Abstract
Why sex is maintained in nature is a fundamental question in biology. Natural genetic transformation (NGT) is a sexual process by which bacteria actively take up exogenous DNA and use it to replace homologous chromosomal sequences. As it has been demonstrated, the role of NGT in repairing deleterious mutations under constant selection is insufficient for its survival, and the lack of other viable explanations have left no alternative except that DNA uptake provides nucleotides for food. Here we develop a novel simulation approach for the long-term dynamics of genome organization (involving the loss and acquisition of genes) in a bacterial species consisting of a large number of spatially distinct populations subject to independently fluctuating ecological conditions. Our results show that in the presence of weak interpopulation migration NGT is able to subsist as a mechanism to reload locally lost, intermittently selected genes from the collective gene pool of the species through DNA uptake from migrants. Reloading genes and combining them with those in locally adapted genomes allow individual cells to readapt faster to environmental changes. The machinery of transformation survives under a wide range of model parameters readily encompassing real-world biological conditions. These findings imply that the primary role of NGT is not to serve the cell with food, but to provide homologous sequences for restoring genes that have disappeared from or become degraded in the local population.
SEXUAL reproduction is a process that brings genomes, or portions of genomes, from different individuals into a common cell, producing a new combination of genes: in eukaryotes, this occurs as a result of fertilization and meiotic recombination; in bacteria, it happens as a result of the acquisition of exogenous DNA. The ubiquity of genetic transfer in bacteria is reflected in the dynamic structure of their genomes, which are constantly being shaped by two opposing forces: selection for shorter length (favoring DNA loss through deletion) and selection for gene function (driving genome loading by the acquisition of exogenous DNA) (Vellai et al. 1999; Mira et al. 2001). The balance of these forces results in most bacteria having highly economized genomes with only a small fraction (∼10%) of noncoding sequences (Vellai et al. 1998; Mira et al. 2001). DNA transfer into the bacterial cell can occur in three ways: (i) transduction by viruses, (ii) conjugation by plasmids, and (iii) natural genetic transformation (NGT) by developing competence, a regulated physiological state in which the bacterial cell is able to take up DNA fragments released by another cell (Avery et al. 1944; Solomon and Grossman 1996). The genetic elements responsible for transduction and conjugation primarily survive as parasites and are located on viral and plasmid DNA. The genes required for competence are, however, located on the bacterial chromosome, placing NGT under the direct control of the cell. While all three mechanisms play a role in rare gene transfer events between bacteria of different species, termed horizontal gene transfer, NGT is the most significant source of active and frequent genetic transfer within a species (for a comparative review of the three processes see Thomas and Nielsen 2005). In bacteria capable of NGT, alleles typically change more frequently by recombination (e.g., 5- to 10-fold in Streptococcus pneumoniae and Neisseria meningitidis) than by mutation (Levin and Bergstrom 2000; Feil et al. 2001; Feil 2004; Thomas and Nielsen 2005). It is this combination of high-throughput genetic mixing among members of the same species and direct cellular control that is responsible for NGT often being referred to as the bacterial analog of meiotic sex in eukaryotes (Maynard Smith et al. 1991; Maynard Smith 1993).
The persistence of NGT raises the same question as the prevalence of meiotic sex (Bernstein et al. 1985; Elena and Lenski 1997; Butlin 2002; Otto and Lenormand 2002): What is the short-term advantage of genetic mixing to the individual? NGT is obviously costly, not just because the machinery of DNA uptake must be maintained, but also because bacteria undergoing transformation face the risk of incorporating defective alleles (Redfield et al. 1997). And while the long-term implications of NGT—for genome adaptation (Cohen et al. 2005) and diversification (Holmes et al. 1999; Vestigian and Goldenfeld 2005)—are clear, the short-term advantage to individuals (an advantage necessary to maintain NGT) remains elusive (Redfield 2001).
In this work we aim to investigate the role of NGT in bacteria and the necessary conditions for its short-term maintenance. The ability to uptake naked DNA through NGT has been detected across a wide phylogenetic spectrum, ranging from archaea through divergent subdivisions of bacteria, including representatives from gram-positive bacteria, cyanobacteria, Thermus spp., green sulfur bacteria, and many other gram-negative bacteria (Thomas and Nielsen 2005). The details of transformation, however, vary widely among bacteria of different species. With the exception of N. gonorrhoeae most naturally transformable bacteria develop time-limited competence in response to specific environmental conditions such as altered growth conditions, nutrient access, cell density (by quorum sensing), or starvation. The conserved ability among a wide range of bacteria to acquire DNA molecules through NGT indicates that the genetic trait is functionally important in the environment, enabling access to DNA as a source of nutrients or genetic information (Thomas and Nielsen 2005).
So far it has been convincingly demonstrated that NGT's role in repairing deleterious mutations under constant selection is insufficient for its survival (Redfield et al. 1997). The lack of other viable explanations has left no alternative except that the uptake of DNA provides nucleotides for food (Redfield 2001). This, however, is difficult to reconcile with the facts that one of the strands is taken up intact (despite the apparent risks of degradation inside the cell) and that highly specific sequences are required for the binding and uptake of DNA in some bacterial species, e.g., N. gonorrhoeae or Haemophilus influenzae (Solomon and Grossman 1996) (even though nucleotides from other sources confer the same nutritional benefits). To understand the relevance of NGT as a vehicle of genetic information one must take into account not only a single population, but also a collection of populations living under diverse and constantly changing ecological conditions, as only these together possess the complete set of genes common to the species (Woese 1998; Ochman et al. 2000). To find the short-term advantage that maintains NGT, we have to consider its role in allowing genetic mixing between populations facing variable selection.
MODEL
The dynamics of genome organization in bacteria:
We view a bacterial species as a metapopulation (Hanski 1998) that is composed of a large number of spatially distinct populations living under varying ecological conditions. Different populations experience selection for different combinations of numerous possible environmental factors [availability of particular metabolites, host recognition (Claverys et al. 2000), and others (Nakamura et al. 2004)]. Most of these factors fluctuate in time over a broad range of timescales (starting from daily weather changes, through seasonal alternations and decades-long host life cycles, up to long-scale climate changes, to mention just a few). The populations are, on one hand, constantly adapting to their own locally changing environments and, on the other hand, connected by weak interpopulation migration that can span large distances (even continents and oceans). What we aim to show is that under these conditions the advantage of NGT may lie in providing locally adapted populations with the ability to respond to environmental changes by importing genes from the collective gene pool of the species through taking up DNA from migrants. This way bacteria can economize their genomes by disposing of genes that are not currently in use in the local environment and picking up those that have just become useful. There is a growing body of experimental evidence showing that such genetic plasticity plays a central role in the adaptation of bacteria, the most well-studied examples being virulence-related genetic diversity (particularly of the genes responsible for capsule composition) in S. pneumoniae (Claverys et al. 2000) and the hypervariable region of Helicobacter pylori (Alm et al. 1999) responsible for different pathophysiologies associated with chronic H. pylori infection in humans.
Modeling a bacterial species:
To test our hypothesis quantitatively, we consider a model of a bacterial species (Figure 1) that is capable of living on 10 different types of food (denoted by A, B,…,J). Individual bacteria can utilize any of these foods only if they have the corresponding model food gene (a, b,…,j, respectively) present in their genomes. Each such model gene is understood to represent the complete group of genes necessary for the utilization of a particular resource type. While under natural conditions there may exist dozens of such resource–gene group pairs, numerical treatment of the model rapidly becomes intractable as this number is increased, forcing us to consider only a limited number of food types and gene groups. The relatively small model genomes used in our simulations (typically a few model genes long) intend to represent the much larger genomes of real bacteria. As a consequence, model gene numbers and model genome lengths have to be suitably rescaled when interpreting the results of the model.
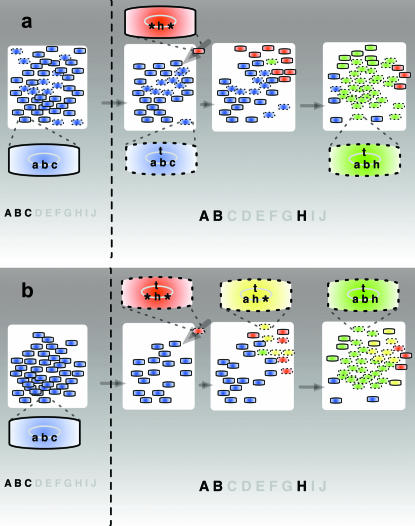
Figure 1.—
Schematic of the two main processes responsible for the spread of NGT. In both scenarios local food types change from ABC to ABH. Bacteria with the most viable genotypes are shown with the relevant model genes indicated inside, and those capable of NGT are emphasized by “perforated” borders. (a) Under rapidly changing conditions the number of bacteria with the t (transformation) model gene usually remains significant (cf. Figure 2b). Newly arrived bacteria possessing the model food gene h (red cells) spread due to a lack of competition, subsequently allowing local bacteria capable of NGT (blue cells with perforation) to successfully adapt by assembling the optimal combination of model genes abh + t (green cells with perforation), and to outcompete all the others, while also spreading the t model gene. (b) Under slowly changing conditions, gene t often disappears from the local population (cf. Figure 2b). It is, however, possible that a competent bacterium possessing gene h (red cells with perforation) arrives from another population. Progeny of this migrant subsequently adapt and outgrow the local bacteria by incrementally (indicated by yellow and then green coloring) assembling the best adapted genome, abh + t (green cells with perforation), which also results in the spread of the model t gene.
At any moment only 3 of the 10 possible food sources are available, but their types are changing in time independently in each population of the metapopulation. This food change is characterized by the common rate Rfood, at which one of the food sources is randomly replaced by another one not currently available in the population (e.g., ABC changes to ABH and then to ADH, etc.). Bacteria reproduce at a rate rr (maximized at 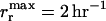 ) that depends on the amount of available food they can utilize and decreases with the number of functional model food genes possessed (to impose genome economization); for details see methods. Bacteria can also be washed out of their populations at a fixed rate rw = 10−2 hr−1, lose any of their functional genes by mutation at a rate rm = 10−4 hr−1 per model gene, and those possessing a functional copy of the model gene for NGT (denoted by t) attempt to incorporate exogenous DNA into their genome from the surrounding medium at a rate rt = 10−3 hr−1 per model gene. Transformation and mutation are considered at the level of model genes (a–j and t). That is to say, a single mutation event in the context of our model encompasses a series of events starting with a deleterious mutation (either a point mutation or a deletion), leading to loss of function and continuing with the subsequent gradual loss of the gene group responsible for the specific function through repeated deletion events. In other words, we consider a given group of genes to be only either present and fully functional or completely absent. We model transformation in a similar fashion: a bacterium may acquire or lose a complete functioning copy of any model gene as a result of a single transformation event. Note that the gene group responsible for transformation (the model t gene) is capable of eliminating even itself by taking up a defective copy from the environment. While the rates of a series of events leading to the acquisition or loss of a complete gene group (a single model gene) responsible for a given function are probably orders of magnitude smaller than those considered in our model, we argue in detail below (see results) that this does not affect the validity of the results obtained.
) that depends on the amount of available food they can utilize and decreases with the number of functional model food genes possessed (to impose genome economization); for details see methods. Bacteria can also be washed out of their populations at a fixed rate rw = 10−2 hr−1, lose any of their functional genes by mutation at a rate rm = 10−4 hr−1 per model gene, and those possessing a functional copy of the model gene for NGT (denoted by t) attempt to incorporate exogenous DNA into their genome from the surrounding medium at a rate rt = 10−3 hr−1 per model gene. Transformation and mutation are considered at the level of model genes (a–j and t). That is to say, a single mutation event in the context of our model encompasses a series of events starting with a deleterious mutation (either a point mutation or a deletion), leading to loss of function and continuing with the subsequent gradual loss of the gene group responsible for the specific function through repeated deletion events. In other words, we consider a given group of genes to be only either present and fully functional or completely absent. We model transformation in a similar fashion: a bacterium may acquire or lose a complete functioning copy of any model gene as a result of a single transformation event. Note that the gene group responsible for transformation (the model t gene) is capable of eliminating even itself by taking up a defective copy from the environment. While the rates of a series of events leading to the acquisition or loss of a complete gene group (a single model gene) responsible for a given function are probably orders of magnitude smaller than those considered in our model, we argue in detail below (see results) that this does not affect the validity of the results obtained.
The frequency of model gene fragments in the surroundings is approximated by that in the living individuals of the population (Cohen et al. 2005). This is consistent with the assumption that the death of bacteria is largely independent of their gene composition and with experiments showing that DNA fragments from lysed bacteria persist for hours to days—as measured in natural transformation assays (Thomas and Nielsen 2005)—a timescale that is too short to allow extracellular DNA to survive the time period between food changes in our model. For details of how the competition of individual bacteria with each possible genotype was modeled see methods.
Migration between the populations is a crucial element of the model, because without it NGT would just futilely reshuffle the existing genes inside the populations. To see this, let us suppose that in a fraction P of a population a certain gene has become defective. Then the frequency of repairing this defective gene by taking up a functional copy from a recently deceased member of the population, rtP(1 − P), is the same as that of accidentally replacing a functional gene by a defective one, rt(1 − P)P. Thus, lacking any short-term advantage, NGT would rapidly disappear, in agreement with the results of Redfield et al. (1997). If, however, we take into account migration, it is able to facilitate the spread of gene t, as illustrated in the context of our model in Figure 1.
Self-consistent migration:
Assuming that bacteria can migrate long distances (such that within this distance a large number of populations exist with independently changing food sources), migration can be taken into account very efficiently in terms of a mean-field approach, commonly used in statistical physics. This means that the genotype distribution averaged over the populations within the migrational range can be well approximated by the time average of the genotype distribution of any single population. In short, spatial and temporal averages are interchangeable. Consequently, it is enough to consider only one population and use its own history to compute the genotype distribution of the arriving migrants. Coupling back, through migration, an ever increasing fraction of a single population's past into its own dynamics results in the convergence of the genotype distribution of the metapopulation to its stationery value in a self-consistent manner, as described in more detail in the following section. The influx of migrants (the number of incoming bacteria per unit time) is defined in the model as the product of the migration rate Rmigr and the average size of the population N (which was in the order of 108 in our simulations).
METHODS
Population dynamics:
Our population dynamics simulations were carried out in a manner that allowed the separate treatment of every bacteria possessing any of the possible model genotypes. The frequency of individual bacteria of each genotype was calculated by solving the 210+1 differential equations describing the number of bacteria n(G) in each of the 210+1 genome states G,
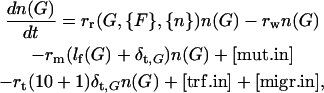 |
(1) |
where lf(G) is the number of functional model food genes in genotype G, and δx,G is equal to unity if G contains an intact model gene x and zero otherwise. n(G) is treated as a continuous variable, but with a lower cutoff at n(G) = 1 to mimic the discrete nature of bacterial populations. This lower cutoff is implemented for each subpopulation with n(G) < 1, by resetting n(G) to 1 with probability n(G) or to 0 with probability 1 − n(G), at each time step of the simulation. The first term on the right-hand side describes the reproduction of bacteria as detailed below. The second term corresponds to the washout of bacteria from the population. The third and fourth terms deal with mutation: the former one considers bacteria with genotype G that undergo mutation, while the latter one [mut.in] is the sum of contributions from all bacteria that mutate into state G. The fifth and sixth terms describe transformation, the former one corresponding to those bacteria in state G that undergo transformation, while the latter one [trf.in] represents the complicated sum of all transformations that lead to state G. The last term [migr.in] corresponding to migration is also described below.
Reproduction rate:
The fitness of individual bacteria with a given genotype G is influenced by available food {F} ≡ F1, F2, F3, as well as by genome length l(G) = lf(G)+ltδt,G, where lt denotes the length of the t gene relative to the model food genes (and is considered to be less than unity, as explained below). Available food types Fi were each considered to be constantly replenished in the environment, each type being characterized by a strength Si corresponding to the number of bacteria the given food type could sustain at the maximum division rate 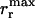 . Each bacterium with a functional copy of model gene fi (corresponding to a given food type Fi) present in its genome (
. Each bacterium with a functional copy of model gene fi (corresponding to a given food type Fi) present in its genome (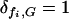 ) received an “equal share”
) received an “equal share” 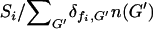 of this food, while those bacteria that did not have an intact copy of the model gene (
of this food, while those bacteria that did not have an intact copy of the model gene (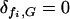 ) in their genome received none. In general the speed at which a given bacterium can divide rr(G, {F}, {n}) is proportional to the total amount of food it can utilize
) in their genome received none. In general the speed at which a given bacterium can divide rr(G, {F}, {n}) is proportional to the total amount of food it can utilize 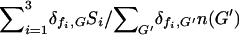 , but also decreases by a factor of 2/(1 + l(G)/lopt) as the total number of intact model food genes present in its genome increases, where lopt = 3 is the optimal genome size. The denominator of the latter factor takes into account that the time necessary for a bacterium to divide has a component proportional to genome length. Bacteria that posses only those three model genes that are necessary for the utilization of the three food types available at a given moment are the ones that divide the fastest (i.e., have the highest fitness), their genomes being the most highly economized. As outlined in model real bacteria may posses several dozen gene groups necessary for the utilization of fluctuating resources, but as we are not able to treat numerically more than a few model gene–food pairs, we have chosen the parameters of the fitness function such that it decreases rapidly as the number of model genes grows (e.g., by 33% if twice the optimal number of model genes is present); that is, we, in this sense, consider each model gene to correspond to several gene groups. In addition, as the gene group responsible for DNA uptake (the t model gene) is only one of several dozen and in reality consists of a relatively small number of genes, we have included it with a smaller relative length (lt = 0.1) in the total genome length. Due to practical constraints, however, the mutation rate of the t gene was considered (except for the data presented in Figure 3c) to be the same as that of the food genes (which corresponds to a 10-fold increase in the mutation rate for the t gene). Therefore, in our simulations the reproduction rate of a bacterium with genotype G and available foods {F} was given by
, but also decreases by a factor of 2/(1 + l(G)/lopt) as the total number of intact model food genes present in its genome increases, where lopt = 3 is the optimal genome size. The denominator of the latter factor takes into account that the time necessary for a bacterium to divide has a component proportional to genome length. Bacteria that posses only those three model genes that are necessary for the utilization of the three food types available at a given moment are the ones that divide the fastest (i.e., have the highest fitness), their genomes being the most highly economized. As outlined in model real bacteria may posses several dozen gene groups necessary for the utilization of fluctuating resources, but as we are not able to treat numerically more than a few model gene–food pairs, we have chosen the parameters of the fitness function such that it decreases rapidly as the number of model genes grows (e.g., by 33% if twice the optimal number of model genes is present); that is, we, in this sense, consider each model gene to correspond to several gene groups. In addition, as the gene group responsible for DNA uptake (the t model gene) is only one of several dozen and in reality consists of a relatively small number of genes, we have included it with a smaller relative length (lt = 0.1) in the total genome length. Due to practical constraints, however, the mutation rate of the t gene was considered (except for the data presented in Figure 3c) to be the same as that of the food genes (which corresponds to a 10-fold increase in the mutation rate for the t gene). Therefore, in our simulations the reproduction rate of a bacterium with genotype G and available foods {F} was given by
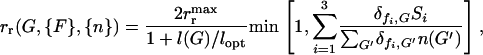 |
(2) |
and we chose the food strength to be Si = 105 for each available food type.
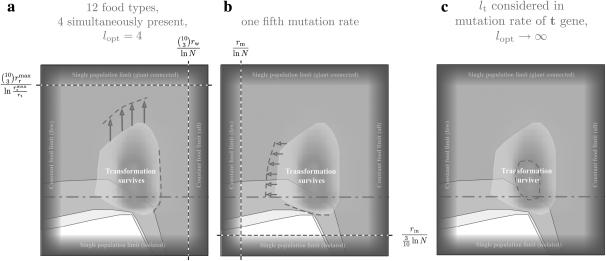
Figure 3.—
Robustness of the proposed mechanism for the survival of NGT. We investigated the robustness of the survival range of NGT by varying several model parameters. (a) We changed the number of possible food types from 10 to 12 and the number simultaneously present from 3 to 4, consequently also setting lopt = 4. The survival range of NGT was found to extend to higher values of the migrational influx in accordance with our predictions, while it only extended to a much smaller degree toward faster food change rates. (b) We changed the mutation rate to one-fifth of that considered in Figure 2. Under these conditions—similar to those in a—the survival range of NGT extends in one direction, toward smaller food change rates, but does not significantly move in the other, toward smaller values of migrational influx. (c) We demonstrate that the t gene survives, although under a more limited set of parameters, even if selection for genome length is absent. During these simulations we gradually approached the limit 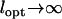 , where selection for genome length disappears. We also increased the mutation rate of food genes by an order of magnitude, while keeping that of the t gene fixed. This way we were able to take into consideration the length of the t gene lt = 0.1 in terms of the relative mutation, while also keeping our simulations tractable.
, where selection for genome length disappears. We also increased the mutation rate of food genes by an order of magnitude, while keeping that of the t gene fixed. This way we were able to take into consideration the length of the t gene lt = 0.1 in terms of the relative mutation, while also keeping our simulations tractable.
Migration:
Since all food types are equivalent and consequently all combinations of these are equally likely, the number of mean-field variables needed to describe the global genotype distribution can be reduced from 210+1 to 10 × 2 corresponding to all genotypes G with 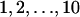 intact metabolic genes, with and without the t gene. To take into account the influx of bacteria from external populations we took the averages of these 10 × 2 types in the population with a sliding-growing time window (always encompassing the last quarter of the simulation) and subsequently calculated the migrational term [migr.in] for each genotype G by multiplying the corresponding mean-field variable with the appropriate combinatorial factor and the migration rate Rmigr.
intact metabolic genes, with and without the t gene. To take into account the influx of bacteria from external populations we took the averages of these 10 × 2 types in the population with a sliding-growing time window (always encompassing the last quarter of the simulation) and subsequently calculated the migrational term [migr.in] for each genotype G by multiplying the corresponding mean-field variable with the appropriate combinatorial factor and the migration rate Rmigr.
RESULTS
Phase diagram for the survival of NGT:
Performing extensive computer simulations for various values of the two main external parameters, the food change rate Rfood and the migration rate Rmigr, we have found that NGT (represented by model gene t) indeed survives under a wide range of parameters, as can be seen in Figure 2a. In these stochastic population dynamics simulations we have numerically followed the time evolution of the number of bacteria in each of the possible 210+1 genotypes (representing the presence or absence of the 10 model food genes and the model transformation gene) in a single population, as detailed in methods.
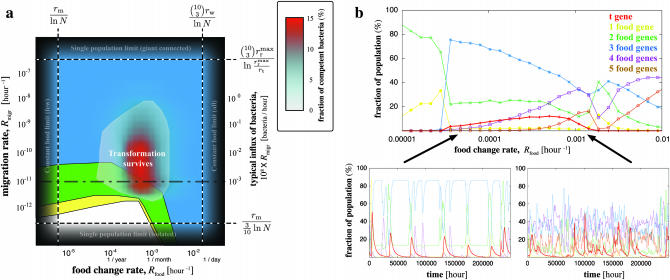
Figure 2.—
Phase diagram for the persistence of NGT. (a) The average percentage of bacteria capable of NGT is shown with color gradient (translucent from white to red) on the Rfood–Rmigr parameter space, with different points corresponding to different possible bacterial metapopulations. The maximum genome size (the number of functional model food genes) in systems where transformation has been turned off is underlain in color code (yellow regions have bacteria with only one functional food gene, green regions have bacteria with at most two, while the blue region corresponds to metapopulations where bacteria exist that can utilize all three food types). The dashed lines indicate the theoretical limits of the persistence of transformation as described in the text. (b) The top shows a slice in the parameter space along the dashed-dotted black line in a, with data points corresponding to the fraction of bacteria with the t gene and the fraction of those having exactly 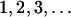 , functional food genes. The bottom displays two illustrative time series from the simulations, corresponding to a low and a high food change rate, as indicated by the arrows.
, functional food genes. The bottom displays two illustrative time series from the simulations, corresponding to a low and a high food change rate, as indicated by the arrows.
Limits for the persistence of NGT:
The limits beyond which NGT cannot persist (dashed straight lines in Figure 2a), because either the temporal or the spatial fluctuations become irrelevant, can be estimated easily. If 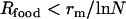 , then the food sources remain unchanged for such a long time that the t gene disappears completely by deletion before it could become beneficial at the next food change. At the other extreme, for
, then the food sources remain unchanged for such a long time that the t gene disappears completely by deletion before it could become beneficial at the next food change. At the other extreme, for 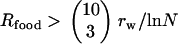 , i.e., when the rate at which any given food combination recurs
, i.e., when the rate at which any given food combination recurs 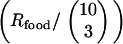 is larger than the rate at which bacteria carrying the corresponding combination of functional genes are completely washed out of the population (
is larger than the rate at which bacteria carrying the corresponding combination of functional genes are completely washed out of the population (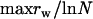 ), transformation cannot confer an advantage through assembling this combination as it is always present in the local population. In other words, the food changes so rapidly that populations effectively experience a constant feeding (with all possible food combinations), and NGT becomes useless.
), transformation cannot confer an advantage through assembling this combination as it is always present in the local population. In other words, the food changes so rapidly that populations effectively experience a constant feeding (with all possible food combinations), and NGT becomes useless.
There are similar constraints on the migration rate as well. Obviously, for very small migration rates, 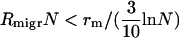 , i.e., when the influx of bacteria with a newly required food gene (
, i.e., when the influx of bacteria with a newly required food gene (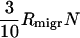 ) is lower than the rate of the complete disappearance of the functional t gene (rm/ln N), migration loses its role in the propagation of NGT; thus, the metapopulation practically falls apart into isolated populations, in which transformation cannot survive. The other limit is
) is lower than the rate of the complete disappearance of the functional t gene (rm/ln N), migration loses its role in the propagation of NGT; thus, the metapopulation practically falls apart into isolated populations, in which transformation cannot survive. The other limit is 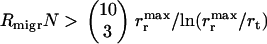 , i.e., when the migration is so intense that after a food change the rate of the arrival of a bacterium with the optimal combination of functional model genes
, i.e., when the migration is so intense that after a food change the rate of the arrival of a bacterium with the optimal combination of functional model genes 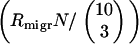 exceeds the rate at which the model gene that has just become beneficial proliferates and subsequently gets incorporated by a member of the original population
exceeds the rate at which the model gene that has just become beneficial proliferates and subsequently gets incorporated by a member of the original population 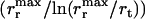 . Then the entire metapopulation effectively becomes a single giant population, in which transformation cannot survive also. Although these are rather crude estimates, which give only the limits outside of which transformation is certain not to survive, we have found nonetheless that within these extremes NGT persists for most parameter values, indicating the robustness of our proposed mechanism.
. Then the entire metapopulation effectively becomes a single giant population, in which transformation cannot survive also. Although these are rather crude estimates, which give only the limits outside of which transformation is certain not to survive, we have found nonetheless that within these extremes NGT persists for most parameter values, indicating the robustness of our proposed mechanism.
Effects of NGT on genome organization:
To demonstrate the dramatic effect of transformation on the genome composition, we take a cut through the parameter space (dashed-dotted line in Figure 2a) and plot the average fraction of bacteria possessing a functional copy of the t gene, as well as the fraction of those having exactly 1, 2,…, functional model food genes in Figure 2b. For a low food change rate, but within the range where NGT survives, most bacteria contain only functional copies of the three necessary food genes, and the average fraction of functional copies of the model gene t is very low as it is used very rarely. Functional copies of the t gene can occasionally disappear from the population and then subsequently be replanted by migrants as illustrated in Figure 1a. For larger food change rates, but still within the survival range of NGT, the t gene becomes beneficial more often, its average fraction increases (except near the other end of the range), and the fraction of bacteria having four or more functional model food genes also increases. Two time series (at a low and a high food change rate) displaying the evolution of these fractions are shown in the bottom of Figure 2b.
Survival range of NGT:
In the model we have chosen numerically tractable values for the rates (rrmax, rw, rm, rt) characterizing a given bacterial species. Some of these rates may be off by up to a few orders of magnitude for certain types of bacteria; this, however, should not fundamentally affect the width of the parameter range where NGT survives (in terms of either Rfood or Rmigr), as this depends primarily on the binomial factor 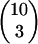 . Moreover, under natural conditions the number of possible food types is usually ≫10, and the number of simultaneously available ones is also >3; thus the survival range of NGT is probably even broader (extending to higher values of Rfood and Rmigr) than what our calculations predict. We have attempted to survey the effect of more realistic, but computationally more demanding parameter sets as presented in Figure 3, a and b, and found that the survival range of NGT indeed became broader. In Figure 3a we increased the number of possible food types from 10 to 12, while also increasing the number present at any one time from 3 to 4, consequently also setting lopt = 4. In Figure 3b we decreased the mutation rate to one-fifth of the value considered previously.
. Moreover, under natural conditions the number of possible food types is usually ≫10, and the number of simultaneously available ones is also >3; thus the survival range of NGT is probably even broader (extending to higher values of Rfood and Rmigr) than what our calculations predict. We have attempted to survey the effect of more realistic, but computationally more demanding parameter sets as presented in Figure 3, a and b, and found that the survival range of NGT indeed became broader. In Figure 3a we increased the number of possible food types from 10 to 12, while also increasing the number present at any one time from 3 to 4, consequently also setting lopt = 4. In Figure 3b we decreased the mutation rate to one-fifth of the value considered previously.
Characterizing transformation by a single rate coefficient rt is also a rather crude simplification, since many bacteria become competent only under certain conditions. Fortunately, the limits for the survival of NGT calculated above depend very weakly on the value of rt. Besides, the parameter range where a better regulated competence will persist is expected to be even larger than it is in our simplified model.
Up to now we have considered only food sources that fluctuate with a single characteristic rate Rfood. To check what happens in more complex situations, we added an equal amount of constant food source (XYZ) to the fluctuating ones. These simulations confirmed that the survival range of NGT remains virtually unaffected, indicating that the ability to lose and reload a few types of intermittently beneficial “operational” genes is advantageous to the bacteria and sufficient to maintain NGT.
Migration between bacterial populations in nature clearly depends on distance and, as a consequence, a wide variety of migration rates are usually present in the metapopulation. Populations close to each other (with strongly correlated environmental fluctuations or with intense interpopulation migration), however, may be grouped together and considered as a large effective population. For NGT to persist it is sufficient that a reasonable number of such “effective” populations exist in the metapopulation, a requirement that does not seem unrealistic in view of the highly varied conditions under which bacteria prevail on Earth.
Finally one very important question remains that we have not addressed so far: Is selection for shorter genome length necessary for the survival of NGT as a mechanism to reload genes? To answer this question we performed simulations where we gradually approached the limit 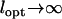 , where selection for genome length disappears. We found that if we took into consideration the length of the t gene lt in terms of the mutation rate, the t gene survived (see Figure 3c). Because smaller mutation rates imply longer convergence times in our simulation, we implemented the value of lt = 0.1, not by decreasing the mutation rate of the t gene, but by increasing those of the food genes by a factor of 10. From these results we can conclude that while selection for shorter genome length substantially increases the size of the parameter range where NGT is able to survive solely as a mechanism to reload genes, it is not indispensable. We may further argue that the relatively small region in Figure 3c where the t gene persists should become much larger for more realistic values of the mutation rate and the number of fluctuating food types (cf. Figure 3, a and b).
, where selection for genome length disappears. We found that if we took into consideration the length of the t gene lt in terms of the mutation rate, the t gene survived (see Figure 3c). Because smaller mutation rates imply longer convergence times in our simulation, we implemented the value of lt = 0.1, not by decreasing the mutation rate of the t gene, but by increasing those of the food genes by a factor of 10. From these results we can conclude that while selection for shorter genome length substantially increases the size of the parameter range where NGT is able to survive solely as a mechanism to reload genes, it is not indispensable. We may further argue that the relatively small region in Figure 3c where the t gene persists should become much larger for more realistic values of the mutation rate and the number of fluctuating food types (cf. Figure 3, a and b).
DISCUSSION
In light of our simulations, we suggest that the existence of NGT is facilitated by its role as a vehicle to reload genes. We argue that the short-term advantage that sustains NGT long enough for its evolutionary effects to emerge, lies, at least in part, in providing mobility to variably selected genes. It allows individuals to reload genes lost from a population—due to long disuse—but still available in the metapopulation, bringing together genes from the collective gene pool of the species with locally adapted genomes. This advantage prevails if spatio-temporal fluctuations (Meyers and Bull 2002) in the environment (imposing variable selection pressure on different populations of the same species) exist in parallel with weak migration between the populations (allowing genetic mixing). Whether or not natural bacterial populations actually experience the kind of population subdivision and interpopulation migration necessary for our model to be applicable remains to be demonstrated experimentally. Some examples that may easily fulfill these conditions, however, readily come to mind; e.g., experience shows that any perishable substance that is a potential food source for bacteria is promptly colonized. One may also consider the intestinal flora of grazing animals, herds of which cover large distances while occasionally encountering each other at locations, such as water sources, where migration may occur between the microbial populations resident in their intestines.
Provided that the above conditions may be rather general, our results compel us to imply that the ability of active DNA uptake may easily have evolved through the gradual specialization of a more general transport mechanism (Woese 1998), driven not by the need to serve the cell with nucleotides for food, but by an advantage conferred through providing homologous sequences for restoring genes eroded by formerly adaptive deletions. In other words NGT is not an accidental by-product of nutrient uptake, but has come into existence to counterbalance gene loss, which inevitably occurs in highly economized genomes under fluctuating selection pressure. It should be emphasized, however, that this does not exclude the advantage transformation confers through enabling access to DNA as a source of nutrients, which most probably helps sustain NGT.
A fundamental long-term effect of genetic mixing by NGT, with important implications for the understanding of the evolution of eukaryotic sex (Maynard Smith 1975; Holmes et al. 1999; Maynard Smith and Szathmáry 2002), is that it prevents bacterial species from falling apart into independently evolving clonal lineages (Dykhuizen and Green 1991), by facilitating genetic mixing between genomes of sufficient homology. From a medical perspective the dynamic nature of bacterial genomes has clear significance for the pressing problem of the rapid spread of antibiotic resistance and other pathogenic traits (Claverys et al. 2000).
Acknowledgments
I.D. acknowledges support from the Hungarian Science Foundation (grant no. OTKA K60665).
References
- Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown et al., 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397: 176–180. [DOI] [PubMed] [Google Scholar]
- Avery, O. T., C. M. MacLeod and M. McCarty, 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J. Exp. Med. 79: 137–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, H., H. C. Byerly, F. A. Hopf and R. E. Michod, 1985. Genetic damage, mutation, and the evolution of sex. Science 229: 1277–1281. [DOI] [PubMed] [Google Scholar]
- Butlin, R., 2002. Opinion–evolution of sex: the costs and benefits of sex: new insights from old asexual lineages. Nat. Rev Genet. 3: 311–317. [DOI] [PubMed] [Google Scholar]
- Claverys, J., M. Prudhomme, I. Mortier-Barriére and B. Martin, 2000. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol. Microbiol. 35: 251–259. [DOI] [PubMed] [Google Scholar]
- Cohen, E., D. Kessler and H. Levine, 2005. Recombination dramatically speeds up evolution of finite populations. Phys. Rev. Lett. 94: 098102. [DOI] [PubMed]
- Dykhuizen, D. E., and C. E. Green, 1991. Combination in Escherichia coli and the definition of biological species. J. Bacteriol. 17: 7257–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S. F., and R. E. Lenski, 1997. Test of synergistic interactions among deleterious mutations in bacteria. Nature 390: 395–398. [DOI] [PubMed] [Google Scholar]
- Feil, E. J., 2004. Small change: keeping pace with microevolution. Nat. Rev. Microbiol. 2: 483–495. [DOI] [PubMed] [Google Scholar]
- Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day et al., 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski, I., 1998. Metapopulation dynamics. Nature 396: 41–49. [Google Scholar]
- Holmes, E. C., R. Urwin and M. C. J. Maiden, 1999. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol. Biol. Evol. 16(6): 741–749. [DOI] [PubMed] [Google Scholar]
- Levin, B. R., and C. T. Bergstrom, 2000. Bacteria are different: observations, interpretations, speculations, and opinions about adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. USA 97: 6981–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith, J., 1975. The Evolution of Sex. Cambridge University Press, Cambridge, UK.
- Maynard Smith, J., 1993. The role of sex in bacterial evolution. J. Hered. 84(5): 326–327. [DOI] [PubMed] [Google Scholar]
- Maynard Smith, J., and E. Szathmáry, 2002. The Major Transitions in Evolution. Oxford University Press, Oxford.
- Maynard Smith, J., C. G. Dowson and B. G. Spratt, 1991. Localised sex in bacteria. Nature 349: 29–31. [DOI] [PubMed] [Google Scholar]
- Meyers, A. M., and J. J. Bull, 2002. Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 17: 551–557. [Google Scholar]
- Mira, A., H. Ochman and N. A. Moran, 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17: 589–596. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., T. Itoh, H. Matsuda and T. Gojobori, 2004. Biased biological function of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36: 760–766. [DOI] [PubMed] [Google Scholar]
- Ochman, H., J. G. Lawrence and E. A. Groisman, 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304. [DOI] [PubMed] [Google Scholar]
- Otto, S. P., and T. Lenormand, 2002. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 3: 252–260. [DOI] [PubMed] [Google Scholar]
- Redfield, J. R., M. R. Schrag and A. M. Dean, 1997. The evolution of bacterial transformation: sex with poor relations. Genetics 146: 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield, R. J., 2001. Do bacteria have sex? Nat. Rev. Genet. 2: 634–639. [DOI] [PubMed] [Google Scholar]
- Solomon, J. M., and A. D. Grossman, 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12: 150–155. [DOI] [PubMed] [Google Scholar]
- Thomas, C. M., and K. M. Nielsen, 2005. Mechanisms of, and barriers to. Horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3: 711–721. [DOI] [PubMed] [Google Scholar]
- Vellai, T., K. Takacs and G. Vida, 1998. A new aspect to the origin and evolution of eukaryotes. J. Mol. Evol. 46: 499–507. [DOI] [PubMed] [Google Scholar]
- Vellai, T., A. L. Kovács, G. Kovács, Cs. Ortutay and G. Vida, 1999. Genome economisation and a new approach to the species concept in bacteria. Proc. R. Soc. Lond. Ser. B 266: 1953–1958. [Google Scholar]
- Vestigian, K., and N. Goldenfeld, 2005. Global divergence of microbial sequences mediated by propagating fronts. Proc. Natl. Acad. Sci. USA 102: 7332–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C., 1998. The universal ancestor. Proc. Natl. Acad. Sci. USA 95: 6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]