Abstract
The arsenal of engineered variants of the GFP [green FP (fluorescent protein)] from Aequorea jellyfish provides researchers with a powerful set of tools for use in biochemical and cell biology research. The recent discovery of diverse FPs in Anthozoa coral species has provided protein engineers with an abundance of alternative progenitor FPs from which improved variants that complement or supersede existing Aequorea GFP variants could be derived. Here, we report the engineering of the first monomeric version of the tetrameric CFP (cyan FP) cFP484 from Clavularia coral. Starting from a designed synthetic gene library with mammalian codon preferences, we identified dimeric cFP484 variants with fluorescent brightness significantly greater than the wild-type protein. Following incorporation of dimer-breaking mutations and extensive directed evolution with selection for blue-shifted emission, high fluorescent brightness and photostability, we arrived at an optimized variant that we have named mTFP1 [monomeric TFP1 (teal FP 1)]. The new mTFP1 is one of the brightest and most photostable FPs reported to date. In addition, the fluorescence is insensitive to physiologically relevant pH changes and the fluorescence lifetime decay is best fitted as a single exponential. The 1.19 Å crystal structure (1 Å=0.1 nm) of mTFP1 confirms the monomeric structure and reveals an unusually distorted chromophore conformation. As we experimentally demonstrate, the high quantum yield of mTFP1 (0.85) makes it particularly suitable as a replacement for ECFP (enhanced CFP) or Cerulean as a FRET (fluorescence resonance energy transfer) donor to either a yellow or orange FP acceptor.
Keywords: Clavularia, fluorescence imaging, fluorescence resonance energy transfer (FRET), genetic fusion, teal fluorescent protein
Abbreviations: FP, fluorescent protein; CCD camera, charge-coupled-device camera; CFP, cyan FP; TFP, teal FP; dTFP, dimeric TFP; ECFP, enhanced CFP; ER, endoplasmic reticulum; EGFP, enhanced GFP; FRET, fluorescence resonance energy transfer; FWHM, full width at half maximum; GFP, green FP; LB, Luria–Bertani; LED, light emitting diode; mECFP, ECFP with the A206K mutation; mTFP, monomeric TFP; ND, neutral density; rmsd, root mean square deviation; YC3.3, yellow cameleon 3.3; YFP, yellow FP
INTRODUCTION
The FPs (fluorescent proteins), including mutants of Aequorea victoria GFP (green FP) [1] and its numerous homologues from Anthozoa corals [2,3], have enabled fluorescence imaging of recombinant fusion proteins to become a popular and widely accessible technique in cell biology research [4,5]. The defining feature of FPs is their remarkable ability to autonomously generate a visible wavelength fluorophore within the confines of their distinctive β-barrel structures [6,7]. Chromophore formation in wild-type Aequorea GFP is a stepwise process that converts a sequence of three amino acids (Ser65, Tyr66 and Gly67) into a unique conjugated system that extends from the side chain of Tyr66 to a five-membered heterocycle formed from main chain atoms [8]. The steric, electrostatic and hydrogen-bonding environment imposed upon the chromophore by the surrounding residues strongly influences the fluorescence properties. This dependence has allowed researchers to engineer Aequorea GFP variants with altered colours, brightness and photostability [1]. For example, in wild-type Aequorea GFP, the ground-state chromophore exists as an equilibrating mixture of the neutral phenol with maximal absorbance at 395 nm and the anionic phenolate with maximal absorbance at 475 nm [9]. Efforts to engineer variants with the ground-state equilibrium shifted towards either the anionic or protonated form of the chromophore have been highly successful and have resulted in popular green fluorescent variants (emission peak at ∼510 nm) such as EGFP (enhanced GFP) [10] and Sapphire [11] respectively. Another particularly useful class of variants that have resulted from protein engineering efforts are the Aequorea GFP-derived YFPs (yellow FPs) that are defined by the T203Y mutation, an anionic phenolate tyrosine-derived chromophore, and an emission peak that is approx. 20 nm red-shifted from that of EGFP [7].
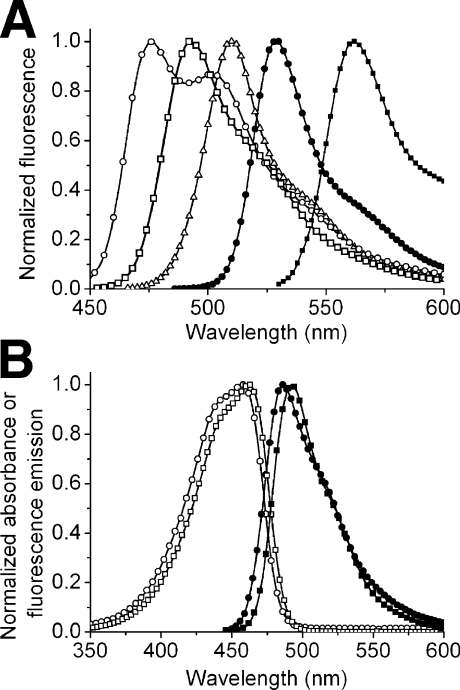
An approach that has proven successful for the engineering of blue-shifted Aequorea GFP variants has been to alter the covalent structure of the chromophore through substitution of other aromatic amino acids for Tyr66. For example, replacing Tyr66 of Aequorea GFP with a tryptophan residue gives an indole-containing chromophore with an emission peak in the cyan region (∼480 nm) of the visible spectrum [11]. Although the original Y66W mutant of Aequorea GFP was only weakly fluorescent, efforts to improve the brightness yielded the widely used variant ECFP [enhanced CFP (cyan FP)] [12,13] and more recently Cerulean [14] and CyPet [15]. Although ECFP has stood the test of time as a useful fluorophore for multicolour labelling and as the preferred FRET (fluorescence resonance energy transfer) donor to a YFP acceptor, its spectral properties are suboptimal [14,16]. Some limitations have been partially addressed in the newer variants. For example, Cerulean is 2-fold brighter than ECFP but it has decreased photostability under arc lamp illumination [14,16]. CyPet exhibits high FRET to the YFP variant YPet [15] but is poorly expressed at 37 °C [16]. Considered as a family, ECFP and its descendents are limited by fluorescence brightness that is less than 50% of the popular YFP variant Citrine [16] and fluorescence lifetimes that are generally multiexponential (although Cerulean is adequately fitted as a single exponential [14]). In addition, variants with tryptophan-derived chromophores have a very broad fluorescence emission with a FWHM (full width at half maximum) of approx. 60 nm (Figure 1A). Other popular FPs with tyrosine-derived chromophores (e.g. EGFP and Citrine) have single-peaked and narrower emission spectra with FWHM of approx. 31 nm.
Figure 1. Absorbance and fluorescence emission spectra of a selection of FPs.
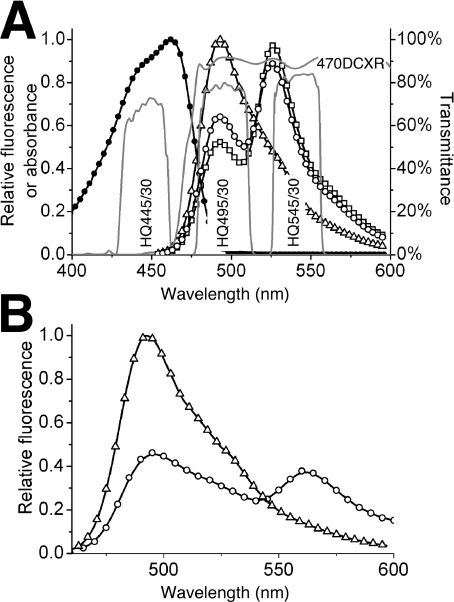
(A) Fluorescence emission spectra of mCerulean (○) [14,34], mTFP1 (□), EGFP (△), Citrine (●) [32] and mOrange (■) [33]. (B) Absorbance (open symbols) and fluorescence emission (filled symbols) spectra of dTFP0.2 (○, ●) and mTFP1 (□, ■).
It had occurred to us that the limitations of the existing CFPs with tryptophan-derived chromophores could be due to intrinsic properties of the chromophore structure. Furthermore, we reasoned that a new FP with a tyrosine-derived chromophore and a fluorescence emission in the cyan region could serve as a superior alternative to the existing CFPs. A number of candidate FPs that fit these general criteria have been cloned from organisms of the phylum Cnidaria in recent years. These proteins include: amFP486 from Anemonia majano [2]; dsFP483 and the very similar DstC1 from Discosoma striata [2,17]; cFP484 from Clavularia sp. [2]; mcCFP and the very similar mc5 from Montastrea cavernosa [18,19]; PdaC1 from Pocillopora damicornis [17]; anm1GFP1 from an unidentified Anthomedusa [20] and MiCy from Acropora sp. [21]. An engineered variant of tetrameric amFP486 known as AmCyan1 (Clontech), and wild-type MiCy (MBL International), are commercially available. The naturally occurring CFPs have emission maxima ranging from 477 to 495 nm, quantum yields ranging from 0.24 to 0.90, and molar absorption coefficients (ϵ) ranging from 24000 to 75000 M−1·cm−1. Despite these promising attributes, the natural CFPs are limited by the fact that they all have quaternary structure ranging from tight dimers in the case of anm1GFP1 and MiCy to obligate trimers or tetramers for all other Anthozoa CFPs reported to date.
While oligomerization of an FP is not particularly relevant to its use as a reporter of gene expression, it can often perturb the proper localization of fusion proteins, particularly when the protein of interest is an oligomer itself [22–25]. The most effective approach for overcoming the problem of FP oligomerization is to use a combination of rational protein engineering and directed evolution to create useful non-oligomerizing variants. Such an approach has been applied to a growing number of FPs including: a red FP from Discosoma sp. [25]; a GFP from Galaxeidae [26]; an orange FP from Fungia concinna [21]; green-to-red photoconvertible FPs from both Lobophyllia hemprichii [27] and Dendronephthya sp. [28]; a photoactivatable GFP from Pectiniidae [29]; and a chromoprotein from Montipora sp. to create a red FP with a large Stokes shift [30]. For Aequorea GFP and its variants, a single point mutation, A206K, is sufficient to abolish the weak dimerization [31]. The application of a monomeric version of MiCy has appeared in the literature [30], but no details on the engineering or properties of this protein have yet been published.
In the present paper, we describe the engineering, optimization and structural characterization of a monomeric variant of cFP484 from Clavularia sp. [2]. The directed evolution of the new FP was guided by selection for blue-shift, photostability and fluorescent brightness. Accordingly, the resulting FP has brightness and photostability that equals or exceeds that of the currently preferred FP variants [16]. We demonstrate that this new FP is an effective and practical FRET donor to both the YFP variant Citrine [32] and the Discosoma red FP-derived mOrange [33].
EXPERIMENTAL
General methods and materials
A synthetic gene library of Clavularia cFP484 variants was commissioned from the DNA Technologies Unit at the NRC (National Research Council) Plant Biotechnology Group (Saskatoon, SK, Canada). The full-length gene library was constructed by PCR-based assembly of overlapping PAGE-purified oligonucleotides that had been synthesized on a Beckman 1000M solid-phase DNA synthesizer. All synthetic DNA oligonucleotides for cloning and construction of subsequent libraries were purchased from Sigma-Genosys Canada or Integrated DNA Technologies. Unless otherwise indicated, Pfu polymerase (Fermentas) was used for all PCR amplifications in the buffer supplied by the manufacturer. PCR products and products of restriction digest were routinely purified using the QIAquick PCR purification kit (Qiagen) according to the manufacturer's protocols. Restriction enzymes were purchased from either Invitrogen or New England Biolabs. The cDNA sequences for all TFP (teal FP) variants and fusion constructs were confirmed by dye terminator cycle sequencing using the DYEnamic ET kit (Amersham Biosciences). Sequencing reactions were analysed at the University of Alberta MBSU (Molecular Biology Service Unit). The proteins mECFP (ECFP with the A206K mutation) and mCerulean have the A206K mutation in addition to their characteristic substitutions [31,34].
Library construction, mutagenesis and genes encoding fusion proteins
The initial synthetic gene library was digested with XhoI and EcoRI and ligated into similarly digested pBAD/His B vector (Invitrogen). Subsequent libraries with saturation mutagenesis at a particular residue were constructed by either by an overlap-extension PCR method [25] or the QuikChange® protocol (Stratagene). Randomly mutated libraries were constructed by error-prone PCR as previously described [25] using Taq polymerase (New England Biolabs) under conditions optimal for three mutations per 1000 bp [35]. Full-length gene libraries resulting from overlap-extension PCR or error-prone PCR were digested with XhoI and EcoRI and ligated into similarly digested pBAD/His B. Regardless of library assembly method, electrocompetent Escherichia coli strain DH10B (Invitrogen) was transformed and plated on to LB (Luria–Bertani)/agar plates supplemented with ampicillin (0.1 mg/ml) and L-arabinose (0.02%). Plates were incubated for 14 h at 37 °C prior to screening.
To construct expression vectors for cameleon variants, the gene encoding YC3.3 (yellow cameleon 3.3) was first inserted into the XhoI and EcoRI sites of the pBAD/His B bacterial expression vector [32]. The cDNAs encoding mECFP, mCerulean and mTFP1 (monomeric TFP1) were each PCR-amplified with primers that added a 5′ XhoI and a 3′ SphI site. To maintain identical linker lengths, the 3′ SphI site was appended immediately after the codon encoding Ala227 of mECFP and mCerulean or after the codon encoding the structurally aligned Arg220 of mTFP1. The purified PCR products were digested and ligated into the similarly digested plasmid containing the YC3.3 gene. Cameleons expressed in E. coli were expressed and purified as described above. To create the mTFP1–YC3.3 mammalian cell expression vector, the full-length gene in pBAD/His B was PCR-amplified with a 5′ primer that appended a HindIII restriction site, a Kozak sequence (gccaccgccATGc, where ATG is the start codon), and the ER (endoplasmic reticulum) targeting sequence of calreticulin (MLLSVPLLLGLLGLAAAD). The 3′ primer appended the ER retention signal (KDEL) followed by an EcoRI restriction site. The PCR product was digested with HindIII/EcoRI and ligated with appropriately digested pcDNA3 (Invitrogen).
The mOrange–mTFP1 fusion gene was created by overlap extension PCR. The gene encoding mOrange was amplified with a 5′ primer that appended an XhoI site and a 3′ primer that appended the sequence for an eight-amino-acid linker after residue 225. In a separate PCR reaction, the gene encoding mTFP1 was amplified with a 5′ primer that added the linker sequence and a 3′ primer that appended an EcoRI site. The two purified PCR products were mixed and re-amplified with the 5′ primer from the mOrange reaction and the 3′ primer from the mTFP1 reaction. The resulting full-length PCR product was digested with XhoI/EcoRI and ligated into similarly digested pBAD/His B vector.
To create the mTFP1–β-actin mammalian expression plasmid, the gene encoding mTFP1 was PCR-amplified with a 5′ primer encoding an NheI site and a 3′ primer encoding an XhoI site. The purified and digested PCR product was ligated into the pEGFP-actin vector (Clontech) that had been previously digested with the same restriction enzymes to excise the EGFP coding sequence. To create the mTFP1–α-tubulin expression vector, an identical procedure was used to replace the EGFP gene in pEGFP-tub (Clontech) with the gene encoding mTFP1.
Library screening
The system for imaging the fluorescence of bacterial colonies grown on a 10 cm Petri dish is a custom-built device that has been described in detail elsewhere [36]. Briefly, the light from a 175 W xenon arc lamp (Sutter) is passed through a 436/20 nm bandpass filter (Chroma) and into a bifurcated fibre optic bundle (Newport). Light exiting the fibre optic bundle illuminates a 10 cm diameter area with an irradiance of approx. 0.04 mW/cm2. For all screening up to the identification of mTFP0.6, colony fluorescence on illuminated plates was viewed through a pair of custom goggles fitted with 3 mm thick GG455 glass (Chroma). When viewed through these goggles, colonies fluorescing at 480–490 nm have a distinctly ‘bluish’ hue and are distinguishable from ‘greenish’ colonies emitting at 500–510 nm. For the identification of mTFP0.7, colony fluorescence was digitally imaged with a Retiga 1300i 12-bit CCD camera (charge-coupled-device camera) (QImaging) fitted with a filter wheel (Sutter) that contains both a 480/40 nm and a 530/30 nm bandpass filter. Through the use of custom macros for Image Pro Plus (Media Cybernetics), images in both emission channels were acquired and the fluorescence intensities of all colonies were individually integrated. Colonies with high 480/530 nm intensity ratios and high brightness at 480 nm were selected for further characterization.
For the identification of mTFP0.8, mTFP0.9 and mTFP1, the screening protocol was modified in order to select for photostability. We equipped six Royal Blue (peak emission at 455 nm) Luxeon V LEDs (light emitting diodes) (Lumileds Lighting) with narrow beam lenses (Fraen) and positioned them to evenly illuminate (55 mW/cm2) the 10 cm dish in the imaging system described above. Through the use of a custom serial port connection, the LEDs could be switched on and off at computer-controlled intervals. An Image Pro Plus macro (Media Cybernetics) was used to automate acquisition and processing. For each plate, fluorescence images were acquired following a series of programmed intervals of intense illumination. Using this system, we could readily identify colonies with decreased propensity to photobleach.
For all screening protocols, colonies with more intense fluorescence, high 480/530 nm intensity ratio, or decreased propensity to photobleach were picked and cultured overnight in 4 ml of LB media containing ampicillin (0.1 mg/ml) and L-arabinose (0.2%). On the following day, 0.1 ml of each culture was dispensed into a 96-well-plate black clear bottom plate (Corning) and the full emission spectra of each variant was measured with a Safire2 plate reader (Tecan). Variants with the most blue-shifted and intense emission peak were used as templates in the subsequent round of library construction.
Protein purification and characterization
To prepare proteins in sufficient quantity for characterization, E. coli strains DH10B or LMG194 were transformed with the pBAD/His B expression vector containing the gene of interest. A single colony was used to inoculate a 4 ml culture that was allowed to grow overnight (37 °C and 225 rev./min) before being diluted into 1 litre of LB media containing ampicillin (0.1 mg/ml) and arabinose (0.2%). The culture was allowed to grow for 12 h before cells were harvested by centrifugation and lysed by a French press. Proteins were purified by Ni-NTA (Ni2+-nitrilotriacetate) chromatography (Amersham). All cameleons and mOrange–mTFP1 fusion constructs were further purified by gel filtration chromatography using a HiLoad 16/60 Superdex 75 pg column (GE Healthcare). Proteins were dialysed into 50 mM Tris (pH 7.5).
The oligomeric structure of mTFP variants was determined by gel filtration chromatography on a HiLoad 16/60 Superdex 75 pg gel filtration column. Samples of the dimeric dTomato and the monomeric mCherry proteins [33] were expressed and purified as described above and used as size standards. The AKTAbasic liquid chromatography system (GE Healthcare) can monitor multiple wavelengths simultaneously. Purified TFP variants were mixed with either dTomato or mCherry and the resulting elution profiles were monitored at both 450 and 550 nm.
Spectroscopy
Absorption spectra were recorded on a DU-800 UV–visible spectrophotometer (Beckman). Quantum yields for TFP variants were measured using fluorescein in 10 mM NaOH as the reference standard [37]. Molar absorption coefficients were measured by the alkali denaturation method [33,38]. All emission spectra were acquired on a QuantaMaster spectrofluorimeter (Photon Technology International) and have been corrected for the instrument response. The photostability of mTFP1 under arc lamp illumination was determined using the method of Shaner et al. [16].
Fluorescence lifetimes were determined on a TimeMaster time-resolved spectrofluorimeter (Photon Technology International) that uses the stroboscopic optical boxcar technique [39]. All lifetimes were determined in 10 mM Tris (pH 8.0) with 1 mM EDTA and 50 mM NaCl at protein concentrations of less than or equal to 0.1 μM. A nitrogen dye laser at 440 nm was used for excitation, and the emission monochromator was set to either 480 nm (for mECFP and mCerulean) or 490 nm for mTFP1. A 480/40 nm bandpass filter was used on the emission channel to further minimize scattered light. Slits were adjusted as necessary to obtain adequate fluorescent intensity when using polarizing filters at ‘magic angle’ conditions or lower concentrations of FP.
To determine the pH-dependence of the fluorescence emission of mECFP, mCerulean and mTFP1, each protein (stock solution of 1 mg/ml in 5 mM Tris, pH 7) was diluted 1:100 in a 96-well-plate black clear bottom plate (Corning) containing 0.1 ml of buffer (100 mM) at pH values ranging from 2 to 9. Full emission spectra at each pH were acquired with a Safire2 plate reader (Tecan). The relative fluorescence at each pH was measured at the peak wavelength.
FRET efficiencies for cameleons and mOrange–mTFP1 fusion constructs were determined by measuring the emission spectra before and after trypsinolysis (50 μg/ml trypsin) under conditions where the linker sequence is cleaved and the FPs remain intact. FRET efficiencies (E) for mECFP and mCerulean donors were calculated using the formula E=1−[(fluorescence at 475 nm before trypsinolysis)/(fluorescence at 475 nm after trypsinolysis)]. For mTFP1 donor with either a Citrine or mOrange acceptor, the fluorescence before and after trypsinolysis was measured at 490 nm.
Protein crystallization and structure determination
Crystals of mTFP1 (dimensions of 1.2 mm×0.2 mm×0.2 mm) grew in 1–2 days by the hanging drop vapour diffusion method. Hanging drops consisted of 2 μl of mTFP1 (48 mg/ml) in 50 mM Hepes (pH 7.9) and 2 μl of the well solution [10% (v/v) ethanol, 65 mM zinc acetate and 0.1 M sodium acetate, pH 5.1]. A single crystal of mTFP1 was swept through paratone-N oil and flash-frozen in liquid nitrogen prior to diffraction data collection at 100 K. Data were collected with a Quantum 315 detector at the Advanced Photon Source (Argonne, IL, U.S.A.) beamline 14 BM-C. Diffraction data were merged and indexed using the HKL suite [40] and the structure was solved by molecular replacement using EPMR (evolutionary programming for molecular replacement) [41] and the co-ordinates from the A chain of EosFP (Protein Data Bank accession code 1ZUX [42]) as the search model. The SHELXL program was used for rigid body, positional and B-factor refinement [43]. Manual model building was performed with COOT [44]. Stereochemical quality of the model was checked with PROCHECK [45], and Figures were created with MOLSCRIPT [46] and RASTER3D [47].
Protein expression and imaging in HeLa cells
All DNA for mammalian cell transfection was purified by a Plasmid Midi kit (Qiagen). HeLa cells were cultured in DMEM (Dulbecco's modified Eagle's medium; Invitrogen) supplemented with 10% (v/v) foetal bovine serum (Sigma) at 37 °C. Cells in 35 mm imaging dishes were transfected with 4 μg of plasmid DNA mixed with 10 μg of poly(ethyleneimine) (linear, molecular mass ∼25000; Polysciences) in 0.5 ml of OptiMEM (Invitrogen) and serum was added after 3 h. Approx. 14–24 h later the medium was exchanged for Hanks balanced salt solution containing no calcium chloride, magnesium chloride, magnesium sulfate or Phenol Red (Invitrogen) and the cells were imaged. HeLa cells expressing mTFP1–β-actin or mTFP1–α-tubulin were imaged using an LSM510 confocal microscope (Zeiss) equipped with a 5 mW 458 nm excitation laser. HeLa cells expressing mTFP1–YC3.3 were imaged with a Zeiss Axiovert 200M epifluorescence inverted microscope equipped with a xenon arc lamp and a monochrome Retiga 2000R 12-bit cooled CCD camera (QImaging). The external excitation filter wheel, excitation shutter and emission filter wheel are controlled through a Lambda 10-3 controller (Sutter). Only dichroic mirrors are housed in the motorized reflector turret. The QED InVivo software package (Media Cybernetics) is used for automated computer control of all microscope hardware and for quantitative image analysis.
RESULTS AND DISCUSSION
Design of a synthetic gene library encoding dimeric variants of cFP484
At the time this project was initiated, the only reported homologues of Aequorea GFP with tyrosine-derived chromophores and emission peaks between approx. 480 and 490 nm were cFP484, amFP486 and dsFP483 [2]. These CFPs each have emission peak shapes and fluorescence brightness comparable with Aequorea GFP but are tetramers and therefore unsuitable for use as non-perturbing genetic fusion partners. A sequence alignment of these three proteins (Supplementary Figure 1 at http://www.BiochemJ.org/bj/400/bj4000531add.htm) revealed that of the 227 structurally aligned residues, there are 78 residues that are conserved in all three proteins and an additional 91 residues that are conserved in two of the three. Considering only those positions of two-thirds conservation, we determined that cFP484 is the variant at 19%, dsFP483 at 34% and amFP486 at 47% of the positions. Thus, of these three proteins, cFP484 is the least divergent from a hypothetical ‘consensus’ sequence. In addition, of these three wild-type proteins, cFP484 has been reported to have the highest fluorescence brightness and the fewest cysteine residues [2]. In consideration of these factors we chose to use the cFP484 amino acid sequence as the starting point for the directed evolution process.
We rationally designed and synthesized a gene library with a theoretical diversity of approx. 5×105 cFP484 variants (Supplementary Table 1 at http://www.BiochemJ.org/bj/400/bj4000531add.htm). Important features of the library included: mammalian codon usage; deletion of 40 non-homologous residues from the N-terminus; addition of the seven N-terminal and seven C-terminal residues of Aequorea GFP; semi-degenerate codons encoding potential ‘tetramer-breaking’ mutations at three external positions; and semi-degenerate codons encoding potential ‘rescuing’ mutations at eleven internal positions. Sequencing of random clones revealed that 12% of the genes had mutations only at desired positions and thus the true library diversity, including unintentional errors introduced during synthesis, was approx. 4×106 variants.
Directed evolution of an mTFP (monomeric TFP)
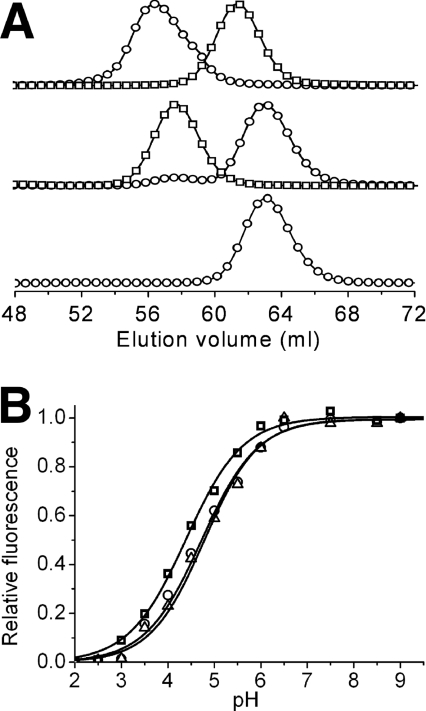
The synthetic gene library was used to transform E. coli and colonies were screened for fluorescence. Fluorescent colonies represented approx. 0.5% of all colonies and were approximately equally divided between colonies that fluoresced at approx. 490 nm (cyan) and colonies that fluoresced at approx. 510 nm (green). We attribute the cyan/green dichotomy in our initial library to the designed arginine/lysine degeneracy at position 70. Of all sequenced variants, the R70K mutation was consistently found in those that fluoresced green but never in those that fluoresced cyan. As will be discussed below, Arg70 is involved in interactions that have since been proposed to be critical for blue-shifting the fluorescence [48]. The brightest CFP identified after extensive screening of the initial library was a dimer with eight mutations relative to wild-type and an emission peak at 486 nm. We have adopted the name ‘teal fluorescent protein’ (TFP), with a preceding ‘d’ for dimeric or ‘m’ for monomeric, and a succeeding numerical identifier, to identify new variants described herein. Following this convention, the dimeric protein identified in our initial screen is dTFP0.1 (dimeric TFP 0.1) and the 170% brighter version resulting from one round of directed evolution is dTFP0.2 (Figure 1B and Table 1; Supplementary Table 2 at http://www.BiochemJ.org/bj/400/bj4000531add.htm). Replacement of dimer interface residues Ser162 and Ser164 with lysine residues produced a monomeric version (Figure 2A), mTFP0.3, which retained only 15% of the brightness of its dimeric precursor. After multiple successive rounds of screening libraries, generated by random or saturation mutagenesis, for variants with improved brightness and high 480/530 nm emission ratio (Supplementary Table 2), we arrived at mTFP0.7 that has fluorescence brightness equivalent to mCerulean (Table 1). However, when we attempted to image an mTFP0.7–β-actin fusion in live HeLa cells by confocal microscopy, we discovered that the fluorescent signal rapidly bleached upon illumination with the 458 nm laser. Further investigation with purified protein revealed that the apparently bleached protein spontaneously recovered its fluorescence over the course of several minutes. We are currently undertaking a detailed investigation of the intriguing photochromic properties of mTFP0.7.
Table 1. Fluorescent properties of CFPs and TFPs.
| Cameleon FRET efficiency (+/−Ca2+) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein | Absorbance (nm) | Emission (nm) | ϵa (×10−3) (M−1·cm−1) | Φb | Brightnessc | pKa | Photostabilityd | − | + |
| cFP484e | 456 | 484 | 35.3 | 0.48 | 17 | n.d.f | n.d. | n.d. | n.d. |
| dTFP0.1 | 456 | 485 | 42 | 0.63 | 26 | n.d. | <1 | n.d. | n.d. |
| dTFP0.2 | 456 | 486 | 60 | 0.68 | 41 | n.d. | <1 | n.d. | n.d. |
| mTFP0.3 | 458 | 488 | 19 | 0.31 | 6 | n.d. | <1 | n.d. | n.d. |
| mTFP0.7 | 453 | 488 | 60 | 0.50 | 30 | 4.0 | <1 | n.d. | n.d. |
| mTFP1 | 462 | 492 | 64 | 0.85 | 54 | 4.3 | 163g/110h | 39% | 49% |
| mECFP | 433/451i | 475/504i | 33/30i | 0.41 | 13/12i | 4.7 | 64h,j | 26% | 39% |
| mCerulean | 433/451i | 475/503i | 43/37i | 0.64 | 27/24i | 4.7 | 36g,j | 30% | 41% |
| EGFPj | 488 | 507 | 56 | 0.60 | 34 | 6 | 174h,j | n.d. | n.d. |
aMolar absorption coefficient.
bQuantum yield.
cProduct of Φ and ϵ×10−3. Values for common FP variants have been previously tabulated [16].
dTime to bleach from an initial emission rate of 1000 photons/s to 500 photons/s.
eValues from [2].
fn.d., not determined.
gMeasured with 10% ND filters.
hMeasured with no ND filters.
iValues for both ‘humps’ of mECFP and mCerulean are provided.
jValues from [16].
Figure 2. Characterization of oligomeric structure and pH-sensitivity.
(A) Gel filtration chromatography elution profile of dTFP0.2 and mTFP1. Detection is at either 450 nm (○) or 550 nm (□). The upper profile is a co-injection of dTFP0.2 and monomeric mCherry [33], the middle profile is dimeric dTomato [33] and mTFP1, and the lower profile is mTFP1 alone. Earlier generations of mTFP had elution times identical with mTFP1 (results not shown). The small 450 nm peak at the dimer elution volume in the middle profile is due to the weak absorbance of dTomato at this wavelength. (B) pH-dependence of the fluorescence emission of mCerulean (○), mTFP1 (□) and mECFP (△).
Directed evolution of mTFP variants with increased photostability
To screen for photostable variants we constructed an array of six 460 nm LEDs (Lumileds, San Jose, CA, U.S.A.) that provides even illumination of a Petri dish with approx. 55 mW/cm2. This irradiance is 1400 times more intense than the excitation used in previous screening and sufficient to photoconvert mTFP0.7 expressed in a colony of E. coli within 20 s. The fluorescence of libraries of mTFP0.7 variants expressed in bacterial colonies was digitally imaged during exposure to intense illumination and colonies with decreased propensity to photobleach were identified. Following several rounds of selection for variants that were photostable, bright and retained a high 480/530 nm emission ratio, we arrived at mTFP1 that has a total of 31 amino acid replacements relative to the wild-type protein (Supplementary Table 2).
In vitro characterization of purified mTFP1 revealed it to be slightly red-shifted (excitation peak at 462 nm and emission peak at 492 nm) from cFP484 (Table 1) and thus the new colour classification of teal rather than cyan is appropriate. A quantum yield of 0.85 and a molar absorption coefficient of 64000 M−1·cm−1 makes mTFP1 comparable in brightness with the popular YFP variants mCitrine [32] and Venus [49] and one of the brightest monomeric FPs of any colour [16]. The fluorescence of mTFP1 is insensitive to physiologically relevant changes in pH (Figure 2B).
To determine the photostability of mTFP1, photobleaching experiments were done on droplets of purified protein suspended in mineral oil following the protocol of Shaner et al. [16] This method is designed to approximate the conditions of a typical wide-field microscopy experiment while rigorously accounting for differences in the spectral properties of the FP as well as the optical properties of the microscope. Using this method, the time for bleaching from an initial emission rate of 1000 photons/s per molecule down to 500 photons/s per molecule (t1/2) was determined to be 163 s when 10% ND (neutral density) filters were used. With no ND filters the t1/2 was 110 s for mTFP1. For the sake of comparison, the t1/2 for Cerulean with 10% ND filters is 36 s and the t1/2 for EGFP with no ND filters is 174 s [16]. As previously noted [16], Cerulean displays an illumination intensity-dependent fast bleaching component that can decrease the intensity to 60% of its initial value within the first few seconds of imaging under typical conditions ([16] and N. Shaner, personal communication). No fast bleaching component was observed for mTFP1.
The fluorescence lifetime decay of mTFP1 (Supplementary Figure 2 at http://www.BiochemJ.org/bj/400/bj4000531add.htm) is unambiguously best fitted as single lifetime (τ=3.2 ns, χ2=1.0). Attempts to fit the mTFP1 lifetime decay data with a double exponential converged to a solution in which τ1=τ2=3.2 ns; strong evidence for the single fluorescent lifetime. Repeating the experiment under ‘magic angle’ conditions had no effect on the observed lifetimes or statistics. Analogous experiments with mECFP and mCerulean resulted in data that were best fitted as double exponentials [for mECFP, τ1=3.7 ns (80%), τ2=1.7 ns (20%), χ2=1.1; for mCerulean, τ1=3.7 ns (80%), τ2=1.9 ns (20%), χ2=0.99]. As previously reported [14], mCerulean could also be adequately fitted with a single exponential (τ=3.5 ns, χ2=1.1), although the residuals were consistently unsatisfactory.
Structural analysis of mTFP1
mTFP1 crystallized in space group C2 with one molecule per asymmetric unit. The data collection, merging and refinement statistics for mTFP1 are excellent (Supplementary Table 3 at http://www.BiochemJ.org/bj/400/bj4000531add.htm). The final model of mTFP1 [R=0.149 (all data); Rfree=0.207; 1.19 Å resolution; 1 Å=0.1 nm] contains all residues from Gly6d to Asn221 (see Supplementary Figure 1 for numbering scheme), 340 water molecules, one acetate ion, one chloride ion and three zinc ions at crystal contacts. PROCHECK analysis of the structure indicates good stereochemistry with 92.8% of the non-glycine and non-proline residues in the ‘most favoured’ regions, 6.1% in the ‘additionally allowed’ regions, 1.1% in the ‘generously allowed’ regions and no residues in the ‘disallowed’ regions of the Ramachandran plot. Careful inspection of the electron density map for the two residues in the generously allowed regions (Lys102 and Glu115) reveals that the modelled conformation for these residues is indeed the best fit for the experimental electron density.
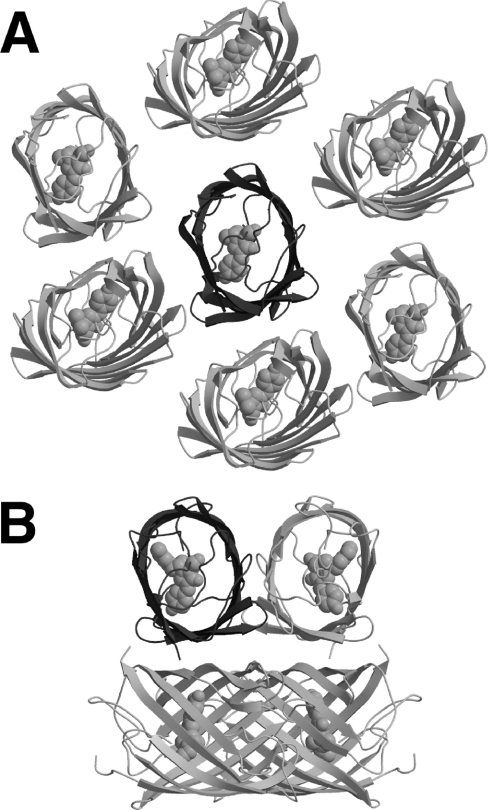
As one would predict on the basis of sequence homology, the fold of mTFP1 is the typical ‘β-can’ motif that is characteristic of Aequorea GFP and it homologues [7]. A comparison with structures in the Protein Data Bank using DALI [50] and DALILITE [51] revealed EosFP (Protein Data Bank code 1ZUX [42]) and dsFP583 (also known as DsRed, Protein Data Bank codes 1G7K [52] and 1GGX [53]) to be the closest structural homologues of mTFP1. EosFP has 66% sequence identity and a 0.6 Å rmsd (root mean square deviation) over 213 aligned C-α atoms, while dsFP583 has 55% sequence identity and a 0.7 Å rmsd over 212 aligned C-α atoms. The structural alignment is only slightly worse for amFP486 (Protein Data Bank code 2A46 [48]), with which mTFP1 has 51% sequence identity and a 0.9 Å rmsd over 213 aligned C-α atoms. The one striking difference between the structure of mTFP1 and those of EosFP, dsFP583 and amFP486 is in the intermolecular interactions observed in the crystal lattice. Shown in Figure 3(A) is the monomer of mTFP1 surrounded by the closest subunits within the crystal lattice. The distance and orientations of these surrounding subunits are quite distinct from those observed for the subunits of the biologically relevant tetramers observed in the crystal structures of other Anthozoa FPs such as amFP486 (Figure 3B). The crystal packing observed for mTFP1 is consistent with the fact that this protein was rationally engineered to be a monomer in solution. It is interesting to note that six (three unique interactions and their symmetrical counterparts) of the most intimate crystal contacts involve surface histidine, glutamic acid and aspartic acid residues bridged by zinc ions. This observation is consistent with the requirement for zinc acetate in the crystallization buffer.
Figure 3. Crystal packing of mTFP1 and amFP486.
(A) Shown in dark grey cartoon representation is the asymmetric unit of mTFP1 (a single polypeptide) crystallized in the monoclinic space group C2. Also shown are the proximal copies of the protein (light grey cartoon) as generated using crystallographic symmetry operations. As expected for a monomeric variant, none of the proximal protein subunits pack in orientations similar to those typically observed in Anthozoa FPs such as the highly homologous amFP486 [48] shown in (B). (B) For amFP486 crystallized in the tetragonal space group I4122 (Protein Data Bank code 2A46), the asymmetric unit contains a single polypeptide (dark grey cartoon), which is shown in the same orientation as mTFP1 in (A) [48]. The additional three subunits (light grey cartoon) of the tightly packed tetrameric biological unit were generated using the crystallographic symmetry operations.
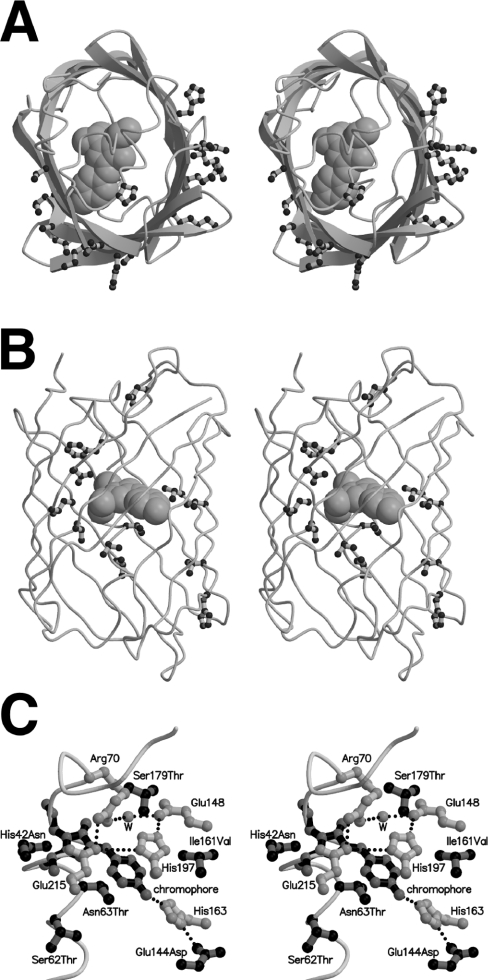
Shown in Figures 4(A) and 4(B) are the locations of all mutations at residues with side chains directed towards either the outside or the inside of the protein structure respectively. Considering only those mutations on the surface of mTFP1 (Figure 4A) and comparing with the subunit orientation in amFP486 (Figure 3B), it is apparent that the mutations cluster in the former protein–protein interfaces. While four of these external mutations were rationally introduced to disrupt the intermolecular interactions at the interface (residues 125, 127, 162 and 164), seven others (residues 123, 141, 158, 182, 186, 216 and 221) were discovered through random mutagenesis and screening. As shown in Figure 4(B), beneficial internal mutations tend to be located in the midsection of mTFP1. With only one exception, individual internal mutations provided only incremental improvements in either the brightness or photostability of the protein, suggesting that the effects were due to slight changes in the side chain packing and internal hydrogen bond network. One particularly dramatic consequence for a single point mutation was the large increase in photostability observed for the N63T mutation that differentiates mTFP0.8 from mTFP0.7. Residue 63 is positioned directly below the chromophore when viewed as in Figure 4(C). Since the mechanism of reversible photobleaching is unknown at this point, it is difficult to speculate on how this replacement promotes the improved photostability. Further investigations of the mechanism of photobleaching are under way.
Figure 4. Location of beneficial mutations in mTFP1.
(A) Stereoview of mTFP1 with the polypeptide backbone in cartoon representation and the chromophore in space filling representation. All residues with fully solvent exposed side chains that are mutated relative to the wild-type cFP484 are shown in ball-and-stick representation. The protein is viewed from the same orientation as the single asymmetric unit shown in Figures 3(A) and 3(B). By comparison with the amFP486 structure shown in Figure 3(B), it is apparent that the external mutations are clustered in the former protein–protein interfaces of the tetrameric wild-type protein. (B) Stereoview of mTFP1 with all residues that are directed towards the interior of the β-barrel and mutated relative to cFP484 shown in ball-and-stick representation. (C) Stereoview of the chromophore region of mTFP1 showing the chromophore (black bonds with grey atoms), all mutations in the immediate vicinity of the chromophore (grey bonds with black atoms), and a selection of residues (i.e. the quadrupole salt-bridge network and His163) that were not mutated but are discussed in the text (white bonds with grey atoms).
The molecular environment of the chromophore is very similar to that observed in amFP486 [48]. In particular, the four residues (Arg70, Glu148, His197 and Glu214) of the critical quadrupole salt-bridge network are conserved both in identity and in conformation (Figure 4C). This quadrupole salt-bridge network has been proposed to be responsible for maintaining a positive charge on His197 that may in turn limit charge transfer in the excited state and thereby give rise to the blue-shift [48]. One major difference in the chromophore environment is the presence of His163 (aligns with an alanine residue in amFP486) that hydrogen-bonds to the phenolate oxygen of the chromophore through the imidazole side chain (Figure 4C). Interestingly, one of the surface mutations identified during selection for photostability, E144D, is hydrogen-bonded to the imidazole side chain of His163 (Figure 4C). A glutamic acid residue at this position (as in the wild-type protein) would probably be unable to adopt a conformation that would allow formation of a similar hydrogen bond. In amFP486, a water molecule substitutes for the imidazole side chain of His163 and makes a similar hydrogen bond to the chromophore.
A surprising aspect of the mTFP1 structure is that the chromophore is significantly non-planar with a CA-CB dihedral ‘tilt’ of 12.7° and a CB-CG dihedral ‘twist’ of −14.3°. Such distortions from planarity have previously been observed for non-fluorescent ‘dark’ states of certain photoswitchable FPs [54]. However, for a protein as brightly fluorescent as mTFP1 the observation of such a distortion is unexpected and apparently inconsistent. Possible explanations for this distorted structure are that it is either an artefact of crystallization or a population averaged mixture of an extremely twisted non-fluorescent chromophore and a planar fluorescent chromophore. Arguing against the first possibility is the fact that the chromophore is completely buried within the β-can structure and crystal contacts with nearby surface residues do not appear to be causing major distortions of the protein main-chain atoms. Arguing against the second possibility is the fact that there is no significant residual electron density in the immediate vicinity of the chromophore and thus no evidence of alternate conformations. Further high-resolution structural studies will be required to address this interesting observation.
mTFP1 as a FRET donor to mCitrine and mOrange
To assess mTFP1's suitability as a FRET donor to mCitrine, analogous versions of YC3.3 [13] containing mECFP, mCerulean or mTFP1 were constructed and their FRET efficiency in the absence and presence of Ca2+ was determined (Table 1 and Figure 5A). All three versions of YC3.3 exhibited similar increases in FRET efficiency upon Ca2+ binding, but the mTFP1–YC3.3 version was notable for its significantly higher efficiencies in both the Ca2+-bound and Ca2+-free states. The trend in experimental FRET efficiencies, mECFP<mCerulean<mTFP1, is consistent with our calculated Ro values of 5.0 nm for mECFP, 5.3 nm for mCerulean and 5.7 nm for mTFP1. It is important to note that the original YC3.3 contained ECFP and had been optimized for maximum FRET difference between the Ca2+-bound and Ca2+-free states. It is therefore unsurprising that switching to an mTFP1 donor (with a different dipole orientation and increased Ro value) results in a slightly decreased dynamic range. This is almost certainly because the protein was empirically optimized [13,55] for interfluorophore distance changes centred on the Ro of ECFP (where there is the strongest dependence of FRET efficiency on distance). If mTFP1–YC3.3 was to be subject to similar systematic optimization, we are confident that the dynamic range could be significantly improved.
Figure 5. mTFP1 as a FRET donor to a YFP or mOrange acceptor.
(A) Shown are the in vitro emission spectra of mTFP1–YC3.3 [13] with no Ca2+ (○), 10 mM Ca2+ (□) and no FRET (△). To obtain the ‘no FRET’ spectrum, the linker between the two FPs was digested with trypsin under conditions where the FPs remain intact. Shown in grey lines is a plot of percentage transmittance versus wavelength for the custom filter set used for imaging of mTFP1–YFP FRET to produce the results shown in Figure 7. Filters are designated with Chroma Technology part numbers. (B) Shown are the in vitro emission spectra for an mTFP1–mOrange fusion protein before (○), and after (△), proteolysis of the linker peptide.
The calculated Ro value (5.7 nm) for mTFP1–mOrange FRET suggested to us that this new monomeric FP pair could compare favourably with the previously reported dimeric MiCy–mKO FRET pair [21]. To experimentally validate mTFP1 as a FRET donor to mOrange, mTFP1 and mOrange were genetically fused with an eight-amino-acid linker. The emission spectra for the purified fusion protein before and after trypsinolysis of the linker are shown in Figure 5(B). The experimental FRET efficiency was calculated to be 53.6%. Relative to EGFP, mTFP1 is a superior FRET donor to mOrange because it is brighter (Table 1) and can be excited at or near its blue-shifted absorbance peak of 462 nm without significant cross-excitation of mOrange. It remains to be seen how mTFP1 and mMiCy will compare in this regard [30].
Live cell imaging of mTFP1 fusions and FRET reporters
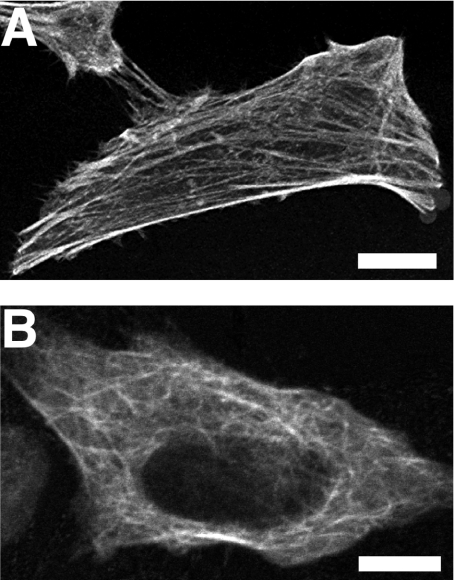
To determine if mTFP1 was suitable for use as non-perturbing fusion partner, we expressed mTFP1–β-actin (Figure 6A) and mTFP1–α-tubulin (Figure 6B) fusions in mammalian cells. Scanning confocal microscopy with a 458 nm laser revealed that both fusion proteins localized correctly. In stark contrast with our results with mTFP0.7–β-actin, there was no significant decrease in fluorescence intensity upon repeated image acquisitions.
Figure 6. Localization of mTFP1 fusion proteins in live HeLa cells.
Confocal fluorescence images of HeLa cells expressing either mTFP1–β-actin (A) or mTFP1–α-tubulin (B). Scale bar, 10 μm in both images.
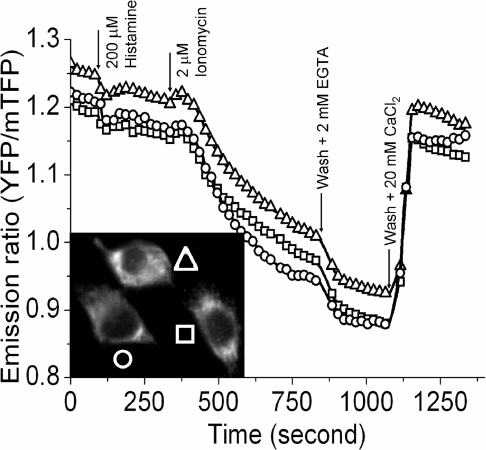
Typical filter sets used to image Cerulean by epi-fluorescence microscopy are adequate but suboptimal for mTFP1. To take advantage of mTFP1's improved brightness, we recommend a 445/30 nm excitation filter, a 470 nm beamsplitter and a 495/30 nm emission filter (Figure 6A and Supplementary Table 3). This combination gives a 2.6-fold increase in fluorescent signal relative to mCerulean imaged using a standard CFP set (e.g. a 436/20 nm excitation filter, a 455 nm beamsplitter and a 480/40 nm excitation filter). We have used our recommended set in combination with a new YFP emission filter (545/30 nm), to demonstrate that mTFP1–YC3.3 can be practically employed in live cell FRET imaging. The gene encoding mTFP1–YC3.3 was cloned into a mammalian expression vector with an N-terminal signal sequence and a C-terminal ER retention peptide [13]. FRET imaging of transiently transfected HeLa cells on a Zeiss Axiovert 200M equipped with a digital CCD camera revealed a typical pattern of ER localization and robust ratiometric responses to induced changes in the free Ca2+ concentration (Figure 7). This result demonstrates that the spectral distinction between mTFP1 and mCitrine is sufficient for these proteins to be used as an advantageous new FRET pair that can be imaged on a standard epi-fluorescence microscope equipped with appropriate bandpass filter sets.
Figure 7. FRET imaging of mTFP1 with a YFP acceptor.
Shown is a plot of the emission ratio versus time for three HeLa cells (see donor fluorescence image in the inset) expressing mTFP1–YC3.3 targeted to the ER. Images in both the donor and acceptor emission channels were acquired every 5 s with identical exposure times (300 ms) and 10% ND filters. Histamine (200 μM) or ionomycin (2 μM) was added at the indicated time points.
Summary
Using a combination of rational protein engineering and directed evolution we have created a new monomeric FP, dubbed mTFP1, that harbours an anionic phenolate-type chromophore chemically identical with that of EGFP. As such, the blue-shifted mTFP1 is the beneficiary of the narrow peak shape, high brightness and high photostability properties that are empirically associated with the EGFP chromophore [16]. A recurring theme in the history of FP engineering is that, given the plethora of applications for which FPs are suitable research tools, no new variant makes all others redundant [5,16,33]. With this in mind, we readily acknowledge that the properties of mTFP1 make it a superior fluorophore for some, but not all, applications. Applications for which mTFP1 offers a clear advantage over existing alternatives include intra- and inter-molecular FRET with a yellow or orange acceptor FP and dual colour imaging in combination with an orange or red FP. For dual colour imaging in combination with a YFP, mTFP1 could provide a brighter fluorescent signal than existing Aequorea-derived CFPs but the bleedthrough into the acceptor emission channel may be only slightly better (or possibly worse) depending on the specific filter combination in place.
In conclusion, relative to monomeric alternatives of similar colour, mTFP1 has a number of favourable attributes that will allow practical FRET imaging at significantly lower intracellular protein concentrations, higher light intensities, and for greater duration than is currently possible. These advantages make mTFP1 an excellent complement to the existing selection of monomeric FPs recommended for use in fluorescence imaging [16].
Online data
Acknowledgments
This research was made possible with financial support from the University of Alberta, CFI (Canada Foundation for Innovation), NSERC (Natural Sciences and Engineering Research Council), and an Alberta Ingenuity New Faculty Award. We are grateful to Linda Reha-Krantz, Dina Tleugabulova, Xue-jun Sun, Ed Kiegle, Glen Loppnow, Sandra Marcus and our Departmental electronics shop for technical assistance and helpful discussion. In addition, we thank the University of Alberta MBSU for analysis of DNA sequencing reactions, David Piston for the gift of mCerulean cDNA and Don Schwab of the NRC Plant Biotechnology Group for gene synthesis. Nathan Shaner and Paul Steinbach at the University of California, San Diego, performed the photobleaching experiments. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR07707. R. E. C. holds a Canada Research Chair in Bioanalytical Chemistry. New FPs that originate from the Campbell laboratory and are described in this paper are covered by a U.S. patent application owned by the University of Alberta.
References
- 1.Tsien R. Y. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Matz M. V., Fradkov A. F., Labas Y. A., Savitsky A. P., Zaraisky A. G., Markelov M. L., Lukyanov S. A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 3.Verkhusha V. V., Lukyanov K. A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J., Campbell R. E., Ting A. Y., Tsien R. Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 5.Chudakov D. M., Lukyanov S., Lukyanov K. A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005;23:605–613. doi: 10.1016/j.tibtech.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Yang F., Moss L. G., Phillips G. N., Jr The molecular structure of green fluorescent protein. Nat. Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 7.Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 8.Cody C. W., Prasher D. C., Westler W. M., Prendergast F. G., Ward W. W. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 9.Chattoraj M., King B. A., Bublitz G. U., Boxer S. G. Ultra-fast excited state dynamics in green fluorescent protein: multiple states and proton transfer. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8362–8367. doi: 10.1073/pnas.93.16.8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormack B. P., Valdivia R. H., Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 11.Heim R., Prasher D. C., Tsien R. Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heim R., Tsien R. Y. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 13.Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., Ikura M., Tsien R. Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 14.Rizzo M. A., Springer G. H., Granada B., Piston D. W. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 2004;22:445–449. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen A. W., Daugherty P. S. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat. Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 16.Shaner N. C., Steinbach P. A., Tsien R. Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 17.Carter R. W., Schmale M. C., Gibbs P. D. Cloning of anthozoan fluorescent protein genes. Comp. Biochem. Physiol. C. 2004;138:259–270. doi: 10.1016/j.cca.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y., Castner E. W., Jr, Lawson C. L., Falkowski P. G. Biophysical characterization of natural and mutant fluorescent proteins cloned from zooxanthellate corals. FEBS Lett. 2004;570:175–183. doi: 10.1016/j.febslet.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 19.Kelmanson I. V., Matz M. V. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: Faviida) Mol. Biol. Evol. 2003;20:1125–1133. doi: 10.1093/molbev/msg130. [DOI] [PubMed] [Google Scholar]
- 20.Shagin D. A., Barsova E. V., Yanushevich Y. G., Fradkov A. F., Lukyanov K. A., Labas Y. A., Semenova T. N., Ugalde J. A., Meyers A., Nunez J. M., et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol. Biol. Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 21.Karasawa S., Araki T., Nagai T., Mizuno H., Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem. J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lauf U., Lopez P., Falk M. M. Expression of fluorescently tagged connexins: a novel approach to rescue function of oligomeric DsRed-tagged proteins. FEBS Lett. 2001;498:11–15. doi: 10.1016/s0014-5793(01)02462-0. [DOI] [PubMed] [Google Scholar]
- 23.Gavin P., Devenish R. J., Prescott M. An approach for reducing unwanted oligomerisation of DsRed fusion proteins. Biochem. Biophys. Res. Commun. 2002;298:707–713. doi: 10.1016/s0006-291x(02)02552-4. [DOI] [PubMed] [Google Scholar]
- 24.Soling A., Simm A., Rainov N. Intracellular localization of Herpes simplex virus type 1 thymidine kinase fused to different fluorescent proteins depends on choice of fluorescent tag. FEBS Lett. 2002;527:153–158. doi: 10.1016/s0014-5793(02)03201-5. [DOI] [PubMed] [Google Scholar]
- 25.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karasawa S., Araki T., Yamamoto-Hino M., Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J. Biol. Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 27.Wiedenmann J., Ivanchenko S., Oswald F., Schmitt F., Rocker C., Salih A., Spindler K. D., Nienhaus G. U. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurskaya N. G., Verkhusha V. V., Shcheglov A. S., Staroverov D. B., Chepurnykh T. V., Fradkov A. F., Lukyanov S., Lukyanov K. A. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 29.Ando R., Mizuno H., Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 30.Kogure T., Karasawa S., Araki T., Saito K., Kinjo M., Miyawaki A. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat. Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 31.Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 32.Griesbeck O., Baird G. S., Campbell R. E., Zacharias D. A., Tsien R. Y. Reducing the environmental sensitivity of yellow fluorescent protein: mechanism and applications. J. Biol. Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 33.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 34.Rizzo M. A., Piston D. W. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys. J. 2005;88:L14–L16. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fromant M., Blanquet S., Plateau P. Direct random mutagenesis of gene-sized DNA fragments using polymerase chain reaction. Anal. Biochem. 1995;224:347–353. doi: 10.1006/abio.1995.1050. [DOI] [PubMed] [Google Scholar]
- 36.Cheng Z., Campbell R. E. Assessing the structural stability of designed β-hairpin peptides in the cytoplasm of live cells. ChemBioChem. 2006;7:1147–1150. doi: 10.1002/cbic.200500540. [DOI] [PubMed] [Google Scholar]
- 37.Brannon J. H., Magde D. Absolute quantum yield determination by thermal blooming-fluorescein. J. Phys. Chem. 1978;82:705–709. [Google Scholar]
- 38.Ward W. W. Biochemical and physical properties of GFP. In: Chalfie M. K. S., editor. Green Fluorescent Protein: Properties, Applications and Protocols. New York: Wiley; 1998. pp. 45–75. [Google Scholar]
- 39.James D. R., Siemiarczuk A., Ware W. R. Stroboscopic optical boxcar technique for the determination of fluorescence lifetimes. Rev. Sci. Instrum. 1992;63:1710–1716. [Google Scholar]
- 40.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Kissinger C. R., Gehlhaar D. K., Fogel D. B. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 42.Nienhaus K., Nienhaus G. U., Wiedenmann J., Nar H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9156–9159. doi: 10.1073/pnas.0501874102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheldrick G. M., Schneider T. R. SHELXL: high-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 46.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 47.Merritt E. A., Bacon D. J. Raster3D: photorealistic molecular graphics. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 48.Henderson J. N., Remington S. J. Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12712–12717. doi: 10.1073/pnas.0502250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 50.Holm L., Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 51.Holm L., Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 52.Yarbrough D., Wachter R. M., Kallio K., Matz M. V., Remington S. J. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution. Proc. Natl. Acad. Sci. U.S.A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wall M. A., Socolich M., Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat. Struct. Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 54.Henderson J. N., Remington S. J. The kindling fluorescent protein: a transient photoswitchable marker. Physiology. 2006;21:162–170. doi: 10.1152/physiol.00056.2005. [DOI] [PubMed] [Google Scholar]
- 55.Miyawaki A., Tsien R. Y. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.