Abstract
In most organisms, meiotic chromosome segregation is dependent on crossovers (COs), which enable pairs of homologous chromosomes to segregate to opposite poles at meiosis I. In mammals, the majority of meiotic chromosome segregation errors result from a lack of COs between homologs. Observations in Homo sapiens and Drosophila melanogaster have revealed a second class of exceptional events in which a CO occurred near the centromere of the missegregated chromosome. We show that in wild-type strains of Saccharomyces cerevisiae, most spore inviability is due to precocious separation of sister chromatids (PSSC) and that PSSC is often associated with centromere-proximal crossing over. COs, as opposed to nonreciprocal recombination events (NCOs), are preferentially associated with missegregation. Strains mutant for the RecQ homolog, SGS1, display reduced spore viability and increased crossing over. Much of the spore inviability in sgs1 results from PSSC, and these events are often associated with centromere-proximal COs, just as in wild type. When crossing over in sgs1 is reduced by the introduction of a nonnull allele of SPO11, spore viability is improved, suggesting that the increased PSSC is due to increased crossing over. We present a model for PSSC in which a centromere-proximal CO promotes local loss of sister-chromatid cohesion.
THE first division of meiosis is distinct from mitosis and meiosis II in that sister chromatids remain associated with each other, while homologous chromosomes segregate to opposite spindle poles. Proper chromosome segregation at meiosis I depends on crossing over, which establishes chromatin bridges (called chiasmata) between homologs (for review, see Page and Hawley 2003; Petronczki et al. 2005). During prometaphase, homologs can become attached to microtubules from the same or opposite spindle poles. Only attachment to opposite poles results in a stable configuration that is maintained until anaphase. Recognition that chromosomes are properly oriented depends on the mechanical tension that results when homologs are pulled toward opposite spindle poles, and this pulling is resisted by chiasmata. Homologs are held together by chiasmata because sister chromatids are glued to each other in regions distal to chiasmata by sister-chromatid cohesion. Cohesion distal to chiasmata is released at anaphase I, as chromosomes begin their poleward movement. Cohesion near the centromere persists until anaphase II when sister chromatids separate and segregate.
The major cause of aneuploid gametes in mammals and Drosophila is a failure to cross over, and thus failure to form a chiasma, between a pair of homologous chromosomes (MacDonald et al. 1994; Hassold et al. 1996; Koehler et al. 1996; Lamb et al. 1996). Failure to cross over results in random segregation, such that homologs are just as likely to move to the same pole as to opposite poles at anaphase I. When nondisjunction at meiosis I is followed by equational chromosome segregation at meiosis II, two of the resulting products lack a copy of the missegregated chromosome and two are disomic, carrying one chromatid from each homolog [referred to as a meiosis I (MI) disome]. These types of nondisjunction events are extremely rare in wild-type budding yeast, presumably because this organism undergoes high levels of meiotic recombination. However, in yeast mutants with reduced levels of crossing over, these events are common (Ross-Macdonald and Roeder 1994; Sym and Roeder 1994). In addition, yeast artificial chromosomes frequently undergo meiosis I nondisjunction because these chromosomes do not recombine as efficiently as normal chromosomes (Ross et al. 1996).
A second type of meiotic missegregation observed in both mammals and Drosophila leads to disomes containing both chromatids from the same homolog (MII disomes). This type of gamete represents ∼22% of aberrant segregation events in humans and ∼6% in flies (Koehler et al. 1996; Lamb et al. 1996; Hassold and Hunt 2001). Rather than being nonrecombinant, these disomes are associated with crossovers (COs) specifically near the centromere. Although the missegregation event occurs at meiosis II, the predisposing event, a centromere-proximal CO, occurs during meiotic prophase. The exact mechanism underlying this type of segregation has been difficult to decipher due to the inability to analyze all four products of a single meiosis.
Meiotic recombination events are distributed nonrandomly throughout the genome. Genetic interference ensures that COs are placed at some distance from each other. Some regions of the genome undergo higher frequencies of recombination than others (Gerton et al. 2000). In general, the regions around centromeres are cold spots for meiotic recombination. This repression of recombination is dependent on a functional centromere and affects both COs and noncrossovers (NCOs) (Lambie and Roeder 1986, 1988). The deficit in recombination near centromeres is due, at least in part, to a reduction in formation of the double-strand breaks that serve to initiate meiotic recombination (Zenvirth et al. 1992; Baudat and Nicolas 1997).
SGS1 is the sole budding-yeast homolog of the RecQ family of helicases (Gangloff et al. 1994; Watt et al. 1995; Miyajima et al. 2000). sgs1 mutants form inviable spores (60–80% spore viability), apparently because they undergo missegregation leading to disomy (Watt et al. 1995). The random pattern of spore inviability observed in tetrads from sgs1 tetrads is inconsistent with a deficiency of COs. In fact, sgs1 mutants have been shown to undergo a 30–40% increase in meiotic crossing over compared to wild type (Rockmill et al. 2003). Consistent with an increase in crossing over, cytological studies indicate that meiotic nuclei from sgs1 contain an increased number of synapsis inititation complexes (SICs) (Rockmill et al. 2003), which are believed to mark the sites of COs (Agarwal and Roeder 2000; Fung et al. 2004). Further evidence that Sgs1 plays an antagonistic role in meiotic crossing over comes from the observation that sgs1 mutants suppress the CO defect in certain meiotic mutants (Jessop et al. 2006).
We find that the major event leading to spore death in wild-type budding yeast is aneuploidy resulting from precocious separation of sister chromatids (PSSC) at meiosis I. The predominant products are disomes containing nonsister chromatids, and these are often associated with crossing over near the centromere. In addition, we show that the sgs1 mutant undergoes the same type of chromosome missegregation events as wild type, and we provide evidence that the decrease in spore viability in sgs1 is due to the increase in crossing over.
MATERIALS AND METHODS
Strains:
Yeast strains are listed in Table 1. Diploids used to detect spores disomic for chromosome III (BR4316, BR4310, and BR4633) are homozygous for insertion of a single CUP1 gene and an arg4-8ts gene at the LEU2 locus on chromosome III (Spector and Fogel 1992). Both genes are deleted from their normal locations on chromosome VIII. The iTHR1, iHYG, and iNAT markers (“i” indicates insertion at an ectopic location) were constructed by transforming with PCR products containing the markers and 55 nt of pericentromeric DNA to each side to place the markers ∼13 kb to the left of CENIII (iTHR1 at CHR3 position 101398), ∼1.3 kb to the right of CENIII (iHYG at CHR3 position 115843), and ∼12 kb to the right of CENIII (iNAT at CHR3 position 125202). Oligonucleotide sequences are available upon request. The iLEU2 marker 23 kb to the right of CENIII was made by transformation with pPGK1 (Clarke and Carbon 1980). The iURA3 and iADE2 markers were made by transformation with plasmids pJC303-4 (Clarke and Carbon 1983) and pCB432 (Chua and Roeder 1997), respectively.
TABLE 1.
Yeast strains
The URA3 heteroalleles present in BR4633 (Table 1, Figure 1C) were derived from Ura+ haploid segregants of the wild-type diploid BR4316 (Table 1, Figure 1A). Replacing the URA3 gene at the centromere with ura3-1 and ura3-stu alleles was achieved by selection on 5-fluoro-orotic acid to select for uracil auxotrophs (Boeke et al. 1984) after transformation with PCR fragments containing these alleles. The ura3-1 allele was amplified from genomic DNA; the ura3-stu allele was amplified from pAZ2a (Rockmill and Roeder 1990).
Figure 1.—
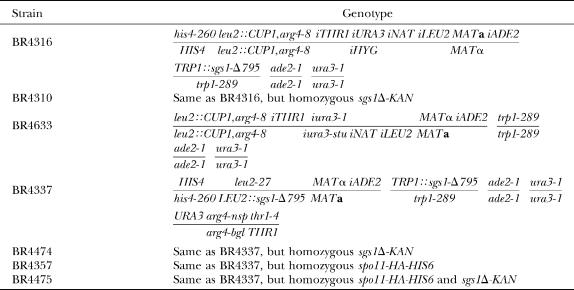
Centromere-proximal crossovers are associated with disomy. (A) Configuration of markers in strains used for the detection and analysis of chromosome III disomy and associated recombination events. (B) Map distances in five intervals from four-spore viable tetrads (converted to random spore data) are compared to map distances found in disomic spores derived from wild type and sgs1-Δ795 (materials and methods). Intervals where map distances in disomic spores are significantly different from map distances in tetrads are indicated by asterisks (*, P < 0.05; **, P ≪ 0.001). (C) Configuration of markers in the strain used to measure COs associated with gene conversion. (D) The frequency of crossing over associated with gene conversion is shown for monosomic Ura+ recombinants and for mating-competent Ura+ disomes. The results from monosomes and disomes are significantly different (P < 0.0001 using chi-square analysis).
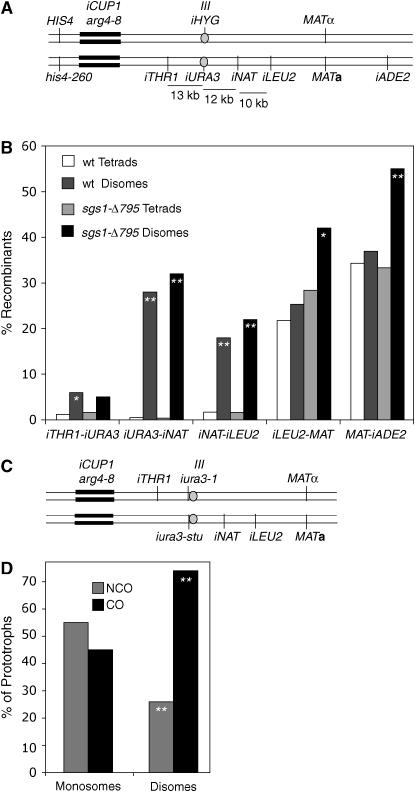
Strains shown in Figure 3A (BR4337, BR4357, BR4474, and BR4475) are diploids in which both haploid parents are isogenic to BR1919-8B (Rockmill and Roeder 1998). Diploids are heterozygous for HIS4 on chromosome III and THR1 on chromosome VIII (constructed by transformation with the corresponding wild-type genes). The iADE2 allele distal to MAT was constructed by transformation with pCB432 (Chua and Roeder 1997). A wild-type copy of LEU2 and a copy of the sgs1-Δ795 truncation allele were inserted at the LEU2 locus by transformation with pRL1 (Mullen et al. 2000). The wild-type TRP1 gene and another copy of the sgs1-Δ795 allele were introduced by transformation with p303 (Rockmill et al. 2003). A wild-type copy of URA3 was introduced upstream of the ARG4 locus by transformation with pMJ177 (Borde et al. 1999). The sgs1-Δ795 strains are homozygous for a complete deletion of the SGS1 open reading frame and carry two insertions of the sgs1-Δ795 allele, one at LEU2 and one at TRP1 (Rockmill et al. 2003). The wild-type strains are identical except that they carry two wild-type copies of SGS1 at the SGS1 locus. The spo11-HA strains were made by transformation with pSK54 (Kee and Keeney 2002). ZIP3-GFP was introduced into haploid segregants that were uracil auxotrophs by transformation with pSA219 (Agarwal and Roeder 2000).
Figure 3.—
SPO11-HA suppresses sgs1-Δ795. (A) Configuration of markers in strains used for analysis of crossing over in various mutant backgrounds. (B) COs were measured in four intervals in wild type (BR4316), sgs1-Δ795 (BR4474), spo11-HA (BR4357), and sgs1-Δ795 spo11-HA (BR4475). Intervals are designated by the color utilized in A. Asterisks denote intervals in which map distances are significantly different from those in sgs1 (P < 0.05). (C) Spread nucleus from sgs1-Δ795 cell at pachytene stained for Zip3-GFP (green) and Zip1 (red) in pachytene. Yellow indicates regions of overlap. Bar, 1 μm. (D) Spread nucleus from sgs1-Δ795 spo11-HA stained as in C. Bar, 1 μm. (E) Zip3-GFP foci were quantified in pachytene-staged chromosome spreads. Asterisks denote strains in which the number of Zip3 foci is significantly different from that of sgs1-Δ795 (P < 0.05 using the t-test). (F) Spore viability. Differences between strains were assessed using the big G-test with spore viability patterns (i.e., four-, three-, two-, one-, and zero-spore viable, not shown). Asterisks denote strains in which spore viability is significantly different from that of sgs1-Δ795.
Genetic analysis:
Map distances were calculated from four-spore viable tetrads and significance (standard error) was calculated using the website: www.groik.com/stahl/. E. Hoffmann and R. Borts supplied a calculator for G-test analysis of spore viability. Comparison of map distances between disomes and tetrads was done by converting tetrad data into random spore data. Thus, map distance was calculated as the number of nonparental ditype (NPD) tetrads plus half the number of tetratype (TT) tetrads divided by the total number of tetrads.
Disomy was measured directly in 1300 tetrads from wild type (BR4316) and 1500 tetrads from an sgs1-Δ795 strain (BR4310). The starting strains are diploids homozygous for a dosage-effect assay that allows chromosome III disomes to be detected (Whittaker et al. 1989). Meiotic segregants that are monosomic for chromosome III are copper sensitive (CuS) and arginine auxotrophic (Arg−), whereas meiotic segregants that are disomic for chromosome III are copper resistant (CuR) and arginine prototrophic (Arg+). Three-spore viable tetrads containing one disomic spore were scored for the segregation of a centromere marker, TRP1, to determine whether the disomic spore had a viable sister. If the sister was viable, then the disomic spore and one other viable spore had the same configuration for TRP1 (i.e., both were Trp+ or Trp−). In addition, disomes were scored for the presence of sister or nonsister chromatids of chromosome III by scoring the centromere markers iURA3 and iHYG. Those that contained both markers were scored as having nonsister chromatids.
HygR Ura+ spores were selected from a pool of purified spores (Rockmill et al. 1991) obtained from wild-type (BR4316) and sgs1-Δ795 (BR4310) diploids. Those testing positive for disomy (i.e., Arg+ and CuR) were scored for homozygosis of recessive markers on chromosome III to assess crossing over. The frequencies of Thr−, NatS, and Leu− spores were doubled to account for the inability to detect the reciprocal products (e.g., LEU2/LEU2).
Ura+ spores were selected from a sporulated wild-type diploid (BR4633) containing URA3 heteroalleles adjacent to CENIII. Recombination between flanking markers was scored directly in 700 Ura+ prototrophs monosomic for chromosome III (i.e., CuS and Arg−). Mating-competent Ura+ disomes (i.e., CuR Arg+) were crossed to a tester strain, and tetrad analysis was carried out to determine the configuration of chromosome III markers. Nonmating disomes were analyzed by PCR to determine heterozygosity of the flanking markers. A set of PCR primers for each marker was used to detect alleles with and/or without the insert (iTHR and iNAT). Mating-competent disomes represent 46% of total disomes.
Cytology:
Meiotic cells were spread and stained as previously described (Rockmill et al. 2003). The number of Zip3 foci was quantitated in nuclei with fully synapsed chromosomes.
RESULTS AND DISCUSSION
Chromosome missegregation in wild type:
Wild-type strains of Saccharomyces cerevisiae produce a low level of inviable spores. To assess the contribution of chromosome missegregation to spore inviability, tetrad analysis was performed using a strain in which meiotic products containing two copies of chromosome III (disomes) can be detected. The starting diploid strain (BR4316, Figure 1A) is homozygous for insertion of the CUP1 gene and the arg4-8ts gene at the LEU2 locus on chromosome III. This strain is sensitive to copper and unable to grow in the absence of arginine at 30°. However, a disomic strain carrying two copies of chromosome III in an otherwise haploid genome is resistant to copper and able to make its own arginine at 30°. Thus, colonies derived from disomic spores can be identified by replica plating to medium containing copper and lacking arginine. The assay is not robust enough to allow disomes to be recovered quantitatively by plating spores on selective medium.
Of 1300 tetrads dissected, 1.5% contained at least one spore disomic for chromosome III. All tetrads containing one or more disomic spores also contained <4 viable spores. To determine whether the observed rate of chromosome missegregation can account for the spore inviability seen in wild type, the predicted level of spore viability was calculated on the basis of the assumption that the frequency of missegregation found for chromosome III is similar for the other 15 chromosome pairs. In 22% (1 − (0.985)16) of meioses, a chromosome is expected to missegregate, rendering at least one meiotic product inviable. Since 1 meiosis results in 4 spores (a tetrad), at least 22 of 400 spores (100 meioses × 4 spores) would be inviable due to aneuploidy, predicting 94.5% spore viability (378/400). The spore viability observed is 91%. Thus, most spore inviability in wild type can be accounted for by aberrant chromosome segregation, if all chromosomes behave like chromosome III.
Missegregation occurs via PSSC:
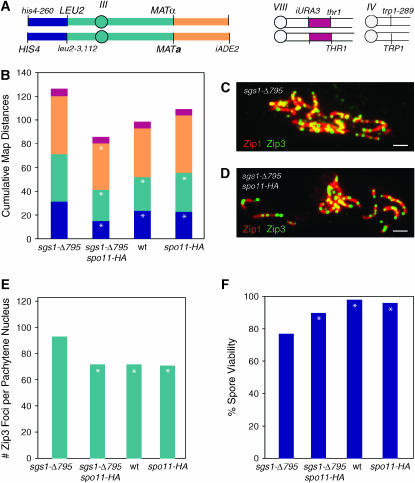
The path of chromosome segregation leading to disomy can be inferred from the analysis of three-spore viable tetrads harboring one disomic spore because all four chromatids are recovered. The segregation of a centromere marker on another chromosome (TRP1 near CENIV) allows the identification of sister spores. In the majority (88%) of three-spore-viable, disome-containing tetrads, the disomic spore has a viable sister spore (Table 2). Furthermore, inspection of the centromere markers on the disomic chromosomes (i.e., iHYG and iURA3, Figure 1A) reveals that most (72%) of the disomes contain nonsister chromosomes (i.e., are Hyg+ Ura+) (Table 2). These observations suggest that for the majority of aberrant meioses, at least one chromosome pair underwent PSSC at meiosis I (Figure 2A).
TABLE 2.
Analysis of disomes recovered from tetrads
| Genotypea | Viable sisterb | Dead sisterb | HygR Ura+ (MI)c | HygR or Ura+ (MII)c |
|---|---|---|---|---|
| Wild type | 8 | 1 | 13 | 5 |
| sgs1-Δ795 | 9 | 1 | 49 | 9 |
Strains analyzed were BR4316 and BR4310.
Tetrads in which three spores were viable and one spore was disomic for chromosome III were analyzed to determine whether the sister spore to the disome was viable.
Disomes recovered from one-, two- and three-spore viable tetrads were scored for containing one or both centromere markers, representing the products of MI and MII missegregation events, respectively.
Figure 2.—
Precocious separation of sister chromatids. (A) PSSC at meiosis I. Shown are two pairs of homologous chromosomes, with each chromosome consisting of two sister chromatids. The larger chromosomes carry the markers used to detect disomes (arg4-8, CUP1, bars with light shading). Circles represent centromeres. At meiosis I, the pair of small chromosomes segregates properly, while one of the large chromosomes undergoes PSSC. At meiosis II, intact pairs of sister chromatids undergo proper sister segregation, but single chromatids segregate randomly. This results in a tetrad in which one spore is disomic, containing nonsister chromatids, and this spore has a viable sister spore. Two spores are euploid, and one is inviable due to aneuploidy. (B) PSSC at meiosis II. Chromosomes segregate properly at meiosis I. At meiosis II, both sister chromatids segregate to the same pole, forming a disomic spore carrying sister chromatids and an inviable sister spore.
COs near centromeres are enriched in disomes:
To analyze a large sample of aneuploid meiotic products, disomic spores were selected from a wild-type diploid (Figure 1A). Disomes were recovered by selecting for both centromere III markers (iURA3 and iHYG) from purified spores. These disomes contain two nonsister chromatids, corresponding to the predominant class of disomes recovered by tetrad analysis. COs in five intervals on chromosome III were scored in these disomes and compared to CO frequencies from four-spore viable tetrads (Tables 3 and 4). Disomes were scored for homozygosis of recessive markers on chromosome III, and this frequency was doubled to account for the inability to detect homozygosis of dominant alleles (e.g., both LEU2/LEU2 and LEU2/leu2 spores are phenotypically Leu+).
TABLE 3.
Tetrad analysis of strains carrying centromere markers
| Genotypea | iTHR1–iURA3 PD:NPD:TTb | cMc | iURA3–iNAT PD:NPD:TTb | cMc | iNAT–iLEU2 PD:NPD:TTb | cMc | iLEU2–MAT PD:NPD:TTb | cMc | MAT–iADE2 PD:NPD:TTb | cMc |
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | 240:0:6 | 1.2 | 909:0:9 | 0.5 | 880:0:32 | 1.8 | 519:4:387 | 22.6 | 343:7:500 | 31.9 |
| sgs1-Δ795 | 202:0:7 | 1.7 | 605:0:5 | 0.4 | 741:0:24 | 1.6 | 220:14:242 | 34.2d | 220:22:218 | 38.0 |
Strains analyzed were BR4316 and BR4310.
PD, parental ditype; NPD, nonparental ditype; TT, tetratype.
Map distances were calculated from four-spore viable tetrads using Perkins' formula (Perkins 1949).
Map distance is significantly different between wild type and mutant.
TABLE 4.
Crossing over in HygR Ura+ MI disomes
| Genotypea | iTHR1–iURA3b (% COc) | iURA3–iNATd (% COc) | iNAT–iLEU2e (% COc) | Total disomes | % CO |
|---|---|---|---|---|---|
| Wild type | 9 (6) | 39 (28) | 26 (18) | 282 | 52 |
| sgs1-Δ795 | 3 (5) | 20 (32) | 14 (22) | 126 | 59 |
Strains analyzed were BR4316 and BR4310.
Disomes that are auxotrophic for threonine represent COs in the iTHR1–iURA3 interval.
To calculate CO frequency, the frequency of recombinants detected was multiplied by two to account for the inability to detect disomes homozygous for dominant alleles.
Disomes that are sensitive to Clonat represent COs in the iURA3–iNAT interval.
Disomes that are auxotrophic for leucine and resistant to Clonat (or sensitive to Clonat and leucine prototrophic) represent COs in the iNAT–iLEU2 interval.
The results in Table 4 and Figure 1B demonstrate a strong, but not absolute, correlation between centromere-proximal recombination events and the occurrence of disomes. For example, among four-spore-viable tetrads (chromosomes properly segregated), the rate of crossing over in the 13-kb iURA3–iNAT interval is 0.5%, but it is 28% in disomes, corresponding to a 56-fold increase. This increase falls off to 10-fold in the adjacent iNAT–iLEU2 interval and 5-fold in the iTHR1–iURA3 interval. Two centromere-distal intervals (iLEU2–MAT and MAT–iADE2) did not display increased COs in disomes. Thus, crossing over in the centromere-proximal region is strongly associated with PSSC. It should be noted, however, that a substantial number of disomes (almost half) do not appear to have undergone a centromere-proximal CO. These disomes may be due to COs that we cannot detect, or they may be unrelated to recombination.
COs, as opposed to NCOs, are preferentially associated with disome formation:
Our data suggest that COs near the centromere interfere with chromosome segregation. However, an alternative possibility is that chromosomes that are prone to missegregate are also more likely to undergo recombination. For example, an altered chromatin structure near the centromere might predispose the chromosome to both events. In this scenario, PSSC should be associated with recombination events in general (including both COs and NCOs), and the relative frequencies of COs vs. NCOs should be the same in both monosomic and disomic spore populations. If, however, COs are preferentially associated with PSSC, then the relative frequency of COs to NCOs should be increased in disomes compared to monosomes.
To distinguish between these possibilities, a diploid was constructed in which mutant URA3 genes are inserted immediately adjacent to CENIII (Figure 1C). Most uracil prototrophs (Ura+) result from gene conversion events within URA3, which may or may not be accompanied by reciprocal exchange between flanking markers. Two different methods were used to measure crossing over in Ura+ disomes. Disomes that were mating competent were crossed to a tester strain, and the configuration of iTHR1 and iNAT markers was determined through the analysis of meiotic progeny (Table 5). Disomes that were nonmaters were analyzed by PCR to score for heterozygosity or homozygosity at iTHR1 and iNAT. By PCR analysis, it is not possible to distinguish disomes containing two recombinant chromatids (class B) from nonrecombinants (class A). These classes can be distinguished by tetrad analysis; thus, mating-competent disomes provide a more complete measure of CO frequency. The frequency of disomes carrying a single recombinant chromatid (classes D and E) is the same in mating-competent disomes as in nonmaters (P = 0.32 for class D; P = 0.41 for class E; P = 1.0 for classes D and E combined), arguing that mating-competent disomes are representative of the population at large. Thus, only mating-competent disomes are considered in the following discussion.
TABLE 5.
Crossing over in disomic Ura+ gene convertants
| Class | Genotype of disome | Matersa (%) | Nonmatersb (%) |
|---|---|---|---|
| A | Het. iTHR1 Het. iNAT (parental configuration)c | 14 (24) | NA |
| B | Het. iTHR1 Het. iNAT (nonparental configuration)d | 20 (34) | NA |
| C | Het. iTHR1 Het. iNAT (configuration unknown)e | NA | 38 (59) |
| D | Hom. iTHR1 Het. iNATf | 14 (24) | 10 (15) |
| E | Het. iTHR1 Hom. iNATf | 9 (16) | 15 (23) |
| F | Hom. iTHR1 Hom. iNATg | 1 (2) | 2 (3) |
| % CO frequency | 74h | >38i (72j) |
Disomic Ura+ gene convertants selected from spores of BR4633 were analyzed for crossing over in the iTHR1-iNAT interval. Het., heterozygous; Hom., homozygous; NA, not applicable.
A total of 58 mating-competent disomes were scored by tetrad analysis.
A total of 65 nonmating disomes were analyzed by PCR.
The two chromatids retained the parental configuration of markers. One carried iTHR1, but not iNAT; the other carried iNAT, but not iTHR1.
Both chromatids displayed a recombinant configuration of markers. One carried both iTHR1 and iNAT; the other chromatid carried neither marker.
Classes A and B cannot be distinguished by PCR analysis.
These classes result from crossing over in the iTHR1–iNAT interval.
This class represents MII disomes carrying sister chromatids.
CO frequency was calculated as the sum of classes B, D, and E divided by the sum of all classes and multiplied by 100.
CO frequency represents the minimum frequency; it is the sum of classes D and E divided by the sum of all classes and multiplied by 100.
CO frequency was calculated on the basis of the assumption that class C contains disomes with the nonparental configuration of markers (i.e., like class C) and that the frequency of these disomes relative to those with the parental configuration (i.e., like class B) is the same as it is among mating-competent disomes. Thus, the CO frequency was calculated as the sum of classes D and E plus 0.59 × class C divided by the sum of all classes and multiplied by 100.
Among Ura+ spores monosomic for chromosome III, 45% had a CO in the CENIII region between iTHR1 and iNAT (Figure 1C). In contrast, among Ura+ spores that were disomic, 74% had undergone a CO in the iTHR1-iNAT region (Table 5; Figure 1C), suggesting that COs (as opposed to NCOs) are more often associated with missegregation.
Among Ura+ disomes, 36% sustained a CO in the iNAT–iLEU interval just to the right of the iTHR1–iNAT interval spanning the centromere (Figure 1C). In total, 88% of disomes had at least one CO in the overall iTHR1–iLEU2 region, supporting the notion that centromere-proximal COs predispose chromosomes to undergo PSSC. In addition, 21% of the disomes sustained two COs in the iTHR1–iLEU2 region. The occurrence of these rare double COs in disomes raises the possibility that multiple recombination events, perhaps even at some distance from the centromere, have an adverse effect on chromosome segregation.
sgs1 mutants display increased COs and increased PSSC:
The sgs1-Δ795 mutant is a deletion of the C-terminal 795 amino acids, which includes the helicase domain (Mullen et al. 2000). The sgs1-Δ795 strains used here carry a deletion of the SGS1 gene at the SGS1 locus; one copy of the sgs1-Δ795 gene is inserted at LEU2 and another at TRP1. The isogenic wild-type strains contain both sgs1-Δ795 inserts, but the wild-type SGS1 gene is present at its normal location on both homologs.
To determine if the spore inviability in sgs1-Δ795 strains is due to PSSC, tetrad analysis was performed on an sgs1-Δ795 strain (BR4310) carrying the chromosome III markers diagrammed in Figure 1A. The frequency of disomes was 3.5%, more than twice the frequency in wild type. This frequency is probably an underestimate, since disomes are particularly unstable in the sgs1-Δ795 background (data not shown), likely leading to a failure to recover a portion of the chromosome III disomes. This rate of disomy formation predicts 86% spore viability, if all chromosomes are equally affected. Spore viability in the sgs1-Δ795 strain is 77%, suggesting that at least half of the spore death occurs via PSSC. In three-spore viable tetrads containing a disome, most disomes (90%) have a viable sister spore and most disomes (84%) contain nonsister chromatids (Table 2), similar to wild type.
To determine whether disomes from sgs1-Δ795 are enriched for crossing over, disomes were selected from an sgs1-Δ795 diploid (BR4310) and examined for CO frequencies on chromosome III. Similar to wild type, disomes from sgs1-Δ795 are enriched for recombination in the centromeric region (Table 4, Figure 1B). Unlike wild type, the centromere-distal intervals also display significant increases in COs in disomic spores. This observation raises the possibility that missegregating chromosomes from sgs1-Δ795 have sustained disproportionate numbers of COs.
In both wild type and sgs1-Δ795, COs in the region to the right of CENIII have a stronger effect on PSSC than those occurring to the left. Interestingly, the region to the right of CENIII is enriched for cohesins and the centromere-binding protein, Sgo1, suggesting that the functional centromere extends beyond the 117-bp CENIII sequence sufficient for centromere activity on a replicating plasmid (Blat and Kleckner 1999; Kiburz et al. 2005). This correspondence between the location of heightened cohesin deposition and the region in which COs predispose to missegregation supports the notion that COs perturb kinetochore function in meiosis, perhaps through disruption of sister-chromatid cohesion.
Reducing COs in sgs1 improves spore viability:
As noted above, crossing over is increased in sgs1-Δ795 strains; it is possible that some of the excess COs occur near centromeres and thus promote PSSC. If the excess COs in sgs1-Δ795 are deleterious, then reducing crossing over should improve spore viability. Spo11 is the endonuclease responsible for catalyzing the double-strand breaks (DSBs) that initiate meiotic recombination (Keeney et al. 1997). In strains homozygous for the tagged SPO11 allele, spo11-HA, the formation of meiotic double-strand breaks is reduced to ∼70% of the wild-type level (Martini et al. 2006, our unpublished data).
Combining the spo11-HA allele with sgs1-Δ795 reduces crossing over to approximately wild-type levels (Table 6, Figure 3B). Accordingly, the number of Zip3 foci is restored to the wild-type level (Figure 3, C–E). Spore viability in the double mutant is significantly increased compared to the sgs1-Δ795 single mutant (Figure 3F), implying that much of the spore death in sgs1-Δ795 mutants is due to excessive meiotic recombination. Taken together with the observations that most chromosome missegregation is due to PSSC and centromere-proximal COs are enriched in disomes, these results suggest that many of the CO events that are deleterious occur near centromeres.
TABLE 6.
Tetrad analysis of spo11-HA diploids
| Genotypea | HIS4–LEU2 PD:NPD:TTb | cMc (foldd) | LEU2–MAT PD:NPD:TTb | cMc (foldd) | MAT–iADE PD:NPD:TTb | cMc (foldd) | ARG4–THR1 PD:NPD:TTb | cMc (foldd) |
|---|---|---|---|---|---|---|---|---|
| Wild type | 218:7:123 | 23.7 (1.0) | 180:5:165 | 27.9 (1.0) | 123:12:228 | 41.3 (1.0) | 298:0:37 | 5.5 (1.0) |
| sgs1-Δ795 | 275:19:187 | 31.3e (1.3) | 193:19:266 | 39.7e (1.4) | 179:34:309 | 49.1 (1.2) | 428:0:60 | 6.1 (1.1) |
| spo11-HA | 379:6:255 | 22.7 (1.0) | 276:10:367 | 32.7 (1.2) | 128:23:278 | 48.5 (1.2) | 584:0:65 | 5.0 (0.9) |
| sgs1-Δ795 spo11-HA | 392:1:160 | 15.0e (0.6) | 318:11:224 | 26.2 (0.9) | 251:24:324 | 39.1 (0.9) | 515:0:61 | 5.3 (1.0) |
Strains analyzed were BR4337, BR4474, BR4357, and BR4475.
PD, parental ditype; NPD, nonparental ditype; TT, tetratype.
Map distances were calculated from four-spore viable tetrads using Perkin's formula (Perkins 1949).
The fold increase in map distance relative to wild type is indicated.
Map distances are significantly different from those in wild type.
These results are consistent with the proposed function for Sgs1 and other RecQ homologs as CO suppressors (Karow et al. 2000; Ira et al. 2003; Wu and Hickson 2003; Hu et al. 2005; Jessop et al. 2006). It is reasonable for vegetative or somatic cells to employ a mechanism to prevent reciprocal recombination events, since loss of heterozygosity can be deleterious and predispose to cancer (Lasko and Cavenee 1991). However, it is less clear why such an activity would be useful during meiosis, where crossing over is essential. One reason, suggested by our results, is that too much crossing over leads to missegregation due to COs near centromeres. In addition, some fraction of recombination events may be aberrant; for example, they might involve interactions between ectopic sequences or they may generate recombination intermediates with unligatable ends. It would be beneficial for the cell to be able to prevent or reverse such interactions. It is possible that aberrant events account for the inability of spo11-HA to restore spore viability to fully wild-type levels in sgs1-Δ795.
Meiotic cells induce about three times as many recombination events as will become COs (Fogel et al. 1979). The phenotype of sgs1 mutants suggests that Sgs1 functions in wild type to reduce the number of COs to the appropriate level. When the number of DSBs is reduced by spo11-HA in an otherwise wild-type background, the level of COs is unchanged (Martini et al. 2006) (Figure 3C). This result suggests the existence of a mechanism to ensure that the wild-type level of COs is attained, despite a reduction in the total number of initiated events, a phenomenon referred to as “CO homeostasis.” Perhaps Sgs1 and CO homeostasis are part of the same mechanism. In strains with a reduced level of DSBs, CO homeostasis may act by downregulating Sgs1 activity. This interaction could explain why the sgs1-Δ795 mutation has little effect in a spo11-HA background.
Disomes containing sister chromatids:
Although most of the disomes recovered from tetrad analysis are composed of nonsister chromatids, a significant number contain sister chromatids (Table 3). Disomes of this type are difficult to recover because they cannot be selected from random spores (they are not HygR Ura+). Nevertheless, information is available from the limited number of disomes detected during tetrad analysis. From wild-type and sgs1 strains combined, a total of 14 disomes containing sister chromatids were recovered; 6 of these experienced a CO in the CENIII region. Thus, centromere-proximal COs are associated with both MI and MII disomes. Segregation could be determined for four tetrads in which three spores were viable. In one tetrad, the chromatids segregated to the same pole at meiosis II, as shown in Figure 2B. Interestingly, in the three remaining tetrads, both pairs of chromosomes III missegregated. One pair segregated to opposite poles at meiosis I as shown in Figure 2A, and the other pair segregated to the same pole at meiosis II as shown in Figure 2B. In all three cases, this resulted in a tetrad in which the disome containing sister chromatids had a viable sister spore.
Meiotic PSSC in other organisms:
As noted in the Introduction, the major source of meiotic aneuploidy in Drosophila and humans results from the failure to cross over, leading to nondisjunction at meiosis I (Koehler et al. 1996; Orr-Weaver 1996; Hassold and Hunt 2001; Lamb et al. 2005). We have shown that this is not the case in yeast, since most meiotic aneuploidy results from PSSC and the missegregating chromatids undergo increased crossing over near the centromere and normal levels of crossing over elsewhere.
A second type of missegregation observed in flies and humans results in disomes containing sister chromatids and is associated with recombination near the centromere (Koehler et al. 1996; Lamb et al. 1996). Two models have been proposed to account for these events (Koehler et al. 1996). The first model suggests that an entanglement of the bivalent resulting from retention of sister-chromatid cohesion prevents segregation at meiosis I. Both homologs go to the same pole at meiosis I and then undergo reductional segregation at meiosis II. This model predicts the formation of two viable meiotic products, both of which are disomes containing sister chromatids. A second model proposes that sister chromatids separate at anaphase I. Nevertheless, both sister chromatids usually continue to the same pole at meiosis I, and this is followed by random segregation at meiosis II. Occasionally, however, the chromatids segregate away from each other at meiosis I to generate disomic gametes containing nonsister chromatids.
The ability to do tetrad analysis in yeast allows us to distinguish between these different patterns of chromosome missegregation. We observed no examples of tetrads predicted by the first model (i.e., two viable disomic spores containing sister chromatids), implying that this model does not apply to yeast. Yeast disomes predominantly contain nonsister chromatids, but a substantial number have sister chromatids. It is likely that the same mechanism underlies both events, since both types of disomes are associated with centromere-proximal COs.
It is attractive to assume that PSSC events associated with centromere-proximal COs occur by the same mechanism in yeast and animals. Our results support a model in which chromatid separation is initiated at meiosis I, but the chromatids can segregate away from each other either at meiosis I or at meiosis II (Figure 2). The difference between PSSC in animals and yeast may simply be the timing of chromatid separation. If chromatids do not fully separate until homologs have already begun to move away from each other at meiosis I, then the two sisters may continue their movement to the same pole. In this case, the resulting disomes would carry sister chromatids, as is most often the case in mammals and flies (Figure 2B). However, if the chromatids separate at or just prior to the metaphase I to anaphase I transition, then the sisters may segregate away from each other at the first division. In this case, the resulting disomes would carry nonsister chromatids, as is usually the case in yeast (Figure 2A).
Perhaps the difference in the timing of sister chromatid separation during PSSC events in yeast vs. animals reflects underlying differences in kinetochore structure. Whereas each kinetochore in budding yeast is attached to a single microtubule, the kinetochores of higher eukaryotes have multiple microtubules attached (Joglekar et al. 2006). Disrupting the monopolar orientation of a chromosome with a compound kinetochore might be more difficult than disrupting a single microtubule attachment.
COs may disrupt centromere structure:
Our studies have revealed that COs are strongly associated with PSSC, suggesting that COs can lead to loss of the pericentric cohesion needed for proper alignment of homologs at metaphase I. Indeed, cytological observations of chiasmata reveal that sister-chromatid cohesion is disrupted at CO sites (e.g., Eijpe et al. 2003; Parra et al. 2004; Kleckner 2006).
A recent model of centromere structure (Bloom et al. 2006) postulates the existence of an intramolecular loop resulting from the chromatid folding back on itself in the region of the centromere. The site of microtubule attachment is located at the tip of the loop, and the loop is stabilized by cohesins. The loop is believed to be dynamic, growing shorter or longer, as more or less of the chromatid is incorporated into the loop. The authors postulate that the dynamic nature of the loop plays a role in the tension-based checkpoint mechanism that ensures proper chromosome segregation. COs that occur close to the centromere might interfere with formation and/or expansion of the loop, thus predisposing the affected chromosome to missegregate.
The correlation between pericentric COs and PSSC found in yeast, as well as in Drosophila and mammals, suggests that limiting the number of COs near the centromere may be a strategy that organisms employ to safeguard against loss of centromeric cohesion.
Acknowledgments
We are grateful to Eva Hoffmann and members of the Roeder laboratory for helpful discussions. We thank Scott Keeney for sharing data and plasmids. This work was supported by the Howard Hughes Medical Institute.
References
- Agarwal, S., and G. S. Roeder, 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. [DOI] [PubMed] [Google Scholar]
- Baudat, F., and A. Nicolas, 1997. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc. Natl. Acad. Sci. USA 94: 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blat, Y., and N. Kleckner, 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98: 249–259. [DOI] [PubMed] [Google Scholar]
- Bloom, K., S. Sharma and N. V. Dokholyan, 2006. The path of DNA in the kinetochore. Curr. Biol. 16: R276–R278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J. D., F. Lacroute and G. R. Fink, 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197: 345–346. [DOI] [PubMed] [Google Scholar]
- Borde, V., T.-C. Wu and M. Lichten, 1999. Use of a recombination reporter insert to define meiotic recombination domains on chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 19: 4832–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua, P. R., and G. S. Roeder, 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 11: 1786–1800. [DOI] [PubMed] [Google Scholar]
- Clarke, L., and J. Carbon, 1980. Isolation of the centromere-linked CDC10 gene by complementation in yeast. Proc. Natl. Acad. Sci. USA 77: 2173–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, L., and J. Carbon, 1983. Genomic substitutions of centromeres in Saccharomyces cerevisiae. Nature 305: 23–28. [DOI] [PubMed] [Google Scholar]
- Eijpe, M., H. Offenberg, R. Jesseberger, E. Revenkova and C. Heyting, 2003. Meiotic cohesin REC8 marks the axial elements of rat synaptonemal complexes before cohesins SMC1β and SMC3. J. Cell Biol. 160: 657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel, S., R. Mortimer, K. Lusnak and F. Tavares, 1979. Meiotic gene conversion: a signal of the basic recombination event in yeast. Cold Spring Harbor Symp. Quant. Biol. 43: 1325–1341. [DOI] [PubMed] [Google Scholar]
- Fung, J. C., B. Rockmill, M. Odell and G. S. Roeder, 2004. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116: 795–802. [DOI] [PubMed] [Google Scholar]
- Gangloff, S., J. P. McDonald, C. Bendixen, L. Arthur and R. Rothstein, 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton, J. L., J. DeRisi, R. Shroff, M. Lichten, P. O. Brown et al., 2000. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97: 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold, T., and P. Hunt, 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2: 280–284. [DOI] [PubMed] [Google Scholar]
- Hassold, T. H., M. Abruzzo, K. Adkins, D. Griffin, M. Merrill et al., 1996. Human aneuploidy: incidence, origin, and etiology. Environ. Mol. Mutagen. 28: 167–175. [DOI] [PubMed] [Google Scholar]
- Hu, Y., X. Lu, E. Barnes, M. Yan, H. Lou et al., 2005. Recq15 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell. Biol. 25: 3431–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira, G., A. Malkova, G. Liberi, M. Foiani and J. E. Haber, 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop, L., B. Rockmill, S. Roeder and M. Lichten, 2006. Synaptonemal complex proteins modulate Sgs1 meiotic anti-crossover activity. PLoS Genet. 2: 1402–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar, A. P., D. C. Bouck, J. N. Molk, K. S. Bloom and E. D. Salmon, 2006. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 5: 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow, J. K., A. Constantinou, J.-L. Li, S. C. West and I. D. Hickson, 2000. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl. Acad. Sci. USA 97: 6504–6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee, K., and S. Keeney, 2002. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney, S., C. N. Giroux and N. Kleckner, 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kiburz, B. M., D. B. Reynolds, P. C. Megee, A. L. Marston, B. H. Lee et al., 2005. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 19: 3017–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner, N., 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115: 175–194. [DOI] [PubMed] [Google Scholar]
- Koehler, K. E., C. L. Boulton, H. E. Collins, R. L. French, K. C. Herman et al., 1996. Spontaneous X chromosome MI and MII nondisjunction events in Drosophila melanogaster oocytes have different recombinational histories. Nat. Genet. 14: 406–414. [DOI] [PubMed] [Google Scholar]
- Lamb, N. E., S. B. Freeman, A. Savage-Austin, D. Pettay, L. Taft et al., 1996. Susceptible chiasmate configurations of chromosome 21 predispose to non-disjunction in both maternal meiosis I and meiosis II. Nat. Genet. 14: 400–405. [DOI] [PubMed] [Google Scholar]
- Lamb, N. E., K. Yu, J. Shaffer, E. Feingold and S. L. Sherman, 2005. Association between maternal age and meiotic recombination for trisomy 21. Am. J. Hum. Genet. 76: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie, E. J., and G. S. Roeder, 1986. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics 114: 769–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambie, E. J., and G. S. Roeder, 1988. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell 52: 863–873. [DOI] [PubMed] [Google Scholar]
- Lasko, D., and W. Cavenee, 1991. Loss of constitutional heterozygosity in human cancer. Annu. Rev. Genet. 25: 281–314. [DOI] [PubMed] [Google Scholar]
- MacDonald, M., T. Hassold, J. Harvey, L. H. Wang, N. E. Morton et al., 1994. The origin of 47,XXY and 47,XXX aneuploidy: heterogeneous mechanisms and role of aberrant recombination. Hum. Mol. Genet. 3: 1365–1371. [DOI] [PubMed] [Google Scholar]
- Martini, E., R. Diaz, N. Hunter and S. Keeney, 2006. Crossover homeostasis in yeast meiosis. Cell 126: 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima, A., M. Seki, F. Onoda, A. Ui, Y. Satoh et al., 2000. Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet. Syst. 75: 319–326. [DOI] [PubMed] [Google Scholar]
- Mullen, J. R., V. Kaliraman and S. J. Brill, 2000. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics 154: 1101–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver, T. L., 1996. Meiotic nondisjunction does the two-step. Nat. Genet. 14: 374–376. [DOI] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: The meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Parra, M. T., A. Viera, R. Gomez, J. Page, R. Benavente et al., 2004. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 117: 1221–1234. [DOI] [PubMed] [Google Scholar]
- Perkins, D. D., 1949. Biochemical mutants in the smut fungus Ustilago maydis. Genetics 34: 607–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki, M., M. F. Siomos and K. Nasmyth, 2005. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112: 423–440. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1990. Meiosis in asynaptic yeast. Genetics 126: 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and G. S. Roeder, 1998. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 12: 2574–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., E. J. Lambie and G. S. Roeder, 1991. Spore enrichment. Methods Enzymol. 194: 146–149. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., J. C. Fung, S. S. Branda and G. S. Roeder, 2003. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13: 1954–1962. [DOI] [PubMed] [Google Scholar]
- Ross, L. O., R. Maxfield and D. Dawson, 1996. Exchanges are not equally able to enhance meiotic chromosome segregation in yeast. Proc. Natl. Acad. Sci. USA 93: 4979–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Macdonald, P., and G. S. Roeder, 1994. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79: 1069–1080. [DOI] [PubMed] [Google Scholar]
- Spector, L. M., and S. Fogel, 1992. Mitotic hyperploidy for chromosome VIII and III in Saccharomyces cerevisiae. Curr. Genet. 21: 309–318. [DOI] [PubMed] [Google Scholar]
- Sym, M., and G. S. Roeder, 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Watt, P. M., E. J. Louis, R. H. Borts and I. D. Hickson, 1995. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81: 253–260. [DOI] [PubMed] [Google Scholar]
- Whittaker, S., B. Rockmill, A. Blechl, D. Maloney, M. Resnick et al., 1989. A genetic system for the detection of meiotic aneuploidy. Mol. Gen. Genet. 215: 10–18. [DOI] [PubMed] [Google Scholar]
- Wu, L., and I. D. Hickson, 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Zenvirth, D., T. Arbel, A. Sherman, M. Goldway, S. Klein et al., 1992. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 11: 3441–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]