Abstract
Dbl family GEFs (guanine nucleotide-exchange factors) for the Rho GTPases almost invariably contain a PH (pleckstrin homology) domain adjacent to their DH (Dbl homology) domain. The DH domain is responsible for GEF activity, and the PH domain plays a regulatory role that remains poorly understood. We demonstrated previously that Dbl family PH domains bind phosphoinositides with low affinity and cannot function as independent membrane targeting modules. In the present study, we show that dimerization of a Dbs (Dbl's big sister) DH/PH domain fragment is sufficient to drive it to the plasma membrane through a mechanism involving PH domain–phosphoinositide interactions. Thus, the Dbs PH domain could play a significant role in membrane targeting if it co-operates with other domains in the protein. We also show that mutations that prevent phosphoinositide binding by the Dbs PH domain significantly impair cellular GEF activity even in chimaeric proteins that are robustly membrane targeted by farnesylation or by the PH domain of phospholipase C-δ1. This finding argues that the Dbs PH domain plays a regulatory role that is independent of its ability to aid membrane targeting. Thus, we suggest that the PH domain plays dual roles, contributing independently to membrane localization of Dbs (as part of a multi-domain interaction) and allosteric regulation of the DH domain.
Keywords: Dbl's big sister (Dbs), guanine nucleotide exchange factor (GEF), membrane targeting, pleckstrin homology (PH) domain, phosphoinositide, Rho
Abbreviations: BS3, bis(sulfosuccinimidyl) suberate; Dbs, Dbl's big sister; DH, Dbl homology; DMEM, Dulbecco's modified Eagle's medium; EGFP, enhanced green fluorescent protein; FBS, fetal bovine serum; FKBP, FK506-binding protein; GEF, guanine nucleotide-exchange factor; GST, glutathione S-transferase; H-Ras, Harvey-Ras; PAK1, p21-activated protein kinase; PBD, p21-binding domain; PH, pleckstrin homology; PLC, phospholipase-C; PtdIns3P, phosphatidylinositol-3-phosphate; PtdIns(4,5)P2, phosphatidylinositol-4,5-bisphosphate; Ras-GRF, Ras guanine-nucleotide releasing factor; RBD, Rho-binding domain; SH3, Src homology 3; SPR, surface plasmon resonance; TTBS, Tris-buffered saline containing 0.1% Triton X-100
INTRODUCTION
Dbl-related GEFs (guanine nucleotide-exchange factors) activate Rho family small GTPases, which control many cellular processes including organization of the actin cytoskeleton [1,2]. Dbl-related GEFs contain a helical DH (Dbl homology) domain that is nearly always followed by a PH (pleckstrin homology) domain [1]. The ∼200 amino acid DH domain harbours the guanine nucleotide-exchange activity, selective for one or several Rho-family GTPases [3,4]. The adjacent 120 amino acid PH domain plays a modulatory role that appears to differ mechanistically across this large family of proteins [5]. With more than 50 Dbl family proteins in the human proteome, and ∼250 PH domains, approx. 20% of known human PH domains are linked directly to a DH domain. Few of these PH domains are well understood.
PH domains are best known for their ability to bind phosphoinositides and to target their host proteins to cellular membranes [6]. Although some PH domains bind strongly and specifically to phosphoinositides, most do so only weakly and with little specificity [7,8]. The binding properties of PH domains from several Dbl family proteins have been analysed [9–11]. Each binds phosphoinositides with an affinity [KD for PtdIns(4,5)P2 (phosphatidylinositol-4,5-bisphosphate)>10 μM] that is thought to be too low to drive membrane targeting independently. With the exception of the Tiam-1 C-terminal PH domain, which shows specificity for PtdIns3P (phosphatidylinositol-3-phosphate) [9,12], all DH-domain-associated PH domains are promiscuous in their phosphoinositide binding. These findings argue that the PH domains of Dbl family proteins do not simply target their host proteins to cellular membranes. Indeed, PH domain mutations that impair phosphoinositide binding did not affect subcellular localization of Dbl family proteins in several reports [12–16], although altered or reduced membrane localization was described in other studies [15,17,18]. In all cases, regardless of whether subcellular localization was altered, the same mutations significantly reduced GEF activity of the Dbl protein in the same cells. Thus, phosphoinositide binding by the PH domain may play a role in GEF activity distinct from (or in addition to) any role in driving membrane association.
Structural studies have shown that the PH domain contributes directly to the interaction of certain Dbl family GEFs with their small GTPase targets [4,5,19–21]. Comparison of Dbs (Dbl's big sister) DH/PH structures with and without bound GTPase [22] suggests a model in which phosphoinositide binding to the PH domain at a membrane leads to a reorientation of the DH and PH domains that maximizes their combined interaction with the membrane-associated Rho-family GTPase. Focusing on Dbs, we have been interested in understanding the relative contributions to the GEF-activating effects of PH domain–phosphoinositide interactions made by allosteric/conformational effects, and of simple PH-domain-mediated membrane targeting.
Dbs was originally identified in a retroviral murine cDNA library screen for genes that transform NIH 3T3 cells [23], and was called Dbs because of its similarity to Dbl. The 1149 amino acid murine Dbs gene product contains a SEC14 domain (residues 82–219), SPEC (spectrin repeat) domains (residues 352–539), a DH domain (residues 623–819), a single PH domain (residues 820–967) and an SH3 (Src homology 3) domain (residues 1060–1108). The DH domain of Dbs activates RhoA and Cdc42 in cellular and in vitro studies [13,15,19,24]. PH domain deletion eliminates the ability of Dbs to transform cells, but addition of the H-Ras (Harvey-Ras) far (farresylation) sequence to the DH domain partially rescues transforming activity [24]. Thus, simple membrane recruitment may play some role in the contribution of the PH domain to Dbs function. We find that the GEF activity of the Dbs DH/PH fragment can be enhanced by targeting it to the plasma membrane by dimerization (to increase PH domain avidity) or by fusion to known membrane-targeting sequences. However, even when Dbs is membrane-targeted, mutations introduced into the PH domain that reduce phosphoinositide binding impair GEF activity. Our results therefore suggest that, while the PH domain may participate in driving membrane localization of Dbs (as one of several co-operating domains), this may be secondary to its role in phosphoinositide-regulated conformational changes that enhance GEF activity.
EXPERIMENTAL
Plasmid constructs and mutagenesis
DNA encoding the murine Dbs DH/PH region (amino acids 623–967) was amplified by PCR and subcloned in between the BamHI and EcoRI sites of pGEX-2TK (Amersham Pharmacia Biotech). The resulting GST (glutathione S-transferase)–DH/PH fusion protein has a PKA (protein kinase A) phosphorylation site between the GST moiety and the DH/PH fragment. To generate a dimerizable DH/PH fragment [fused to EGFP (enhanced green fluorescent protein)], the FKBPF36V (FK506-binding protein) sequence was fused to the Dbs DH/PH coding sequence, and the resulting fusion (Figure 1A) was subcloned between the BglII and EcoRI sites of pEGFP-C1 (Clontech, Palo Alto, CA, U.S.A.). DNA encoding the rat PLC-δ1 (phospholipase C-δ1) PH domain (amino acids 11–127) was subcloned between the BglII and EcoRI sites of pEGFP-C1 to generate an EGFP–PLC-δ1-PH domain fusion, and was also fused to the C-terminus of pEGFP–DH/PH-FKBPF36V to produce the fusion protein EGFP–DH/PH-FKBPF36V–PLC-δ1-PH. A fusion protein with the PLC-δ1 PH domain linked directly to the C-terminus of an EGFP–Dbs DH fusion protein (containing amino acids 623–824 from Dbs) was also generated. This is EGFP–Dbs DH–PLC-δ1-PH. As an alternative mode of membrane targeting, the coding sequence of H-Ras1 (amino acids 170–189), corresponding to its far signal, was fused to the C-terminus of pEGFP–DH/PH (using the EcoRI site) to generate the fusion protein EGFP–DH/PH–far.
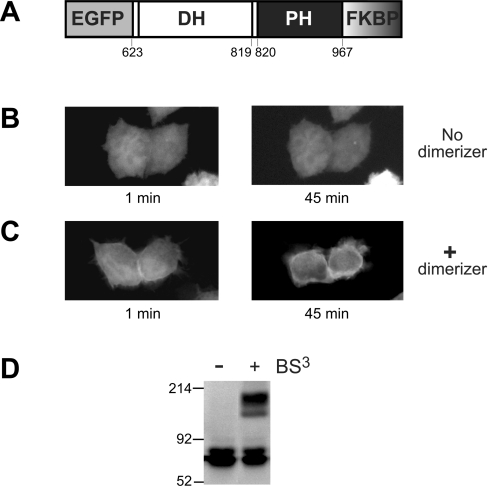
Figure 1. The DH/PH fragment of Dbs can be driven to the plasma membrane by dimerization.
(A) FKBPF36V was fused to the C-terminus of an EGFP–Dbs DH/PH fragment to give the EGFP–DH/PH–FKBPF36V fusion protein schematized here. Amino acid numbers corresponding to domain boundaries are given. HeLa cells transiently transfected with the EGFP–DH/PH–FKBPF36V construct (containing the wild-type Dbs PH domain) were either untreated (B) or treated (C) with the AP20187 dimerizer at a final concentration of 200 nM (see Experimental section), and live cells were visualized by fluorescence microscopy after the indicated times. (D) AP20187 addition promotes dimerization of the EGFP–DH/PH–FKBPF36V fusion protein as assessed by cross-linking with BS3 (see Experimental section). Whole-cell lysates were subjected to treatment with BS3, and analysed by SDS/PAGE and immunoblotting with an anti-GFP antibody.
PH domain mutations were made in the GST–DH/PH, EGFP–DH/PH-FKBPF36V, EGFP–DH/PH-FKBPF36V–PLC-δ1-PH and EGFP–DH/PH–far proteins. All mutations were generated using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene). Amino-acid replacements made in the PH domain β1/β2 loop (PH*; K849Q, K851Q, R855Q and K857Q) and β2 strand (R861Q) are depicted in Figure 2(A). Constructs expressing the PBD (p21-binding domain; amino acids 67–150) of human PAK1 (p21-activated kinase-1) [25] and the RBD (Rho-binding domain; amino acids 7–89) of murine Rhotekin [26] were provided kindly by Dr Channing Der (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, NC, U.S.A.) and Dr Xiang-Dong Ren (Department of Dermatology, Stony Brook University, Stony Brook, NY, U.S.A.) respectively.
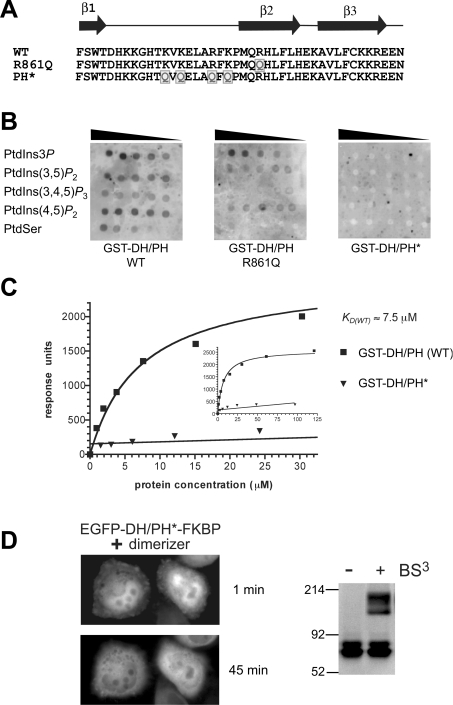
Figure 2. Mutations in the PH domain of Dbs abolish its weak PtdIns(4,5)P2 binding.
(A) The sequence from strand β1 to β3 of the Dbs PH domain is shown, corresponding to the region of PH domains known to interact with phosphoinositides [32]. Mutations (to glutamine) were introduced into the β2 strand (R861Q) or at four basic residues (K849, K851, R855, and K857) in the β1/β2 loop (PH*) of the Dbs PH domain as indicated by the grey boxed Qs. (B) 32P-Labelled GST–DH/PH (10 μg) was used to probe nitrocellulose filters spotted with serial 2-fold dilutions of PtdIns3P, PtdIns(3,5)P2, PtdIns(3,4,5)P3, PtdIns(4,5)P2 and phosphatidylserine (PtdSer) as marked (beginning at 2 mg/ml). (C) GST–DH/PH and GST–DH/PH* proteins were assessed for binding to PtdIns(4,5)P2 using SPR as described [8]. GST–DH/PH or GST–DH/PH* at a series of concentrations were flowed over a Biacore sensor chip containing 3% (mol/mol) PtdIns(4,5)P2 in dioleoylphosphatidylcholine. Steady-state binding signals are plotted against protein concentration, with the best-fit to a simple 1:1 binding curve superimposed. The KD (app) value for binding of (dimeric) GST–DH/PH (wild-type) was approx. 7.5 μM. The inset curve demonstrates saturation of PtdIns(4,5)P2 binding by GST–DH/PH. GST–DH/PH* gave no significant binding signal at any concentration tested. Curves are representative of at least three independent experiments. (D) Introduction of PH* mutations (which prevent phosphoinositide binding as shown in B and C) abolished the ability of the EGFP–DH/PH*–FKBPF36V to translocate to the plasma membrane upon addition of AP20187. Details are as described for Figures 1(B) and 1(C). BS3 cross-linking experiments were performed as in Figure 1(D) and show that AP20187 induces robust EGFP–DH/PH*–FKBPF36V dimerization.
Overlay analysis of phosphoinositide binding
The Dbs GST–DH/PH and GST–DH/PH* fusion proteins were expressed in Escherichia coli, purified, labelled with 32[P]Pi, and used in lipid-overlay experiments exactly as described previously [7,12], with serial 2-fold dilutions of lipids [starting with 2 mg/ml in 1:1 (v/v) chloroform/methanol containing 0.1% HCl] spotted on to a nitrocellulose membrane.
SPR (surface plasmon resonance)
SPR studies of phosphoinositide binding were performed exactly as described previously [8], using a Biacore X.
Activated Cdc42- and RhoA-pulldown assays
Pull-down assays to assess cellular levels of activated Cdc42 and RhoA were performed as described previously [25,26]. HeLa cells were seeded in DMEM (Dulbecco's modified Eagle's medium) and 10% (v/v) FBS (fetal bovine serum) at 8×106 cells per 10 cm diameter plate. After 15 h, cells were transfected with 20 μg of the relevant DNA using Lipofectamine™ 2000 (Invitrogen). After 4 h, cells were washed twice in PBS and resuspended in DMEM with 0.1% FBS for a further 16 h. Cells were then lysed in ice-cold lysis buffer [25 mM Hepes, pH 7.5, 150 mM NaCl, 1% (v/v) Nonidet P40, 0.25% sodium deoxycholate, 10% (w/v) glycerol, 10 mM MgCl2 and 1 mM PMSF] and spun at 16000 g in a microcentrifuge at 4 °C for 4 min. Supernatant (lysate) was retained and the total protein concentration in each sample was measured using the bicinchoninic acid protein assay (Pierce, Rockford, IL, U.S.A.). Total protein (70 μg) from the lysate was saved for Western blotting controls (to assess total Cdc42, RhoA, and Dbs expression levels). For activated Cdc42-pulldown assays, 600 μg of total protein was incubated with 30 μg of PAK1 PBD bound to glutathione–agarose (Sigma) for 1 h at 4 °C. Agarose beads were washed twice in 4 °C lysis buffer, resuspended in SDS/PAGE sample buffer [62.7 mM Tris/HCl, pH 6.8, 2% (w/v) SDS, 10% (w/v) glycerol and 0.01% Bromophenol Blue], and boiled for 3 min prior to analysis by immunoblotting with mouse anti-Cdc42 (610929 clone 44; BD Transduction Laboratories, Lexington, KY, U.S.A.) at 1:300 dilution in TTBS (Tris-buffered saline containing 0.1% Triton X-100) with 5% (w/v) dried milk powder (for PAK1 PBD pull-down or whole-cell lysate samples). For activated RhoA-pulldown assays, 1 mg of total protein was incubated with 30 μg of GST–RBD bound to glutathione–agarose (Sigma) for 1 h at 4 °C. Agarose beads were washed twice in cold lysis buffer (without Nonidet P40), resuspended in SDS/PAGE sample buffer, and boiled for 3 min prior to analysis by immunoblotting. Mouse anti-RhoA (sc-179; Santa Cruz Laboratories; Santa Cruz, CA, U.S.A.) at 1:250 dilution in TTBS containing 5% (w/v) dried milk powder was used for Western blotting of RBD pull-down or whole-cell lysate samples. Mouse anti-GFP (B34; Covance Research Products, Berkeley, CA, U.S.A.) was used at 1:10000 dilution in TTBS containing 5% (w/v) dried milk powder for Western blots of whole-cell lysate samples to assess levels of GFP fusion protein expression.
In vitro nucleotide-exchange assays
Nucleotide-exchange assays were performed exactly as described previously [19], monitoring incorporation of N-methylanthraniloyl-GTP (Molecular Probes) into Cdc42 or RhoA expressed and purified from E. coli. Exchange assay reaction mixtures contained 20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol and 100 μM N-methylanthraniloyl-GTP, plus 2 μM GTPase (Cdc42 or RhoA).
Fluorescence microscopy
HeLa cells were plated at 3.75×105 cells per 13-mm-diameter dish and transfected after 12 h with 2 μg of the relevant pEGFP-C1 derivative using Lipofectamine™ 2000 (Invitrogen). After 4 h, cells were washed twice in PBS and resuspended in DMEM with 0.1% FBS for a further 16 h. Living cells were then visualized using a Leica DM-IRB inverted fluorescence microscope and 0.1 μm z-sections were collected and processed using OpenLab deconvolution software.
For in vivo dimerization experiments, cells in fresh DMEM containing 10% (v/v) FBS in a 13-mm-diameter dish were placed in a 37 °C incubation block attached to a Leica DM-IRB inverted fluorescence microscope 14 h after transfection, and the AP20187 dimerizer (ARIAD Pharmaceuticals Inc., Cambridge, MA, U.S.A.) was added to a final concentration of 200 nM in the medium (from a 100 μM stock solution in ethanol). Cells were monitored for changes in localization of the EGFP-tagged FKBPF36V fusion proteins by fluorescence microscopy beginning 1 min after the addition of AP20187 and every 15 min thereafter. As a control, HeLa cells transfected with parental pEGFP-C1 were treated with dimerizer as described above and observed for changes in localization of EGFP.
Chemical cross-linking studies
HeLa cell lysates were treated with the covalent cross-linker BS3 [bis(sulfosuccinimidyl) suberate]. The cells were incubated for 15 h after transfection with 20 μg of the relevant expression construct, and then the AP20187 dimerizer was added to a final concentration of 200 nM. After a further 3 h, cells were washed in cold PBS containing dimerizer, and lysed in 20 mM phosphate buffer, pH 7.5, containing 150 mM NaCl, 1% (v/v) Triton X-100, 10 μg/ml aprotinin, 0.5 mM PMSF and dimerizer. The lysate was centrifuged at 16000 g for 10 min and BS3 was added to the supernatant (from a 10 mM stock in 20 mM phosphate buffer, pH 7.5) to a final concentration of 0.25 mM for 30 min at room temperature (22 °C). The cross-linking reaction was quenched with 20 mM glycine, pH 9.5, for 15 min, and the lysate was analysed by SDS/PAGE and immunoblotting with an anti-GFP antibody as described above (see Figure 1D).
RESULTS
In agreement with the argument that the PH domain of Dbs does not function as a simple independent membrane-targeting module [5], we found that an EGFP fusion protein containing the isolated Dbs DH/PH fragment (residues 623–967) is entirely cytoplasmic, with no evidence for defined subcellular localization (Figure 1B). However, this does not rule out the possibility that the Dbs PH domain nonetheless contributes to membrane association in a functionally important way, perhaps by co-operating with other membrane-targeting domains within the same protein (or in an oligomer). For example, the PH domain could co-operate with the N-terminal Sec14 domain in Dbs, also reported to bind phosphoinositides [27], to drive the protein to phosphoinositide-rich areas of the plasma membrane. Similarly, if Dbs forms oligomers, as reported for Dbl, Ras-GRF 1/2 (Ras guanine-nucleotide-releasing factor 1/2) and Cool-2 [28–30], multiple PH domains within the same oligomer could co-operate with one another to drive high-avidity (multivalent) membrane recruitment, as demonstrated previously for the PH domain of dynamin [31,32].
Dimerization can drive translocation of the Dbs DH/PH fragment to the plasma membrane
To determine whether the Dbs PH domain can promote membrane association as part of a multi-domain interaction, we generated an artificial chemically-inducible dimer of the DH/PH fragment and investigated its subcellular localization. We appended an F36V mutant of FK506 binding protein-12 to the C-terminus of an EGFP–DH/PH fusion to give the EGFP–DH/PH–FKBPF36V protein in Figure 1(A). A cell-permeant dimeric analogue of FK506 (AP20187) binds bivalently to two FKBP molecules (with a KD≈1 nM), and induces efficient dimerization of this fusion protein [33]. Corvera et al. [34] have used this approach to increase the avidity of FYVE domain interactions with PtdIns3P-containing endosomal membranes.
The EGFP–DH/PH–FKBPF36V fusion protein was completely diffuse and cytoplasmic when transiently expressed in HeLa cells without added dimerizer (Figure 1B). However, when AP20187 dimerizer was added to a final concentration of 200 nM (see Experimental section), plasma membrane association was discerned within 1 min in approx. 30% of transfected cells (Figure 1C), and this intensified significantly over the next 45 min. AP20187 treatment had no influence on the distribution of EGFP in HeLa cells expressing EGFP alone (results not shown). To confirm that 200 nM AP20187 induces efficient dimerization of the EGFP–DH/PH–FKBPF36V fusion protein, we incubated lysates from AP20187-treated cells with the bifunctional cross-linking agent BS3. An anti-GFP Western blot (Figure 1D) showed that EGFP–DH/PH–FKBP dimers were only cross-linked following treatment of cells with AP20187. Sedimentation equilibrium analytical ultracentrifugation studies (results not shown) confirmed that an E. coli expressed Dbs DH/PH fragment was entirely monomeric at concentrations up to 10 μM. Thus, dimerization of the Dbs DH/PH fragment is sufficient to drive it to the plasma membrane of HeLa cells.
Membrane targeting of the dimerized Dbs DH/PH fragment is PH-domain-mediated
To determine whether the membrane targeting seen in Figure 1(C) required PH domain–phosphoinositide interactions, we made mutations to disrupt PtdIns(4,5)P2 binding by the Dbs PH domain (Figure 2A). Arg861 in the β2 strand of the Dbs PH domain [19] corresponds to a critical and reasonably well-conserved residue in the lipid-binding site, at which mutations usually abolish PH domain phosphoinositide binding [32]. Rossman et al. [13] showed previously that the R861Q mutation impairs phosphoinositide binding significantly. In our lipid-overlay studies using an E. coli-produced GST–DH/PH fusion protein, we found that significant residual phosphoinositide binding remained in the R861Q mutant (Figure 2B). We therefore replaced four basic residues in the β1/β2 phosphoinositide-binding loop (K849, K851, R855 and K857) with glutamine to generate the PH* variant. GST–DH/PH* showed no detectable phosphoinositide binding in lipid overlay studies (Figure 2B), and gave no significant PtdIns(4,5)P2-binding signal when analysed in SPR studies (Figure 2C), even when injected at 100 μM.
As shown in Figure 2(D), the EGFP–DH/PH*–FKBPF36V fusion protein does not become membrane associated upon addition of the AP20187 dimerizer. This finding suggests that dimerizer-induced membrane translocation of the wild-type EGFP–DH/PH–FKBPF36V fusion is driven primarily by its PH domain, and requires both the ability of the PH domain to bind phosphoinositides and its dimerization. It therefore seems reasonable to argue that the Dbs PH domain could contribute to membrane targeting of full-length Dbs in a functionally important way by co-operating with other domains in Dbs or with other PH domains in a Dbs oligomer. When fused to GST, which dimerizes with high affinity [35], the Dbs DH/PH fragment binds to lipid vesicles containing 3% (mol/mol) PtdIns(4,5)P2 with a KD (app) of 7.5 μM as assessed by SPR studies (Figure 2C). This is in the range where membrane targeting should be evident in cellular studies [32], particularly if DH domain interactions with Cdc42 or RhoA can also contribute. By contrast, we could not detect PtdIns(4,5)P2 binding by the monomeric DH/PH fragment in SPR studies at the concentrations tested here (results not shown). We reported previously [31] a similar observation for the dynamin PH domain. In that case, the phosphoinositide head-group binding by a monomeric PH domain had a KD value in the 1–2 mM range [36,37], and dimerization mediated by GST brought the KD for PtdIns(4,5)P2 in membranes to approx. 20 μM [31]. Thus, the phosphoinositide-binding characteristics of the Dbs and dynamin PH domains are rather similar. In both cases, mutations that impair the very low-affinity phosphoinositide binding also impair cellular function of the respective protein [13,15,38–40]. Dbs PH therefore represents another example in which low-affinity phosphoinositide binding by a PH domain appears to be functionally relevant, possibly through its involvement in multivalent inter-aticactions with membranes [32].
PH domain mutations do not affect solubility or intrinsic GEF activity of DH/PH fragments
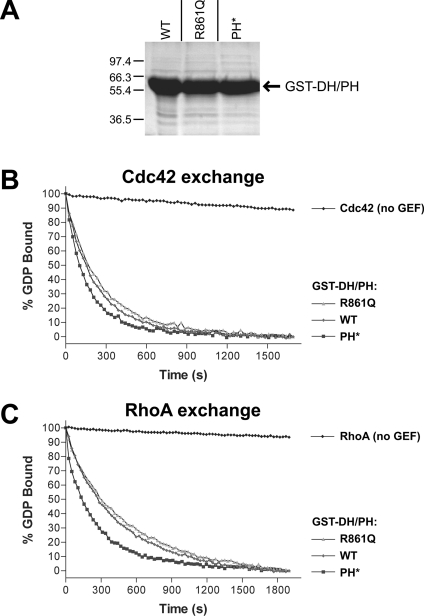
The mutations detailed in Figure 2(A) do not appear to cause misfolding, insolubility or instability of the expressed polypeptides. Each mutant was expressed well (and solubly) in E. coli at 37 °C with similar yields, as shown in Figure 3(A). In addition, to confirm that the PH domain mutations do not affect intrinsic GEF activity, we assayed the Cdc42- and RhoA-exchange activity of each GST–DH/PH fusion protein in vitro, using fluorescence spectroscopy to monitor N-methylanthraniloyl-GTP binding (see Experimental section). As shown in Figures 3(B) and 3(C), the intrinsic ability of the Dbs DH/PH fragment to promote nucleotide exchange on soluble GTPases was not impaired by PH domain mutations. Since the observed in vitro GEF activity of the Dbs DH/PH fragment requires that the PH domain is correctly folded [19], these observations argue that the PH* mutations reduce phosphoinositide binding without impairing overall structure.
Figure 3. PH domain mutations do not impair solubility or in vitro GEF activity of the Dbs DH/PH fragment.
(A) A Coomassie Blue-stained SDS/PAGE gel of E. coli-expressed wild-type (WT), R861Q and PH* forms of GST–DH/PH after one-step purification with glutathione–agarose beads is shown. Expression levels, solubility and stability appeared identical for the three proteins. In vitro GTP/GDP exchange on Cdc42 (B) and RhoA (C) was measured for wild-type (WT) and mutated GST–DH/PH proteins as described in the Experimental section. The data were fitted as single exponential decays, giving kobs for Cdc42 of 0.15×10−3 s−1 (no GEF), 4.29×10−3 s−1 (wild-type), 3.72×10−3 s−1 (R861Q) and 6.02×10−3 s−1 (PH*). Rates for RhoA were 0.31×10−3 s−1 (no GEF), 2.25×10−3 s−1 (wild type), 1.97×10−3 s−1 (R861Q) and 3.99×10−3 s−1 (PH*). Each experiment was performed twice with identical results.
Membrane targeting of the Dbs DH/PH fragment promotes Rho GTPase activation
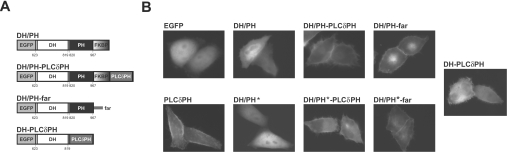
In the dimerizer studies described in Figure 1(C), we noticed AP20187-dependent formation of membrane protrusions that resemble filopodia in cells with membrane-translocated EGFP–DH/PH–FKBPF36V. This might reflect Cdc42 activation by the membrane-translocated DH/PH fragment, which in turn leads to filopodia formation [2,41]. We were not able to detect Cdc42 or RhoA activation reproducibly using biochemical assays in dimerizer-treated cells expressing EGFP–DH/PH–FKBPF36V. Reasoning that this might reflect relatively weak membrane association (and GEF activity) of dimerized EGFP–DH/PH–FKBPF36V, we generated alternative constructs in which more robust plasma membrane-targeting domains were fused to a monomeric Dbs DH/PH fragment. In one set of constructs, we fused the PLC-δ1-PH domain to the C-terminus of EGFP–DH/PH–FKBPF36V (to give DH/PH–PLCδ1PH in Figure 4A), maintaining the FKBPF36V moiety as a ‘spacer’ between the Dbs DH/PH fragment and the PLC-δ1-PH domain. PLC-δ1-PH binds with high affinity to PtdIns(4,5)P2 [32], and is targeted robustly to the plasma membrane of mammalian cells [42]. In another approach, to drive PtdIns(4,5)P2-independent plasma membrane targeting, we fused the H-Ras far sequence to the C-terminus of the Dbs DH/PH fragment (to give DH/PH–far in Figure 4A), as described for plasma membrane targeting of other proteins [43,44]. We also generated an EGFP fusion protein with the PLC-δ1 PH domain linked directly to the C-terminus of the Dbs DH domain (DH–PLCδ PH in Figure 4A), effectively replacing the Dbs PH domain with PLC-δ1-PH. For the remainder of the manuscript, all chimaeric proteins discussed are fused to EGFP, and will be referred to according to the convention shown in Figure 4A (with PH* denoting the presence of the PH* mutations shown in Figure 2A). Fluorescence microscopy studies (Figure 4B) demonstrated that fusing PLC-δ1-PH or the H-Ras plasma-membrane-targeting signal promotes significant plasma membrane localization of the EGFP–DH/PH fusions (similar to that seen with EGFP–PLC-δ1-PH), regardless of the presence of PH* mutations. By contrast, the chimaeric DH/PH and DH/PH* showed no evidence at all of concentration at the plasma membrane, and were indistinguishable from EGFP alone.
Figure 4. Membrane targeting of the Dbs DH/PH fragment.
(A) Schematic representation of the chimaeric constructs employed for membrane targeting of DH/PH and DH/PH*. To target the Dbs DH/PH fragment with PLC-δ1-PH, the PLC-δ1 PH domain was fused to the C-terminus of the EGFP–DH/PH–FKBPF36V chimaera shown in Figure 1(A), using the FKBP moiety as a spacer between the two PH domains. For targeting the DH/PH fragment by farnesylation (far), the farnesylation sequence of H-Ras was fused directly to the C-terminus of the EGFP–DH/PH fusion. To analyse the isolated DH domain, PLC-δ1-PH was fused directly to its C-terminus. Constructs including the Dbs PH domain were made with both the wild-type and PH* versions of the PH domain. The nomenclature used in the text for each construct is noted above its scheme. (B) Membrane localization of each EGFP chimaera was assessed by fluorescence microscopy of transiently-transfected live HeLa cells. No significant plasma membrane localization is seen for EGFP, or EGFP–DH/PH (wild-type or PH*). All fusion proteins containing PLC-δ1-PH or the far signal were robustly targeted to the plasma membrane. Structures resembling filopodia were seen in more than 60% of cells (n=150) exhibiting membrane localization of a DH-domain-containing fragment (DH/PH–PLC δ1 PH, DH/PH*–PLC δ1 PH, DH/PH–far, DH/PH*–far and DH–PLC δ1 PH).
We assessed levels of activation of endogenous RhoA and Cdc42 in HeLa cells transiently expressing the membrane-targeted Dbs chimaeric DH/PH shown in Figure 4. We used the RBD of Rhotekin to selectively precipitate activated RhoA from transfected cells [26]. To determine levels of Cdc42 activation, we used PAK1 PBD to selectively precipitate GTP-bound Cdc42 [25]. Parallel immunoblots of GTP-bound and total RhoA or Cdc42 then allowed an estimation of the relative proportion of the small GTPase that had been activated. Although expression of the chimaeric DH/PH enhanced RhoA (Figure 5) and Cdc42 (Figure 6) activation to some extent in HeLa cells, membrane targeting of the DH/PH fragment by fusion to PLC-δ1-PH (in DH/PH–PLCδ PH) or far (in DH/PH–far) led to a significant further increase (compare lanes 5 and 7 with lanes 3 in Figures 5 and 6). Thus, membrane targeting appears to be sufficient to promote GEF activity of the Dbs DH/PH fragment in a cellular context, consistent with the appearance of filopodia in cells with a membrane-targeted dimeric DH/PH fragment. Expression of each membrane-targeted DH/PH-containing chimaera in Figure 4(B) also led to the appearance of structures resembling filopodia that were absent in cells expressing EGFP or an EGFP–PLC-δ1-PH fusion protein.
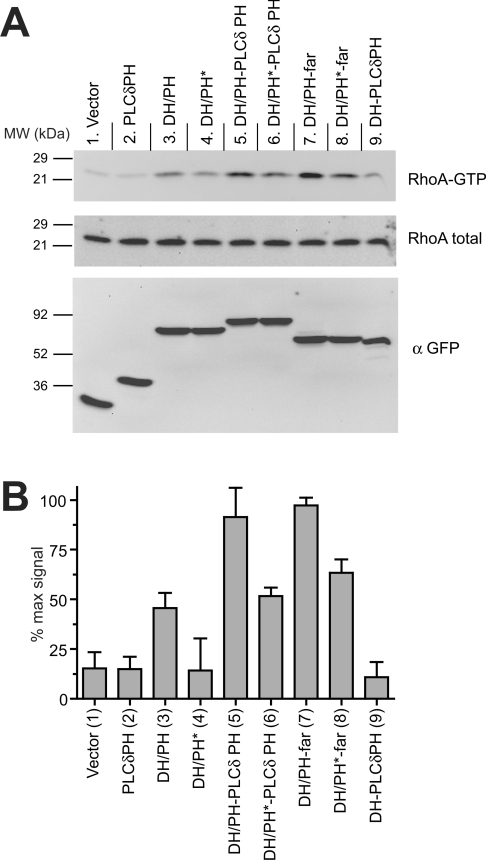
Figure 5. RhoA activation by membrane-targeted Dbs DH/PH.
(A) Representative immunoblots for RhoA activation experiments. HeLa cells were transfected with the indicated construct (see Figure 4A), and the active pool of (GTP-bound) RhoA was specifically isolated from serum-starved cells by affinity chromatography using GST–Rhotekin RBD as described in the Experimental section. The top and middle panels were immunoblotted with an antibody to RhoA, and represent (top) RhoA protein precipitated with GST–Rhotekin RBD (GTP-bound, active) and (middle) whole-cell lysate (total RhoA). The bottom panel represents whole-cell lysate immunoblotted with an antibody against GFP for comparison of expression levels of the different chimaeric Dbs DH/PH. (B) Immunoblots from at least three independent experiments were quantified using Kodak ImageStation software. The normalized intensity of each band was evaluated as a percentage of the maximum RhoA activation in that experiment, and results are presented as means±S.D.
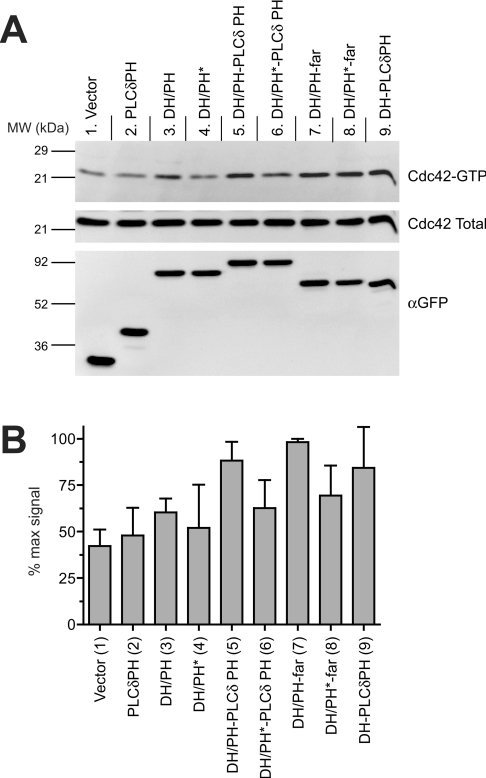
Figure 6. Cdc42 activation by membrane-targeted Dbs DH/PH.
(A) Representative immunoblots for Cdc42 activation experiments. Experiments were performed as for Figure 5(A), but using GST–PAK PBD to specifically isolate activated Cdc42. Top and middle panels were immunoblotted with an antibody to Cdc42, and represent activated (top) and total (middle) Cdc42 respectively. The bottom panel represents whole-cell lysate immunoblotted with an antibody against GFP for assessing Dbs fragment expression levels. (B) Quantification of Cdc42 activation experiment, as described in the legend to Figure 5(B).
PH domain mutations impair in vivo GEF activity even in the context of membrane-targeted Dbs fragments
If the primary role of phosphoinositide binding by the Dbs PH domain is to promote membrane targeting, wild-type and PH* mutants of otherwise membrane-targeted Dbs DH/PH fragments should be equivalent in their cellular GEF activity. Contrary to this notion, however, PH*-mutated Dbs DH/PH fragments showed significantly reduced apparent RhoA-GEF activity even when membrane-targeted by PLC-δ1-PH (Figure 5; compare lane 5 with lane 6) or farnesylation (Figure 5; compare lane 7 with lane 8). The PH* mutations do not appear to alter membrane targeting of DH/PH–PLC-δ1-PH or DH/PH–far (Figure 4B), yet, in an already membrane-targeted context, the PH* mutations significantly reduce GEF activity. This observation suggests a specific role for PH domain–phosphoinositide interactions in regulating Dbs activity at the membrane. Similar observations were also made for Cdc42 activation (Figure 6), although the effect of the PH* mutation on GEF activity of the chimaeric DH/PH–far was less clear. The PH* mutation also reduced RhoA and Cdc42 activation by the chimaeric DH/PH, which may reflect a reduction in its low level of membrane targeting.
Finally, by analysing cells expressing DH–PLCδPH, we tested the cellular GEF activity of the membrane-targeted DH domain alone (fused to PLC-δ1-PH without including the Dbs PH domain). As seen in Figures 5 and 6, lanes 9, DH–PLC-δ1-PH expression led to robust Cdc42 activation (Figure 6), but had only a small effect on RhoA activation (Figure 5). Interestingly, comparing lane 6 with lane 9 in Figure 6 suggests that a phosphoinositide-binding-defective PH domain may actually have a negative influence on the ability of the DH domain to activate Cdc42 (but not RhoA).
DISCUSSION
We conclude from these findings that the Dbs PH domain has the capacity to modify subcellular localization of Dbs, but only as one component of a multidomain interaction. If full-length Dbs forms oligomers, as reported for Dbl, Ras-GRF and Cool-2 [28–30], multiple copies of the Dbs PH domain could co-operate in targeting these oligomers to the plasma membrane. More likely, the Dbs PH domain might contribute to membrane association of the whole protein through co-operation with one or more other membrane-targeting domains within Dbs. In addition to an SH3 domain at its C-terminus that could bind to proline-rich regions of a membrane-localized protein, Dbs contains a SEC14 domain at the N-terminus. The SEC14 domain, first identified in yeast in a phosphatidylinositol transfer protein [45], has been shown to bind phospholipids in vitro and in vivo [45,46]. The SEC14 domains of Dbl, Ost (the rat orthologue of Dbs) and Dbs were recently reported to bind to phosphoinositides and to contribute significantly to the subcellular localization of these proteins [27,47]. In either case, the fact that only two PH domains are required for membrane targeting (i.e. a DH/PH dimer is significantly membrane-associated) argues that the Dbs PH domain can contribute a significant amount of binding energy to membrane association of the whole protein.
To investigate whether PtdIns(4,5)P2 binding by the Dbs DH/PH fragment plays a role independent of its ability to promote membrane targeting, we analysed Cdc42 and RhoA activation in HeLa cells by mutants defective in PtdIns(4,5)P2 binding that were robustly membrane targeted by PLC-δ1-PH or a farnesyl group. Activation of both small GTPases was reduced as a result of these mutations, even when made in the context of a strongly membrane-targeted DH/PH-containing protein. This observation supports the hypothesis that phosphoinositide binding by the Dbs PH domain plays an important role beyond its contributions to membrane targeting, in agreement with previous arguments [13,15]. Interestingly, when the isolated DH domain from Dbs was plasma membrane targeted by direct fusion to PLC-δ1-PH, RhoA activation was minimal (Figure 5), but Cdc42 activation was significant (Figure 6). Crystallographic studies show that Cdc42 and RhoA interact directly with both the DH and PH domains of Dbs [4,19]. The fact that the PH domain appears more important for RhoA than for Cdc42 activation may indicate that PH domain contacts play a greater role in assisting RhoA exchange than Cdc42 exchange, perhaps because of stronger PH domain–RhoA interactions or weaker DH domain–RhoA interactions. It is therefore interesting that the crystal structure of the Dbs DH/PH fragment bound to RhoA shows at least one additional hydrogen bond between the PH domain β3/β4 loop and the RhoA switch 2 region (i.e. between Tyr883 of the PH domain and Arg68 of RhoA) that is not present in the Cdc42 complex [4].
Crystal structures are now available for the Dbs DH/PH fragment alone [22], in complex with Cdc42 [19] and in complex with RhoA [4]. Analysis of the GTPase-free structure indicates that the spatial relationship between the PH and DH domains is not fixed, and suggests that phosphoinositide binding to the PH domain may restrict conformational heterogeneity so that the relationship between DH and PH domains is ideal for optimal productive interactions of both domains with Cdc42 and RhoA [22]. In the context of this model, our results suggest (Figure 7) that RhoA activation requires its interaction with both the DH and PH domains, which is optimized by PtdIns(4,5)P2 binding. Association with the DH domain alone is not sufficient. For Cdc42 activation, DH domain interactions may dominate (and may be sufficient on their own). PtdIns(4,5)P2 binding may relieve a negative influence of the PH domain on Cdc42–DH domain interactions, in addition to optimizing the observed Cdc42–PH domain interactions.
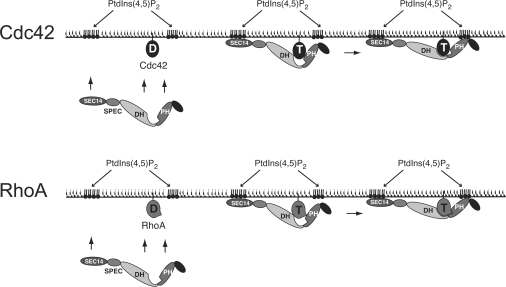
Figure 7. Schematic model for the role of the Dbs PH domain in regulating GEF activity.
Membrane recruitment of Dbs is promoted by the co-operation of multiple domains, including the Sec14 domain [27], possibly the SH3 domain, plus PH domain–phosphoinositide interactions, PH domain–GTPase (Cdc42 or RhoA) interactions, and DH domain–GTPase (Cdc42 or RhoA) interactions. At the plasma membrane, optimal binding to the GTPase target requires ligation of the PH domain by PtdIns(4,5)P2 in order to relieve possible steric hindrance to DH domain interactions and maximize interactions between the GTPase and the PH domain. In the case of Cdc42, relief of inhibition may play a more important role, as discussed in the text. Our results suggest that the PH domain interactions may be more important in the case of RhoA, as indicated in the Figure. D, GDP bound to Cdc42 or RhoA; T; GTP-bound state.
In the Dbl family of proteins, which accounts for a significant fraction of all PH-domain-containing proteins, the phosphoinositide-binding properties and subcellular localization of isolated PH domains argues that they contribute to, but do not dominate or drive, membrane targeting. Once at the membrane surface, however, PH domain binding to phosphoinositides appears to stabilize a DH/PH conformation that is optimal for Rho-family GTPase activation, presumably restricting activation of these GTPases to phosphoinositide-containing membranes.
Acknowledgments
We thank M. Yohe for assistance with in vitro GEF assays, members of the M. A. L. laboratory for valuable discussions, and Dr Channing Der and Dr Xiang-Dong Ren for constructs. This work was supported in part by National Institutes of Health Grants R01-GM56846 (to M. A. L.), F31-GM65066 (to M. A. B.), and R01-GM62299 (to J.S.). K. L. R. is supported by postdoctoral fellowship PF-05-129-01-GMC from the American Cancer Society.
References
- 1.Whitehead I. P., Campbell S., Rossman K. L., Der C. J. Dbl family proteins. Biochim. Biophys. Acta. 1997;1332:F1–F23. doi: 10.1016/s0304-419x(96)00040-6. [DOI] [PubMed] [Google Scholar]
- 2.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 3.Hart M. J., Eva A., Zangrilli D., Aaronson S. A., Evans T., Cerione R. A., Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalysed by a common domain on the dbl oncogene product. J. Biol. Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 4.Snyder J. T., Worthylake D. K., Rossman K. L., Betts L., Pruitt W. M., Siderovski D. P., Der C. J., Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat. Struct. Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- 5.Rossman K. L., Sondek J. Larger than Dbl: new structural insights into RhoA activation. Trends Biochem. Sci. 2005;30:163–165. doi: 10.1016/j.tibs.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Lemmon M. A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- 7.Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., Lemmon M. A. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- 8.Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol. Cell. 2004;13:677–688. doi: 10.1016/s1097-2765(04)00083-8. [DOI] [PubMed] [Google Scholar]
- 9.Snyder J. T., Rossman K. L., Baumeister M. A., Pruitt W. M., Siderovski D. P., Der C. J., Lemmon M. A., Sondek J. Quantitative analysis of the effect of phosphoinositide interactions on the function of Dbl family proteins. J. Biol. Chem. 2001;276:45868–45875. doi: 10.1074/jbc.M106731200. [DOI] [PubMed] [Google Scholar]
- 10.Han J., Luby-Phelps K., Das B., Shu X., Xia Y., Mosteller R. D., Krishna U. M., Falck J. R., White M. A., Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J., Chen R. H., Corblan-Garcia S., Cahill S. M., Bar-Sagi D., Cowburn D. The solution structure of the pleckstrin homology domain of human SOS1. A possible structural role for the sequential association of diffuse B cell lymphoma and pleckstrin homology domains. J. Biol. Chem. 1997;272:30340–30344. doi: 10.1074/jbc.272.48.30340. [DOI] [PubMed] [Google Scholar]
- 12.Baumeister M. A., Martinu L., Rossman K. L., Sondek J., Lemmon M. A., Chou M. M. Loss of phosphatidylinositol 3-phosphate binding by the C-terminal Tiam-1 pleckstrin homology domain prevents in vivo Rac1 activation without affecting membrane targeting. J. Biol. Chem. 2003;278:11457–11464. doi: 10.1074/jbc.M211901200. [DOI] [PubMed] [Google Scholar]
- 13.Rossman K. L., Cheng L., Mahon G. M., Rojas R. J., Snyder J. T., Whitehead I. P., Sondek J. Multifunctional roles for the PH domain of Dbs in regulating Rho GTPase activation. J. Biol. Chem. 2003;278:18393–18400. doi: 10.1074/jbc.M300127200. [DOI] [PubMed] [Google Scholar]
- 14.Palmby T. R., Abe K., Der C. J. Critical role of the pleckstrin homology and cysteine-rich domains in Vav signaling and transforming activity. J. Biol. Chem. 2002;277:39350–39359. doi: 10.1074/jbc.M202641200. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes E. J., Karnoub A. E., Booden M. A., Der C. J., Campbell S. L. Critical role of the pleckstrin homology domain in Dbs signaling and growth regulation. J. Biol. Chem. 2003;278:21188–21196. doi: 10.1074/jbc.M211792200. [DOI] [PubMed] [Google Scholar]
- 16.Kubiseski T. J., Culotti J., Pawson T. Functional analysis of the Caenorhabditis elegans UNC-73B PH domain demonstrates a role in activation of the Rac GTPase in vitro and axon guidance in vivo. Mol. Cell. Biol. 2003;23:6823–6835. doi: 10.1128/MCB.23.19.6823-6835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellanger J.-M., Estrach S., Schmidt S., Briancon-Marjollet A., Zugasti O., Fromont S., Debant A. Different regulation of the trio Dbl-homology domains by their associated PH domains. Biol. Cell. 2003;95:625–634. doi: 10.1016/j.biolcel.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Vanni C., Mancini P., Gao Y., Ottaviano C., Guo F., Salani B., Torrisi M. R., Zheng Y., Eva A. Regulation of proto-Dbl by intracellular membrane targeting and protein stability. J. Biol. Chem. 2002;277:19745–19753. doi: 10.1074/jbc.M111025200. [DOI] [PubMed] [Google Scholar]
- 19.Rossman K. L., Worthylake D. K., Snyder J. T., Siderovski D. P., Campbell S. L., Sondek J. A crystallographic view of interactions between Dbs and Cdc42: PH domain-assisted guanine nucleotide exchange. EMBO J. 2002;21:1315–1326. doi: 10.1093/emboj/21.6.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derewenda U., Oleksy A., Stevenson A. S., Korczynska J., Dauter Z., Somlyo A. P., Otlewski J., Somlyo A. V., Derewenda Z. S. The crystal structure of RhoA in complex with the DH/PH fragment of PDZRhoGEF, an activator of the Ca(2+) sensitization pathway in smooth muscle. Structure. 2004;12:1955–1965. doi: 10.1016/j.str.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Kristelly R., Gao G., Tesmer J. J. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J. Biol. Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- 22.Worthylake D. K., Rossman K. L., Sondek J. Crystal structure of the DH/PH fragment of Dbs without bound GTPase. Structure. 2004;12:1078–1086. doi: 10.1016/j.str.2004.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Whitehead I., Kirk H., Kay R. Retroviral transduction and oncogenic selection of a cDNA encoding Dbs, a homolog of the Dbl guanine nucleotide exchange factor. Oncogene. 1995;10:713–721. [PubMed] [Google Scholar]
- 24.Whitehead I. P., Lambert Q. T., Glaven J. A., Abe K., Rossman K. L., Mahon G. M., Trzaskos J. M., Kay R., Campbell S. L., Der C. J. Dependence of Dbl and Dbs transformation on MEK and NF-κB activation. Mol. Cell. Biol. 1999;19:7759–7770. doi: 10.1128/mcb.19.11.7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benard V., Bohl B. P., Bokoch G. M. Characterization of Rac and Cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J. Biol. Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 26.Ren X. D., Kiosses W. B., Schwartz M. A. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostenko E. V., Mahon G. M., Cheng L., Whitehead I. P. The Sec14 homology domain regulates the cellular distribution and transforming activity of the Rho-specific guanine nucleotide exchange factor, Dbs. J. Biol. Chem. 2004;280:2807–2817. doi: 10.1074/jbc.M411139200. [DOI] [PubMed] [Google Scholar]
- 28.Anborgh P. H., Qian X., Papageorge A. G., Vass W. C., DeClue J. E., Lowy D. R. Ras-specific exchange factor GRF: oligomerization through its Dbl homology domain and calcium-dependent activation of Raf. Mol. Cell. Biol. 1999;19:4611–4622. doi: 10.1128/mcb.19.7.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feng Q., Baird D., Cerione R. A. Novel regulatory mechanisms for the Dbl family guanine nucleotide exchange factor Cool-2/α-Pix. EMBO J. 2004;23:3492–3504. doi: 10.1038/sj.emboj.7600331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu K., Debreceni B., Bi F., Zheng Y. Oligomerization of DH domain is essential for Dbl-induced transformation. Mol. Cell. Biol. 2001;21:425–437. doi: 10.1128/MCB.21.2.425-437.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein D. E., Lee A., Frank D. W., Marks M. S., Lemmon M. A. The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J. Biol. Chem. 1998;273:27725–27733. doi: 10.1074/jbc.273.42.27725. [DOI] [PubMed] [Google Scholar]
- 32.Lemmon M. A., Ferguson K. M. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 2000;350:1–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Strasser V., Fasching D., Hauser C., Mayer H., Bock H. H., Hiesberger T., Herz J., Weeber E. J., Sweatt J. D., Pramatarova A., et al. Receptor clustering is involved in Reelin signaling. Mol. Cell. Biol. 2004;24:1378–1386. doi: 10.1128/MCB.24.3.1378-1386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayakawa A., Hayes S. J., Lawe D. C., Sudharshan E., Tuft R., Fogarty K., Lambright D., Corvera S. Structural basis for endosomal targeting by FYVE domains. J. Biol. Chem. 2004;279:5958–5966. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- 35.Tudyka T., Skerra A. Glutathione S-transferase can be used as a C-terminal, enzymatically active dimerization module for a recombinant protease inhibitor, and functionally secreted into the periplasm of Escherichia coli. Protein Sci. 1997;6:2180–2187. doi: 10.1002/pro.5560061012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salim K., Bottomley M. J., Querfurth E., Zvelebil M. J., Gout I., Scaife R., Margolis R. L., Gigg R., Smith C. I., Driscoll P. C., et al. Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 1996;15:6241–6250. [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng J., Cahill S. M., Lemmon M. A., Fushman D., Schlessinger J., Cowburn D. Identification of the binding site for acidic phospholipids on the PH domain of dynamin: implications for stimulation of GTPase activity. J. Mol. Biol. 1996;255:14–21. doi: 10.1006/jmbi.1996.0002. [DOI] [PubMed] [Google Scholar]
- 38.Achiriloaie M., Barylko B., Albanesi J. P. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol. Cell. Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee A., Frank D. W., Marks M. S., Lemmon M. A. Dominant-negative inhibition of receptor-mediated endocytosis by a dynamin-1 mutant with a defective pleckstrin homology domain. Curr. Biol. 1999;9:261–264. doi: 10.1016/s0960-9822(99)80115-8. [DOI] [PubMed] [Google Scholar]
- 40.Vallis Y., Wigge P., Marks B., Evans P. R., McMahon H. T. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 41.Nobes C. D., Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 42.Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell. Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aronheim A., Engelberg D., Li N., al-Alawi N., Schlessinger J., Karin M. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell. 1994;78:949–961. doi: 10.1016/0092-8674(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 44.Logan S. K., Falasca M., Hu P., Schlessinger J. Phosphatidylinositol 3-kinase mediates epidermal growth factor-induced activation of the c-Jun N-terminal kinase signaling pathway. Mol. Cell. Biol. 1997;17:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bankaitis V. A., Malehorn D. E., Emr S. D., Greene R. The Saccharomyces cerevisiae SEC14 gene encodes a cytosolic factor that is required for transport of secretory proteins from the yeast Golgi complex. J. Cell Biol. 1989;108:1271–1281. doi: 10.1083/jcb.108.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cockcroft S. Phosphatidylinositol transfer proteins: a requirement in signal transduction and vesicle traffic. BioEssays. 1998;20:423–432. doi: 10.1002/(SICI)1521-1878(199805)20:5<423::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 47.Ueda S., Kataoka T., Satoh T. Role of the Sec14-like domain of Dbl family exchange factors in the regulation of Rho family GTPases in different subcellular sites. Cell. Signalling. 2004;16:899–906. doi: 10.1016/j.cellsig.2004.01.007. [DOI] [PubMed] [Google Scholar]