Abstract
The establishment of silent chromatin requires passage through S-phase, but not DNA replication per se. Nevertheless, many proteins that affect silencing are bona fide DNA replication factors. It is not clear if mutations in these replication factors affect silencing directly or indirectly via deregulation of S-phase or DNA replication. Consequently, the relationship between DNA replication and silencing remains an issue of debate. Here we analyze the effect of mutations in DNA replication factors (mcm5-461, mcm5-1, orc2-1, orc5-1, cdc45-1, cdc6-1, and cdc7-1) on the silencing of a group of reporter constructs, which contain different combinations of “natural” subtelomeric elements. We show that the mcm5-461, mcm5-1, and orc2-1 mutations affect silencing through subtelomeric ARS consensus sequences (ACS), while cdc6-1 affects silencing independently of ACS. orc5-1, cdc45-1, and cdc7-1 affect silencing through ACS, but also show ACS-independent effects. We also demonstrate that isolated nontelomeric ACS do not recapitulate the same effects when inserted in the telomere. We propose a model that defines the modes of action of MCM5 and CDC6 in silencing.
THE position-dependent repression of genes via distal nonpromoter regulatory elements is referred to as “silencing.” In Saccharomyces cerevisiae, the mating-type (HM) loci HML and HMR, the subtelomeric portion of the chromosomes, and the rRNA genes are the main genome regions where silencing has been observed and extensively studied (reviewed in Rusche et al. 2003).
At HML and HMR silencing is established through two specialized silencer elements that flank each of these loci (Rusche et al. 2003). These elements contain binding sites for Rap1p and Abf1p, which act as transcriptional activators/repressors in other contexts (Eisenberg et al. 1988; Biswas and Biswas 1990; Planta et al. 1995), as well ARS consensus sites (ACS), which bind the origin recognition complex (ORC) (Labib and Diffley 2001; Lei and Tye 2001). It is believed that the juxtaposition of Rap1p, Abf1p, and ORC creates a high local concentration of the main components of silenced chromatin, the SIR proteins, which eventually leads to the nucleation and spreading of the silent chromatin state (Rusche et al. 2003).
At telomeres, gene expression is reversibly silenced to produce either active or completely repressed chromatin, a phenomenon referred to as telomeric position effect (TPE) (Tham and Zakian 2002; Rusche et al. 2003). Mutations in the regulators of silencing at HM, the SIR genes, the ORC genes, ABF1, and RAP1, also have an effect on TPE (Aparicio et al. 1991; Pryde and Louis 1999; Roy and Runge 2000). On the other hand, genes involved in nonhomologous end joining, telomere maintenance, or histone methylation (HDF1 and HDF2, MRE11, XRS2, RAD50, COMPASS) as well as localization at the nuclear periphery affect silencing at telomeres, but not at the HM loci (Boulton and Jackson 1998; Laroche et al. 1998; Fisher and Zakian 2005; Mueller et al. 2006). Hence, silencing of the telomeres is mediated by similar, but not identical mechanisms to these employed at the mating loci. The growing list of factors engaged in silencing also includes the histone acetyl transferase NuA4 (Babiarz et al. 2006; Clarke et al. 2006), the histone chaperone CAF-1 (Tamburini et al. 2006), cohesin (Suter et al. 2004; Chang et al. 2005), SUM1 (Irlbacher et al. 2005), and SCP160 (Marsellach et al. 2006).
Unlike the HM loci, subtelomeric regions do not contain apparent strong silencers. At these positions the telomeric (TG1–3) repeats, which contain binding sites for Rap1p (Gilson 1989; Marcand et al. 1996; Grunstein 1997), cooperate with Abf1p-binding sites (ABF1-BS) and ACS, which are found in the core X- and Y′-subtelomeric elements. ABF1-BS and ACS do not act as silencers on their own and hence they are referred to as protosilencers (Boscheron et al. 1996; Fourel et al. 1999; Pryde and Louis 1999; Lebrun et al. 2001). In the core X- and Y′-elements, protosilencers coreside with sequences that have chromatin partitioning or antisilencing activities that are referred to as subtelomeric antisilencing regions (STARs) (Fourel et al. 1999, 2004; Pryde and Louis 1999). Thus, silencing at telomeres seems to be regulated by a complex assembly of proto- and antisilencers that is not so well understood.
A most puzzling issue regarding gene silencing is its link to DNA replication. The establishment of silent chromatin requires passage through S-phase (Miller and Nasmyth 1984), but does not require DNA replication (Dubey et al. 1991; Kirchmaier and Rine 2001; Li et al. 2001). Furthermore, robust silencing at HMR does not take place until M-phase (Lau et al. 2002). Nevertheless, many of the genes that affect silencing both at telomeres and at HM loci encode for bona fide DNA replication factors such as ORC2, ORC5, minichromosome maintenance (MCM)5, MCM10, CDC44(RF-C), CDC45, and POL30(PCNA) (Ehrenhofer-Murray et al. 1995, 1999; Dillin and Rine 1997; Fox et al. 1997; Dziak et al. 2003; Suter et al. 2004; Liachko and Tye 2005).
We have shown that a mutation in MCM5 (mcm5-461) derepresses multiple natural subtelomeric genes as well as reporter cassettes inserted next to telomeres (Dziak et al. 2003). To clarify the mechanism by which the mcm5-461 allele disturbs silencing, we attempted to identify natural subtelomeric sequences that confer its effect. We also compared the effect of mcm5-461 to that of mutations in several other replication genes. We show that replication factor mutants differentially affect silencing at telomeres. Interestingly, we show that nontelomeric ACS does not recapitulate the effects observed with natural telomeric ACS.
MATERIALS AND METHODS
Yeast strains and plasmids:
The strains used in this study are shown in supplemental Table 4 at http://www.genetics.org/supplemental/. All experiments were performed at the permissive temperatures as stated in the original studies in supplemental Table 4.
The GF2, GF3, GF6, GF9, GF10, GF44, GF46, and GF61 constructs are described in Fourel et al. (1999). URA3-tel (pURTEL) was described in Gottschling et al. (1990). ARS1-URA3-tel and URA3-ARS1-tel were produced by inserting the BamHI/EcoRI fragment of pARS1wt (Marahrens and Stillman 1992) in the indicated positions in pURTEL. ARS1ΔACS-URA3-tel, ARS1ΔABF1-URA3-tel, GF6ΔACS, and GF44ΔACS were generated by site-directed mutagenesis of ACS and ABF1-BS in ARS1-URA3-tel, GF6, and GF44, respectively.
Yeast transformation and fluoroorotic acid resistance assays:
Integrating constructs were produced by restriction digestion of the GF plasmids as described in Fourel et al. (1999) or by PCR. Cells were transformed and colonies were selected on SC/−Ura plates. For each measurement of percentage of fluoroorotic acid resistance (%FOAR) three colonies were streaked on SC/FOA plates to confirm the variegated expression and then inoculated in 3 ml SC medium and diluted 1000 times into a fresh SC medium for 3 consecutive days. Serial 1:10 dilutions were prepared for each culture and 5-μl aliquots were spotted on SC and SC/FOA plates. The %FOAR cells was acquired as the average number of cells on SC/FOA plates divided by the average number of cells on SC plates. The numbers presented in supplemental Tables 1–3 at http://www.genetics.org/supplemental/ are average values ± standard error of n triplicate measurements with each strain/construct combination. Average values, error measurement, and ratios between %FOAR in different strains were calculated in Microsoft Excel.
Replication timing was estimated by the transient hemimethylation assay (Friedman et al. 1995). DamI methylase-expressing yeast strains were generated by transforming cells with XhoI-linearized pDP6-Dam1 (Hoekstra and Malone 1985). Transformants expressing DamI methylase and their genomic DNAs were tested for sensitivity to DpnI. Cells at OD600 = 0.3–0.4 were arrested at G2/M in YPD medium containing 15 μg/ml nocodazole [Sigma (St. Louis) M1404] for 3 hr. An aliquot of arrested cells was collected and the extent of arrest was determined by FACS and by microscope (>90% large budded, not shown). The cells were released from the arrest by resuspension in fresh media after washing twice with H2O. Samples were harvested at 0, 25, 35, 45, 55, 65, 75, 85, 95, 105, and 125 min. Genomic DNA was isolated, digested with EcoRI or XhoI and DpnI, respectively, separated on 0.8% agarose gels, and transferred to Nylon membranes. The membranes were probed with a PCR-generated probe to ARS305 or with a 1.3-kb XhoI fragment Y′-probe (Brevet et al. 2003), respectively. Signals were acquired by a Typhoon phosphorimager (Amersham Biosciences, Arlington Heights, IL) and analyzed with Kodak ID image analysis software.
RESULTS
Experimental strategy:
The evaluation of the expression of a reporter gene (most frequently URA3) inserted in the subtelomeric region of a chromosome (most frequently in the VII-L telomere) is a routine assay for telomere position effect (TPE). At these positions URA3 shows variegated expression—it is either fully transcribed or completely repressed. The proportion of cells that do not express URA3 is used as an indirect measure of the extent of subtelomeric silencing. Because URA3 expression renders the cells sensitive to FOA, the proportion of nonexpressing cells is assessed as % FOAR.
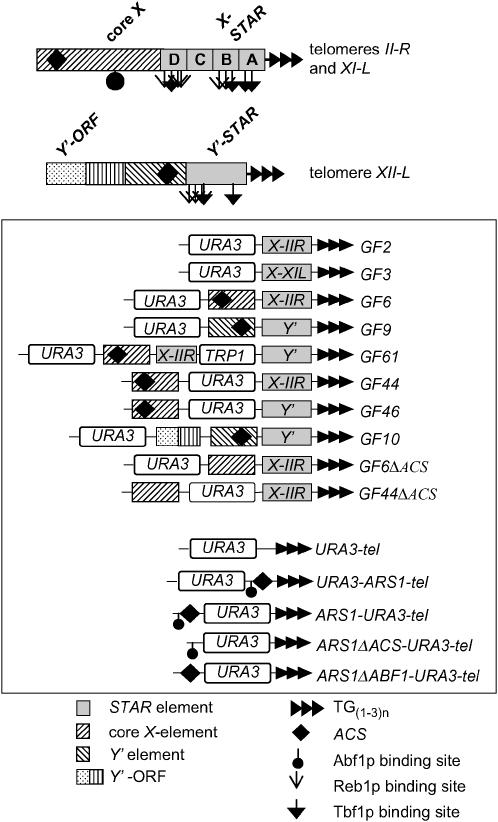
To identify sequences that confer the effect of mcm5-461 and other alleles on TPE, we selected nine previously published URA3 reporter cassettes (Fourel et al. 1999) (Figure 1). These cassettes contain a telomere repeat [TG(1–3)n] and different combinations of natural Y′- or core X- or STAR elements. For cross-reference, we kept the names of the constructs (GF2, GF3, etc.) as in the original article (Fourel et al. 1999). In Figure 1 we show the positions of ACS and the binding sites of Reb1p, Tbf1p, and Abf1p in the building blocks of these constructs.
Figure 1.—
Diagrams of the constructs used in this study. Maps (not to scale) of the core X-element at the II-R and XI-L telomeres and the Y′-element at the XII-L telomere are shown at the top. The positions of STAR, the core X- and Y′-elements, the telomeric TG1–3 repeats, ACS, and the binding sites for Abf1p, Reb1p, and Tbf1p are indicated by different patterns and the symbols shown below. The combinations of these elements and the position of the URA3 reporter in the different constructs are shown in the middle.
All these constructs were integrated in the VII-L telomere of mcm5-461, mcm5-1, orc2-1, orc5-1, cdc45-1, cdc7-1, and cdc6-1 and the corresponding isogenic wild-type strains. Variegated expression was confirmed by streaking the cells on SC/−Ura and SC/FOA plates. Subsequently, three colonies (per experiment) were picked and grown for >30 generations in nonselective medium to reach an equilibrium of variegated expression and to allow for the complete loss of any nonintegrated cassettes. Serial dilutions of these cultures were then spotted on SC and SC/FOA plates and the average %FOAR was measured. Initial experiments showed that the three wild-type strains (W303, mcm5-461∷MCM5, and mcm5-1∷MCM5) impose similar levels of URA3 repression in identical constructs (Figure 2). GF10 showed about two times higher repression in mcm5-461∷MCM5 (Figure 2). While we do not understand the reason for this dissimilarity, it appears on a background of comparable patterns of repression between all other constructs in the three strains (Figure 2). Hence, our reporter cassettes are subject to only minor strain-specific variations. We postulate that any significant variation in the repression of these constructs in mutant strains should be attributed to the mutant alleles.
Figure 2.—
Reporter constructs display similar repression in different wild-type strains. The GF series of constructs in Figure 1 were integrated in the wild-type W303 and the merodipolid mcm5-461∷MCM5 and mcm5-1∷MCM5 strains and %FOAR was measured in (n) triplicate measurements. Average %FOAR cells for the different strain/construct combinations are plotted. Data are from supplemental Table 1 at http://www.genetics.org/supplemental/.
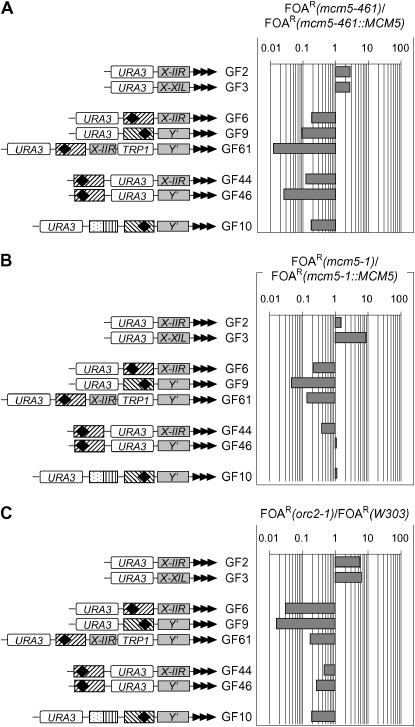
Next, we calculated the average %FOAR for each strain/construct combination in several (n) independent experiments (supplemental Table 1 at http://www.genetics.org/supplemental/). Finally, the ratio of %FOAR in the mutant strain vs. the %FOAR in the isogenic wild-type strain was calculated and plotted to represent the effect of different mutations on the silencing of URA3 in different constructs (Figure 3, A–G). In these graphs, values below one signify derepression of URA3 in the mutant strain, while values above one signify increased repression.
Figure 3.—
Differential silencing of URA3 in mutant strains. The GF constructs in Figure 1 were inserted in the VII-L telomere of mcm5-461, mcm5-461∷MCM5, mcm5-1, mcm5-1∷MCM5, W303, orc2-1, orc5-1, cdc45-1, cdc7-1, and cdc6-1 strains and %FOAR was determined for each construct/strain combination. The graphs show the ratio of %FOAR in the mutant vs. %FOAR in the isogenic wild-type strain. Data are from supplemental Table 1 at http://www.genetics.org/supplemental/. The diagrams from Figure 1 are shown on the left. Values above one indicate increased repression of URA3 in this construct in the mutant relative to the isogenic wild-type strain. Values below one indicate derepression. The following ratios are shown: (A) %FOAR mcm5-461 vs. %FOAR mcm5-461∷MCM5; (B) %FOAR mcm5-1 vs. %FOAR mcm5-1∷MCM5; (C) %FOAR orc2-1 vs. %FOAR W303; (D) %FOAR orc5-1 vs. %FOAR W303; (E) %FOAR cdc45-1 vs. %FOAR W303; (F) %FOAR cdc7-1 vs. %FOAR W303; (G) %FOAR cdc6-1 vs. %FOAR W303.
orc2-1, mcm5-461, and mcm5-1 perturb silencing predominantly via ACS-containing core X- and Y′-elements:
We noted significant similarities in the effects of mcm5-461, mcm5-1, and orc2-1 (Figure 3, A–C). These mutations caused derepression of URA3 in the constructs, which harbor the ACS-containing core X- and Y′-elements (GF6, GF9, GF61, GF44, GF46, and GF10). This effect was stronger in GF6, GF9, and GF61, in which the ACS-containing element was juxtaposed to URA3 toward the telomere. The constructs, in which the ACS-containing element was positioned next to URA3 toward the centromere (GF44, GF46) or was separated from URA3 by additional Y′-sequences (GF10) showed a weaker effect in mcm5-461 and orc2-1 and almost no effect in mcm5-1 (Figure 3, A–C). The results indicate that in these constructs URA3 is exposed to ACS-emitted silencing signals, which are compromised in orc2-1, mcm5-461, and mcm5-1.
The two constructs that contained no ACS plus a STAR between the telomere and URA3 (GF2, GF3) showed a moderate increase in silencing in mcm5-461, mcm5-1, and orc2-1 (Figure 3, A–C). These data are consistent with the idea that a general genomewide decrease in ACS-mediated silencing could release factors that can engage in telomere-mediated silencing. The alternative interpretation is that mcm5-461, mcm5-1, and orc2-1 have a direct positive effect on the STARs in constructs that are missing ACS. We favor the former idea because the MCM and ORC complexes associate with subtelomeric ACS (Wyrick et al. 2001) (M. A. Rehman, unpublished data) and because we have no reasonable model to explain how they could work on ACS-less chromatin.
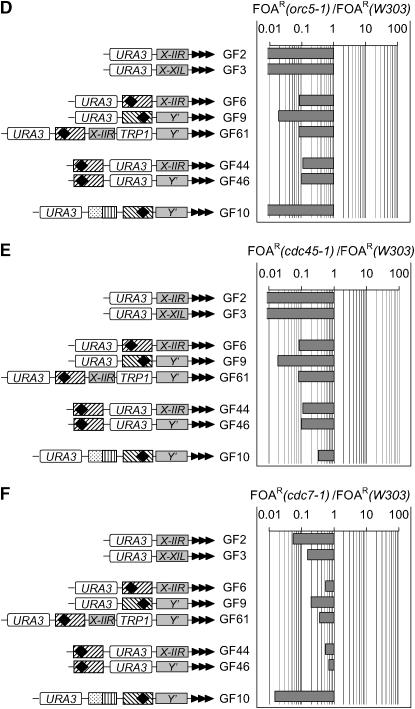
orc5-1, cdc45-1, and cdc7-1 show ACS-independent effects:
orc5-1, cdc45-1, and cdc7-1 caused significant derepression of URA3 in all constructs (Figure 3, D–F). Remarkably, strongest derepression was observed in the ACS-less constructs (GF2, GF3) and the construct with the greatest distance between ACS and URA3 (GF10). These data do not exclude a role of these alleles via ACS; however, they strongly imply that orc5-1, cdc45-1, and cdc7-1 profoundly affect another step in silencing that is ACS independent.
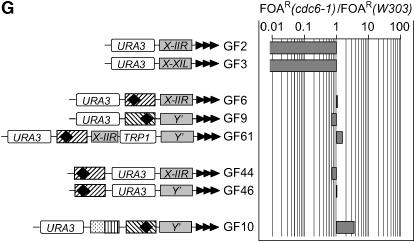
cdc6-1 does not affect silencing via ACS:
cdc6-1 showed strong derepression of URA3 in the ACS-less constructs (GF2, GF3) while the ACS-containing constructs were unaffected (Figure 3G). The results suggest that the cdc6-1 allele does influence silencing, but this effect is independent of ACS.
cdc6-1, orc5-1, cdc45-1, and cdc7-1 strongly to moderately decreased the repression of URA3 in the GF2 and GF3 constructs, which contain only a STAR element next to the telomere (Figure 1). Considered separately, these data point to the possibility that the four wild-type genes counter the effect of STARs; however, the mechanism of this action remains completely unknown.
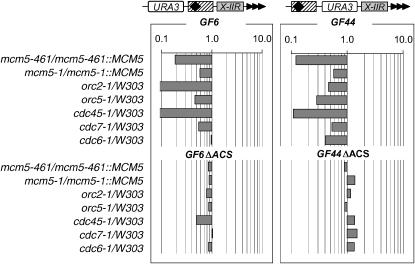
The deletion of ACS precludes the effect of the mutations:
To evaluate the dependence of the observed effects on ACS, we destroyed ACS in two of the tested constructs, GF6 and GF44. They differ in the position of the X-element and URA3 relative to the telomere (Figure 1). We inserted these constructs in the VII-L telomere and performed FOAR assays (supplemental Table 2 at http://www.genetics.org/supplemental/). We estimated the effect of each mutation by dividing the %FOAR cells in the mutants by the %FOAR cells in the corresponding wild-type strains (Figure 4). Consistent derepression (although at varying magnitude) of URA3 in GF6 and GF44 in all mutants relative to wild-type strains was observed except for GF6 in cdc6-1 (Figure 4, top graphs). The destruction of ACS strongly reduced (GF6ΔACS) or eliminated (GF44ΔACS) these effects with residual derepression still notable for GF6ΔACS in cdc45-1 and cdc7-1 (Figure 4, bottom graphs). These results clearly show that the effects of all mutants (with the exception of cdc6-1) are mediated by ACS. In conjunction with Figure 3, the data in Figure 4 provide the conclusion that orc5-1, cdc45-1, and cdc7-1 affect silencing through the ACS, but also though another unknown mechanism.
Figure 4.—
Deletion of ACS in GF6 and GF44 abolishes the effect of mutations. The ratio of %FOAR in mutant vs. %FOAR in isogenic wild-type strains as indicated on the left was calculated from the numbers in supplemental Table 2 at http://www.genetics.org/supplemental/ and plotted. Each series of bars represents the calculations for the constructs shown at the top.
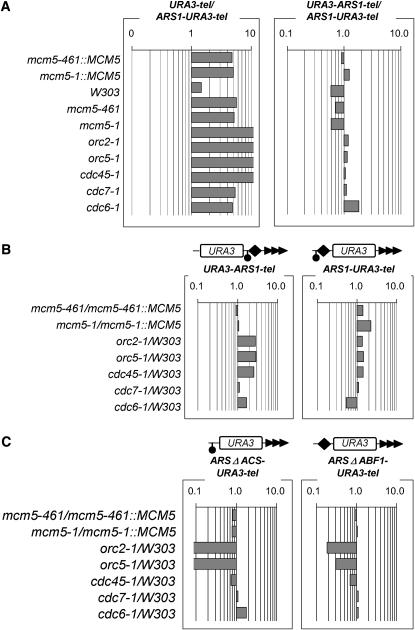
Nontelomeric ACS does not recapitulate the effects of core X- and Y′-ACS:
The ACS sequences within the X- and the Y′-elements are the obvious candidates that convey the effect of mutations in DNA replication factors on TPE. We therefore tested if isolated nontelomeric ACS could recapitulate these effects. We constructed five reporter cassettes containing URA3, a telomeric repeat, and the genomic ARS1 (encompassing ACS through ABF1-BS) (Marahrens and Stillman 1992) (Figure 1). We must note that the consensus ACS and ABF1-BS in the X-element and in ARS1 are not identical. URA3-ARS1-tel and ARS1-URA3-tel differ in the position of ARS1 and URA3 relative to the telomere (Figure 1). In ARS1ΔABF1-URA3-tel and ARS1ΔACS-URA3-tel the ABF1-BS and ACS were destroyed by site-directed mutagenesis. URA3-tel does not contain ARS1 (Figure 1). These five constructs were integrated in the VII-L telomere of all mutant and isogenic wild-type strains and TPE assays were performed (supplemental Table 3 at http://www.genetics.org/supplemental/). As expected, ARS1 acted as a protosilencer in all strains as exemplified by the moderate to strong increase in repression in ARS1-URA3-tel relative to URA3-tel (Figure 5A). We also show that the position of ARS1 has little effect on URA3 in all mutants tested (Figure 5B).
Figure 5.—
Isolated nontelomeric ACS does not recapitulate the effects of mutations in the GF constructs. (A) The ratio of %FOAR in URA-tel vs. ARS1-URA3-tel and in URA3-ARS1-tel vs. ARS1-URA3-tel in the strains shown on the left was calculated and plotted. Data are from supplemental Table 3 at http://www.genetics.org/supplemental/. (B and C) The ratio of %FOAR in mutant vs. %FOAR in wild-type strains as indicated on the left was calculated and plotted from the numbers in supplemental Table 3. Each series of bars represents the calculations for the constructs shown at the top.
The impact of mutations in replication factors on the repression of these constructs was assessed by dividing the %FOAR in the mutant strains by %FOAR in the isogenic wild-type strains (Figure 5B). Surprisingly, despite the fact that the net effect of ACS in URA3-ARS1-tel and ARS1-URA3-tel is increased silencing (supplemental Table 3 at http://www.genetics.org/supplemental/), the mutations had very little or even a weak derepression effect in these constructs (Figure 5B). Even more, the destruction of ACS in ARS1-URA3-tel strongly decreased repression in orc2-1 and orc5-1 cells, while the other mutants were indifferent to the omission of this element (Figure 5C). Corresponding, but weaker derepression was observed upon deletion of ABF1-BS (Figure 5C), arguing that the effect of these mutations was only partially mediated by ACS. For comparison, orc2-1 and orc5-1 did not affect repression in the GF6ΔACS and GF44ΔACS constructs (Figure 4) and most mutations reduced repression in GF6 and GF44 (Figure 4). The overall impression from these experiments was that isolated nontelomeric ACS responds quite differently to mutations in replication factors as compared to “natural” telomeric ACS.
Repression in the GF constructs does not correlate with deregulation of the cell cycle or the replication timing of telomeres:
In an attempt to correlate the results obtained so far to a foreseeable deregulation of cell cycle or DNA replication, we directly compared the cell cycle distribution, minichromosome stability, and the replication timing of telomeres in the mutant strains.
Cell cycle distribution was evaluated in exponentially growing cells in rich medium at the permissive temperatures. We noted a moderate increase in the fraction of G2/M cells in cdc6-1 and mcm5-461 and a more substantial increase in cdc45-1 (supplemental Figure 1 at http://www.genetics.org/supplemental/). There was also a moderate increase in the fraction of S-cells in cdc7-1. Apart from these differences, we did not note a correlation between the effect of these mutations on the cell cycle (supplemental Figure 1) and that on telomeric silencing (Figure 3). All strains demonstrated significant instability of minichromosomes (data not shown), but again failed to draw a parallel to the pattern of silencing.
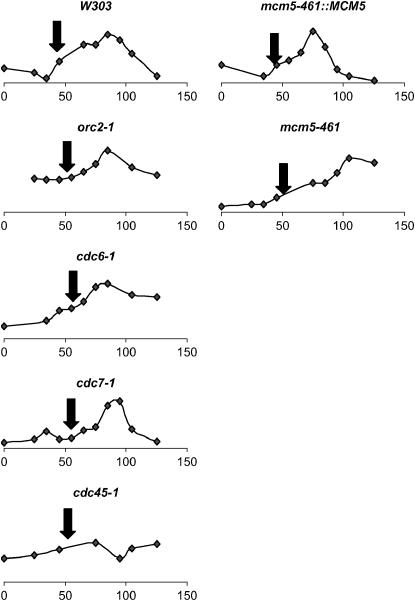
Replication timing of telomeres in the different mutants was estimated by the transient hemimethylation assay (Friedman et al. 1995) in nocodazole-synchronized cultures. Briefly, the DamI methylase gene was introduced in all strains and the cells were arrested in mitosis by exposure to 15 μg/ml nocodazole. The cells were then released and aliquots were taken at different time points. The genomic DNA from these samples was isolated and digested with DpnI. Because DpnI digests yeast DNA only if it is methylated by DamI and because newly replicated DNA is not immediately methylated by DamI, the detection of DpnI-resistant genomic DNA indicates the time when this DNA is replicated (Friedman et al. 1995). We evaluated the timing of replication of two genomic loci, the early firing ARS305 and telomeres (Figure 6). In all strains ARS305 replicated between 45 and 55 min after the release from mitotic arrest. In most strains subtelomeric Y′-DNA was replicated ∼90 min after the release from mitotic arrest. In mcm5-461 the Y′-DNA was replicated 100 min after the release. Notably, cdc45-1 did not display a distinct peak of hemimethylated DNA during the cell cycle, thus suggesting untimed replication of subtelomeric DNA (Figure 6).
Figure 6.—
Replication timing of telomeric DNA in different mutants. Each graph represents the relative abundance of DpnI-resistant telomeric DNA in nocodazole synchronized cells that express DamI methylase. The name of each strain is shown at the top of each graph. Time (minutes) after release of mitotic block is shown at the bottom. The arrow indicates the peak of abundance of DpnI-resistant ARS305 DNA. Details are provided in materials and methods.
Taken together, these data revealed no correlation between the effect of the mutations on silencing on the one hand (Figure 3) and that on cell cycle, minichromosome stability, or replication timing of telomeres on the other hand.
DISCUSSION
DNA replication factors differentially regulate subtelomeric protosilencers:
The understanding of the relationship between silencing and DNA replication is a challenge. The establishment of silent chromatin requires a passage through S-phase (Miller and Nasmyth 1984), but the requirement for DNA replication per se has been ruled out (Dubey et al. 1991; Kirchmaier and Rine 2001; Li et al. 2001; Sharma et al. 2001). Nevertheless, the list of DNA replication factors that affect silencing has been steadily growing (Ehrenhofer-Murray et al. 1995, 1999; Dillin and Rine 1997; Fox et al. 1997; Dziak et al. 2003; Suter et al. 2004; Liachko and Tye 2005). In many instances it is not clear if mutations in these factors affect silencing directly or indirectly through effects on DNA replication or cell cycle. Here we provide conclusive evidence regarding the mode of action of two of these genes and details on the roles of others.
First, we show that the mcm5-461 and mcm5-1 alleles share a high level of similarity with orc2-1. Previously, disruption of silencing at multiple genomic loci by the orc2-1 allele was conclusively demonstrated (Rusche et al. 2003; Ramachandran et al. 2006). mcm5-461 and mcm5-1 as well as orc2-1 reduced repression in the constructs, which contain URA3 next to ACS-containing core X- or Y′-elements (Figure 3, A–C). In support of ACS-dependent function, we show that in mcm5-461, mcm5-1, and orc2-1 the deletion of ACS in GF6 and GF44 abolished the effect of these mutations (Figure 4). Altogether, we present evidence that, similar to ORC2, MCM5 is directly involved in ACS-mediated silencing at telomeres. Most, if not all of the effects of these two genes on silencing are via ACS. In constructs lacking ACS (GF2 and GF3) these mutations produced a moderate increase in silencing (Figure 3, A–C). These results are consistent with a genomewide deficiency in the ACS-mediated silencing, which in turn releases and redirects silencing factors to the telomere. However, the proposed scenario may not be universal. For example, a telomeric URA3 reporter without a STAR element (URA-tel) was actually derepressed in orc2-1 and orc5-1, but not in mcm5-461 mutants (supplemental Table 3 at http://www.genetics.org/supplemental/). Hence, the response of repressed genes at ACS-independent locations may vary in different mutants.
Our second finding is that CDC6 (or at least the cdc6-1 allele) regulates silencing independently of ACS. Cdc6p is essential for the loading of the MCM complex on ORC-bound ACS at origins of replication (Blow and Dutta 2005). The cdc6-1 allele impairs DNA replication (Weinreich et al. 2001; Tsuyama et al. 2005). However, it selectively caused derepression only in the ACS-less GF2 and GF3 constructs, while having negligible effects on the ACS-containing constructs (Figure 3G). In agreement with our results, derepression of genes in the nontelomeric DAL gene cluster on chromosome IX in the cdc6-1 mutant occurs independently of several nearby ACS (W. Burhans, personal communication). This behavior of cdc6-1 contrasts with the derepression of the same genes by the orc2-1 mutation, which is ACS dependent (Ramachandran et al. 2006). The experiments in Figure 3G and this unrelated set of data reinforce the conclusion that cdc6-1 affects silencing, however, in an ACS-independent fashion.
Our data do not allow unambiguous conclusions about the modes of action of ORC5, CDC45, and CDC7. orc5-1, cdc45-1, and cdc7-1 decreased silencing in all constructs regardless of the presence of ACS (Figure 3, D–F). The trivial interpretation of these data is that, like cdc6-1, the three alleles do not affect silencing via ACS. However, the deletion of ACS in GF6 and GF44 reduced repression in these mutants (Figure 4), arguing in favor of ACS-dependent function. The most likely scenario is that orc5-1, cdc45-1, and cdc7-1 act via ACS, but the specificity of this action is masked by effects on other ACS-independent process(es). We can only speculate on the nature of these ACS-independent processes. For example, a recent article shows extensive crosstalk between DNA replication/silencing factors and genes, which regulate sister chromatid cohesion or histone acetylation (Suter et al. 2004). It is possible that orc5-1, cdc45-1, and cdc7-1 disturb the links to cohesins or some unknown feature of the maintenance of the silent chromatin state in addition to decreasing the efficiency of silencing emitted by ORC/MCM complexes on ACS.
ACS are not created equal:
We attempted to recapitulate the effects of mutations in replication factors in a more defined experimental system. We used isolated ACS and ABF1-BS derived from ARS1 as opposed to the ACS and ABF1-BS in the subtelomeric core X- and Y′-elements. We stress that the consensus ACS and ABF1-BS in ARS1 differ from the ones in core X. The ARS1 containing constructs (Figure 5) and the core X containing GF6 and GF44 (Figure 4) provide a basis for comparison between these two types of ACS. We noted that the telomere- and ARS1-derived ACS display extensive dissimilarities in terms of telomeric silencing (Figures 4 and 5). The results suggest that subtelomeric ACS are not functional replicas of origin ACS and their action as protosilencers is not subject to the same regulatory mechanisms. This idea is in agreement with Palacios DeBeer et al. (2003), who demonstrate that origin and HM silencer ACS differ in their affinity for the ORC and that this difference reflects the preferential function of the corresponding ACS in silencing or origin firing. We propose that an additional functional dissimilarity of these two types of ACS is their dependence on the cdc6-1 allele (Figure 3) (Weinreich et al. 2001; Tsuyama et al. 2005). It is possible that the surrounding sequence of core X- and Y′-elements influences the ACS silencing specificity. More focused work is needed to address in detail how origin and subtelomeric ACS differ.
Concluding remarks:
There is an extensive overlap of proteins, which work in both replication and silencing. Many of these bind ACS or regulate the function of ACS-bound proteins (Rusche et al. 2003). In DNA replication, these proteins control origin licensing and firing (Rusche et al. 2003). It remains unclear how the same proteins work in silencing. Here we show that ACS-related factors require different subtelomeric elements for their silencing effects and are not uniform ACS-dependent repressors. We support the idea that at least some of these factors work directly on the silenced domains rather than affecting them indirectly via DNA replication or the cell cycle.
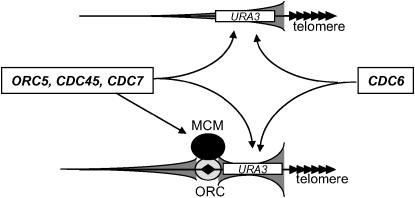
We propose that the MCM proteins (or at least Mcm5p) are recruited to subtelomeric ACS, where together with the ORC they build up a protosilencer (Figure 7). The protosilencer activity is directly influenced by Orc5p, Cdc45p, and Cdc7p. However, these three proteins also modulate silencing independently of ACS via other processes (Figure 7). Finally, Cdc6p does not participate in the regulation of protosilencers. Still, Cdc6p can control silencing via some unknown ACS-independent mechanism. Future studies should address how these genes are involved in telomeric silencing.
Figure 7.—
A model for ACS-mediated silencing at yeast telomeres. The array of solid triangles represents the telomeric repeat. The solid diamond represents ACS. The oval with light shading over ACS represents the ORC. The solid oval represents MCM proteins. URA3 is shown as a rectangle. The elongated cones with dark shading represent areas and levels of repression. In the absence of protosilencers the only silencing element is the telomere. Its activity is independent of ORC and MCM proteins. Subtelomeric ACS protosilencers require the binding of ORC and MCM proteins.
Acknowledgments
We thank J. Rine, B. Stillman, P. Pasero, B. Duncker, and S. Jacobson for the generous donation of strains and plasmids. This study was supported by a grant to K.Y. from National Science and Engineering Research Council (Canada) and to E.G. from the Ligue Nationale Contre le Cancer (France).
References
- Aparicio, O. M., B. L. Billington and D. E. Gottschling, 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279–1287. [DOI] [PubMed] [Google Scholar]
- Babiarz, J. E., J. E. Halley and J. Rine, 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20: 700–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas, S. B., and E. E. Biswas, 1990. ARS binding factor I of the yeast Saccharomyces cerevisiae binds to sequences in telomeric and nontelomeric autonomously replicating sequences. Mol. Cell. Biol. 10: 810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow, J. J., and A. Dutta, 2005. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell. Biol. 6: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscheron, C., L. Maillet, S. Marcand, M. Tsai-Pflugfelder, S. M. Gasser et al., 1996. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 15: 2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Boulton, S. J., and S. P. Jackson, 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17: 1819–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brevet, V., A. S. Berthiau, L. Civitelli, P. Donini, V. Schramke et al., 2003. The number of vertebrate repeats can be regulated at yeast telomeres by Rap1-independent mechanisms. EMBO J. 22: 1697–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C. R., C. S. Wu, Y. Hom and M. R. Gartenberg, 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19: 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, A. S., E. Samal and L. Pillus, 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 17: 1744–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin, A., and J. Rine, 1997. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics 147: 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, D. D., L. R. Davis, S. A. Greenfeder, L. Y. Ong, J. G. Zhu et al., 1991. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell. Biol. 11: 5346–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziak, R., D. Leishman, M. Radovic, B. K. Tye and K. Yankulov, 2003. Evidence for a role of MCM (mini-chromosome maintenance)5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J. Biol. Chem. 278: 27372–27381. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray, A. E., M. Gossen, D. T. Pak, M. R. Botchan and J. Rine, 1995. Separation of origin recognition complex functions by cross-species complementation. Science 270: 1671–1674. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray, A. E., R. T. Kamakaka and J. Rine, 1999. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics 153: 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg, S., C. Civalier and B. K. Tye, 1988. Specific interaction between a Saccharomyces cerevisiae protein and a DNA element associated with certain autonomously replicating sequences. Proc. Natl. Acad. Sci. USA 85: 743–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, T. S., and V. A. Zakian, 2005. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair 4: 1215–1226. [DOI] [PubMed] [Google Scholar]
- Fourel, G., E. Revardel, C. E. Koering and E. Gilson, 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18: 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., F. Magdinier and E. Gilson, 2004. Insulator dynamics and the setting of chromatin domains. BioEssays 26: 523–532. [DOI] [PubMed] [Google Scholar]
- Fox, C. A., A. E. Ehrenhofer-Murray, S. Loo and J. Rine, 1997. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science 276: 1547–1551. [DOI] [PubMed] [Google Scholar]
- Friedman, K. L., M. K. Raghuraman, W. L. Fangman and B. J. Brewer, 1995. Analysis of the temporal program of replication initiation in yeast chromosomes. J. Cell Sci. 19(Suppl.): 51–58. [DOI] [PubMed] [Google Scholar]
- Gilson, E., 1989. Multipurpose role for RAP1, a yeast DNA looping protein. Res. Microbiol. 140: 433–438. [DOI] [PubMed] [Google Scholar]
- Gottschling, D. E., O. M. Aparicio, B. L. Billington and V. A. Zakian, 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63: 751–762. [DOI] [PubMed] [Google Scholar]
- Grunstein, M., 1997. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 9: 383–387. [DOI] [PubMed] [Google Scholar]
- Hoekstra, M. F., and R. E. Malone, 1985. Expression of the Escherichia coli dam methylase in Saccharomyces cerevisiae: effect of in vivo adenine methylation on genetic recombination and mutation. Mol. Cell. Biol. 5: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irlbacher, H., J. Franke, T. Manke, M. Vingron and A. E. Ehrenhofer-Murray, 2005. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 19: 1811–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier, A. L., and J. Rine, 2001. DNA replication-independent silencing in S. cerevisiae. Science 291: 646–650. [DOI] [PubMed] [Google Scholar]
- Labib, K., and J. F. Diffley, 2001. Is the MCM2–7 complex the eukaryotic DNA replication fork helicase? Curr. Opin. Genet. Dev. 11: 64–70. [DOI] [PubMed] [Google Scholar]
- Laroche, T., S. G. Martin, M. Gotta, H. C. Gorham, F. E. Pryde et al., 1998. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 8: 653–656. [DOI] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16: 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun, E., E. Revardel, C. Boscheron, R. Li, E. Gilson et al., 2001. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics 158: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei, M., and B. K. Tye, 2001. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 114: 1447–1454. [DOI] [PubMed] [Google Scholar]
- Li, Y. C., T. H. Cheng and M. R. Gartenberg, 2001. Establishment of transcriptional silencing in the absence of DNA replication. Science 291: 650–653. [DOI] [PubMed] [Google Scholar]
- Liachko, I., and B. K. Tye, 2005. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics 171: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens, Y., and B. Stillman, 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823. [DOI] [PubMed] [Google Scholar]
- Marcand, S., S. W. Buck, P. Moretti, E. Gilson and D. Shore, 1996. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap 1 protein. Genes Dev. 10: 1297–1309. [DOI] [PubMed] [Google Scholar]
- Marsellach, F. X., D. Huertas and F. Azorin, 2006. The multi-KH-domain protein of Saccharomyces cerevisiae Scp160p contributes to the regulation of telomeric silencing. J. Biol. Chem. 281: 18227–18235. [DOI] [PubMed] [Google Scholar]
- Miller, A. M., and K. A. Nasmyth, 1984. Role of DNA replication in the repression of silent mating type loci in yeast. Nature 312: 247–251. [DOI] [PubMed] [Google Scholar]
- Mueller, J. E., M. Canze and M. Bryk, 2006. The requirement for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics 173: 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer, M. A., U. Muller and C. A. Fox, 2003. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 17: 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta, R. J., P. M. Goncalves and W. H. Mager, 1995. Global regulators of ribosome biosynthesis in yeast. Biochem. Cell Biol. 73: 825–834. [DOI] [PubMed] [Google Scholar]
- Pryde, F. E., and E. J. Louis, 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18: 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran, L., D. T. Burhans, P. Laun, J. Wang, P. Liang et al., 2006. Evidence for ORC-dependent repression of budding yeast genes induced by starvation and other stresses. FEMS Yeast Res. 6: 763–776. [DOI] [PubMed] [Google Scholar]
- Roy, N., and K. W. Runge, 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10: 111–114. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516. [DOI] [PubMed] [Google Scholar]
- Sharma, K., M. Weinberger and J. A. Huberman, 2001. Roles for internal and flanking sequences in regulating the activity of mating-type-silencer-associated replication origins in Saccharomyces cerevisiae. Genetics 159: 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter, B., A. Tong, M. Chang, L. Yu, G. W. Brown et al., 2004. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini, B. A., J. J. Carson, J. G. Linger and J. K. Tyler, 2006. Dominant mutants of the Saccharomyces cerevisiae ASF1 histone chaperone bypass the need for CAF-1 in transcriptional silencing by altering histone and Sir protein recruitment. Genetics 173: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham, W. H., and V. A. Zakian, 2002. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene 21: 512–521. [DOI] [PubMed] [Google Scholar]
- Tsuyama, T., S. Tada, S. Watanabe, M. Seki and T. Enomoto, 2005. Licensing for DNA replication requires a strict sequential assembly of Cdc6 and Cdt1 onto chromatin in Xenopus egg extracts. Nucleic Acids Res. 33: 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich, M., C. Liang, H. H. Chen and B. Stillman, 2001. Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle. Proc. Natl. Acad. Sci. USA 98: 11211–11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick, J. J., J. G. Aparicio, T. Chen, J. D. Barnett, E. G. Jennings et al., 2001. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360. [DOI] [PubMed] [Google Scholar]