Abstract
The critical role of IL-5 (interleukin-5) in eosinophilic inflammation implicates it as a therapeutic target for allergic diseases. The aim of the present study was to elucidate the molecular basis for the involvement of reversible histone acetylation in IL-5 transcriptional regulation. We provide evidence that HDAC4 (histone deacetylase 4) and p300, a known HAT (histone acetyltransferase), reversibly controlled the activity of the IL-5 promoter in vivo and in vitro, with a concurrent alteration of histone H3 acetylation status at the promoter regions. The nucleo-cytoplasmic shuttling of HDAC4 was shown to play an important role in the suppressive function of HDAC4 in IL-5 gene expression. Point mutation and reporter ChIP (chromatin immunoprecipitation) studies determined that the four transcription factors binding on the IL-5 promoter, i.e. C/EBPβ (CAAT/enhancer-binding protein β), GATA3 (GATA binding protein 3), NFAT (nuclear factor of activated T cells) and YY1 (Yin and Yang 1), were essential for the recruitment of HDAC4. Consistent with these observations, HDAC4 was found to form protein complexes with GATA3 and YY1, and to co-exist in the nuclei with GATA3. We propose that the unique regulatory mechanism of IL-5 gene transcription involves the reversible histone modification catalysed by HDAC4 and p300, which are recruited by the transcription factors. The dynamic balance in IL-5 transcriptional regulation is achieved through interactions among HATs/HDACs, histones and transcription factors. These data contribute to understanding the molecular mechanisms of IL-5 regulation, which is crucial to the development of new therapeutic strategies for IL-5-related allergic diseases.
Keywords: chromatin remodelling, histone acetyltransferase, histone deacetylase, histone modification, interleukin-5
Abbreviations: CBP, CREB (cAMP-response-element-binding protein)-binding protein; CaMK, calcium/calmodulin-dependent protein kinase; C/EBPβ, CCAAT/enhancer-binding protein β; ChIP, chromatin immunoprecipitation; CoIP, co-immunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole; GATA3, GATA binding protein 3; GFP, green fluorescent protein; GM-CSF, granulocyte macrophage colony-stimulating factor; HAT, histone acetyltransferase; HDAC, histone deacetylase; IL, interleukin; HDACi, HDAC inhibitor; HEK, human embryonic kidney; NaBu, sodium butyrate; NFAT, nuclear factor of activated T cells; RE, regulatory element; RT, reverse transcriptase; TRITC, tetramethylrhodamine β-isothiocyanate; TSA, trichostatin A; Th2, T helper 2; YY1, Yin and Yang 1
INTRODUCTION
IL-5 (interleukin-5), an important cytokine primarily synthesized by activated Th2 (T helper 2) cells [1], is involved uniquely in control of the growth, differentiation and activation of eosinophils [2], which are the major determinants of the tissue damage observed in allergic disorders [3]. Apparently, a full elucidation of the regulatory mechanisms of IL-5 synthesis may be helpful for seeking effective strategies for the management of allergic diseases.
The expression of the IL-5 gene, in common with many other cytokine genes, has been demonstrated to be regulated primarily at the transcriptional level [4]. Differentiation of naive CD4+ T cells into Th2 cells is accompanied by chromatin remodelling including hyperacetylation of histones in the nucleosomes associated with the Th2 cytokine gene loci, such as IL-5, IL-4, IL-13 and GM-CSF (granulocyte macrophage colony-stimulating factor) genes [5], in which histone hyperacetylation of the IL-5 gene displays significantly delayed kinetics, suggesting a distinct remodelling mechanism for the IL-5 gene locus. Three regulatory elements (REI, II and III) of the hIL-5 promoter have been identified [6], among which the REI (−80 bp to −45bp) and REII (−123 bp to −92 bp) regions are proven as the activating elements [7], whereas the REIII region (−170 bp to −130 bp) appears to function as a negative regulatory element and is less well described. Besides these, Schwenger et al. [8] identified another negative regulatory element (−459 bp to −447 bp) at the distal region of the IL-5 promoter, suggesting a complicated regulatory mechanism for IL-5 gene transcription. A variety of transcription factors, for instance, C/EBPβ (CCAAT/enhancer-binding protein β), GATA3 (GATA binding protein 3), NFAT (nuclear factor of activated T cells), YY1 (Yin and Yang 1), AP1 (activating protein 1) and Ets1 (E twenty-six 1) have been reported to bind to the 5′-flanking region of the human IL-5 gene and play distinguishing roles on IL-5 gene expression [9–15]. Although the functions of the individual factors were described in some detail, the precise molecular basis for the co-ordination between multiple factors in determination of IL-5 gene expression has not been well illustrated.
The histone acetylation/deacetylation modification has emerged as a major form of epigenetic mechanism that regulates the expression of genes in eukaryotic cells [16]. Acetylation of nucleosomal histones in vivo is a dynamic, reversible process governed by the opposing activities of HATs (histone acetyltransferases) and HDACs (histone deacetylases) [17]. Active gene transcription is tightly associated with hyperacetylation of histones, whereas hypoacetylation is correlated with reduced transcription or gene silencing [18]. Besides histones, HATs and HDACs may also act on non-histone proteins such as transcription factors. The recruitment of HATs or HDACs by specific transcription factors enables these chromatin-modifying enzymes to regulate specific sets of target genes. HDACs that have been described in mammals [17] are grouped into three classes according to their structures, expression patterns and catalytic mechanisms [19]. Class I HDACs are expressed primarily in the nucleus and are involved in gene silencing of both specific genes and entire chromosomal domains [17]. In contrast, Class II HDACs possess the capability of active nucleo-cytoplasmic shuttling, which is suggested to be regulated by CaMK (Ca2+/calmodulin-dependent protein kinase)-dependent phosphorylation [20]. In fact, Class II HDACs display unique responses to the various signalling molecules and cell-type specific regulation of different sets of target genes. The activities of HDACs of both classes can be specifically inhibited by HDACis (HDAC inhibitors). Class III HDACs are proteins relating to the yeast NAD+-dependent deacetylase Sir2 and their regulatory mechanisms are specific and to date are not well understood.
Although alterations in chromatin structure and histone acetylation are implicated as playing important roles in gene regulation of the Th2 gene loci [21], their impact on the IL-5 promoter are still disputable. It has recently been reported that TSA (trichostatin A), one of the HDACs, induced a T cell-suppressive effect and decreased expression of IL-5 [22]. In contrast, work from another group suggested that glucocorticoid-induced repression of IL-5 expression could be relieved by treatment of transfected cells with TSA, and this repression could be augmented by HDAC1 co-transfection [23]. A variety of transcription factors binding to the IL-5 promoter have been shown to be capable of recruiting p300/CBP [CREB (cAMP-response-element-binding protein)-binding protein], one of the known HATs [24–26]. Moreover, YY1 was confirmed to exert repressive roles through recruiting and interacting with Class I HDACs [27], suggesting that the transcription of the IL-5 gene may be tightly related to histone acetylation. Despite these studies to date, a number of important issues regarding roles of acetylation modification on IL-5 gene regulation still remain to be elucidated.
In the present study, we demonstrate that the reversible acetylation catalysed by p300 and HDAC4 affected the IL-5 promoter activity as well as the IL-5 mRNA level in vitro and in vivo, which was accompanied by alterations in acetylation status of histones at the IL-5 promoter. The nucleo-cytoplasmic shuttling of HDAC4 may play an important role. Moreover, we show that transcription factors C/EBPβ, NFAT, GATA3 and YY1 were involved in the recruitment of HDAC4 at the IL-5 promoter. We conclude that the coordinated action of p300 and HDAC4 play significant roles in maintenance of the dynamic balance of IL-5 production in vivo.
EXPERIMENTAL
Plasmids and site-directed mutagenesis
Plasmids expressing human HDAC1–6 (fused to the FLAG-epitope) were gifts from Dr W. C. Greene (Gladstone Institute of Virology and Immunology, San Francisco, CA, U.S.A.) and Dr E. Seto (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, U.S.A.). The p300 expression plasmid was provided by Dr J. Boys (The Institute of Cancer Research, London, U.K.). The constructs expressing C/EBPβ, NFAT (the permanently active mutant, NFATp[S>A]), GATA3–GFP and YY1 were kindly supplied by Dr S. Akira (Research Institute of Microbial Diseases, Osaka University, Japan), Dr A. Goldfeld (Department of Pathology, Harvard Medical School, Boston, MA, U.S.A.), Dr J. Zhu (National Institute of Allergy and Infections Diseases, National Institutes of Health, U.S.A.), Dr K. Murphy (Department of Pathology and Center for Immunology, Washington University School of Medicine, U.S.A.) and Dr E. Bonneloy (Régulation de la Transcription et Maladies Génétiques, CNRS UPR 2228, Université René Descartes, France) respectively. pCaMKIV and its inactive mutant pCaMKIV-ΔdCT were provided by Dr J. Xie (Department of Physiology, Faculty of Medicine, University of Manitoba, Canada). The construct pGL3-IL-5-luc containing the human IL-5 promoter (−529 bp to+16 bp) was amplified as previously described [28]. Constructs of the mutated IL-5 promoter for binding sites of C/EBPβ, NFAT, GATA3 and YY1 were generated by an overlap extension PCR procedure [29]. The mutated sequences were designed as follows: C/EBPβ (−235 bp): AAT to GAC; NFAT (−110 bp): GAA to TCG; GATA3 (−70 bp): CTATCT to GTCGAC; GATA3 (−152 bp): GATA to CTTA; GATA3 (−400 bp): GATTAATC to CTTTAAAG; YY1 (−90 bp and −450 bp): CCAT to TCGC. All of the constructs were sequenced to confirm the correct mutations before use.
Cell culture, HDACi treatment, transfection and luciferase assays
Jurkat cells and HEK (human embryonic kidney)-293T cells were maintained in RPMI-1640 and IMDM (Iscove's modified Dulbecco's medium; Gibco) respectively, containing 10% FBS (foetal bovine serum), penicillin (100 units/ml) and streptomycin (100 μg/ml). For the treatments with HDACi, cells were treated with various concentrations of TSA (Sigma) or NaBu (sodium butyrate; Sigma) for 24 h. PMA (20 ng/ml; Sigma) and ionomycin (1 mM; Sigma) were added 6 h before harvesting for T cell-activation (for Jurkat cells). Transient transfections of HEK-293T cells and Jurkat cells were performed using the conventional calcium phosphate-DNA precipitation method and electroporation respectively. Transfected cells were analysed for luciferase activity using a Promega dual-luciferase reporter assay system. The Renilla luciferase control plasmid pREP7-RLuc was co-transfected in each experiment for normalization. RLA (relative luciferase activity) was calculated with the reporter activity of pGL3-IL-5-luc plasmid alone as 1 unless noted. All the results represent the means±S.D. based on at least three independent experiments.
Total RNA isolation and reverse transcription
Total cellular RNA was extracted using a total RNA isolation system (Promega) according to the manufacturer's instructions. RNA (1 μg) per sample was reverse transcribed to cDNA in a total volume of 20 μl using a RT (reverse transcriptase) reaction kit (Promega).
Quantitative real-time PCR
Real-time PCR was performed using an ABI Prism® 7700 sequence detection system (PE Applied Biosystems) according to the manufacturer's instruction and SYBR® Premix Ex Taq (Takara) as a DNA-specific fluorescent dye. Expression of β-actin was used as an internal control with the primers 5′-TCGTGCGTGACATTAAGGAG-3′ (sense) and 5′-ATGCCAGGGTACATGGTGGT-3′ (antisense). PCR was carried out for 45 cycles of 95 °C for 15 s and 60 °C for 60 s. Primer sequences for detection of IL-5 mRNA expression were synthesized as 5′-CTGCCTACGTGTATGCCATCC-3′ (sense) and 5′-CATTGGCTATCAGCAGAGTTCG-3′ (antisense). All the reactions were repeated at least three times. Data were analysed using the software provided by ABI Company (Applied Biosystems) and calculated by the 2−ΔΔCt method.
Antibodies, immunofluorescence and flow cytometric analysis
Antibodies used in the present study were anti-IL-5 (Chemicon), anti-acetyl histone H3 and H4 (Upstate), anti-FLAG (Sigma), anti-GFP (green fluorescent protein; Chemicon), anti-YY1 (Santa Cruz Biotechnology) and anti-actin (Santa Cruz Biotechnology). For immunofluorescence, HEK-293T cells were grown on coverslips and transfected with FLAG–HDAC4, GFP–GATA3, pCaMKIV or pCaMKIV-ΔdCT expression plasmids. The harvested cells were fixed, permeabilized and subsequently incubated with rabbit anti-FLAG antibody. After staining with a TRITC (tetramethylrhodamine β-isothiocyanate)-conjugated goat anti-rabbit secondary antibody (Zhongshan) and DAPI (4′,6-diamidino-2-phenylindole) DNA dye, cells were washed and imaged under a fluorescence microscope (Nikon).
Protein isolation, Western blotting and CoIP (co-immunoprecipitation) assay
Western blotting was performed using HEK-293T cells transfected with the FLAG–HDAC4 plasmid. Cells (2×107) were scraped from the coverslips and after washing were lysed with lysis buffer [10 mM Tris/HCl (pH 8.0), 150 mM NaCl, 0.5% Nonidet P-40 and protease inhibitor cocktail] for 10 min at 4 °C. Total cell extracts were obtained using sonication and centrifugation. Following SDS/PAGE (15% gels) and transfer to a nitrocellulose membrane, the samples were incubated with anti-FLAG or anti-actin antibodies. The expression of actin was used as an internal control. Interactions between HDAC4 and transcription factors were confirmed by CoIP assay. Total cell extracts from FLAG–HDAC4 and GFP–GATA3 or YY1 co-transfected cells were pre-cleared with salmon sperm DNA/protein A–agarose beads (50% bead slurry; Upstate). Anti-GFP or anti-YY1 antibodies were added for immunoprecipitation. Immunoprecipitates were then subjected to SDS/PAGE followed by transfer on to a nitrocellulose membrane. After incubation with an anti-FLAG antibody, followed by a HRP (horseradish peroxidase)-conjugated goat anti-rabbit secondary antibody (1:100000; Sigma) samples were detected using the AEC detection method.
ChIP (chromatin immunoprecipitation) assay
Transfected HEK-293T cells were cross-linked with 1% formaldehyde (final concentration) after washing. Cells were lysed with lysis buffer [50 mM Tris/HCl (pH 8.1), 10 mM EDTA, 1% SDS and protease inhibitor cocktail] and sonicated on ice, then pre-cleared with protein A–agarose. Following immunoprecipitation with rabbit anti-acetyl-H3/H4 or anti-FLAG antibodies, protein complexes were immunoprecipated and washed in turn with low salt, high salt, lithium chloride buffer and TE buffer [10 mM Tris (pH 8.0) and 1 mM EDTA]. After elution and reverse crosslinking, the purified DNA was resuspended in TE buffer. DNA samples (2 μl) were then amplified by PCR and real-time PCR. Primer pairs for the IL-5 promoter were as follows: for IL-5p, 5′-AGGAGATCTTTTTAGTCACTGGCAACA-3′ (sense) and 5′-CGTCTCGAGGGCAAAGAAAGTGCATAG-3′ (antisense); for IL-5-d1, 5′-TTTAAGATATAAGGCATTGGAA-3′ (sense) and 5′-GCAAAGAAAGTGCATAGTACAA-3′ (antisense); for IL-5-d2, 5′-TATTAACCCAAAGATTCCTTC-3′ (sense) and 5′-GTTTCCAATGCCTTATATCTTA-3′ (antisense); for IL-5-d3, 5′-GCCACAGTCATAGTAGAACATAGC-3′ (sense) and 5′-GGAATCTTTGGGTTAATACATCA-3′ (antisense); and for IL-5-d4, 5′-ACCTTCCCTCTTTATCTTCA-3′ (sense) and 5′-TAAGGTAGACCACTAAACAGAAT-3′ (antisense). The primers used in the reporter ChIP assay were 5′-AAATGTGGGGCAATGATGTA-3′ (sense) and 5′-GCGGGCCTTTCTTTATGT-3′ (antisense) (a part of the IL-5 promoter region).
RESULTS
HDAC4 repressed the IL-5 promoter activity and p300 counteracted the effect of HDAC4
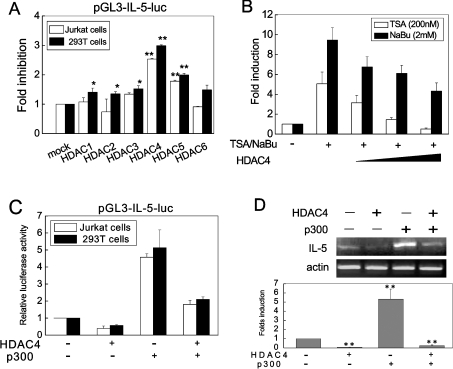
Our previous work revealed that TSA and NaBu, the two known HDACis, enhanced the IL-5 promoter activity (S. Han, J. Lu, Y. Zhang, C. Cheng, L. Li, L. Han and B. Huang, unpublished work). This was consistent with our previous report showing that p300/CBP increased not only the activity of the IL-5 promoter, but also the endogenous IL-5 mRNA level. The HAT activity of p300/CBP was necessary for this function [28]. To verify that histone deacetylation truly plays a repressive role in IL-5 transcription, HEK-293T cells were co-transfected with expression plasmids of the six HDACs (HDAC1–6) and the reporter gene plasmid pGL3-IL-5-luc, or transfected with the reporter gene plasmid alone. The results showed that the six HDACs tested exerted distinct repressive effects on the IL-5 promoter activity, among which HDAC4 had a much more prominent effect on IL-5 repression, bringing about a nearly 3-fold repression of the promoter activity (Figure 1A). We also repeated the same transfection experiments in Jurkat cells, which expressed IL-5 in vivo on activation, and obtained similar results (Figure 1A). Meanwhile, transfection of HDAC inhibitors, TSA or NaBu-treated cells with HDAC4 resulted in a diminished IL-5 mRNA level (Figure 1B), implicating an antagonizing effect of HDAC4 on the functions of these inhibitors. These data strongly suggested that HDACs played important roles in transcriptional repression of the IL-5 gene. We further tested the effects of p300 and HDAC4 on IL-5 gene regulation and the results indicated that overexpression of HDAC4 induced a decrease of the IL-5 promoter activity in both HEK-293T cells and Jurkat cells, whereas ectopic expression of p300 stimulated the IL-5 promoter activation (Figure 1C). An intriguing phenomenon was observed when HDAC4 and p300 were co-transfected, i.e. the suppression of IL-5 transcriptional activity by HDAC4 was moderated by co-transfection with p300. In contrast, the promotive role of p300 on the luciferase activity of the IL-5 promoter was reduced by co-transfection with HDAC4 (Figure 1C). This effect was confirmed by the detection of the endogenous IL-5 mRNA levels in cells transfected with HDAC4 and p300. Real-time PCR showed that the IL-5 mRNA level was reduced by overexpression of HDAC4 and was elevated by p300 transfection (Figure 1D), indicating the antagonistic effect between HDAC4 and p300 in control of IL-5 gene expression. All these data demonstrated that the reversible acetylation catalysed by HDAC4 and p300 exerted critical functions in the transcriptional regulation of the IL-5 gene.
Figure 1. The reporter activity and mRNA level of the IL-5 gene are reversibly regulated by HDAC4 and p300.
(A) Jurkat cells and HEK-293T cells were transfected with pGL3-IL-5-luc plasmid together with HDAC constructs expressing HDAC1–6 respectively. Luciferase activities were determined and normalized to Renilla activity 24 h after transfection. For T cell activation, Jurkat cells were treated with PMA and ionomycin 6 h before harvest. Results are shown as fold repression relative to that of the cells transfected without HDAC plasmid and are the means±S.D. from at least three individual experiments. (B) HEK-293T cells were transfected with HDAC4 plasmid for 6 h followed by treatment with HDAC for 24 h. Cells were harvested for detection of mRNA expression by real-time PCR. Data represent means±S.D. of three individual experiments. (C) The pGL3-IL-5-luc construct was co-transfected with HDAC4, p300 or both into Jurkat cells or HEK-293T cells and luciferase activities were calculated relative to that of the cells with pGL3-IL-5-luc transfection alone. Values are means±S.D. (n=3). (D) HDAC4 and p300 were transiently transfected into HEK-293T cells, and the mRNA level was estimated by RT-PCR and real-time PCR analysis and expressed as fold induction compared with that of the cells transfected with empty vector (n=3), **P<0.01, *P<0.05.
The acetylation level of histone H3 at the IL-5 promoter was reversibly altered by HDAC4 and p300 transfection
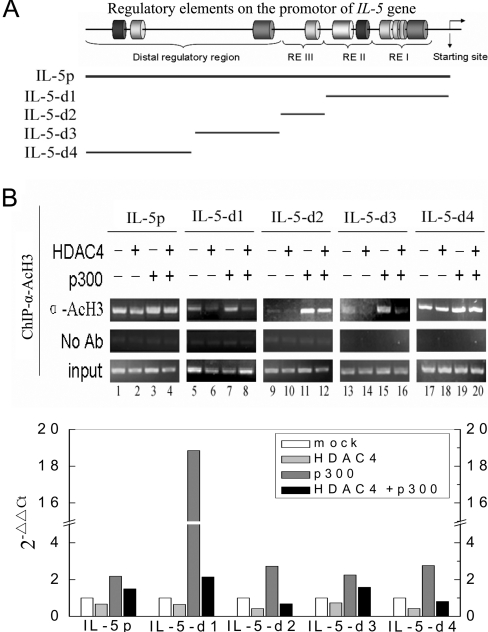
HDACs were thought to be responsible for the hypoacetylation of core histones, especially histones H3 and H4 at promoter regions. For ChIP assays we designed a series of primers corresponding to five individual regions in the IL-5 promoter (Figure 2A), in an attempt to detect changes of the acetylation level of histones at the IL-5 promoter. The chromatin fragments from HEK-293T cells co-transfected with HDAC4 and p300 plasmids were immunoprecipitated with antibodies against acetylated histone H3 and H4. Following the isolation of precipitated DNA, distinct regions of the IL-5 promoter were amplified. The results showed that the acetylation level of H3 was markedly influenced by the transfection of HDAC4 and p300 (Figure 2B). Specifically, in cells transfected with HDAC4, decreased accumulations of acetyl-H3 were observed in all of the regions of IL-5 promoter examined. In contrast, transfection of p300 led to an elevation of the acetylation level of H3 at the IL-5 promoter. Meanwhile, cells co-transfected with both HDAC4 and p300 exhibited higher levels of acetyl-H3 compared with HDAC4-transfected cells but lower levels of acetyl-H3 compared with p300-transfected cells (Figure 2B), indicating the antagonistic roles between HDAC4 and p300 in modifying the status of histone acetylation at the same region of the IL-5 promoter. Meanwhile, no obvious changes in the acetylation level of histone H4 were detected in the parallel experiments (results not shown).
Figure 2. ChIP assay for detection of acetylated histones on the IL-5 promoter.
(A) Diagram of the 5′-flanking region of the IL-5 gene showing regulatory elements and binding sites of transcription factors. Lines indicate the five regions of the IL-5 promoter amplified by specific primers in ChIP analysis. (B) HEK-293T cells were cross-linked with formaldehyde 24 h after transfection. DNA was sheared and immunoprecipitated with an anti-acetyl histone H3 antibody. After reversing the cross-linking, the DNA was amplified using primers specific for the regions indicated in (A). PCR products were resolved on a 1.5% agarose gel (upper panel). α-AcH3, anti-acetylated histone H3; Ab, antibody. Results from the real-time PCR are shown in the lower panel. Input indicates DNA prior to immunoprecipitation. Experiments were performed in triplicate.
HDAC4 was recruited to the IL-5 promoter regions and its nucleo–cytoplasm shuttling was important to its function
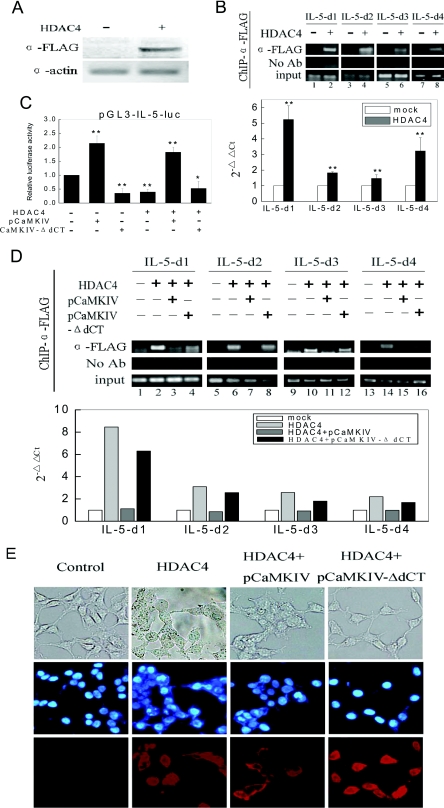
To further investigate the mechanisms of HDAC4 acting on IL-5 gene regulation, the recruitment of HDAC4 to the IL-5 promoter was checked using the ChIP assay. First, we confirmed the ectopic expression of FLAG–HDAC4 in HEK-293T cells by Western blotting (Figure 3A). For ChIP experiments, the immunoprecipitated DNA was prepared from HEK-293T cells transfected with the FLAG–HDAC4 construct and incubated with an anti-FLAG antibody. As shown in Figure 3(B), binding of HDAC4 was detected at all of the four regions in the IL-5 promoter, though the intensity of the HDAC4 binding varied (Figure 3B). Among these, HDAC4 recruitment appeared to be more intensive at the region containing REI and REII (IL-5-d1) (Figure 3B).
Figure 3. Recruitment of HDAC4 to the IL-5 promoter region.
(A) Western blot analysis of whole-cell extracts from HEK-293T cells. HEK-293T cells were transfected with FLAG–HDAC4 for 24 h and harvested. Whole-cell extracts were separated by SDS/PAGE and blotted with an anti-FLAG antibody. The expression of exogenous HDAC4 was detected using the ECL® staining method. (B) ChIP assays of the IL-5 promoter region. HEK-293T cells transfected with FLAG–HDAC4 were harvested and cross-linked. After sonication, cells were immunoprecipitated with an anti-FLAG antibody. DNA was amplified with primers specific for four regions of the IL-5 promoter. The recruitment of HDAC4 in each region was measured by PCR and real-time PCR. (C) HEK-293T cells were transfected with FLAG–HDAC4, or co-transfected with expression plasmids encoding CaMKIV or CaMKIV-ΔdCT (inactive mutant), as indicated, along with pGL3-IL-5-luc reporter plasmid. Values represent the relative luciferase activity (±S.D.) compared with that of the reporter plasmid transfected alone (n=3), **P<0.01, *P<0.05. (D) Alteration of HDAC4 recruitment to the IL-5 promoter. Cells were transfected with FLAG–HDAC4 plasmid or together with pCaMKIV or pCaMKIV-ΔdCT constructs as indicated. An anti-FLAG antibody was used for immunoprecipitation. The recruitment of HDAC4 was measured by PCR and real-time PCR. Experiments were performed in triplicate. (E) Subcellular localization of exogenous HDAC4 in HEK-293T cells. HEK-293T cells were plated on to glass slides and transfected with empty vector, FLAG–HDAC4 alone, FLAG–HDAC4 plus CaMKIV and FLAG–HDAC4 plus CaMKIV-ΔdCT. Following transfection (48 h), cells were fixed and permeabilized. The localization of HDAC4 was shown by immunofluorescence of FLAG visualized by TRITC staining (red), and the nuclei were counterstained with DAPI (blue). Top panel, phase contrast images of the cells; middle panel, cells stained with DAPI; bottom panel, cells stained with TRITC.
As a member of the Class II HDACs, HDAC4 possesses the capability of nucleo–cytoplasm shuttling. The subcellular localization of HDAC4 is suggested to be regulated by CaMKIV. Through phosphorylation, HDAC4 binds to a partner protein 14-3-3 and this leads to efficient nuclear export [20]. In order to further confirm the directly repressive role of HDAC4, we examined the influence of overexpression of pCaMKIV on the activity of the IL-5 promoter. We showed that CaMKIV increased the IL-5 promoter activity, and this elevation was moderated by transfection of pCaMKIV-ΔdCT, which was the inactive mutant of pCaMKIV (Figure 3C). Furthermore, pCaMKIV enhanced IL-5 promoter activity even when co-transfected with HDAC4, whereas the mutant pCaMKIV-ΔdCT did not (Figure 3C). In ChIP assays aiming to test the influence of pCaMKIV on the recruitment of HDAC4 to the IL-5 promoter, we found that CaMKIV decreased the binding of HDAC4 in IL-5 promoter regions, whereas pCaMKIV-ΔdCT did not (Figure 3D). Meanwhile, results from immunofluorescence studies suggested that HDAC4 was expressed both in the nuclei and in the cytoplasm of HEK-293T cells. pCaMKIV transfection induced shuttling of HDAC4 out of the nuclei whereas pCaMKIV-ΔdCT did not have this effect (Figure 3E), which was consistent with the results observed in the ChIP assay. These data demonstrated that the suppression of IL-5 transcription by HDAC4 was direct and the nucleo–cytoplasm shuttling of HDAC4 was essential for its repressive role in IL-5 gene transcription.
HDAC4 was recruited by the transcription factors bound to the IL-5 promoter and the integrity of the binding sites was essential to HDAC4 function
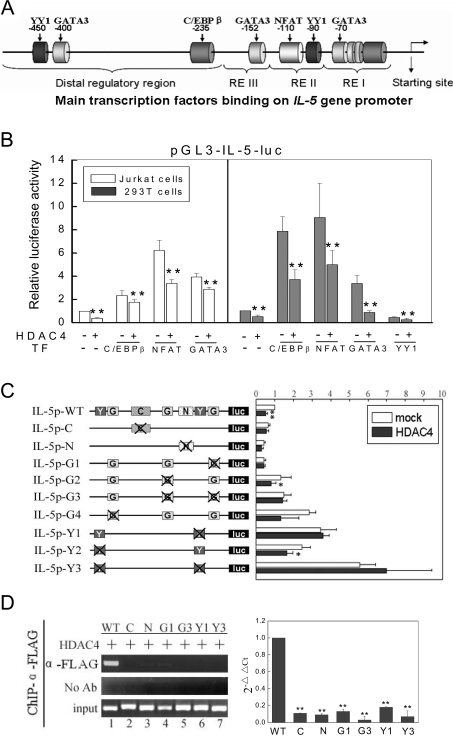
As co-repressors of transcription, HDACs do not contain canonical DNA-binding domains and they are usually recruited to specific chromatin regions by specific transcription factors. We were therefore interested in finding out whether the cis-elements at the IL-5 promoter region were responsible for the recruitment of HDAC4. We first showed that transcription factors that bind to the IL-5 promoter (Figure 4A), i.e. C/EBPβ, NFAT and GATA3 promoted the activity of the IL-5 reporter gene (Figure 4B), which was coincident with the results performed in Jurkat cells, while YY1 played a repressive role (Figure 4B). When HDAC4 was co-transfected, the effects of all the four factors on IL-5 promoter activity were restrained, implying that the suppressive role of HDAC4 probably relied on its recruitment by these factors to the promoter. For testing this assumption, we generated site-directed mutations for the binding sites for all of the four transcription factors at the IL-5 promoter (see the Experimental section). Meanwhile, we also generated double-site mutations of GATA3 (−70 bp and −152 bp) and YY1 (−90 bp and −450 bp) for further confirmation (Figure 4C). Through transfection, the activities of the IL-5 promoter were diminished when the binding sites for C/EBPβ, NFAT and GATA3 (−70 bp) were mutated, whilst the mutations of binding sites for GATA3 (−400 bp) and YY1 (−90 bp and −450 bp) led to an apparent increase in IL-5 promoter activity (Figure 4C) compared with the wild-type promoter. IL-5p-G2 (a mutant of the GATA3 binding site at −152 bp) didn't show any obvious change (Figure 4C). Unexpectedly, IL-5p-G3, the double site mutant of GATA3 (−70 bp and −152 bp) showed slightly increased luciferase activity compared with the wild-type promoter, which was conflictive with the results gained from that of single mutants. Despite this, the repressive roles of HDAC4 were all abolished when HDAC4 was co-transfected with these mutants (Figure 4C). These data provided evidence that the integrity of the transcription factor binding sites was essential for the recruitment and the suppressive role of HDAC4 on the IL-5 promoter. Moreover, the role of transcription factors in recruitment of HDAC4 to the IL-5 promoter was further confirmed by reporter ChIP assay using an anti-FLAG antibody. Transfection of mutant reporters of the IL-5 promoter abolished the binding of HDAC4 on the IL-5 promoter (Figure 4D), indicating the indispensable role of these factors on recruitment of HDAC4.
Figure 4. The recruitment of HDAC4 by transcription factors to the IL-5 promoter.
(A) Schematic diagram of the IL-5 gene promoter showing the binding sites of transcription factors. (B) HDAC4 repressed transcription factor-directed luciferase activities of the IL-5 promoter. Jurkat cells and HEK-293T cells were transfected with the luciferase reporter plasmid pGL3-IL-5-luc together with expression plasmids of C/EBPβ, NFAT, GATA3 or YY1, with or without HDAC4 co-transfection. Luciferase activities were measured 24 h after transfection and standardized against Renilla activity from a co-transfected control vector. Values represent means±S.D. from at least three separate experiments. **P<0.01, *P<0.05. (C) Site-directed mutation studies of the IL-5 promoter. Diagrams of constructs containing the IL-5 promoter are shown with each of the transcription factor binding sites represented as filled boxes. ‘G’ represents the binding site of GATA3, ‘Y’ represents the binding site of YY1, ‘N’ represents the binding site of NFAT and ‘C’ represents the binding site of C/EBPβ. The crossed boxes denote the mutated sites for the indicated transcription factors. The wild-type IL-5 promoter reporter plasmid and its mutants were transfected with or without HDAC4. Relative luciferase activities were normalized against Renilla activity. Data represent means±S.D. (n=3). (D) Reporter ChIP assay for detection of the recruitment of HDAC4 to the mutant IL-5 promoter. The wild-type and mutant reporter plasmids of the IL-5 promoter were co-transfected with FLAG–HDAC4 and immunoprecipitated using an anti-FLAG antibody. The recruitment of HDAC4 was analysed by PCR and real-time PCR using primers amplifying the specified promoter regions.
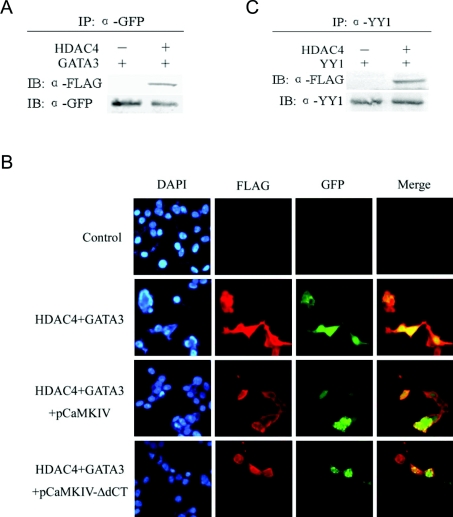
GATA3 and YY1 recruited and interacted with HDAC4 in vivo
Finally, we tested the interactions between HDAC4 and the factors GATA3 and YY1 using CoIP assays. Whole-cell extracts from HEK-293T cells transfected with GFP–GATA3 and FLAG–HDAC4, or GFP–GATA3 alone were immunoprecipitated with an anti-GFP antibody after harvest. Following immunoblotting with an anti-FLAG antibody, we detected a band in cells co-transfected with HDAC4 and GATA3, whereas cells transfected with GATA3 alone did not display this band (Figure 5A), suggesting that HDAC4 could interact with GATA3 in vivo. Additionally, an anti-GFP antibody was used in a blotting assay to ensure the equal amount of plasmids in transfection. This interaction was further verified using immunofluorescence. In HEK-293T cells co-transfected with HDAC4 and GATA3, it can be seen that HDAC4 and GATA3 co-existed in the nuclei (Figure 5B). Co-transfection of pCaMKIV caused the transport of HDAC4 out of the nuclei, which consequently prevented GATA3 from interacting with HDAC4 in the nucleus; meanwhile co-transfection of pCaMKIV-ΔdCT did not have this effect (Figure 5B). These data suggested that GATA3 could recruit HDAC4. Similarly, the interaction between YY1 and FLAG–HDAC4 in vivo was also demonstrated with a CoIP assay using an anti-YY1 antibody (Figure 5C).
Figure 5. Interactions of HDAC4 with GATA3 and YY1 in vivo.
(A) GFP–GATA3 was co-transfected with FLAG–HDAC4 into HEK-293T cells for 24 h. Whole-cell extracts were prepared and immunoprecipitated with an anti-GFP antibody and incubated with protein A–agarose beads. Immunoprecipitates were subjected to immunoblotting analysis with an anti-FLAG antibody and detected using ECL® (upper panel). The blot was re-probed with an anti-GFP antibody to confirm that GATA3 was successfully immunoprecipitated (lower panel). (B) The co-localization of GATA3 and HDAC4 in HEK-293T cells. HEK-293T cells were plated on to glass slides and transfected with empty vector, GFP–GATA3 plus FLAG–HDAC4, GFP–GATA3 plus FLAG–HDAC4 and pCaMKIV, GFP–GATA3 plus FLAG–HDAC4 and pCaMKIV-ΔdCT. The cells were fixed in formaldehyde and stained with an anti-FLAG antibody followed by a TRITC secondary antibody, and visualized under a fluorescence microscope. HDAC4 is immunostained in red and GATA3 is in green. The nuclei were counterstained with DAPI (blue). The merged images were created with Simple PCI software (Cimaging). (C) YY1 exists in a protein complex with HDAC4 in vivo. A construct expressing YY1 was co-transfected with FLAG–HDAC4 into HEK-293T cells for 24 h. Whole-cell lysates were immunoprecipitated with an anti-YY1 antibody and blotted with an anti-FLAG antibody (upper panel). The lower panel shows the blot of transfected YY1 using an anti-YY1 antibody.
To conclude, experimental data in the present study demonstrate that HDAC4 plays an important role in the transcriptional repression of IL-5. Working together with p300, it reversibly regulates the IL-5 promoter activity. This regulation is achieved through the alteration of the acetylation status of histone H3 at the IL-5 promoter. Moreover, HDAC4 can be recruited to the IL-5 promoter through interactions with specific transcription factors, and this recruitment is dependent on the integrity of the binding sites for these factors on the IL-5 promoter. The CaMKIV-mediated nucleo–cytoplasm shuttling of HDAC4 exerts important roles in the suppression of the IL-5 gene. The unique regulation of IL-5 transcription is the result of interactions between HATs and HDACs with histones and non-histone proteins.
DISCUSSION
The IL-5 gene is commonly described as being co-ordinately regulated with other Th2 cytokines, such as IL-4, IL-13 and GM-CSF. Considering the unique control of eosinophilia by IL-5, such coordinate regulation would be contradictory. In fact, the biological specificity of eosinophilia and its control by IL-5 suggests a unique and independent control of IL-5 regulation. To date, the specific regulation of IL-5 has still not been fully studied. Lavender et al. [23] showed that TSA treatment caused a partial relief of the repressive role of glucocorticoid on IL-5 gene expression. Glucocorticoids such as dexamethasone are common medicines in asthma therapy; their effects mediate the repression of a number of cytokine genes, such as IL-4, IL-5, IL-13 and GM-CSF. However, use of dexamethasone broadly represses Th2 cytokine expression. In addition, glucocorticoid is shown to specifically inhibit the inducible binding of factors to the REII region on the IL-5 promoter, which is not sufficient to interpret the specific modification of IL-5.
Among the six human HDACs tested in the present study, HDAC4 was found to be much more effective in repressing the transcriptional activity of the IL-5 promoter (Figure 1A). HDAC1–6 represent the enzymes belonging to two different classes, which have distinct features in structure and exert different functions. Compared with Class I members, Class II HDACs (HDAC4, 5, 6 and 7) display more specific responses to various signalling molecules. Such specificities allow specific target genes to be regulated by individual Class II HDAC members in a type-specific pattern. We have demonstrated that overexpression of HDAC4 resulted in histone hypoacetylation of the IL-5 promoter (Figure 2B) and consequently in a drastic reduction of IL-5 activity (Figures 1A and 1C), suggesting that members of Class II HDACs might be more effective in inhibiting transcription of the IL-5 gene. Moreover, HDAC4 could moderate the promotive effects of TSA and NaBu on IL-5 mRNA expression in a dose-dependent manner, further confirming that the elevated role of HDACis could be attributed to suppressing the activity of HDAC4. Furthermore, we show that p300 and HDAC4 can function as counteracting regulators to reversibly acetylate and deacetylate histones, thus changing the chromatin structure of the IL-5 promoter and subsequently modulating transcription of IL-5. In ChIP assays we demonstrated that pCaMKIV induced HDAC4 release from the promoter (Figures 3D and 3E) and led to an elevation of IL-5 activity (Figure 3C), indicating that the nucleo–cytoplasm shuttling of HDAC4 was an important mechanism for IL-5 suppression.
As co-repressors, HDACs require specific transcription factors for directing them to target DNA elements for regulatory functions. Our luciferase reporter and reporter ChIP assays based on mutation analysis revealed that the repressive role of HDAC4 might involve the co-operation between HDAC4 and four transcription factors (Figures 4B and 4C). Among these, GATA3 exhibited a dual effect on the IL-5 promoter activity, that is, the binding sites at −70 bp and −152 bp seemed to function as activating elements whilst that at −400 bp was a repressive element (Figure 4C), which is consistent with the conclusion drawn by Schwenger et al. [10]. However, the double mutant of GATA3 (−70 bp and −152 bp) led to a slight increase of luciferase activity compared with the wild-type promoter (Figure 4C). The underlying mechanism of this phenomenon is unclear.
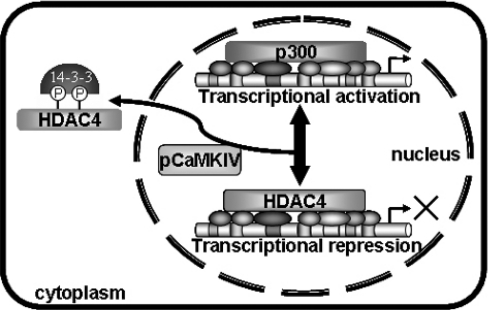
We tested the interactions between HDAC4 and two transcription factors GATA3 and YY1 respectively. Our CoIP assays suggested that HDAC4 interacted with GATA3 in vivo (Figure 5A). In addition, GATA3 existed in nuclei together with HDAC4 (Figure 5B). It has been shown that other members of the GATA family, GATA1 and GATA2, are able to interact with HDACs through their zinc-finger domains [30–32]. Moreover, GATA3 has also been previously demonstrated to interact with and be acetylated by p300 [26], making it an apparent target for HDAC action. Similarly, we showed that YY1 was able to bind to HDAC4 in vivo (Figure 5C). YY1 is known to interact with Class I HDACs both in vitro and in vivo [27]. Up to now, there has been no evidence suggesting the interaction of YY1 with any of the Class II HDAC members. On the other hand, Class II HDACs have been shown to be able to interact with Class I HDACs through the homologous domains [33]. Nevertheless, our data support that these transcription factors are potential molecules for recruiting HDAC4-containing repressor complexes on the IL-5 promoter. GATA3 plays a prominent role in Th2 cytokine production and in the maintenance of continuously remodelled open chromatin at the Th2-specific cytokine loci, including IL-4, IL-13, IL-5 and GM-CSF [34]. Results from the present study confirmed the participation of GATA3 in HDAC4-mediated IL-5 regulation. However, the complicated regulation of IL-5 may require a co-ordinated action of other factors specific to the IL-5 promoter. In the present study, we describe experimental results to show the recruitment of HDAC4 by GATA3 and YY1, although we could not rule out the possibility that the interaction between HDAC4 and the transcription factors might be indirect. Additionally, it is also intriguing to deduce that the other two transcriptional factors, C/EBPβ and NFAT, could probably interact with HDAC4, because both of them are substrates of p300. However, further data will be needed. The co-operation between GATA3 and other factors, such as C/EBPβ, NFAT and YY1 can also partly explain the diverse roles of GATA3 on IL-5 and IL-4 transcription [35]. It is therefore likely that GATA3 responds to Th2-specific transcription type, while the unique regulation of IL-5 is achieved through the co-ordination of factors binding at the IL-5 promoter. Meanwhile, the reversible modification of IL-5 activity catalysed by p300 and HDAC4 is also essential. Alterations in the expression of IL-5 are associated with allergic disorders, such as asthma, eczema and allergic rhinitis. The role of eosinophilia in allergic disorders also implicates IL-5 as a potential target for therapy. Despite the extensive research focused on the regulation of Th2 cell type cytokines, the understanding of the unique molecular mechanisms of IL-5 action still remain incomplete. The hyperacetylation of histones and chromatin remodelling taking place at Th2 type cytokine loci during Th2 differentiation [36] implicates the involvement of HATs and HDACs in this regulatory process, although the biochemical basis for these alterations has not been well illustrated. The modification by HDAC4 and p300 on IL-5 transcriptional modification manifested in the present study provides an insight into the mechanisms of the reversible regulation of IL-5 expression. It appears that a balanced acetylation status of the histones at the IL-5 promoter collaborated by HDAC4 and p300 plays a role in expression of this gene. Based on these experimental data, a picture is emerging in which multiple regulatory factors interact to favourably alter chromatin structure that allows proper transcription of the IL-5 gene. In this process, co-ordinated actions between the specific transcription factors and the reversible chromatin modifying enzymes play critical roles. We therefore propose a hypothesized model of IL-5 regulation (Figure 6). In resting cells, a status of hypoacetylation on the histones of the IL-5 promoter promotes basal expression of IL-5, which is maintained by HDAC4 recruited by transcription factors. After activation, HDAC4 is phosphorylated by CaMKIV, which provides binding sites for 14-3-3 proteins and promotes nuclear export of HDAC4. At this time, p300 is recruited on the promoter by transcription factors. Histones are acetylated and the chromatin structure is remodelled to an open status that permits transcriptional elevation and protein expression. In fact, the normal production of IL-5 may be dependent on a subtle balance of the activities of HATs and HDACs. However, the signalling molecules that drive the process still need to be further elucidated. These studies will help to achieve a better understanding of the control of IL-5 expression, and finally benefit the development of new therapeutic strategies for allergic diseases.
Figure 6. Proposed model showing roles of HDAC4 and p300 in regulation of the IL-5 gene.
For basal expression, HDAC4 is recruited by transcription factors bound on the IL-5 promoter resulting in hypoacetylation of histones at the regulatory elements of the IL-5 promoter, and finally leading to repression of gene transcription. Stimulation of cells with some molecules causes activation of CaMK-dependent signalling, which drives phosphorylation of HDAC4 by CaMKIV. Phosphorylated HDAC4 exports out of the nucleus, thus allowing p300 to be recruited by transcription factors resulting in activation of IL-5 gene transcription.
Acknowledgments
We are grateful to Dr W. C. Greene (Gladstone Institute of Virology and Immunology, San Francisco, CA, U.S.A., Dr E. Seto (H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, U.S.A.), Dr J. Boys (The Institute of Cancer Research, London, U.K.), Dr S. Akira (Research Institute of Microbial Diseases, Osaka University, Japan), Dr A. Goldfeld, Dr J. Zhu, Dr K. Murphy, Dr E. Bonneloy and Dr J. Xie for kindly providing FLAG–HDAC1–6, p300, C/EBPβ, NFATp (S>A), GATA3–GFP, YY1, pCaMKIV and pCaMKIV-ΔdCT expression plasmids. This work was supported by grants from the National Natural Science Foundation of China (30571698), the National Basic Research Program of China (2005CB522404), and the Program for Changjiang Scholars and Innovative Research Team (PCSIRT) in Universities (IRT0519).
References
- 1.Campbell H. D., Tucker W. Q., Hort Y., Martinson M. E., Mayo G., Clutterbuck E. J., Sanderson C. J., Young I. G. Molecular cloning, nucleotide sequence, and expression of the gene encoding human eosinophil differentiation factor (interleukin 5) Proc. Natl. Acad. Sci. U.S.A. 1987;84:6629–6633. doi: 10.1073/pnas.84.19.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi Y., Hayashi Y., Sugama Y., Miura Y., Kasahara T., Kitamura S., Torisu M., Mita S., Tominaga A., Takatsu K., Suda T. Highly purified murine interleukin 5 (IL-5) stimulates eosinophil function and prolongs in vitro survival. IL-5 as an eosinophil chemotactic factor. J. Exp. Med. 1988;167:1737–1742. doi: 10.1084/jem.167.5.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin L. B., Kita H., Leiferman K. M., Gleich G. J. Eosinophils in allergy: role in disease, degranulation, and cytokines. Int. Arch. Allergy Immunol. 1996;109:207–215. doi: 10.1159/000237239. [DOI] [PubMed] [Google Scholar]
- 4.Crabtree G. R. Contingent genetic regulatory events in T lymphocyte activation. Science. 1989;243:355–361. doi: 10.1126/science.2783497. [DOI] [PubMed] [Google Scholar]
- 5.Avni O., Lee D., Macian F., Szabo S. J., Glimcher L. H., Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 6.Stranick K. S., Zambas D. N., Uss A. S., Egan R. W., Billah M. M., Umland S. P. Identification of transcription factor binding sites important in the regulation of the human interleukin-5 gene. J. Biol. Chem. 1997;272:16453–16465. doi: 10.1074/jbc.272.26.16453. [DOI] [PubMed] [Google Scholar]
- 7.Cousins D. J., Richards D., Kemeny D. M., Romagnani S., Lee T. H., Staynov D. Z. DNase I footprinting of the human interleukin-5 gene promoter. Immunology. 2000;99:101–108. doi: 10.1046/j.1365-2567.2000.00947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwenger G. T., Mordvinov V. A., Karlen S., D'Ercole M., Sanderson C. J. Identification of two novel palindromic regulatory elements in the murine interleukin-5 promoter. Mol. Immunol. 1998;35:149–158. doi: 10.1016/s0161-5890(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 9.Li-Weber M., Giaisi M., Krammer P. H. Roles of CCAAT/enhancer-binding protein in transcriptional regulation of the human IL-5 gene. Eur. J. Immunol. 2001;31:3694–3703. doi: 10.1002/1521-4141(200112)31:12<3694::aid-immu3694>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 10.Schwenger G. T., Fournier R., Kok C. C., Mordvinov V. A., Yeoman D., Sanderson C. J. GATA-3 has dual regulatory functions in human interleukin-5 transcription. J. Biol. Chem. 2001;276:48502–48509. doi: 10.1074/jbc.M107836200. [DOI] [PubMed] [Google Scholar]
- 11.De Boer M. L., Mordvinov V. A., Thomas M. A., Sanderson C. J. Role of nuclear factor of activated T cells (NFAT) in the expression of interleukin-5 and other cytokines involved in the regulation of hemopoetic cells. Int. J. Biochem. Cell Biol. 1999;31:1221–1236. doi: 10.1016/s1357-2725(99)00069-2. [DOI] [PubMed] [Google Scholar]
- 12.Schwenger G. T., Fournier R., Hall L. M., Sanderson C. J., Mordvinov V. A. Nuclear factor of activated T cells and YY1 combine to repress IL-5 expression in a human T-cell line. J. Allergy Clin. Immunol. 1999;104:820–827. doi: 10.1016/s0091-6749(99)70293-9. [DOI] [PubMed] [Google Scholar]
- 13.Mordvinov V. A., Schwenger G. T., Fournier R., De Boer M. L., Peroni S. E., Singh A. D., Karlen S., Holland J. W., Sanderson C. J. Binding of YY1 and Oct1 to a novel element that downregulates expression of IL-5 in human T cells. J. Allergy Clin. Immunol. 1999;103:1125–1135. doi: 10.1016/s0091-6749(99)70188-0. [DOI] [PubMed] [Google Scholar]
- 14.Schwenger G. T., Kok C. C., Arthaningtyas E., Thomas M. A., Sanderson C. J., Mordvinov V. A. Specific activation of human interleukin-5 depends on de novo synthesis of an AP-1 complex. J. Biol. Chem. 2002;277:47022–47027. doi: 10.1074/jbc.M207414200. [DOI] [PubMed] [Google Scholar]
- 15.Blumenthal S. G., Aichele G., Wirth T., Czernilofsky A. P., Nordheim A., Dittmer J. Regulation of the human interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J. Biol. Chem. 1999;274:12910–12916. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 17.Cress W. D., Seto E. Histone deacetylases, transcriptional control, and cancer. J. Cell. Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 18.Sterner D. E., Berger S. L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grozinger C. M., Schreiber S. L. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X., Ito A., Kane C. D., Liao T. S., Bolger T. A., Lemrow S. M., Means A. R., Yao T. P. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 2001;276:35042–35048. doi: 10.1074/jbc.M105086200. [DOI] [PubMed] [Google Scholar]
- 21.Van Lint C., Emiliani S., Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J. H., Oh S. W., Kang M. S., Kwon H. J., Oh G. T., Kim D. Y. Trichostatin A attenuates airway inflammation in mouse asthma model. Clin. Exp. Allergy. 2005;35:89–96. doi: 10.1111/j.1365-2222.2004.02006.x. [DOI] [PubMed] [Google Scholar]
- 23.Jee Y. K., Gilmour J., Kelly A., Bowen H., Richards D., Soh C., Smith P., Hawrylowicz C., Cousins D., Lee T., Lavender P. Repression of interleukin-5 transcription by the glucocorticoid receptor targets GATA3 signaling and involves histone deacetylase recruitment. J. Biol. Chem. 2005;280:23243–23250. doi: 10.1074/jbc.M503659200. [DOI] [PubMed] [Google Scholar]
- 24.Kovacs K. A., Steinmann M., Magistretti P. J., Halfon O., Cardinaux J. R. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biol. Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Rodriguez C., Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP) J. Exp. Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamagata T., Mitani K., Oda H., Suzuki T., Honda H., Asai T., Maki K., Nakamoto T., Hirai H. Acetylation of GATA-3 affects T-cell survival and homing to secondary lymphoid organs. EMBO. J. 2000;19:4676–4687. doi: 10.1093/emboj/19.17.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W. M., Inouye C., Zeng Y., Bearss D., Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu C. Y., Lu J., Tan J., Li L., Huang B. Q. Human interleukin-5 expression is synergistically regulated by histone acetyltransferase CBP/p300 and transcription factors C/EBP, NF-AT and AP-1. Cytokine. 2004;27:93–100. doi: 10.1016/j.cyto.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 30.Watamoto K., Towatari M., Ozawa Y., Miyata Y., Okamoto M., Abe A., Naoe T., Saito H. Altered interaction of HDAC5 with GATA-1 during MEL cell differentiation. Oncogene. 2003;22:9176–9184. doi: 10.1038/sj.onc.1206902. [DOI] [PubMed] [Google Scholar]
- 31.Hong W., Nakazawa M., Chen Y. Y., Kori R., Vakoc C. R., Rakowski C., Blobel G. A. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO. J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozawa Y., Towatari M., Tsuzuki S., Hayakawa F., Maeda T., Miyata Y., Tanimoto M., Saito H. Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood. 2001;98:2116–2123. doi: 10.1182/blood.v98.7.2116. [DOI] [PubMed] [Google Scholar]
- 33.Fischle W., Dequiedt F., Hendzel M. J., Guenther M. G., Lazar M. A., Voelter W., Verdin E. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 34.Yamashita M., Ukai-Tadenuma M., Miyamoto T., Sugaya K., Hosokawa H., Hasegawa A., Kimura M., Taniguchi M., DeGregori J., Nakayama T. Essential role of GATA3 for the maintenance of type 2 helper T (Th2) cytokine production and chromatin remodeling at the Th2 cytokine gene loci. J. Biol. Chem. 2004;279:26983–26990. doi: 10.1074/jbc.M403688200. [DOI] [PubMed] [Google Scholar]
- 35.Takemoto N., Arai K., Miyatake S. Cutting edge: the differential involvement of the N-finger of GATA-3 in chromatin remodeling and transactivation during Th2 development. J. Immunol. 2002;169:4103–4107. doi: 10.4049/jimmunol.169.8.4103. [DOI] [PubMed] [Google Scholar]
- 36.Agarwal S., Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]