (green fluorescent protein) and tau–CFP (cyan fluorescent protein)
A tau–GFP fusion protein plasmid and a tau–CFP fusion protein plasmid (provided by Professor E. Mandelkow and Professor E. M. Mandelkow) were transfected in HT22 using TransFectin™ (BioRad). One day before transfection, 5×104 cells were seeded into glass bottom Petri dishes (35 mm diameter). Inhibitor or oxidant treatment was performed 1 day after transfection. Tau distribution was investigated by CLSM (confocal laser-scanning microscopy) using a Zeiss LSM 510 confocal laser-scanning microscope.
Measurement of tau turnover and tau immunoprecipitation
For endogenous protein labelling, cells were seeded at a density of 3×104 cells/cm2 and incubated with [3H]lysine-supplemented medium at a concentration of 46.25 kBq/ml (1.25 μCi/ml) for 72 h. Prior to the experiments, cells were washed twice in culture medium to remove any remaining unincorporated radioactivity, and all experiments were performed either in medium, or in PBS, without any additional radioactive substance. To immunoprecipite the tau protein, cells were harvested and centrifuged at 300 g for 10 min. The cells were washed twice with cold PBS (PAA) and centrifuged at 10000 g for 10 min. The pellet was resuspended in 500 μl PBS and homogenized using a syringe. The cells were lysed by freeze-thawing cycles and the protein content was determined. Immunoprecipitation of the tau protein was performed according to Zwilling et al. [33]. Briefly, cell lysates were first pre-adsorbed on 10% Protein A–Sepharose, equilibrated 1:1 with Tris/NaCl buffer containing 50 mM Tris/HCl (pH 8.0), 150 mM NaCl and 0.1% BSA. A primary antibody reaction was then performed with 100 μg of lysate protein and 4 μg of the Tau-5 antibody directed against the phosphorylation-independent epitope in the centre of the tau protein, in a total volume of 2000 μl of Tris/NaCl buffer, for 2 h at 4 °C. For secondary antibody reactions, 4 μg of HRP (horseradish peroxidase)-conjugated sheep anti-(mouse IgG) (0.5 μg/μl stock) was added and incubated for 2 h at 4 °C. Precipitation was achieved by addition of 40 μl of Protein A–Sepharose for 16 h at 4 °C. Controls were incubated only with the secondary antibody and Protein A–Sepharose. The Sepharose was then centrifuged (12000 g for 30 sec) and washed five times with Tris-buffered saline (50 mM Tris and 150 mM NaCl). Sepharose-bound proteins were separated by SDS/PAGE (12% gels) and stained with Coomassie Blue. The stained bands were excised and radioactivity was measured by scintillation counting. Control samples, containing secondary antibody plus Protein A–Sepharose, or Protein A–Sepharose alone, produced no radioactive bands in the gels.
Immunoblot analyses
Normally, proteins were extracted from cells by gentle shaking in lysis buffer [1 mM DTT (dithiothreitol) in 50 mM Tris/HCl and 25 mM NaCl] on ice, followed by centrifugation at 14000 g for 30 min at 4 °C. For some reactions, the pellet was resuspended in lysis buffer [250 mM sucrose, 25 mM Hepes, 10 mM MgCl2·6H2O, 1 mM EDTA and 1 mM DTT (pH 7.8)], lysed by freeze–thawing cycles, and centrifuged at 10000 g for 10 min. In both cases, supernatants were removed for protein determination. Identical amounts of total protein were boiled in loading buffer followed by electrophoretic separation, and transfer on to PVDF membranes (Amersham). Afterwards the membranes were incubated with primary and secondary antibodies, and finally analysed for immunoreactivity using a POD chemiluminescence kit (NEN). Pre-stained precision molecular mass markers (Bio-Rad) were used to estimate the apparent molecular masses of the protein bands. The relative absorbances were quantified using TabLab software.
ELISA for detection of protein oxidation
The protein carbonyl content was determined in cell lysates (4 mg/ml in lysis buffer with 1 mM butylated hydroxytoluene) by an ELISA as described previously by Buss et al. [34] with the modifications described by Sitte et al. [35]. Primary dinitrophenyl rabbit IgG antiserum (Sigma) and a secondary monoclonal POD-conjugated anti-(rabbit IgG) (Sigma) were used as the detection system. Development was performed with o-phenylene diamine and H2O2.
Immunodetection of oxidized tau
Tau oxidation in cell lysates was assayed by increasing carbonylation of the tau protein, following immunoprecipitation and derivatization with 2,4-dinitrophenylhydrazine. Proteins were separated by SDS/PAGE, transferred to PVDF membranes and then incubated with an antibody directed against the derivatized carbonyl group. Oxidized tau was visualized using the POD chemiluminescence kit (NEN) and BioMax films (Kodak). The relative optical densities were quantified using TabLab software.
Proteasome inhibition
To inhibit proteasome activity, cells were pre-incubated for 1 h at 37 °C, with medium containing the selective proteasome inhibitor MG132, at a concentration of 0.5 μM. MG132 is known to inhibit proteasome reversibly and, indeed, we observed increases in activity after 2 h. Therefore the treatment was changed to a 1 h pre-incubation and up to 24 h continuous treatment with MG132 at a concentration of 10 μM.
Proteasome activity
The activity of proteasome was determined using the artificial fluorogenic peptide suc-LLVY-MCA (Bachem), whose degradation reflects the chymotrypsin-like activity of this multicatalytic proteinase complex. Cells were washed twice with PBS and lysed by repeated freeze–thaw cycles in 1 mM DTT. Lysates were centrifuged at 10000 g for 30 min and protein concentrations were determined using a BCA Kit (Pierce). Cell lysates (10 μl) were incubated with 80 μl proteolysis buffer [0.15 M sucrose, 25 mM Hepes (pH 7.8), 20 mM MgCl2, 1 mM EDTA and 1 mM DTT] containing 10 μl suc-LLVY-MCA (2 mM stock solution in DMSO). This solution was incubated for 30 min at 37 °C and the reaction was stopped by addition of an equal volume of ice-cold 96% ethanol. Fluorescence determinations were carried out at an excitation wavelength of 380 nm and an emission wavelength of 440 nm, using free MCA as a standard, and background fluorescence (without suc-LLVY-MCA) was subtracted.
OA and CsA treatment
The PP1/PP2A inhibitor OA or the PP2B inhibitor CsA was added to washed cells in DMEM supplemented with 10% foetal calf serum, 1% glutamine and 0.35% glucose. In various experiments, cells were incubated for 1, 3, 6 or 24 h with 0.5 μM OA or 5 μM CsA.
Combined treatment with OA and H2O2
After pre-incubation with OA at 0.5 μM for 3 h, the medium was replaced by PBS containing 1 mM H2O2 and cells were incubated for a further 30 min. The cells were then washed with warm PBS and either harvested or cultivated further with normal DMEM for up to 24 h.
Cell viability
Cells were collected and stained with 0.4% (w/v) Trypan Blue dye. The number of viable cells was determined by counting in a Neubauer cytometer.
RESULTS AND DISCUSSION
Proteasome-dependent tau turnover
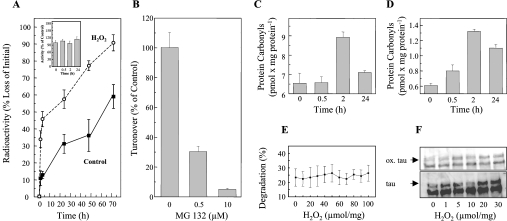
Our experiments indicate that the proteasome is primarily responsible for tau degradation (Figure 1). As demonstrated in Figure 1(A), the half-life of tau is approx. 60 h in HT22 cells, and tau degradation is blocked by the proteasome inhibitor MG132 (Figure 1B). These results confirm and extend the findings of Shimura et al. [16], Petrucelli et al. [36] and David et al. [17], firmly establishing the proteasome-dependency of tau turnover.
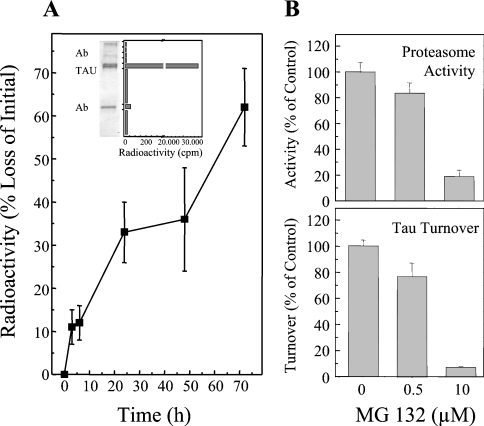
Figure 1. Measurement of tau turnover by the proteasome in HT22 neuronal cells.
Cells were labelled with [3H]lysine for 72 h. Tau was then immunoprecipitated and analysed by electrophoresis. To measure the radioactivity in the tau proteins the resulting gel was cut into bands and the bands were analysed by scintillation counting. The immunoblot inset in (A) shows that such a radioactivity profile contains one major band which is the tau protein as revealed by immunoblotting. Turnover of the tau protein was measured by this technique, and the results are presented in the main part of (A). (B) The inhibition of proteasome activity, as measured by cleavage of the suc-LLVY-AMC peptide substrate, by MG132. (C) Inhibition of cellular tau degradation by the proteasome determinations.
Next, we examined the capacity of purified 20 S proteasome to degrade the tau protein in vitro (Figure 2A). For these experiments, we expressed human tau (htau40 gene) in E. coli, purified the recombinant protein, and incubated it with purified 20 S proteasome to measure proteolysis. For some experiments purified tau was first phosphorylated by incubation with GSK-3β (pTS199/S202 in Figure 2A), in order to test the effects of phosphorylation on proteolytic susceptibility. Importantly, proteasome was not exposed to the phosphorylating system at any time. Tau was rapidly degraded by the 20 S proteasome, but pTS199/S202 (phospho-tau) was degraded approx. 50% slower. Since the experiment of Figure 2(A) only reports the effects of tau phosphorylation by GSK-3β, and this kinase is only one of several that may phosphorylate the tau protein, we decided to try to hyperphosphorylate tau in intact cells (OA inhibition of phosphatases) and then lyse the cells and measure tau degradation by added proteasome. As shown in Figure 2(B), tau phosphorylation greatly increased after HT22 cells were exposed to OA: both pTS199/S202 and pTT231 increased to a very similar extent to that shown in Figure 3(A). Importantly, when the cells were lysed and purified proteasome was added, the degradation of pTS199/S202 was inhibited by approx. 62%, and the degradation of pTT231 was inhibited by approx. 78%, relative to native tau (please note the different y-axis scales in Figures 2A and 2B). Thus from Figures 2(A) and 2(B), we can reasonably conclude that phosphorylation of tau makes the protein more resistant to proteasomal degradation. As far as we are aware, this negative effect of phosphorylation on tau degradation by the 20 S proteasome is a novel finding that has not previously been discussed in the literature.
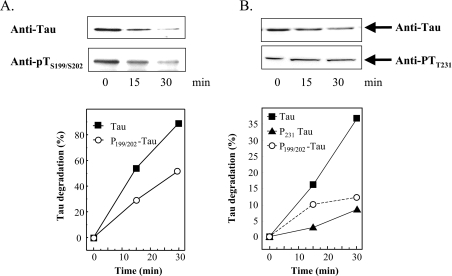
Figure 2. Tau phosphorylation inhibits tau degradation by the proteasome.
(A) Human tau was expressed in E. coli in a pET19b-plasmid (Invitrogen). Tau was purified as described by Lee et al. [37] and Keck et al. [15]. GSK3β was purchased from Biomol. Tau was phosphorylated according to Leclerc et al. [38] by adding GSK3β to purified tau (1:450 w/w) in the presence of 1 mM ATP, and incubating overnight at 30 °C. Control tau was also incubated overnight, but without GSK3β. Both non-phosphorylated tau and phospho-tau were incubated for up to 30 min with purified red blood cell 20 S proteasome (180:1, w/w) in vitro as described previously [39]. The 20 S proteasome was also purified as previously described [39]. Immunoblots, using anti-tau or anti-phospho-tau (Anti-pTS199/S202) antibodies, were performed (example shown at the top of A) and quantified (bottom of A) as described in the Materials and methods section. (B) HT22 cells were incubated for 3 h with 0.5 μM OA. The cells were then lysed and incubated with purified 20 S proteasome (180:1, w/w). The levels of two phosphorylated forms of tau, pTS199/202 and pTT231, were significantly increased by OA treatment; see Figure 3 for full details. Although tau was rapidly degraded, both pTS199/S202 and pTT231, were much more resistant to degradation by proteasome [note that the y-axis of (B) has a much smaller scale than that of (A)]. Values are means for five experiments for which S.E.M. were always less than 5%.
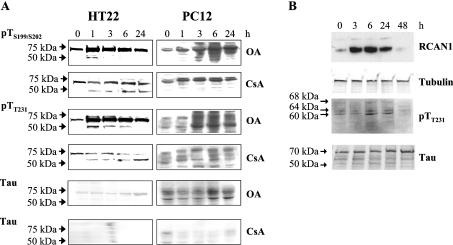
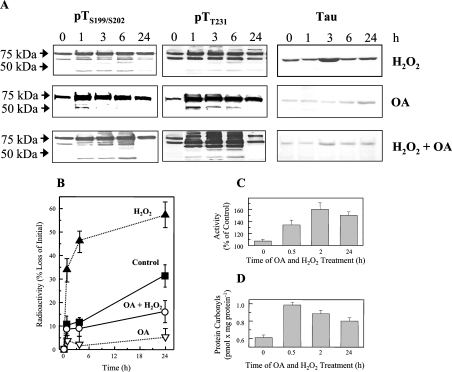
Figure 3. Tau phosphorylation induced by the phosphatase inhibitors OA and CsA, or by overexpression of the RCAN1 gene.
(A) HT22 and PC12 cells were treated with 0.5 μM OA or 5 μM CsA, and were incubated for 1–24 h. Cells were then lysed and analysed by immunoblotting with the anti-tau antibody, or the two phosphospecific antibodies anti-tau pTS199/S202 or anti-tau pTT231. (B) RCAN1 was overexpressed in PC12 cells, using the a RCAN1 transgene tet-off system [10,32], as described in the Materials and methods section. Equal amounts of total protein from each sample were loaded and tubulin detection was used to control loading levels. X-ray films were quantified using IPLab software (Scanalytics) and adjusted according to the loading. Phosphorylated tau protein (pTau) was detected using the pTT231 antibody, that specifically recognizes tau phosphorylated at Thr231. pTau protein levels were elevated approx. 88% after 6 h of RCAN1 overexpression. The elevation was statistically significant (P≤0.05), as tested by the Student's t test (one population).
Tau phosphorylation mediated by phosphatase inhibitors and RCAN1
We next turned our attention to various mechanisms for altering the phosphorylation status of tau in intact cells, so that these conditions could be used to further test how phosphorylation may regulate tau turnover. Using our two cell lines we first tested hyperphosphorylating conditions. Both neuronal cell lines were incubated with 0.5 μM OA, to inhibit PP1 and PP2A, or 5 μM CsA, to inhibit PP2B. The PC12 cell line was used to overexpress a RCAN1 transgene, using the tet-off gene expression system [10,32]. We found that OA treatment, CsA treatment and induction of the RCAN1 transgene all resulted in increased tau phosphorylation, although to differing extents and with a different pattern for each agent (Figure 3; compare the zero time point, in all cases, with 1, 2, 3, 6 and 24 h of inhibitor treatment). Of the three conditions used to induce tau hyperphosphorylation, the PP1/PP2A inhibitor OA was clearly the most potent (Figure 3A).
Tau hyperphosphorylation inhibits degradation by the proteasome
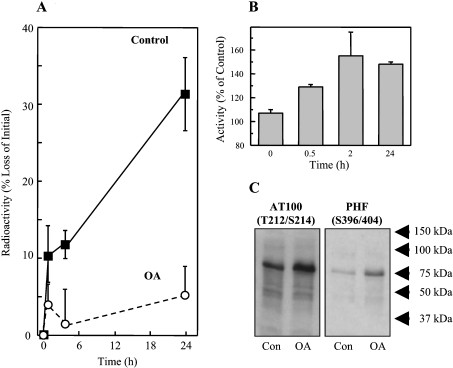
Since OA was most effective in inducing tau phosphorylation, we used this agent to test the effect of phosphorylation on tau turnover in intact cells (Figure 4). OA caused a dramatic decline in time-dependent tau degradation (Figure 4A) accompanied by an increase in tau reactivity with antibodies that normally react with the hyperphosphorylated tau proteins found in neurofibrillary tangles (AT100) and PHFs (Figure 4C). Importantly, this depression of tau turnover was not accompanied by a decline in actual proteasomal activity: in fact, proteasome activity actually increased by almost 60% during the first 2 h of OA treatment (Figure 4B). Thus the negative effect of phosphorylation on tau degradation seen in vitro in Figures 2(A) and 2(B), is also observed in intact cells.
Figure 4. Tau hyperphosphorylation and decreased turnover after treatment of HT22 neuronal cells with OA.
(A) HT22 cells were labelled with [3H]lysine for 72 h, and then treated for 3 h with 0.5 μM OA (or used as controls). Tau was then immunoprecipitated and analysed for radioactivity to calculate tau degradation, as described in Figure 1. (B) Cell lysates were tested for proteasome activity before, and at various time points after, OA treatment, using degradation of the fluorogenic substrate suc-LLVY-MCA as described in the Materials and methods section. The data in both (A) and (B) represent the means±S.E.M. for three independent determinations. (C) Immunoreactivity of HT22 cell lysates to the AT100 and PHF-1 (hyperphosphorylated tau) antibodies in control cells, and after treatment with OA.
To our knowledge, no-one has previously reported that tau hyperphosphorylation directly inhibits its degradation by the proteasome. Tau hyperphosphorylation, tau aggregation and tau accumulation all occur in important neurodegenerative diseases, such as Alzheimer's disease. Thus the direct inhibition of proteasome-dependent tau degradation by tau hyperphosphorylation may have important implications for human disease. Previously, Litersky and Johnson [40] and Wang et al. [41] described the degradation of tau by calpains, and reported an inhibitory effect of phosphorylation. Thus tau phosphorylation may be a common mechanism to inhibit tau degradation by a variety of intracellular proteases. Zhang et al. [42] used various antibodies, such as PHF-1, to examine proteasome-dependent degradation of tau isolated from rat brain, but they did not study the effects of phosphorylation. Conflicting indirect effects of phosphorylation on ubiquitin-mediated tau degradation by the 26 S proteasome have been reported. Shimura et al. [16] reported improved ubiquitinylation of tau after phosphorylation, and suggested that CHIP [C-terminus of the Hsc (heat-shock cognate) 70-interacting protein] mediates tau ubiquitinylation in a phosphorylation dependent manner. Petrucelli et al. [36], however, reported exactly the opposite results, indicating that tau phosphorylation diminishes ubiquitination and 26 S proteasomal degradation. If, as we suggest here, tau degradation is largely conducted by the 20 S proteasome in a process that is directly inhibited by tau phosphorylation, such indirect effects of tau phosphorylation on its degradation by the 26 S proteasome may be of minor importance.
Oxidative stress increases tau turnover by indirect means
Our group has found that mild oxidative modification significantly increases the proteolytic susceptibility of many proteins, and increases their degradation by the 20 S proteasome [28,29,39,43,46]. Exceptions to this rule are proteins with little or no secondary/tertiary structure, such as α-casein, which do not undergo oxidation-induced unfolding. We have shown previously [27,30] that increased recognition and degradation of oxidatively modified proteins is the consequence of an oxidation-induced unfolding process, involving the exposure of (previously shielded) hydrophobic moieties on the surface of the unfolded protein. Such hydrophobic ‘patches’ appear to be favoured substrates for the 20 S proteasome. Since the tau protein has little or no secondary or tertiary structure [44], we hypothesized that tau oxidation would have only minor effects on its degradation. Somewhat surprisingly, therefore, H2O2 significantly increased the turnover of the tau protein in intact HT22 cells (Figure 5A). In actuality, the entire increase in tau proteolysis, following cellular H2O2 treatment, occurred in the first 2 h, after which time both H2O2 treated and control cells exhibited identical rates of tau degradation (Figure 5A). Importantly, actual proteasome activity was not affected by the H2O2 treatment (Figure 5A inset). Furthermore, the turnover of tau during cellular oxidative stress (as in untreated cells) was almost completely blocked by the proteasome inhibitor MG132 (Figure 5B).
Figure 5. Influence of oxidative stress on tau turnover in HT22 neuronal cells.
(A) To measure tau turnover in HT22 cells, intracellular proteins were first radiolabelled with [3H]lysine for 72 h in a pulse–chase procedure (see the Materials and methods section). Cells were then diluted, and resuspended in PBS (pH 7.4) and treated for 30 min with 1 mM H2O2 (or used as controls). Tau was immunoprecipitated and analysed for radioactivity as described in Figure 1. The insert in (A) shows proteasome activity after H2O2 treatment, measured by degradation of the fluorogenic substrate suc-LLVY-AMC as described in the Materials and methods section. (B) The effect of the proteasome inhibitor MG132 on tau turnover after cell treatment with H2O2. (C) and (D) The oxidation status of total cellular proteins (C) or immunoprecipitated tau protein (D) measured by an ELISA technique (see the Materials and methods). (E) The degradation of isolated tau protein by the 20 S proteasome in vitro after exposure of tau (only) to various concentrations of H2O2. The data in (A)–(E) represent the means±S.E. of five independent determinations. (F) The effect of in vitro tau oxidation by H2O2 as reflected by the immunoblot determination of protein carbonyls (ox. tau), or as detected with an anti-tau antibody (tau).
We next tested the effects of oxidative stress on overall protein oxidation and tau oxidation in intact HT22 cells. Although H2O2 treatment may not be as powerful an inducer of protein carbonyls as other oxidation products, such as HNE [47–50], it does cause carbonylation of many cellular proteins [50–53], and it is certainly of both physiological and pathological significance in vivo. The overall level of protein carbonylation in HT22 cells and the level of tau carbonylation were both significantly increased by 2 h of exposure to H2O2 (Figures 5C and 5D). We have previously shown that such oxidized proteins are usually rapidly removed from the intracellular protein pool by proteolysis [45,47–53]. As shown in Figure 5(C), the carbonyl-containing (oxidized) proteins induced by H2O2 2 h after treatment, were almost completely degraded after 24 h. In contrast with these results, for the general pool of cellular proteins, less than one-third of the oxidized (i.e. carbonyl-containing) tau protein produced by cellular H2O2 treatment was degraded after 24 h (Figure 5D). It is also very interesting to note that the extent of tau oxidation was actually 8-fold lower than that seen for the general intracellular protein pool (compare the 2 h time points in Figures 5C and 5D). Thus it appears that tau may be significantly less sensitive to H2O2 oxidation in intact cells than are the majority of cellular proteins, although other oxidation products such as HNE may cause more extensive tau carbonylation and may be more affected by phosphate [47–50]. Under our conditions, however, it seems unlikely that tau oxidation could explain the increased degradation of tau seen in H2O2 treated cells (Figure 5A). We therefore decided to test the proteolytic susceptibility of tau and oxidized tau, in a simple in vitro system, with purified 20 S proteasome.
Purified tau exhibited significant susceptibility to degradation by the purified 20 S proteasome in vitro, but direct exposure to various concentrations of H2O2 (from low to high) neither increased nor decreased its proteolytic susceptibility (Figure 5E). Furthermore, direct in vitro exposure to various concentrations of H2O2 only induced a very moderate degree of oxidative modification of the tau protein, as judged by immunoblot determination of carbonyl groups (Figure 5F). In trying to reconcile the various results of Figure 5, it seems that tau is only mildly susceptible to oxidative modification, both in intact cells, and in vitro (Figures 5D–5F). Oxidatively modified tau showed no increase in proteolytic susceptibility in vitro (Figure 5E) yet cells exposed to oxidative stress exhibited an initial increase in tau degradation (Figure 5A). Although tau degradation during cellular oxidative stress was proteasome-dependent (Figure 5B), it was not the result of increased proteasome activity (Figure 5A inset). Therefore the increased turnover of tau during cellular oxidative stress seems to be a regulatory phenomenon, and not the result of conformational changes in the protein due to oxidation, or alterations in proteasome activity. These results led us to more carefully consider the potential effects of oxidative stress on tau phosphorylation, as a possible explanation of altered proteolytic turnover (see below).
Influence of oxidative stress on PP1- and RCAN1-mediated tau phosphorylation
H2O2 causes Cdk5 activation, resulting in decreased tau phosphorylation due to (phosphorylation-dependent) inactivation of the PP1 inhibitor I-2 [14]. On the other hand RCAN1 is induced during adaptation to oxidative stress, where it causes increased tau phosphorylation via inhibition of calcineurin [10,32] and induction of GSK3β [54]. Thus Cdk5 and RCAN1 can actually oppose one another during oxidative stress; depending on the cell type, and both the intensity and duration of the stress [24–26].
Therefore we next tested for changes in tau phosphorylation following the same H2O2 cell treatment used in Figures 5(A)–5(D). Our results show no significant increase in tau phosphorylation under these specific oxidative stress conditions, although subtle changes in the pattern of pTS199/S202 and pTT231 could be observed (Figure 6). The PP1/PP2A inhibitor OA caused extensive tau phosphorylation in our cells, whereas the PP2B antagonist CsA was far less effective (Figure 3A). Together with the failure of H2O2 to increase overall tau phosphorylation (Figure 6), these results suggest that PP1/PP2A are much more significant tau phosphatases in HT22 cells than is PP2B. Considering the previously mentioned results of Kins et al. [12] and Gong et al. [13], PP1 actually seems to be the most crucial for accumulation of oxidized, phosphorylated tau. To further explore this possibility we next treated cells with both H2O2 and OA. As shown in Figure 6(A), the combination of H2O2 and OA induced significant tau phosphorylation, and with a rather different pattern than that seen with either H2O2 or OA alone.
Figure 6. Tau turnover during hyperphosphorylation and oxidation in HT22 neuronal cells.
HT22 cells were used as controls, were exposed to 1.0 mM H2O2 alone, or to 0.5 μM OA alone, or to the combination of 1.0 mM H2O2 plus 0.5 μM OA, as described in the Materials and methods section. At various time points between 0 and 24 h, cells were harvested, lysed and analysed by immunoblotting (A) using anti-tau, or the anti-phospho-tau antibodies indicated, as described in Figure 3. Tau turnover (B) was measured as in Figure 1(A), and proteasome activity, using the suc-LLVY-AMC fluorogenic peptide substrate (C) was measured as in Figure 4(B). The extent of tau oxidation (D) following H2O2 plus OA treatment was measured by ELISA, as in Figure 5(D). The data in (B)–(D) represent the means±S.E.M. for three independent determinations.
If our interpretations thus far are correct, then exposure of HT22 cells to the combination of H2O2 and OA should cause a decrease in tau degradation compared with control cells or H2O2 treated cells, but an increase in comparison with OA alone. As shown in Figure 6(B), these exact results were, in fact, observed following the joint H2O2 and OA treatment. Importantly, the decline in tau degradation caused by combined H2O2 and OA treatment was not due to any inhibition of the proteasome, since proteasome activity actually increased in these experiments (Figure 6C). Furthermore, the joint H2O2 and OA treatment still caused only mild tau oxidation (Figure 6D) to an extent very similar to that seen with H2O2 alone in Figure 5(D).
The results of Figure 6 suggest that an oxidized but (hyper)phosphorylated tau is a poor substrate for proteasomal degradation. This is in agreement with the accumulation of hyperphosphorylated tau proteins, simultaneous with the oxidizing conditions of chronic inflammation, seen during Alzheimer's disease, suggesting that pathologically enhanced phosphorylation of the tau protein may be sufficient to cause accumulation of the protein inside neurons.
Toxicity of hyperphosphorylated tau
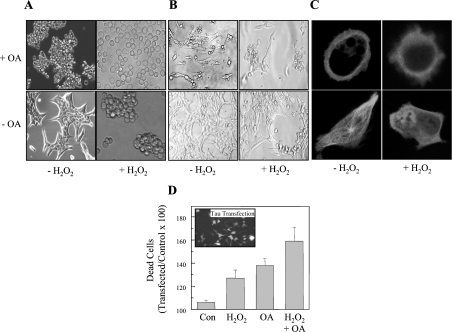
If accumulation of the tau protein might be caused by hyperphosphorylation of the protein, the question arises whether the phosphorylation status of the tau protein also modulates the toxicity of the accumulating intracellular tau aggregates. As judged by microscopy, treatment with OA and H2O2 leads to severe changes in cellular structure (Figure 7A). As expected the most severe effects were observed following the combined treatment of H2O2 plus OA.
Figure 7. Toxicity of tau aggregates.
A day after transfection with tau–CFP, HT22 cells were used as controls, or were exposed to 1.0 mM H2O2 alone for 30 min, or to 0.5 μM OA alone for 3 h, or to the combination of 1.0 mM H2O2 plus 0.5 μM OA, as described in the legend to Figure 6. (A) Light microscopy images are shown of control and treated cells. (B) The distribution of tau–CFP directly after each of the treatment procedures, as detected by CLSM. To measure the effect of tau phosphorylation and oxidation on cell viability, HT22 cells were transfected with tau–GFP (C) The effect of H2O2 and OA on the viability of both transfected (fluorescent) cells, and non-transfected cells was measured. To estimate the role of tau in the cell survival, we calculated the ratio between the viability of transfected cells and the viability of non-transfected cells (×100), and these results (means±S.E.M., n=3) are reported in (D) as percentage of dead cells.
Since we used a tau–CFP construct, to generate a tau–CFP fusion protein inside the cells, we also investigated transfected cells under our various treatment conditions by CLSM. In Figure 7(B) one can see that in control cells the tau–CFP fusion protein was located in fibrils throughout the cytoplasm. Treatment with either OA or H2O2 was accompanied by a decline of these fibrillar structures and, especially in the case of OA, by a more rounded cell shape. The strongest effects were, again, achieved following the double treatment of H2O2 plus OA.
Quantification of cell viability was measured using cells transfected with a tau–GFP construct, in comparison with non-transfected cells (Figure 7C). These experiments indicated that transfection with tau–GFP alone (no other treatment) had minimal effects on cell viability (approx. 5% mortality), whereas OA caused 37% cell death, and H2O2 caused 28% death. Treatment with H2O2 plus OA caused an additive 58% loss of cell viability. Our results indicate that an accumulation of hyperphosphorylated and oxidized tau protein is highly toxic to neurons, and we suggest that this may well be an important contributor to neuronal damage in Alzheimer's disease.
Summary and conclusions
Our results indicate that turnover of the tau protein in neurons depends on proteolytic degradation by the proteasome. Hyperphosphorylation of tau inhibits proteasome-dependent proteolysis and causes tau accumulation. Tau can be phosphorylated by several kinases and dephosphorylated by PP1, PP2A, and PP2B. PP1 is normally repressed by I-2, but Cdk5 can phosphorylate I-2, dissociate it from PP1, and derepress (‘activate’) PP1, leading to dephosphorylation of tau. Alternatively, RCAN1 can inhibit calcineurin and induce GSKβ, resulting in increased tau phosphorylation.
The tauopathies (including Alzheimer's disease) typically involve an accumulation of hyperphosphorylated tau, under the oxidizing conditions of chronic inflammation. Adaptation to oxidative stress induces RCAN1 and GSK3β, thereby increasing tau phosphorylation [10], whereas oxidative stress itself seems to decrease tau phosphorylation via activation of Cdk5 [21]. The complex interactions between tau and its phosphorylases (and their inhibitors/activators), and the effects of phosphorylation or dephosphorylation on tau degradation by the proteasome, are shown in Figure 8. In the present paper, oxidative stress alone transiently stimulated a short burst of tau degradation, even though the tau protein was only minimally oxidized. We suspect that a regulatory mechanism, perhaps involving a rearrangement of tau phosphorylation by PP1 (mediated by Cdk5), may explain this transient effect. We next attempted to model the tau hyperphosphorylation and oxidative stress conditions of tauopathies by combined treatment of HT22 neurons with the PP1/PP2A inhibitor OA and H2O2. During the combined treatment, phosphorylation strongly inhibited tau degradation, almost completely blocking the stimulation of tau degradation seen with H2O2 alone. Thus whatever the mechanism of transiently increased tau proteolysis caused by H2O2 may be, it would appear to be relatively unimportant (compared with tau phosphorylation) for the tauopathies. Exposure of cells to OA plus H2O2 caused cell rounding, aggregation of tau in the periphery and loss of cell viability. We conclude that conditions of tau hyperphosphorylation and chronic inflammation may significantly inhibit tau turnover by the proteasome, thereby promoting tau accumulation and aggregation, and contributing to cellular toxicity and neurodegeneration.
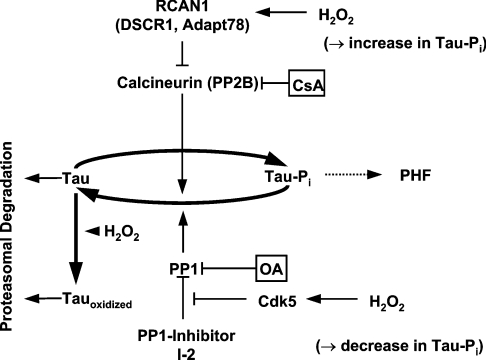
Figure 8. Influence of phosphorylation on tau degradation by the proteasome during oxidative stress and following adaptation to oxidative stress.
Oxidative stress-mediated activation of Cdk5 leads to phosphorylation of the PP1 inhibitor I-2. I-2 then dissociates from PP1 and leads to PP1 ‘activation’ (de-repression). Consequently PP1 is able to dephosphorylate tau. On the other hand adaptation to oxidative stress leads to an increased tau phosphorylation by inducing RCAN1, a PP2B inhibitor, and GSK3β (a tau kinase). CsA inhibits PP2B, whereas OA inhibits PP1 and PP2A. Therefore these inhibitors increase the phosphorylation level of tau by different pathways, in different patterns, and to different extents. Phosphorylation of tau results in decreased tau degradation by the proteasome, and may lead to the formation of PHF. Tau is only mildly sensitive to direct oxidation, and mild oxidation does not alter the proteolytic susceptibility of the tau protein.
Acknowledgments
This work was supported by DFG grants, SFB 507 and SFB575 to T. G., and by NIH-NIEHS (National Institutes of Health–National Institutes of Environmental Health Sciences) grant #ES 03598 to K. J. A. D. Special thanks are extended to Professor E. Mandelkow and Professor E. M. Mandelkow for provision of the tau–CFP and htau40 plasmid.
References
- 1.Goedert M., Spillantini M. G., Jakes R., Rutherford D., Crowther R. A. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron. 1989;3:519–526. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 2.Brich J., Shie F.-S., Howell B. W., Li R., Tus K., Wakeland E. K., Jin L.-W., Mumby M., Churchill G., Herz J., Cooper J. A. Genetic modulation of tau phosphorylation in the mouse. J. Neurosci. 2003;23:187–192. doi: 10.1523/JNEUROSCI.23-01-00187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gustke N., Steiner B., Mandelkow E. M., Biernat J., Meyer H. E., Goedert M., Mandelkow E. The Alzheimer-like phosphorylation of tau protein reduces microtubule binding and involves Ser-Pro and Thr-Pro motifs. FEBS Lett. 1992;307:199–205. doi: 10.1016/0014-5793(92)80767-b. [DOI] [PubMed] [Google Scholar]
- 4.Biernat J., Gustke N., Drewes G., Mandelkow E. M., Mandelkow E. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11:153–163. doi: 10.1016/0896-6273(93)90279-z. [DOI] [PubMed] [Google Scholar]
- 5.Jameson L., Frey T., Zeeberg B., Dalldorf F., Caplow M. Inhibition of microtubule assembly by phosphorylation of microtubule-associated proteins. Biochemistry. 1980;19:2472–2479. doi: 10.1021/bi00552a027. [DOI] [PubMed] [Google Scholar]
- 6.Lindwall G., Cole R. D. Phosphorylation affects the ability of tau protein to promote microtubule assembly. J. Biol. Chem. 1984;259:5301–5305. [PubMed] [Google Scholar]
- 7.Ishiguro K., Shiratsuchi A., Sato S., Omori A., Arioka M., Kobayashi S., Uchida T., Imahori K. Glycogen synthase kinase 3β is identical to tau protein kinase I generating several epitopes of paired helical filaments. FEBS Lett. 1993;325:167–172. doi: 10.1016/0014-5793(93)81066-9. [DOI] [PubMed] [Google Scholar]
- 8.Baum L., Seger R., Woodgett J. R., Kawabata S., Maruyama K., Koyama M., Silver J., Saitoh T. Overexpressed tau protein in cultured cells is phosphorylated without formation of PHF: implication of phosphoprotein phosphatase involvement. Mol. Brain Res. 1995;34:1–17. doi: 10.1016/0169-328x(95)00111-5. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T., Zhong J., Iqbal K., Trenkner E., Grundke-Iqbal I. The regulation of phosphorylation of tau in SY5Y neuroblastoma cells: the role of protein phosphatases. FEBS Lett. 1998;426:248–254. doi: 10.1016/s0014-5793(98)00346-9. [DOI] [PubMed] [Google Scholar]
- 10.Ermak G., Harris C. D., Davies K. J. A. The DSCR1 (Adapt78) isoform 1 protein calcipressin 1 inhibits calcineurin and protects against acute calcium-mediated stress damage, including transient oxidative stress. FASEB J. 2002;16:814–824. doi: 10.1096/fj.01-0846com. [DOI] [PubMed] [Google Scholar]
- 11.Goedert M., Jakes R., Qi Z., Wang J. H., Cohen P. Protein phosphatase 2A is the major enzyme in brain that dephosphorylates tau protein phosphorylated by proline-directed protein kinases or cyclic AMP-dependent protein kinase. J. Neurochem. 1995;65:2804–2807. doi: 10.1046/j.1471-4159.1995.65062804.x. [DOI] [PubMed] [Google Scholar]
- 12.Kins S., Crameri A., Evans D. R., Hemmings B. A., Nitsch R. M., Gotz J. Reduced protein phosphatase 2A activity induces hyperphosphorylation and altered compartmentalization of tau in transgenic mice. J. Biol. Chem. 2001;276:38193–38200. doi: 10.1074/jbc.M102621200. [DOI] [PubMed] [Google Scholar]
- 13.Gong C. X., Wegiel J., Lidsky T., Zuck L., Avila J., Wisniewski H. M., Grundke-Iqbal I., Iqbal K. Regulation of phosphorylation of neuronal microtubule-associated proteins MAP1b and MAP2 by protein phosphatase-2A and -2B in rat brain. Brain Res. 2000;853:299–309. doi: 10.1016/s0006-8993(99)02294-5. [DOI] [PubMed] [Google Scholar]
- 14.Agarwal-Mawal A., Paudel H. K. Neuronal Cdc2-like protein kinase (Cdk5/p25) is associated with protein phosphatase 1 and phosphorylates inhibitor-2. J. Biol. Chem. 2001;276:23712–23718. doi: 10.1074/jbc.M010002200. [DOI] [PubMed] [Google Scholar]
- 15.Keck S., Nitsch R., Grune T., Ullrich O. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J. Neurochem. 2003;85:115–122. doi: 10.1046/j.1471-4159.2003.01642.x. [DOI] [PubMed] [Google Scholar]
- 16.Shimura H., Schwartz D., Gygi S. P., Kosik K. S. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J. Biol. Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 17.David D. C., Layfield R., Serpell L., Narain Y., Goedert M., Spillantini M. G. Proteasomal degradation of tau protein. J. Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 18.Goedert M. The significance of tau and alpha-synuclein inclusions in neurodegenerative diseases. Curr. Opin. Genet. Dev. 2001;11:343–351. doi: 10.1016/s0959-437x(00)00200-8. [DOI] [PubMed] [Google Scholar]
- 19.Maccioni R. B., Munoz J. P., Barbeito L. The molecular bases of Alzheimer's disease and other neurodegenerative disorders. Arch. Med. Res. 2001;32:367–381. doi: 10.1016/s0188-4409(01)00316-2. [DOI] [PubMed] [Google Scholar]
- 20.Pfeiffer J. M., Askew E. W., Roberts D. E., Wood S. M., Benson J. E., Johnson S. C., Freedman M. S. Effect of antioxidant supplementation on urine and blood markers of oxidative stress during extended moderate-altitude training. Wilderness Environ. Med. 1999;10:66–74. doi: 10.1580/1080-6032(1999)010[0066:eoasou]2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Zambrano C. A., Egana J. T., Nunez M. T., Maccioni R. B., Gonzalez-Billault C. Oxidative stress promotes tau dephosphorylation in neuronal cells: the roles of Cdk5 and PP1. Free Radical Biol. Med. 2004;36:1393–1402. doi: 10.1016/j.freeradbiomed.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Butterfield D. A. Amyloid β-peptide (1–42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer's disease brain. Free Radical Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 23.Perry G., Roder H., Nunomura A., Takeda A., Friedlich A. L., Zhu X., Raina A. K., Holbrook N., Siedlak S. L., Harris P. L., Smith M. A. Activation of neuronal extracellular receptor kinase (ERK) in Alzheimer disease links oxidative stress to abnormal phosphorylation. NeuroReport. 1999;10:2411–2415. doi: 10.1097/00001756-199908020-00035. [DOI] [PubMed] [Google Scholar]
- 24.Shea T. B., Prabhakar S., Ekinci F. J. β-Amyloid and ionophore A23187 evoke tau hyperphosphorylation by distinct intracellular pathways: differential involvement of the calpain/protein kinase C system. J. Neurosci. Res. 1997;49:759–768. doi: 10.1002/(SICI)1097-4547(19970915)49:6<759::AID-JNR10>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 25.Xie H. Q., Johnson G. V. Calcineurin inhibition prevents calpain-mediated proteolysis of tau in differentiated PC12 cells. J. Neurosci. Res. 1998;53:153–164. doi: 10.1002/(SICI)1097-4547(19980715)53:2<153::AID-JNR4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Hartigan J. A., Johnson G. V. Transient increases in intracellular calcium result in prolonged site-selective increases in Tau phosphorylation through a glycogen synthase kinase 3β-dependent pathway. J. Biol. Chem. 1999;274:21395–21401. doi: 10.1074/jbc.274.30.21395. [DOI] [PubMed] [Google Scholar]
- 27.Pacifici R. E., Kono Y., Davies K. J. A. Hydrophobicity as the signal for selective degradation of hydroxyl radical-modified hemoglobin by the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1993;268:15405–15411. [PubMed] [Google Scholar]
- 28.Grune T., Reinheckel T., Joshi M., Davies K. J. A. Proteolysis in cultured liver epithelial cells during oxidative stress. Role of the multicatalytic proteinase complex, proteasome. J. Biol. Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- 29.Grune T., Reinheckel T., Davies K. J. A. Degradation of oxidized proteins in K562 human hematopoietic cells by proteasome. J. Biol. Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- 30.Grune T., Merker K., Sandig G., Davies K. J. A. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem. Biophys. Res. Commun. 2003;305:709–718. doi: 10.1016/s0006-291x(03)00809-x. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg S. G., Davies P., Schein J. D., Binder L. I. Hydrofluoric acid-trated tau-PHF proteins display the same biochemical properties as normal tau. J. Biol. Chem. 1992;267:564–569. [PubMed] [Google Scholar]
- 32.Ermak G., Cheadle C., Becker K. G., Harris C. D., Davies K. J. DSCR1 (Adapt78) modulates expression of SOD1. FASEB J. 2004;18:62–69. doi: 10.1096/fj.03-0451com. [DOI] [PubMed] [Google Scholar]
- 33.Zwilling S., Konig H., Wirth T. High mobility group protein 2 functionally interacts with the POU domains of octamer transcription factors. EMBO J. 1995;14:1198–1208. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buss H., Chan T. P., Sluis K. B., Domigan N. M., Winterbourn C. C. Protein carbonyl measurement by a sensitive ELISA method. Free Radical Biol. Med. 1997;23:361–366. doi: 10.1016/s0891-5849(97)00104-4. [DOI] [PubMed] [Google Scholar]
- 35.Sitte K., Merker K., Grune T. Proteasome-dependent degradation of oxidized proteins in MRC-5 fibroblasts. FEBS Lett. 1998;440:399–402. doi: 10.1016/s0014-5793(98)01495-1. [DOI] [PubMed] [Google Scholar]
- 36.Petrucelli L., Dickson D., Kehoe K., Taylor J., Snyder H., Grover A., De Lucia M., McGowan E., Lewis J., Prihar G., et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Human Mol. Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 37.Lee V. M., Wong J., Trojanovski J. Q. Purification of paired helical filament tau and normal tau from human brain tissue. Methods Enzymol. 1999;309:81–89. doi: 10.1016/s0076-6879(99)09008-4. [DOI] [PubMed] [Google Scholar]
- 38.Leclerc S., Garnier M., Hoessel R., Marko D., Bibb J. A., Snyder G. L., Greengard P., Biernat J., Wu Y. Z., Mandelkow E. M., et al. Indirubins inhibit glycogen synthase kinase-3β and CPK5/p25 two protein kinases involved in abnormal tau phosphorylation in Alzheimer's disease: a property common to most cyclin-dependent kinase inhibitors? J. Biol. Chem. 2001;276:251–260. doi: 10.1074/jbc.M002466200. [DOI] [PubMed] [Google Scholar]
- 39.Reinheckel T., Sitte N., Ullrich O., Kuckelkorn U., Davies K. J., Grune T. Comparative resistance of the 20S and 26S proteasome to oxidative stress. Biochem. J. 1998;335:637–642. doi: 10.1042/bj3350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litersky J. M., Johnson G. V. W. Phosphorylation by cAMP-dependent protein kinase inhibits the degradation of tau by calpain. J. Biol. Chem. 1992;267:1563–1568. [PubMed] [Google Scholar]
- 41.Wang J.-Z., Gong C. X., Zaidi T., Grundke-Iqbal I., Iqbal K. Dephosphorylation of Alzheimer paired helical filaments by protein phosphatase-2A and -2B. J. Biol. Chem. 1995;270:4854–4860. doi: 10.1074/jbc.270.9.4854. [DOI] [PubMed] [Google Scholar]
- 42.Zhang J. Y., Liu S. J., Li H. L., Wang J.-Z. Microtubule-associated protein tau is a substrate of ATP/Mg2+-dependent proteasome protease system. J. Neural Transm. 2005;112:547–555. doi: 10.1007/s00702-004-0196-x. [DOI] [PubMed] [Google Scholar]
- 43.Shringarpure R., Grune T., Davies K. J. Protein oxidation and 20S proteasome-dependent proteolysis in mammalian cells. Cell Mol. Life Sci. 2001;58:1442–1450. doi: 10.1007/PL00000787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mandelkow E., Song Y. H., Schweers O., Marx A., Mandelkow E. M. On the structure of microtubules, tau, and paired helical filaments. Neurobiol. Aging. 1995;16:347–354. doi: 10.1016/0197-4580(95)00026-b. [DOI] [PubMed] [Google Scholar]
- 45.Ernst A., Stolzing A., Sandig G., Grune T. Protein oxidation and the degradation of oxidized proteins in the rat oligodendrocyte cell line OLN 93-antioxidative effect of the intracellular spin trapping agent PBN. Mol. Brain Res. 2004;122:126–132. doi: 10.1016/j.molbrainres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Shaw M., Cohen P., Alessi D. R. The activation of protein kinase B by H2O2 or heat shock is mediated by phosphoinositide 3-kinase and not by mitogen-activated protein kinase-activated protein kinase-2. Biochem. J. 1998;336:241–246. doi: 10.1042/bj3360241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Q., Smith M. A., Avila J., DeBernardis J., Kansal M., Takeda A., Zhu X., Nunomura A., Honda K., Moreira P. I., et al. Alzheimer-specific epitopes of tau represent lipid peroxidation-induced conformations. Free Radical Biol. Med. 2005;38:746–754. doi: 10.1016/j.freeradbiomed.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Dozza B., Smith M. A., Perry G., Tabaton M., Strocchi P. Regulation of glycogen synthase kinase-3β by products of lipid peroxidation in human neuroblastoma cells. J. Neurochem. 2004;89:1224–1232. doi: 10.1111/j.1471-4159.2004.02413.x. [DOI] [PubMed] [Google Scholar]
- 49.Santa-Maria I., Hernandez F., Martin C. P., Avila J., Moreno F. J. Quinones facilitate the self-assembly of the phosphorylated tubulin binding region of tau into fibrillar polymers. Biochemistry. 2004;43:2888–2897. doi: 10.1021/bi035345j. [DOI] [PubMed] [Google Scholar]
- 50.Takeda A., Smith M. A., Avila J., Nunomura A., Siedlak S. L., Zhu X., Perry G., Sayre L. M. In Alzheimer's disease, heme oxygenase is coincident with Alz50, an epitope of tau induced by 4-hydroxy-2-nonenal modification. J. Neurochem. 2000;75:1234–1241. doi: 10.1046/j.1471-4159.2000.0751234.x. [DOI] [PubMed] [Google Scholar]
- 51.Rudeck M., Volk T., Sitte N., Grune T. Ferritin oxidation in vitro. Implication of iron release and degradation by the 20S proteasome. IUBMB Life. 2000;49:451–456. doi: 10.1080/152165400410317. [DOI] [PubMed] [Google Scholar]
- 52.Lasch P., Petras T., Ullrich O., Backmann J., Naumann D., Grune T. Hydrogen peroxide induced structural alterations of RNase A. J. Biol. Chem. 2001;276:9492–9502. doi: 10.1074/jbc.M008528200. [DOI] [PubMed] [Google Scholar]
- 53.Bota D., Davies K. J. A. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism Nat. Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 54.Ernak G., Harris C. D., Battocchio D., Davies K. J. A. RCAN1 (DSCR1 or Adapt 7) stimulates expression of GSK-3β. FEBS J. 2006;273:2100–2109. doi: 10.1111/j.1742-4658.2006.05217.x. [DOI] [PubMed] [Google Scholar]