Abstract
All identified membrane fusion proteins are transmembrane proteins. In the present study, we explored the post-mitotic reassembly of the NE (nuclear envelope). The proteins that drive membrane rearrangements in NE assembly remain unknown. To determine whether transmembrane proteins are prerequisite components of this fusion machinery, we have focused on nuclear reconstitution in a cell-free system. Mixing of soluble interphase cytosolic extract and MV (membrane vesicles) from amphibian eggs with chromatin results in the formation of functional nuclei. We replaced MV and cytosol with protein-free phosphatidylcholine LS (liposomes) that were pre-incubated with interphase cytosol. While later stages of NE assembly yielding functional nucleus did not proceed without integral proteins of MV, LS-associated cytosolic proteins were sufficient to reconstitute membrane targeting to the chromatin and GTP-dependent lipid mixing. Binding involved LS-associated A-type lamin, and fusion involved Ran GTPase. Thus in contrast with post-fusion stages, fusion initiation in NE assembly, like membrane remodelling in budding and fission, does not require transmembrane proteins.
Keywords: liposome, membrane–chromatin binding, membrane fusion, nuclear envelope assembly, Ran GTPase
Abbreviations: Cyt-LS, liposomes with bound cytosolic proteins; DOPC, dioleoylphosphatidylcholine; FRAP, fluorescence recovery after photobleaching; FRET, fluorescence resonance energy transfer; GTP[S], guanosine-5′-[γ-thio]triphosphate; LPC, lysophosphatidylcholine; LS, liposomes; MV, membrane vesicle; MWB, membrane wash buffer; NBD, 7-nitrobenz-2-oxa-1,3-diazole; NE, nuclear envelope; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PLC, phospholipase C; SNARE, soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor; TMD, transmembrane domain
INTRODUCTION
The NE (nuclear envelope) of metazoa consists of an outer membrane which is continuous with the endoplasmic reticulum and an inner membrane which is supported by the lamina, a network of intermediate filaments [1]. Reformation of the NE around the segregated chromosomes at the end of mitosis involves many steps, all of which have been studied in the amphibian cell-free system [2,3]. The MV (membrane vesicles) bind to decondensed chromatin and undergo a GTP-dependent fusion [4]. This fusion reaction and the later steps of NE formation, including insertion of nuclear pore complexes and further chromatin swelling, involve Ran GTPase [5]. The later steps of NE formation involve insertion of nuclear pore complexes and further chromatin swelling.
Protein fusogens that initiate membrane fusion at the early stage of tightly controlled NE assembly are yet to be identified [1,6,7]. Proteins that merge membranes during enveloped virus entry [8–10], SNARE (soluble N-ethylmaleimide-sensitive fusion protein attachment protein receptor)-dependent intracellular fusion [11–13], and proteins that are thought to drive cell fusion in development [14], are anchored in the membranes by one or more TMDs (transmembrane domains). Both for viral and for SNARE-dependent fusion, the length and the sequence of the TMD of the fusion proteins are of importance for their fusogenic activity [15,16]. Moreover, diverse fusion reactions appear to involve interactions between the TMDs of the proteins involved and between TMD and other domains of the same fusion protein [17–19]. As all known protein fusogens are transmembrane proteins, we asked the question ‘are transmembrane proteins required for fusion at the early stages of NE assembly’?
EXPERIMENTAL
Lipids, antibodies and recombinant proteins
All lipids were purchased from Avanti Polar Lipids. The monoclonal anti-histone PAN antibody (MAB3422) and the monoclonal anti-lamin A/C antibody (MAB3211) were obtained from Chemicon. The polyclonal Ran antibody (ab4781) was purchased from Abcam. Anti-Gp210 and anti-ribophorin were a gift from D. Forbes (Division of Biological Sciences, University of California at San Diego, CA, U.S.A.). Recombinant Cdk1/cyclin B was obtained from Upstate.
In vitro nuclear assembly
Xenopus MV, soluble cytosolic extract and sperm chromatin were prepared as in [3]. Nuclei were assembled as previously described [20] and were detected as round structures approximately 20 μm in diameter with a smooth rim, readily distinguishable from the rough surface of chromatin with bound but unfused vesicles. The assembled nuclei excluded 70 kDa dextran and actively imported a substrate containing a nuclear localization signal.
Liposome preparation
Large unilamellar vesicles were formed by extrusion through 100 nm filters. Lipid compositions were 100% DOPC [dioleoylPC (phosphatidylcholine)], 97 mol% DOPC and 3 mol% rhodamine-DOPE (PE is phosphatidylethanolamine) or 98.5 mol% DOPC with 0.6 mol% rhodamine-DOPE and 0.85 mol% NBD-DOPE (7-nitrobenz-2-oxa-1,3-diazole-dioleoylPE). In some experiments, we also formed LS (liposomes) from a 1-palmitoyl, 2-oleoyl-PC/1,2-dioleoyl phosphatidylserine/NBD-PE/rhodamine-PE mixture in an 82:15:1.5:1.5 molar ratio.
Preparation of Cyt-LS (liposomes with bound cytosolic proteins)
LS (10 μl of a 1 mg/ml suspension) were incubated with 40 μl of cytosol (20–30 mg/ml of total protein) for 1 h at 4 °C. To isolate LS with bound cytosolic proteins from the remaining unbound cytosolic proteins, we mixed the sample with 250 μl of 75% sucrose in MWB [membrane wash buffer; 250 mM sucrose, 50 mM KCl, 2.5 mM MgCl2, 50 mM Hepes (pH 8.0), 1 mM dithiothreitol, 0.5 mM ATP, aprotonin at 1μg/ml and leupeptin at 1μg/ml] and overlaid it with 150 μl of 40% sucrose in MWB and 300 μl of 25% sucrose in MWB. After an 18 h centrifugation at 41000 rev./min (SW55Ti, Beckman) at 4 °C, Cyt-LS were collected as the 150 μl fraction at the top of the gradient. We measured the distribution of lipids and proteins in five 150 μl fractions from the top to the bottom of the gradient using rhodamine fluorescence and the Bradford assay respectively. Generally, Cyt-LS, prepared as described above, bind to chromatin and fuse on its surface in a manner that mimics MV binding and fusion. We did not perform extensive optimization of the Cyt-LS preparation and functional activity of Cyt-LS varied between preparations.
Chromatin-binding assay
We incubated 10 μl of LS for 1 h at room temperature (22–23 °C) with 1 μl of decondensed sperm chromatin that was prepared in a 30 min incubation with 10 μl of heat-inactivated cytosol. Chromatin associated with LS was pelleted with protein G–agarose beads coated with anti-histone PAN antibodies. As an alternative method, biotinylated chromatin was also pulled down with streptavidin-coated magnetic beads. After two washes with MWB, we measured fluorescence of chromatin-associated LS on a spectrofluorimeter.
Fusion assays
Cyt-LS fusion was observed under a fluorescent microscope using the FRET (fluorescence resonance energy transfer) assay. In this assay, 10 μl of Cyt-LS labelled with rhodamine- and NBD-tagged PE and 10 μl of unlabelled Cyt-LS were mixed in the presence of decondensed chromatin. The samples were incubated for 1 h at room temperature before analysis under the microscope. FRET, detected as rhodamine emission at approx. 585 nm resulting from NBD excitation at approx. 470 nm, decreases when the average spatial separation of the probes increases upon fusion of labelled and unlabelled membranes. We also monitored Cyt-LS fusion, using the lipid-dequenching assay. In this assay, 10 μl of Cyt-LS labelled with rhodamine-tagged PE and 10 μl of unlabelled Cyt-LS were preincubated with decondensed chromatin (1 μl) and 1 mM GTP for 15 min at 4 °C prior to being resuspended in MWB prewarmed to room temperature. The increase in the fluorescent signal at λemission=590 nm (λexcitation=550 nm) resulting from lipid mixing was continuously recorded with a spectrofluorimeter. In some experiments, we added 1 mM GTP[S] (guanosine-5′-[γ-thio]triphosphate) to the preincubation mix. The level of lipid mixing at the end of the recording (t=30 min) was taken as the final extent.
FRAP (fluorescence recovery after photobleaching) experiments
An LSM510 Zeiss confocal microscope with a 100× objective was used at 22 °C for FRAP experiments. A circular area of the membrane (2–6 μm in diameter) of the reformed nucleus was bleached and monitored for fluorescence recovery for 5–20 s. Images were analysed with ImageJ software. For the time course of fluorescence recovery, the fluorescence of the bleached area immediately before and after bleaching was considered to be 1 and 0 respectively. Data were fitted by the following equation:
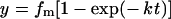 |
where fm represents the mobile fraction of the fluorophores in the bleached region and k is proportional to the diffusion coefficient. The diffusion coefficient was estimated with the equation D=0.32kr2, where r is the radius of the photobleached area.
Biochemistry methods
Depletions of Ran and type-A lamin were performed with the corresponding antibodies immobilized on protein G–agarose beads. After several washes with PBS, the antibody–bead complexes were incubated with 50 μl of extract overnight at 4 °C. After depletion, extracts were incubated with LS for binding and fusion analysis. Mock-treated extracts were incubated with protein G–agarose beads alone.
RESULTS
Transmembrane proteins are not required for targeting membranes to the chromatin surface
To explore the involvement of cytosolic proteins in MV binding to the surface of the chromatin and in fusion itself, we replaced MV with LS that were formed from DOPC with 3 mol% of rhodamine-tagged PE as a fluorescent probe. Like MV, LS bound to condensed chromatin in the absence of cytosol (Figures 1A and 1B). In contrast with MV, LS bound to decondensed chromatin only after a 30 min incubation in cytosol (Figures 1C–1F). This finding suggested that protein-free LS acquire the ability to bind to decondensed chromatin from the cytosol.
Figure 1. Cytosol-dependent binding of LS (fluorescence of the lipid probe in B, D and F) at the surface of sperm chromatin (fluorescence of the DNA probe Hoechst 33342 in A, C and E).
After binding of LS to condensed chromatin (A and B), cytosol was applied, and binding was monitored 5 min (C and D) and 30 min (E and F) later. Scale bar=10 μm.
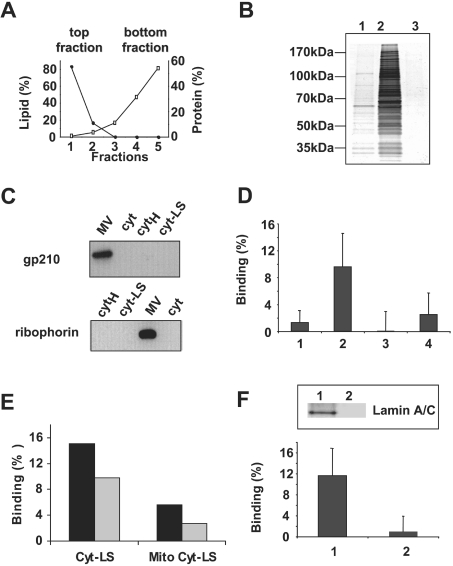
To simplify the experimental system, we removed cytosolic factors that would not associate with our LS. We incubated LS with cytosol and then isolated LS with bound cytosolic proteins from the remaining unbound cytosolic proteins by flotation through a sucrose density gradient. Like protein-free LS, Cyt-LS were recovered at the top of the gradient, as evidenced by the distribution of lipid fluorescence (Figure 2A). While most of the proteins were recovered at the bottom of the gradient (Figures 2A and 2B, lane 2), silver staining allowed us to detect a small amount of protein at the top of the gradient that was recruited by LS (Figure 2B, lane 1). No proteins were observed at the top of the gradient in the experiments in which we applied cytosol that was not pre-incubated with LS (Figure 2B, lane 3). The cytosol used in the present study was separated from most of the membranes by ultracentrifugation. The lack of any remaining MV in our Cyt-LS preparation was further verified by the finding that the Cyt-LS fraction contained neither gp210, an integral protein component of nuclear pores, nor ribophorin, an integral protein of the rough endoplasmic reticulum membrane (Figure 2C), in amounts detectable by Western blotting.
Figure 2. LS-associated cytosolic proteins mediate binding between LS and chromatin.
(A) Distribution of proteins (□) and LS lipids (●) along the centrifugation gradient after LS incubation with cytosol. (B) The amount of protein and number of protein bands detected by silver staining at the top of the gradient (Cyt-LS, lane 1) were much lower than those for the bottom fraction of the gradient (lane 2). No protein was detected in the top fraction in the absence of LS (lane 3). (C) Neither Cyt-LS nor cytosol contains proteins associated with MV. Antibodies against gp210 and ribophorin each recognized a single protein band (at approx. 210 and 72 kDa respectively), in MV but not in cytosol, heat-inactivated cytosol (CytH) or in Cyt-LS. (D) Binding to decondensed chromatin of the mixture of protein-free LS and cytosol separately floated through sucrose gradient (1, n=10), of Cyt-LS (2, n=10), of Cyt-LS that were prepared with heat-inactivated cytosol (3, n=3) and of trypsin-treated Cyt-LS (4, n=6) is quantified as the percentage of LS fluorescence associated with the chromatin pellet. Binding observed for protein-free LS in the absence of cytosol is subtracted in each experiment. Means±S.D. are shown. Values of bars 1, 3 and 4 are statistically different from bar 2 as verified by paired two-tailed t test. (E) Cyt-LS binding to decondensed chromatin was assayed for LS prepared with either interphase or mitotic cytosol. Results of two independent experiments are shown as black and grey bars. (F) Cyt-LS binding was measured in the presence (1) or in the absence (2) of type-A lamin. Means±S.D. are shown (n=3). The degree of depletion was analysed by Western blotting on LS pretreated with mock-depleted cytosol (inset, lane 1) and LS pretreated with type-A lamin-depleted cytosol (inset, lane 2).
To quantify binding between LS and chromatin, we measured the fluorescence of the LS co-precipitated with chromatin. Cyt-LS bound to chromatin that was decondensed by the addition of heat-inactivated cytosol (Figure 2D, bar 2). Binding was strongly inhibited for trypsin-treated Cyt-LS (Figure 2D, bar 4); for LS that were pretreated with heat-inactivated cytosol (Figure 2D, bar 3); and when top fractions of separately floated protein-free LS and cytosol were applied to decondensed chromatin (Figure 2D, bar 1). These findings indicate that cytosolic proteins on LS mediated the interactions between Cyt-LS and chromatin. Moreover, Cyt-LS binding to chromatin was inhibited when interphase cytosol used to prepare Cyt-LS was pretreated with a recombinant form of stabilized cyclin B to initiate mitotic events (Figure 2E). To find out whether Cyt-LS bound to the decondensed chromatin by biologically relevant mechanisms, we explored the involvement of lamins, proteins that are implicated in MV targeting to chromatin in vitro [21]. We identified the lamin A/C among the proteins associated with Cyt-LS (Figure 2F, inset lane 1). Moreover, chromatin binding was inhibited for the Cyt-LS prepared with laminimmunodepleted cytosol (Figure 2F, inset lane 2). Thus lamins appear to be involved in Cyt-LS binding to decondensed chromatin.
In brief, binding between Cyt-LS and decondensed chromatin involves LS-associated proteins present in interphase cytosol, including lamins. This indicates that transmembrane proteins are not required for targeting membranes to the chromatin surface.
Liposome-associated cytosolic proteins mediate GTP- and chromatin-dependent lipid mixing between LS
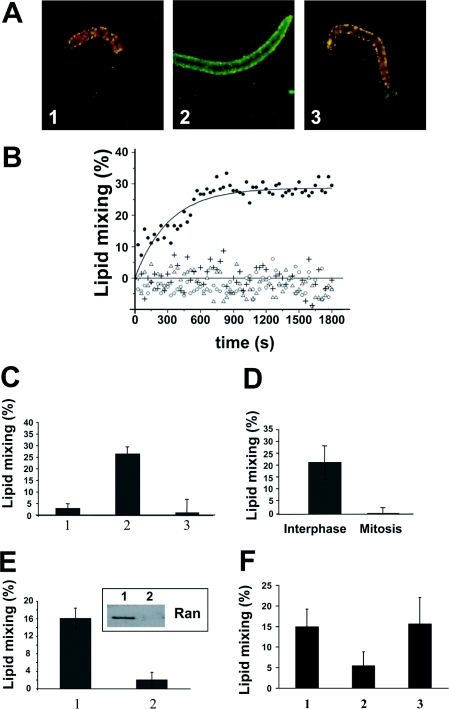
The binding of Cyt-LS to decondensed chromatin allowed us to study the fusion stage of NE assembly. Decondensed chromatin incubated with unlabelled Cyt-LS and Cyt-LS labelled with rhodamine- and NBD-tagged lipids acquired yellow colouration due to an efficient FRET between the fluorescent probes in the labelled Cyt-LS (Figure 3A, panel 1). GTP application changed the colour of the chromatin structures from yellow to green (Figure 3A, panel 2). This change reflected the loss of FRET upon lipid mixing between labelled and unlabelled Cyt-LS and the resulting dilution of the fluorescent probes. Note that lipid mixing between two labelled Cyt-LS does not dilute fluorescent probes. Thus decondensed chromatin incubated with labelled Cyt-LS in the absence of unlabelled Cyt-LS did not develop green colouration after application of 1 mM GTP (Figure 3A, panel 3). No yellow-to-green change in the colouration of the chromatin caused by GTP-dependent fusion between labelled and unlabelled Cyt-LS was observed in the presence of 1 mM GTP-γS (results not shown).
Figure 3. Chromatin-, GTP- and cell cycle-dependent lipid mixing between Cyt-LS.
(A) Lipid mixing between NBD and rhodamine-labelled and unlabelled Cyt-LS at the surface of the decondensed chromatin was observed by fluorescence microscopy as a decrease in FRET in the presence of GTP (2) but not in its absence (1). Each image is a superposition of images excited with a 470-nm laser and detected with NBD (green) and rhodamine (red) filters. Changes in the chromatin colouration from yellow to green reflects the loss of FRET. No change in FRET was observed in the control experiment, in which all Cyt-LS were labelled (3). (B) Lipid mixing between Cyt-LS was assayed by spectrofluorimetry as rhodamine dequenching in the presence of GTP (○), decondensed chromatin (+), and both decondensed chromatin and GTP (●). △ shows a control experiment with both decondensed chromatin and GTP, in which Cyt-LS were replaced with protein free-LS. (C) Final extents of lipid mixing between Cyt-LS in the presence of GTP and decondensed chromatin were measured with (3) and without (2) GTP[S]. (1) Final extent of lipid mixing between protein-free LS in the presence of GTP and decondensed chromatin. Means±S.D. are shown (n=3). (D) Final extents of lipid mixing in the presence of GTP and decondensed chromatin for Cyt-LS prepared either with interphase or mitotic cytosol. Means±S.D. are shown (n=3). (E) Final extents of lipid mixing in the presence of GTP and decondensed chromatin for Cyt-LS prepared either with mock-treated cytosol (1) or with cytosol immunodepleted from Ran GTPase (2). The presence of Ran on the Cyt-LS was verified by Western blotting of the Cyt-LS prepared either with mock-depleted cytosol or with Ran-depleted cytosol (inset, lanes 1 and 2 respectively). Means±S.D. are shown (n=3). (F) Final extents of lipid mixing in the presence of GTP and decondensed chromatin between Cyt-LS were assayed in the absence (1) or presence (2) of 10 nM lauroyl LPC. Dilution of the sample in LPC-free MWB reversed LPC inhibition (3). Means±S.D. are shown (n=5). The value of bar 2 is statistically different from the values of both bar 1 and 3 as verified by a paired two-tailed t test.
GTP- and chromatin-dependent fusion between Cyt-LS was also observed with a 96-well plate assay and Cyt-LS labelled with a self-quenching concentration of rhodamine-tagged PE. In this case, the dilution of the fluorescent lipid upon lipid mixing between labelled and unlabelled Cyt-LS was detected as an increase in fluorescence in the presence of both decondensed chromatin and 2 mM GTP (Figure 3B). No lipid mixing was observed when decondensed chromatin was replaced with condensed chromatin (Figure 3B) or in the presence of 1 mM GTP[S] (Figure 3C; compare 2 with 3). Similarly, lipid mixing was not observed when the Cyt-LS pretreated with interphase cytosol were replaced with Cyt-LS pretreated with heat-inactivated cytosol or with interphase cytosol converted to a mitotic state by addition of cyclin B (Figure 3D). This result is consistent with an earlier study in which membranes do not support NE assembly in mitotic extract [22].
The dependence of lipid mixing on GTP and its inhibition by GTP[S] pointed to a functional importance of GTP hydrolysis in this reaction. We tested whether Cyt-LS fusion involved the small GTPase Ran, which plays an important role in NE assembly [5,23]. Indeed, Ran was found on Cyt-LS, as indicated on the immunoblot by the 28 kDa band (Figure 3E, inset lane 1). To explore the functional role of Ran, we immunodepleted the protein from the cytosol and verified that Cyt-LS pretreated with Ran-depleted cytosol no longer carried the Ran protein (Figure 3E, inset lane 2). Lipid mixing was strongly inhibited in the absence of Ran (Figure 3E, bars 1 and 2), confirming the direct involvement of Ran or a Ran-binding protein in Cyt-LS fusion on decondensed chromatin.
In brief, lipid mixing between Cyt-LS bound to decondensed chromatin reconstitutes many properties of the biologically relevant fusion of MV in NE assembly, including dependencies on the cell cycle, the presence of decondensed chromatin and Ran-dependent GTP-hydrolysis. However, since cytosol-dependent leakage of the aqueous dye encapsulated into liposomes hindered the application of the content mixing assays (results not shown), it remains possible that cytosolic proteins associated with Cyt-LS merged only contacting monolayers of the fusing membranes, and thus that they hemifuse rather than fuse the membranes. In viral and SNARE-dependent fusion, effective transition from hemifusion to complete fusion depends on the transmembrane domain of the fusion protein [16,24,25], and the number of available fusion proteins [16,26], and might require additional proteins [27]. Alternatively, lipid mixing in our system represents complete but leaky fusion.
The LS used in these experiments were formed from a lipid (PC) that, in contrast with fusogenic lipids such as PE, favours a bilayer structure and does not promote non-specific fusion [28]. Another fusogenic lipid, diacylglycerol, a product of PLC (phospholipase C) activity, has been implicated in NE assembly in sea urchin egg preparation [29]. However, since lipid mixing between Cyt-LS proceeds only at the surface of decondensed chromatin, it is unlikely that this lipid mixing is based on the PLC-generated change in the lipid composition of the LS. In addition, there are no known eukaryotic PLC that hydrolyse PC [29]. Lipid mixing between Cyt-LS did not appear to be limited to any particular lipid composition and was also observed with very similar efficiency (results not shown) for LS from the PC and phosphatidylserine mixture often used in experiments on SNARE-mediated proteoliposome fusion [30]. While some compositions supported lipid mixing between Cyt-LS, LPC (lysoPC) inhibited this fusion reaction (Figure 3F, 1 and 2). LPC is known to inhibit diverse biological fusion reactions, including viral and intracellular fusion, by inhibiting bending of the monolayers of the fusing membranes into hemifusion intermediates [31]. As for viral fusion and exocytosis, LPC inhibition of lipid mixing between Cyt-LS was reversible: LPC removal by dilution of the samples in LPC-free buffer restored the fusion competency (Figure 3F, bar 3).
Assembly of functional nuclei by fusion between MV and liposomes
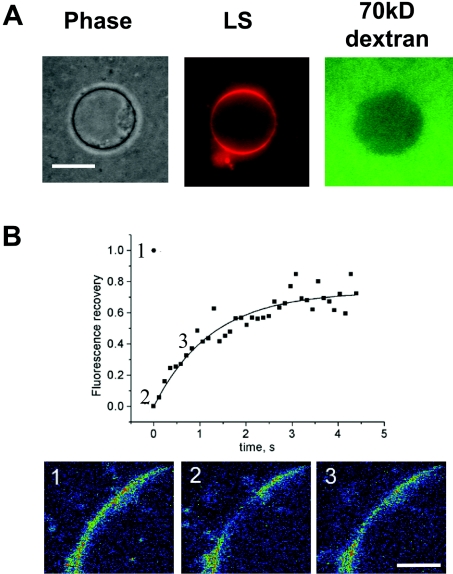
The early stages of nucleus assembly, including docking and fusion of MV at the chromatin surface, are followed by the assembly of nuclear pore complexes, additional fusion events, and further chromatin swelling. While neither Cyt-LS nor LS in the presence of cytosol formed nuclei on their own (results not shown), they were able to join MV in the assembly of a functional nucleus. We first incubated rhodamine-tagged LS and cytosol with chromatin and then added MV. This yielded fluorescent nuclei morphologically indistinguishable from those observed in the absence of LS (Figure 4A). To find out whether the development of this NE from LS and MV involves fusion of the LS at the surface of the chromatin, we examined the lateral mobility of the lipid molecules at the fluorescent rim of these nuclei (Figure 4B, panel 1). Very rapid and complete recovery of FRAP of an approx. 2–6 μm-diameter patch at the surface of the chromatin (Figure 4B, panels 2 and 3) indicated that lipids freely diffuse over distances much exceeding approx. 100 nm diameter of our LS [32]. Thus lipids were distributed in the unified bilayers at the chromatin surface rather than limited in their diffusion, as expected for the bound LS or, in the case of LS hemifusion, in which only the outer monolayers of the adjacent LS would merge and the inner monolayers would remain distinct.
Figure 4. LS fuse with MV during NE formation.
(A) Nuclear reconstitution reactions were performed in the presence of rhodamine-tagged LS. Formation of fluorescently labelled nuclear membranes was assessed at 2 h. The integrity of the NE was verified by exclusion of fluorescein isothiocyanate-70-kDa dextran. Scale bar=10 μm. (B) FRAP experiments indicate that a patch of a nuclear membrane approx. 6 μm in diameter recovers within several seconds (D=2.3×10−8 cm2/s). Images at the bottom of the graph were taken before (1), immediately after (2), and 830 ms after (3) the bleach. The solid line shows a single exponential fit of the experimental data. Scale bar=5 μm.
DISCUSSION
In the present study we reconstituted early stages of NE assembly in an experimental system based on protein-free LS. Several in vitro studies have implicated transmembrane proteins in MV binding to chromatin [33–35] and naked DNA [36]. However, our findings that cytosolic proteins mediate LS binding to the decondensed chromatin and that lamin-immunodepletion of the cytosol blocks this binding emphasize the role of soluble proteins. These soluble proteins could contribute to the initial binding of membranes to chromatin and be the basis for more specific interactions between transmembrane proteins and chromatin [36]. Soluble lamins might interact with membranes either directly, possibly via a 15-carbon isoprene lipid [37], or indirectly, via membrane-associated proteins. The reports on the role of lamins in NE assembly and, in particular, on the effects of their biochemical depletion have been somewhat contradictory [38,39]. This inconsistency between different studies suggests that MV might carry some amount of membrane-associated lamins [40]. Ran, a protein that like lamins was found on the Cyt-LS, has been also implicated in the binding of membranes to the chromatin surface [41]. However, since the LS that were pretreated with mitotic cytosol still carried Ran (results not shown) but did not bind to the decondensed chromatin, it appears that Ran on its own is insufficient for binding.
The mechanisms by which Ran GTPase controls fusion between MV and fusion between Cyt-LS at the surface of the decondensed chromatin remain to be understood. One may hypothesize that GTP hydrolysis by Ran, directly or indirectly bound to Cyt-LS, activates membrane-associated cytosolic proteins that mediate fusion. To date the only identified target of Ran in NE assembly is the importin β1 protein which, when added in excess, blocks NE membrane fusion. The blockage is released by addition of RanQ69L, a non-hydrolysable form of Ran [20]. Our finding that early stages of NE assembly can be reconstituted in the absence of transmembrane proteins agrees with recent reports that demonstrate that vertebrate transmembrane nucleoporins (gp210, POM121 and Ndc1) act at later stages of NE formation [7,42]. Our results are also consistent with a recent demonstration that soluble Nup155 protein and/or proteins associated with it play an important role in MV fusion in NE assembly [43]. While chromatin-targeting and early fusion stages proceeded without transmembrane proteins, LS did not form a full-size nucleus. The involvement of transmembrane proteins at the later stages of NE assembly might reflect the existence of a checkpoint that coordinates fusion steps with the formation of nuclear pores and their dilation [42,44].
In conclusion, in the present study we reconstituted early stages of post-mitotic reassembly of NE with cytosolic proteins and liposomal membranes lacking transmembrane proteins. Our results show the importance of cytosolic regulation of membrane targeting to the chromatin. Like MV, which lose their chromatin-targeting activity when membrane-associated soluble proteins are removed in a high salt buffer [45], liposomes in their binding to chromatin require the association of soluble proteins. Our results suggest that, in contrast with transmembrane proteins involved in viral and intracellular SNARE-dependent fusion [9–12], proteins that mediate membrane fusion in NE assembly lack transmembrane domains. While unexpected, the finding that membrane anchoring via transmembrane domains is not a prerequisite for fusion proteins is in line with the known ability of peripheral membrane proteins to drive membrane remodelling in budding and fission [46]. As MV fusion in ER assembly [47] and in early stages of NE assembly [4], initiation of Cyt-LS fusion required only proteins that are tightly associated with membranes. The simplicity of the developed experimental system and the limited number of proteins which are tightly associated with Cyt-LS and which thus serve as components of the minimal binding and fusion machinery provide a promising approach to the identification of the key fusion proteins that control post-mitotic recompartmentalization of divided cells.
Acknowledgments
We thank Evgenia Leikina (Laboratory of Cellular and Molecular Biophysics, National Institute of Child Health and Human Development, NIH, Bethesda, MD, U.S.A.), Benjamin Podbilewicz (Department of Biology Technion, Israel Institute of Technology, Haifa, Israel), Katherine L. Wilson (The Department of Cell Biology, Johns Hopkins University School of Medicine, Baltimore, MD, U.S.A.), Elena Zaitseva (Laboratory of Cellular and Molecular Biophysics, National Institute of Child Health and Human Development, NIH, Bethesda, MD, U.S.A.) and Joshua Zimmerberg (Laboratory of Cellular and Molecular Biophysics, National Institute of Child Health and Human Development, NIH, Bethesda, MD, U.S.A.) for valuable discussions. We are also grateful to Tom Sargent (Laboratory of Molecular Genetics, National Institute of Child Health and Human Development, NIH, Bethesda, MD, U.S.A.) and Elena Zaitseva for very helpful experimental advice. Special thanks are due to Douglass Forbes (Division of Biological Sciences, University of California at San Diego, CA, U.S.A.) and Valerie Delmar (Division of Biological Sciences, University of California at San Diego, CA, U.S.A.) for providing reagents and critical comments on the manuscript. This research was supported by the Intramural Research Program of the NIH (National Institutes of Health), the NICHD (National Institute of Child Health and Human Development).
References
- 1.Hetzer M., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- 2.Lohka M. J., Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 3.Newport J., Dunphy W. Characterization of the membrane binding and fusion events during nuclear envelope assembly using purified components. J. Cell Biol. 1992;116:295–306. doi: 10.1083/jcb.116.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman A. L., Delannoy M. R., Wilson K. L. GTP hydrolysis is required for vesicle fusion during nuclear envelope assembly in vitro. J. Cell Biol. 1992;116:281–294. doi: 10.1083/jcb.116.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hetzer M., Bilbao-Cortes D., Walther T. C., Gruss O. J., Mattaj I. W. GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell. 2000;5:1013–1024. doi: 10.1016/s1097-2765(00)80266-x. [DOI] [PubMed] [Google Scholar]
- 6.Harel A., Forbes D. J. Importin β: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 7.Mansfeld J., Guttinger S., Hawryluk-Gara L. A., Pante N., Mall M., Galy V., Haselmann U., Muhlhausser P., Wozniak R. W., Mattaj I. W., et al. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 2006;22:93–103. doi: 10.1016/j.molcel.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Blumenthal R., Clague M. J., Durell S. R., Epand R. M. Membrane fusion. Chem. Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 9.Earp L. J., Delos S. E., Park H. E., White J. M. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 2005;285:25–66. doi: 10.1007/3-540-26764-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kielian M., Rey F. A. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson M. B., Chapman E. R. Fusion pores and fusion machines in Ca(2+)-triggered exocytosis. Annu. Rev. Biophys. Biomol. Struct. 2006;35:135–160. doi: 10.1146/annurev.biophys.35.040405.101958. [DOI] [PubMed] [Google Scholar]
- 12.Jahn R., Lang T., Sudhof T. C. Membrane fusion. Cell. 2003;112:519–533.. doi: 10.1016/s0092-8674(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 13.Mayer A. Membrane fusion in eukaryotic cells. Annu. Rev. Cell Dev. Biol. 2002;18:289–314. doi: 10.1146/annurev.cellbio.18.032202.114809. [DOI] [PubMed] [Google Scholar]
- 14.Mohler W. A., Shemer G., del Campo J. J., Valansi C., Opoku-Serebuoh E., Scranton V., Assaf N., White J. G., Podbilewicz B. The type I membrane protein EFF-1 is essential for developmental cell fusion. Dev. Cell. 2002;2:355–362. doi: 10.1016/s1534-5807(02)00129-6. [DOI] [PubMed] [Google Scholar]
- 15.Armstrong R. T., Kushnir A. S., White J. M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 2000;151:425–438. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Y., Zhang F., Su Z., McNew J. A., Shin Y. K. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 17.Melikyan G. B., Markosyan R. M., Roth M. G., Cohen F. S. A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell. 2000;11:3765–3775. doi: 10.1091/mbc.11.11.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamm L. K. Hypothesis: spring-loaded boomerang mechanism of influenza hemagglutinin-mediated membrane fusion. Biochim. Biophys. Acta. 2003;1614:14–23. doi: 10.1016/s0005-2736(03)00159-7. [DOI] [PubMed] [Google Scholar]
- 19.Han X., Wang C. T., Bai J., Chapman E. R., Jackson M. B. Transmembrane segments of syntaxin line the fusion pore of Ca2+-triggered exocytosis. Science. 2004;304:289–292. doi: 10.1126/science.1095801. [DOI] [PubMed] [Google Scholar]
- 20.Harel A., Chan R. C., Lachish-Zalait A., Zimmerman E., Elbaum M., Forbes D. J. Importin β negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 2003;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass J. R., Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J. Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- 23.Hughes M., Zhang C., Avis J. M., Hutchison C. J., Clarke P. R. The role of the ran GTPase in nuclear assembly and DNA replication: characterisation of the effects of Ran mutants. J. Cell Sci. 1998;111:3017–3026. doi: 10.1242/jcs.111.20.3017. [DOI] [PubMed] [Google Scholar]
- 24.Kemble G. W., Danieli T., White J. M. Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 25.Giraudo C. G., Hu C., You D., Slovic A. M., Mosharov E. V., Sulzer D., Melia T. J., Rothman J. E. SNAREs can promote complete fusion and hemifusion as alternative outcomes. J. Cell Biol. 2005;170:249–260. doi: 10.1083/jcb.200501093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chernomordik L. V., Frolov V. A., Leikina E., Bronk P., Zimmerberg J. The pathway of membrane fusion catalyzed by influenza hemagglutinin: restriction of lipids, hemifusion, and lipidic fusion pore formation. J. Cell Biol. 1998;140:1369–1382. doi: 10.1083/jcb.140.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reese C., Heise F., Mayer A. Trans-SNARE pairing can precede a hemifusion intermediate in intracellular membrane fusion. Nature. 2005;436:410–414. doi: 10.1038/nature03722. [DOI] [PubMed] [Google Scholar]
- 28.Brugger B., Nickel W., Weber T., Parlati F., McNew J. A., Rothman J. E., Sollner T. Putative fusogenic activity of NSF is restricted to a lipid mixture whose coalescence is also triggered by other factors. EMBO J. 2000;19:1272–1278. doi: 10.1093/emboj/19.6.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barona T., Byrne R. D., Pettitt T. R., Wakelam M. J., Larijani B., Poccia D. Diacylclycerol induces fusion of nuclear envelope membrane precursor vesicles. J. Biol. Chem. 2005;280:41171–41177. doi: 10.1074/jbc.M412863200. [DOI] [PubMed] [Google Scholar]
- 30.Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Sollner T. H., Rothman J. E. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 31.Chernomordik L. V., Kozlov M. M. Membrane hemifusion: crossing a chasm in two leaps. Cell. 2005;123:375–382. doi: 10.1016/j.cell.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 32.Hinterdorfer P., Baber G., Tamm L. K. Reconstitution of membrane fusion sites. A total internal reflection fluorescence microscopy study of influenza hemagglutinin-mediated membrane fusion. J. Biol. Chem. 1994;269:20360–20368. [PubMed] [Google Scholar]
- 33.Pyrpasopoulou A., Meier J., Maison C., Simos G., Georgatos S. D. The lamin B receptor (LBR) provides essential chromatin docking sites at the nuclear envelope. EMBO J. 1996;15:7108–7119. [PMC free article] [PubMed] [Google Scholar]
- 34.Segura-Totten M., Kowalski A. K., Craigie R., Wilson K. L. Barrier-to-autointegration factor: major roles in chromatin decondensation and nuclear assembly. J. Cell Biol. 2002;158:475–485. doi: 10.1083/jcb.200202019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gant T. M., Harris C. A., Wilson K. L. Roles of LAP2 proteins in nuclear assembly and DNA replication: truncated LAP2β proteins alter lamina assembly, envelope formation, nuclear size, and DNA replication efficiency in Xenopus laevis extracts. J. Cell Biol. 1999;144:1083–1096. doi: 10.1083/jcb.144.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulbert S., Platani M., Boue S., Mattaj I. W. Direct membrane protein–DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 2006;173:469–476. doi: 10.1083/jcb.200512078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber K., Plessmann U., Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- 38.Gant T. M., Wilson K. L. Nuclear assembly. Annu. Rev. Cell Dev. Biol. 1997;13:669–695. doi: 10.1146/annurev.cellbio.13.1.669. [DOI] [PubMed] [Google Scholar]
- 39.Lopez-Soler R. I., Moir R. D., Spann T. P., Stick R., Goldman R. D. A role for nuclear lamins in nuclear envelope assembly. J. Cell Biol. 2001;154:61–70. doi: 10.1083/jcb.200101025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lourim D., Krohne G. Membrane-associated lamins in Xenopus egg extracts: identification of two vesicle populations. J. Cell Biol. 1993;123:501–512. doi: 10.1083/jcb.123.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilbao-Cortes D., Hetzer M., Langst G., Becker P. B., Mattaj I. W. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 2002;12:1151–1156. doi: 10.1016/s0960-9822(02)00927-2. [DOI] [PubMed] [Google Scholar]
- 42.Antonin W., Franz C., Haselmann U., Antony C., Mattaj I. W. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 43.Franz C., Askjaer P., Antonin W., Iglesias C. L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I. W. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 2005;24:3519–3531. doi: 10.1038/sj.emboj.7600825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen M., Feinstein N., Wilson K. L., Gruenbaum Y. Nuclear pore protein gp210 is essential for viability in HeLa cells and Caenorhabditis elegans. Mol. Biol. Cell. 2003;14:4230–4237. doi: 10.1091/mbc.E03-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vigers G. P., Lohka M. J. A distinct vesicle population targets membranes and pore complexes to the nuclear envelope in Xenopus eggs. J. Cell Biol. 1991;112:545–556. doi: 10.1083/jcb.112.4.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corda D., Hidalgo Carcedo C., Bonazzi M., Luini A., Spano S. Molecular aspects of membrane fission in the secretory pathway. Cell. Mol. Life Sci. 2002;59:1819–1832.. doi: 10.1007/PL00012508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]