Abstract
The plant hormone auxin can be regulated by formation and hydrolysis of amide-linked indole-3-acetic acid (IAA) conjugates. Here, we report the characterization of the dominant Arabidopsis iaa–leucine resistant3 (ilr3-1) mutant, which has reduced sensitivity to IAA–Leu and IAA–Phe, while retaining wild-type responses to free IAA. The gene defective in ilr3-1 encodes a basic helix-loop-helix leucine zipper protein, bHLH105, and the ilr3-1 lesion results in a truncated product. Overexpressing ilr3-1 in wild-type plants recapitulates certain ilr3-1 mutant phenotypes. In contrast, the loss-of-function ilr3-2 allele has increased IAA–Leu sensitivity compared to wild type, indicating that the ilr3-1 allele confers a gain of function. Microarray and quantitative real-time PCR analyses revealed five downregulated genes in ilr3-1, including three encoding putative membrane proteins similar to the yeast iron and manganese transporter Ccc1p. Transcript changes are accompanied by reciprocally misregulated metal accumulation in ilr3-1 and ilr3-2 mutants. Further, ilr3-1 seedlings are less sensitive than wild type to manganese, and auxin conjugate response phenotypes are dependent on exogenous metal concentration in ilr3 mutants. These data suggest a model in which the ILR3/bHLH105 transcription factor regulates expression of metal transporter genes, perhaps indirectly modulating IAA-conjugate hydrolysis by controlling the availability of metals previously shown to influence IAA–amino acid hydrolase protein activity.
THE phytohormone auxin is an essential mediator of many facets of plant development. Plants employ several strategies in addition to de novo synthesis to precisely regulate indole-3-acetic acid (IAA) levels, including forming and hydrolyzing conjugates that act as storage forms of IAA. Amide-linked conjugates identified in Arabidopsis seedlings include IAA–Leu, IAA–Ala, IAA–Asp, IAA–Glu (Tam et al. 2000; Kowalczyk and Sandberg 2001), and several IAA–peptide conjugates (Bialek and Cohen 1992; Walz et al. 2002).
Arabidopsis screens have revealed mutants specifically resistant to root growth inhibition caused by IAA–amino acid conjugates (reviewed in Woodward and Bartel 2005b). Through these screens, genes modulating IAA-conjugate sensitivity have been identified, including those encoding the amidohydrolases IAA–Leu resistant (ILR)1 (Bartel and Fink 1995) and IAA–Ala resistant (IAR)3 (Davies et al. 1999) that cleave IAA–amino acid conjugates to release the active hormone. IAA–amino acid resistance screens have also uncovered the predicted membrane protein IAR1 (Lasswell et al. 2000), the pyruvate dehydrogenase E1α subunit homolog IAR4 (LeClere et al. 2004), and the novel protein ILR2 (Magidin et al. 2003).
Triple-mutant seedlings deficient in three IAA-conjugate hydrolases (ILR1, IAR3, and ILL2) have reduced responsiveness to exogenous IAA conjugates and free IAA, display low-auxin phenotypes, and have decreased IAA levels compared to wild type, indicating that hydrolysis of endogenous IAA–amino acid conjugates by these enzymes contributes free IAA to the auxin pool during germination (Rampey et al. 2004). The hydrolases active on IAA–amino acids have putative N-terminal signal sequences and C-terminal ER retrieval signals (Bartel and Fink 1995; Davies et al. 1999), suggesting localization in the ER lumen or an ER-derived compartment. The IAA-conjugate hydrolase genes are expressed in overlapping but distinct patterns not only during germination, but also at other growth stages (Rampey et al. 2004). IAR3 (Titarenko et al. 1997; Sasaki et al. 2001) and ILR1 (Zimmermann et al. 2004) transcripts are induced by jasmonic acid (JA), suggesting that these genes might play roles in JA conjugate hydrolysis or that IAA release may be JA inducible. However, proteins controlling hydrolase gene expression have not been identified.
In addition to transcriptional regulation, hydrolase activity may be controlled post-translationally via the availability of metal cofactors, because in vitro assays have shown that hydrolase activity requires Mn2+ or Co2+ (Bartel and Fink 1995; Davies et al. 1999; LeClere et al. 2002). The findings that several genes with roles in metal transport appear to regulate conjugate responsiveness suggest that the metal microenvironment affects hydrolase activity. For example, ILR2 appears to inhibit an unknown metal transporter (Magidin et al. 2003). The ilr2 mutant is resistant to the inhibitory effects of IAA–amino acid conjugates as well as Mn2+ and Co2+ on root elongation, and ilr2 seedling microsomes transport more Mn2+ than wild type (Magidin et al. 2003).
The IAA-conjugate-resistant iar1 mutant is defective in a predicted metal transporter with seven apparent transmembrane domains and many His-rich regions (Lasswell et al. 2000). The mouse IAR1 homolog ZIP7/KE4 transports zinc from the Golgi apparatus into the cytoplasm (Huang et al. 2005) and complements the iar1 mutant (Lasswell et al. 2000), suggesting that IAR1 might efflux metals from a subcellular compartment, perhaps removing inhibitory metals from the compartment in which the hydrolases reside (Lasswell et al. 2000).
Here, we describe the isolation and characterization of ilr3-1, a dominant mutation that confers resistance to IAA–Leu, IAA–Phe, and Mn2+. The gene defective in ilr3-1 encodes a basic helix-loop-helix (bHLH) leucine zipper transcription factor, bHLH105. We recapitulated several aspects of ilr3-1 phenotypes in wild-type seedlings by overexpressing an ilr3-1 mutant cDNA. Microarray and quantitative real-time PCR analyses identified five genes, including three encoding putative metal transporters, with decreased expression in ilr3-1 seedlings compared to wild type. Indeed, metal accumulation is altered in ilr3 mutants and the phenotypes of gain- and loss-of-function ilr3 mutant alleles depend on exogenous iron concentration, suggesting a role for ILR3/bHLH105 in metal homeostasis and reinforcing the importance of metal homeostasis for auxin metabolism.
MATERIALS AND METHODS
Plant materials and growth conditions:
Plants from the Columbia (Col-0), Wassilewskija (Ws-1), and Landsberg erecta (Ler) accessions were used. For phenotypic assays, seeds were surface sterilized (Last and Fink 1988) and grown aseptically on plant nutrient medium containing 0.5% (w/v) sucrose (PNS) (Haughn and Somerville 1986) unless indicated otherwise and solidified with 0.6% agar. Seedlings were grown on medium alone, medium supplemented with indicated concentrations of IAA, IAA-l-amino acid conjugates (Aldrich, Milwaukee) or other hormones (from 0.1-, 1-, or 100-mm stocks in 100% ethanol), Basta [glufosinate ammonium (Crescent Chemical, Augsburg, Germany) from a 50-mg/ml stock in 25% (v/v) ethanol], or kanamycin (from a filter-sterilized 25-mg/ml stock in H2O). Media supplemented with metals (filter-sterilized 100 mm MnCl2, 500 mm CoCl2, 2 m CaCl2, 20 mm CdCl2, 500 mm ZnSO4 stocks in H2O) did not contain sucrose. Plates were sealed with gas-permeable Leukopor surgical tape (LecTec, Minnetonka, MN). Plates were incubated under yellow long-pass filters to slow the breakdown of indolic compounds (Stasinopoulos and Hangarter 1990) with constant illumination (25–45 μE m−2 sec−2) at 22°. Plants transferred to soil (Metromix 200; Scotts, Marysville, OH) were grown at 22°–25° under Cool White fluorescent bulbs (Sylvania, Danvers, MA) with continuous illumination.
Plants grown for inductively coupled plasma–mass spectrometry (ICP–MS) analysis were seeded (n = 12) into 20-row plastic trays, stratified for 3 days at 4°, and allowed to grow for 5 weeks at 19°–22° under 90 μE m−2 sec−1 of photosynthetically active light provided by fluorescent bulbs (10 hr light/14 hr dark). The growth medium was Sunshine Mix LB2 (Carl Brehob & Son, Indianapolis) spiked with As, Cd, Co, Li, Ni, Pb, and Se (Lahner et al. 2003). Plants were watered twice per week with 1/4 type 2 Hoaglands (Lahner et al. 2003) in which the normal Fe was replaced with 0.5–30 μm Fe–N,N′-Di(2-hydroxybenzyl) ethylenediamine-N,N′-diacetic acid monohydrochloride hydrate (Fe–HBED). Fe–HBED was prepared by mixing HBED (Strem Chemicals, Newburyport, MA) with an equimolar amount of iron (III) nitrate, brought to pH 6.0 with KOH.
Mutant isolation and positional cloning:
The ilr3-1 mutant was isolated as described previously (Bartel and Fink 1995; Davies et al. 1999) from the progeny of Col-0 seed mutagenized via fast-neutron bombardment (60 Gy). ilr3-1 was outcrossed to the Ws and Ler accessions for recombination mapping. F2 seeds were plated on 30 μm IAA–Leu, seedlings displaying wild-type sensitivity were selected for the mapping population, and the genome was examined for an area with linkage to the Ws or Ler parental ecotypes. The ilr3-1 mutation was localized to chromosome 5 using published markers (Konieczny and Ausubel 1993; Bell and Ecker 1994), markers posted on The Arabidopsis Information Resource (http://www.arabidopsis.org/), and the following new markers (see supplemental Table 1 at http://www.genetics.org/supplemental/ for primer sequences), including several dCAPs markers (Michaels and Amasino 1998; Neff et al. 1998): MBA10-2 and MBA10-3 yield a 171-bp product with an altered nucleotide in MBA10-3 that creates a BamHI site in Ws but not Col-0; F6N7-1 and F6N7-3 yield a 183-bp product with an altered nucleotide in F6N3-3 that creates a PvuII site in Col-0 but not Ws; ILL3-5B and ILL3-16 cut with NdeI yield a 360-bp product in Col-0 and 260- and 100-bp products in Ws; MDK4-4 and MDK4-5 yield an ∼500-bp product with polymorphisms identified by sequencing with MDK4-4; At5g54510-5 and At5g54510-6 yield an ∼1.1-kb product with polymorphisms identified by sequencing with At5g54510-6; MRB17-17 and MRB17-18 yield an ∼1.1-kb product with polymorphisms identified by sequencing with MRB17-18; MRB17-23 and MRB17-24 yield an ∼1.1-kb product with polymorphisms identified by sequencing with MRB17-23; and K5F14-2 and K5F14-3 yield a 151-bp product with an altered nucleotide in K5F14-3 that creates a DdeI site in Ws but not in Col-0.
One candidate gene within the ilr3-1 mapping region, At5g54680, was sequenced using ilr3-1 mutant genomic DNA. DNA was isolated from a homozygous ilr3-1 line backcrossed three times and At5g54680 was PCR amplified with the following oligonucleotides: MRB17-27 and MRB17-28, MRB17-29 and MRB17-30, and MRB17-31 and MRB17-32 (supplemental Table 1 at http://www.genetics.org/supplemental/). The resulting products were sequenced directly with the corresponding oligonucleotides (SeqWright Laboratories, Houston).
The 859-bp region from chromosome 4 inserted in the At5g54680/ILR3 gene in the ilr3-1 mutant included 280 bp upstream of At4g22180 and the first 579 bp of the predicted At4g22180 coding sequence. At4g22180 is a hypothetical gene that lacks introns and is the third of three adjacent putative F-box genes (At4g22165, At4g22170, and At4g22180) on chromosome 4 that lack EST evidence for expression. No rearrangement of the sequence occurred upon insertion. We used PCR analysis with oligonucleotides flanking this region on chromosome 4 to determine that the At4g22180 locus was intact in the backcrossed ilr3-1 mutant.
ilr3-2 is a sequence-indexed Arabidopsis T-DNA insertion mutant (SALK_004997) isolated by the Salk Institute Genomic Analysis Laboratory (Alonso et al. 2003) that we obtained from the Arabidopsis Biological Resource Center (ABRC) (Ohio State University, Columbus, OH). The position of the T-DNA insertion in ilr3-2 was verified using PCR analysis. PCR amplification with ILR3-12 and ILR3-13 (supplemental Table 1 at http://www.genetics.org/supplemental/) yielded a 689-bp product from wild-type genomic DNA, whereas amplification with MRB17-28 and LB1-Salk, a modified version of LBb1 (http://signal.salk.edu), yielded an ∼400-bp product from ilr3-2 genomic DNA. This product was sequenced, revealing that the T-DNA is located at position 186 of ILR3 (where 1 is the A position of the initiator ATG).
ilr1-5 is a mutant in the Col-0 accession isolated by screening progeny of γ-irradiated seeds on 50 μm IAA–Leu for auxin conjugate-resistant root elongation as previously described (Bartel and Fink 1995). The mutant contains a C-to-T mutation at nucleotide 1309 of ILR1 (where 1 is the initiator ATG) that replaces a Thr residue with an Ile. The ilr1-5 mutant was backcrossed to Col-0 five times prior to analysis.
Reporter gene analysis:
A 1.8-kb potential ILR3 regulatory region (including −1863 to −1 bp from the ILR3 initiator ATG) was amplified from purified Col-0 DNA with Triplemaster polymerase mix (Eppendorf AG, Hamburg, Germany) using the oligonucleotides ILR3–GUS-1 and ILR3–GUS-2, and the resulting product was cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA). The insert of the resulting plasmid was sequenced to verify the absence of PCR-derived mutations. The ILR3 promoter fragment was removed from this plasmid with HindIII and BamHI and ligated into pBI101.2 (Jefferson et al. 1987) cut with the same enzymes to give pBI101.2-ILR3-prom, which was electroporated (Ausubel et al. 1999) into Agrobacterium tumefaciens strain GV3101 (Koncz et al. 1992) for transformation into Col-0 plants (Clough and Bent 1998). Transgenic lines containing the ILR3 promoter–β-glucuronidase (GUS) construct were plated on medium containing 12 μg/ml kanamycin. Progeny of kanamycin-resistant T1 plants were grown for 1–8 days, and GUS localization was observed after staining for 4 hr with 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β-d-glucuronide as previously described (Bartel and Fink 1994). Thirty-seven- to 44-day-old adult plant parts were stained for 12–15 hr. The six independent transgenic lines that were observed had similar staining patterns with variable intensities.
ILR3 and ilr3-1 cDNA Isolation:
Col-0 and ilr3-1 seeds were surface sterilized (Last and Fink 1988) and plated on filter paper on 150-mm plates containing PNS. Seedlings were grown for 7 days at 22° in yellow-filtered light. RNA was isolated using RNeasy Mini Kits (QIAGEN, Valencia, CA), and 1 μg of total Col-0 or ilr3-1 RNA was reverse transcribed with SuperScript III (Invitrogen) with oligonucleotide (supplemental Table 1 at http://www.genetics.org/supplemental/) ILR3-5 for ILR3 and ILR3-6 for ilr3-1. Each cDNA was PCR amplified with Triplemaster polymerase using oligonucleotides ILR3-4 and ILR3-5 for ILR3 and ILR3-4 and ILR3-6 for ilr3-1. PCR products were purified and cloned into the pCR4-TOPO vector. The inserts of the resulting plasmids, TOPO-ILR3 and TOPO-ilr3-1, were sequenced to verify the absence of PCR-derived mutations.
Overexpression analysis:
The ILR3 and ilr3-1 cDNAs were removed from TOPO-ILR3 and TOPO-ilr3-1 with SalI and NotI and ligated into 35SpBARN (LeClere and Bartel 2001) cut with XhoI and NotI. The resulting plasmids, 35S–ILR3 and 35S–ilr3-1, were sequenced using vector-derived oligonucleotides, 35S-F and NOS-R (LeClere and Bartel 2001), and transformed (Clough and Bent 1998) into Col-0. T1 plants containing each construct were selected on PN containing 10 μg/ml Basta, and homozygous plants were identified in subsequent generations by following segregation of Basta resistance. For 35S–ILR3, nine transgenic lines were obtained, and two lines (D1 and K6) were arbitrarily selected for further study. For 35S–ilr3-1, only three transgenic lines were obtained, and two of these (C1 and E3) were arbitrarily selected for further study.
Microarray analysis:
Col-0 and ilr3-1 seeds were plated on filter paper overlaid on 150-mm plates containing PNS. Seedlings were grown for 7 days at 22° in yellow-filtered light. After 7 days, seedlings were frozen in liquid N2. Total RNA was isolated from three biological replicates of each genotype using RNeasy Mini Kits (QIAGEN), and 30–40 μg of total RNA from each sample was sent to the laboratory of Thomas McKnight at Texas A&M University where mRNA from Col-0 and ilr3-1 samples was converted to cDNA and amplified to produce biotin-labeled cRNA. The cRNA was hybridized to Affymetrix ATH1 Arabidopsis whole-genome (∼22,000 genes) microarray chips and analyzed with Microarray Suite 5.0 (Affymetrix). Transcripts with detectable signals (P < 0.05) on all three Col-0 chips or all three ilr3-1 chips or both (n = 14,065) were analyzed further in Excel (Microsoft) and are displayed in Figure 7 and supplemental Table 2 at http://www.genetics.org/supplemental/. For these transcripts, a two-tailed t-test assuming unequal variance was performed to test for significant differences between the three wild-type samples and three ilr3-1 samples.
Figure 7.—
Global transcript analysis in the ilr3-1 mutant. RNA prepared from three biological replicates of 7-day-old Col-0 (wild type) and ilr3-1 seedlings was analyzed using Affymetrix ATH1 Arabidopsis microarray chips. A scatter plot of normalized mean signal intensities for 14,065 transcripts with detectable signals on all three Col-0 or all three ilr3-1 (or both) chips is shown. The 507 transcripts with significant differences between ilr3-1 and wild type (P ≤ 0.05 in two-tailed t-tests assuming unequal variance) are shown as solid dots; genes with insignificant differences between ilr3-1 and wild type (P > 0.05) are shaded. The six transcripts with >2.5-fold reduction in ilr3-1 relative to wild type are labeled with gene names.
Quantitative real-time PCR analysis:
Total RNA was isolated from 7-day-old seedlings as described above. For each sample, 0.3 μg total RNA was treated with DNase I (Amplification Grade; Roche Applied Science, Indianapolis, IN) and reverse transcribed in a 20-μl volume using 200 units of SuperScript III (Invitrogen) according to the manufacturer's recommendations. For IAR3, ILR1, and ILR2, each reaction contained a mixture of the reverse primers (IAR3-QPCR-R, ILR1-QPCR-R, and ILR2-QPCR-R) at a final concentration of 2 μm; all other reactions contained 2 μm random hexamers. The resulting cDNAs were diluted to 100 μl with H2O. Gene-specific primers and probes were selected using Primer Express Software (Applied Biosystems, Foster City, CA) and are listed in supplemental Table 1 at http://www.genetics.org/supplemental/. GAPDHβ (supplemental Table 1) and APRT (Woodward and Bartel 2005a) primers and probes were used as endogenous controls. Probes were from Applied Biosystems and were 5′-labeled with 6-FAM (5-carboxyfluorescein) and 3′-labeled with MGBNFQ (minor groove binder/nonfluorescent quencher).
Quantitative real-time PCR was performed in triplicate or duplicate for each reverse transcription reaction using 10 μl diluted cDNA (from 30 ng total RNA) per PCR amplification. Each 25-μl reaction contained TaqMan Universal PCR Master Mix (Applied Biosystems), the appropriate forward and reverse primers (0.5 μm each), and the corresponding probe (0.2 μm). PCR conditions were 2 min at 50°, 10 min at 95°, and 40 cycles of 95° for 15 sec and 60° for 1 min. Amplification was monitored in real time utilizing the ABI Prism 7000 sequence detection system software. Template levels were normalized to APRT cDNA amplification using the comparative CT method (ABI Prism 7700 sequence detection system user bulletin no. 2, http://www.appliedbiosystems.com).
Ionomic analysis:
Medium-age rosette leaves (generally two leaves from opposite sides of the plant) from 5-week-old plants were harvested for ionomic analysis. Approximately 3 mg dry weight of each plant was sampled into Pyrex tubes (16 × 100 mm) and dried at 92° for 20 hr. After cooling, 7 of 108 samples from each tray were weighed. All samples were digested with 0.7 ml concentrated nitric acid (OmniTrace, VWR) and diluted to 6.0 ml with 18 MΩ water. Elemental analysis was performed with an ICP–MS (Elan DRCe; Perkin-Elmer, Norwalk, CT) for Li, B, Na, Mg, P, K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, and Cd. Ten samples from each run were retained and rerun as a unit at the end of the experiment to facilitate cross-tray comparisons. All samples were normalized to calculated weights, as determined with an iterative algorithm using the best-measured elements, the weights of the seven weighed samples, and the solution concentrations, implemented in Microsoft Excel (Lahner et al. 2003).
RESULTS
Dominant IAA–leucine resistance is conferred by mutation of a bHLH–leucine zipper transcription factor:
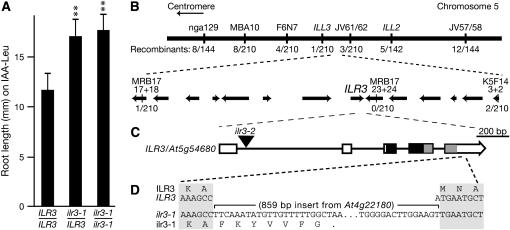
The ilr3-1 mutant was isolated from progeny of fast neutron-bombarded Arabidopsis seeds as an individual resistant to root elongation inhibition by exogenous IAA–Leu (Figure 1A). We identified the gene defective in ilr3-1, using a map-based positional cloning strategy. Because the IAA–Leu resistance phenotype of ilr3-1 is dominant (Figure 1A), plants from an F2 outcrossed population displaying wild-type IAA–Leu sensitivity, and therefore lacking the ilr3-1 mutation, were selected for mapping with PCR-based markers (Bell and Ecker 1994; http://www.arabidopsis.org). We mapped the defective gene to an interval on the bottom arm of chromosome 5 containing 12 annotated genes (Figure 1B). None of these genes were similar to genes known to be important for auxin conjugate or metal responses. We sequenced a gene encoding a basic helix-loop-helix leucine zipper (bHLH–ZIP) transcription factor in this region, At5g54680/bHLH105, and found in the last exon of ilr3-1 a single-base-pair deletion accompanied by an 859-bp insertion from chromosome 4. This insertion results in a premature stop codon 21 bp into the insertion in the At5g54680 coding sequence (Figure 1D). We sequenced ilr3-1 RT–PCR products to confirm the presence of the predicted ilr3-1 mRNA (data not shown). We used PCR to confirm that the region on chromosome 4 from which the ilr3-1 insertion originated was intact in our backcrossed ilr3-1 line and therefore did not contribute to ilr3-1 phenotypes.
Figure 1.—
ILR3 encodes a bHLH–leucine zipper transcription factor. (A) IAA–leucine resistance in ilr3-1 is dominant. Root lengths of wild-type Col-0 (ILR3/ILR3), F1 seedlings from an ilr3-1 cross to Col-0 (ilr3-1/ILR3), and homozygous ilr3-1 (ilr3-1/ilr3-1) grown for 8 days on medium containing 20 μm IAA–Leu and 0.5% sucrose are shown. Bars represent means plus standard deviations, n ≥ 10. **, P < 0.001 in a comparison with wild type using a two-tailed t-test assuming unequal variance. (B) Recombination mapping localized ilr3-1 on chromosome 5 (thick line) between markers MRB17-17+18 and K5F14-3+2. DNA markers are shown above the line, and the number of recombinants over the number of chromosomes scored is shown below. Annotated genes in this region are represented by arrows, with arrowheads depicting the direction of transcription. (C) ILR3 contains five exons (boxes) separated by four introns (lines). The ILR3–bHLH domain (solid rectangle) and the leucine zipper domain (shaded rectangle) are both present in ilr3-1. The location of the ilr3-2 T-DNA insertion is indicated by the triangle. (D) The 859-bp insertion in ilr3-1 begins after position 1529 (where 1 is the A of the initiator ATG) in the fifth exon. RT–PCR analysis indicates that the ilr3-1 coding sequence includes seven codons after the insertion begins before a stop codon occurs. Nucleotides 1523–1538 in ILR3 are shown.
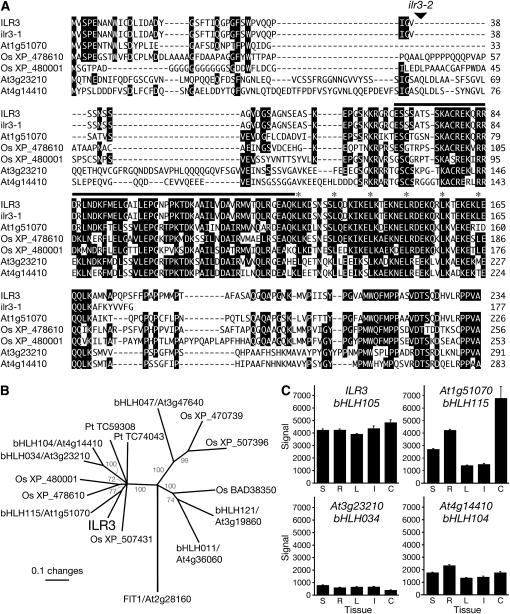
The ILR3 gene encodes a 234-amino-acid protein similar to characterized bHLH proteins from animals and plants. bHLH proteins constitute one of the largest families of transcription factors in Arabidopsis, with 162 members (Bailey et al. 2003; Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003). Only 7 of these members, including ILR3/bHLH105, contain a canonical leucine zipper domain (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003), which follows the bHLH domain (Figure 2A). Basic domains are responsible for DNA binding, while HLH and leucine zipper domains allow dimerization. All of these domains remain present in the predicted ilr3-1 protein. The insertion in ilr3-1 removes the C-terminal 64 amino acids; this C-terminal domain is highly conserved in the ILR3 subfamily (Figure 2; Toledo-Ortiz et al. 2003).
Figure 2.—
The ILR3 family in plants. (A) Alignment of ILR3, ilr3-1, and related proteins. ILR3 is a predicted transcription factor with a leucine zipper motif directly following the bHLH domain (solid rectangle). Leu residues in the zipper are indicated with asterisks. ILR3 and related bHLH–leucine zipper transcription factors in Arabidopsis and rice (Os) were aligned using the Megalign program (DNAStar, Madison, WI) ClustalW method with gap penalty 10.0 and gap length penalty 0.20, using the Gonnet series protein weight matrix. Residues conserved in at least four proteins have solid shading. (B) Phylogenetic tree of the ILR3 family. Sequences corresponding to ILR3 amino acids 62–176 (the bHLH–leucine zipper region) were aligned with bHLH–ZIP proteins from Arabidopsis, Oryza sativa (Os, listed with GenBank accession numbers), and Pinus taeda (Pt, from the TIGR gene index database; Quackenbush et al. 2001) as described in A. The unrooted phylogram was generated using PAUP 4.0b (Swofford 2001). The bootstrap method was performed for 1000 replicates with a distance optimality criterion, and all characters were weighted equally. Bootstrap values are in gray type at the tree nodes. Arabidopsis FIT1 is a bHLH protein without a leucine zipper included for comparison. (C) ILR3 and homolog tissue expression profiles. Bars represent average expression signal from Genevestigator (Zimmermann et al. 2004) compiled microarray data plus standard errors. Expression data are shown for entire seedlings (S; n = 320 chips), roots (R; n = 187), rosettes (L; n = 576), inflorescences (I; n = 139), and cell suspension (C; n = 42) of wild-type Col-0.
ILR3 is expressed throughout plant development:
To characterize ILR3 expression, we analyzed compiled microarray data present on Genevestigator (Zimmermann et al. 2004) and found robust ILR3 expression in all tissues and growth stages analyzed (Figure 2C and data not shown). We also noted that ILR3 may be somewhat more highly expressed than the three most closely related Arabidopsis bHLH genes, bHLH115, bHLH034, and bHLH104 (Figure 2C). To examine ILR3 expression within tissues, we generated plants in which ILR3 upstream sequences drove expression of the GUS reporter gene. In agreement with the Genevestigator data, ILR3–GUS was widely expressed throughout development. In particular, we detected ILR3–GUS in seedling primary and lateral root tips (Figure 3, A–C and E), vasculature (Figure 3D), and stipules (Figure 3F). In mature plants, ILR3–GUS was abundant in rosette and cauline leaf vasculature (Figure 3, G and H), hydathodes (Figure 3I), and stem vasculature (Figure 3M). In reproductive tissues, ILR3–GUS activity was apparent in sepal vasculature, anthers, pollen grains (Figure 3, J–L), siliques (Figure 3N), funicules (Figure 3O), and the abscission zones between siliques and pedicels (Figure 3P).
Figure 3.—
Expression of an ILR3–GUS fusion. (A–F) ILR3–GUS accumulates in seedling root tips, vasculature, and stipules. Seedlings were grown under continuous white light for 2 (A), 3 (B and C), or 8 (D–F) days prior to histochemical staining for GUS activity. (G–P) ILR3–GUS accumulates in tissues of all major plant organs. Parts shown are from 37- to 44-day-old adult plants grown in soil under continuous light stained for GUS activity. (A, B, D–H, J, K, and M–O) Bars, 1 mm; (C, I, L, and P) bars, 100 μm.
Phenotypic characterization of ilr3-1:
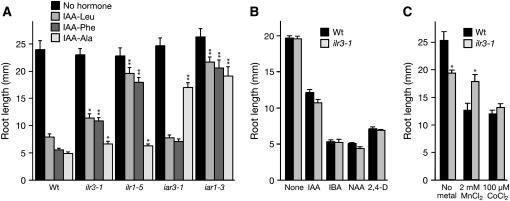
Phenotypic analyses after three backcrosses to wild type revealed that ilr3-1 roots were less sensitive than wild type to both IAA–Leu and IAA–Phe but responded normally to IAA–Ala, a conjugate resistance profile similar to the amidohydrolase mutant ilr1 (Figure 4A). This profile contrasts with iar3, a mutant defective in another amidohydrolase, which is preferentially IAA–Ala resistant (Davies et al. 1999), and the putative metal transporter mutant iar1, which displays decreased sensitivity to several conjugates (Lasswell et al. 2000).
Figure 4.—
Altered ilr3-1 responses to IAA conjugates and metals. (A) ilr3-1 responses to IAA–amino acid conjugates. Root lengths of wild-type Col-0 (Wt), ilr3-1, ilr1-5, iar3-1, and iar1-3 seedlings grown for 8 days on unsupplemented medium with 0.5% sucrose or medium containing 20 μm IAA–Leu, IAA–Phe, or IAA–Ala and 0.5% sucrose are shown. (B) ilr3-1 has wild-type responses to free auxins. Root lengths of Col-0 (Wt) and ilr3-1 seedlings grown for 8 days on medium without hormones or media containing 10 nm IAA, 10 μm IBA, 300 nm NAA, or 60 nm 2,4-D and 0.5% sucrose are shown. (C) Root lengths of Col-0 (Wt) and ilr3-1 seedlings grown for 8 days in yellow-filtered light at 22° on medium without sucrose containing no supplemental metal, 2 mm MnCl2, or 100 μm CoCl2. Error bars indicate standard errors of the means (n ≥ 10). Two-tailed t-tests assuming unequal variance were performed to compare mutant and wild-type values; *, P < 0.01; **, P < 0.001.
Although ilr3-1 roots displayed reduced sensitivity to certain IAA conjugates, the mutant responded like wild type to primary root elongation inhibition caused by natural and synthetic auxins including IAA, indole-3-butyric acid (IBA), 1-naphthaleneacetic acid (NAA), and 2,4-dichlorophenoxyacetic acid (2,4-D) (Figure 4B). ilr3-1 hypocotyl length resembled wild type, and ilr3-1 seedlings had wild-type numbers of lateral roots (data not shown). We did not note aberrant phenotypes of ilr3-1 plants grown in soil. Thus, ilr3-1 is specifically resistant to certain IAA–amino acid conjugates rather than generally compromised in auxin responses.
Because other IAA-conjugate response mutants suggest a link between metal homeostasis and IAA-conjugate sensitivity (Lasswell et al. 2000; Magidin et al. 2003), we tested ilr3-1 seedlings for root growth responses on medium supplemented with Mn2+, Co2+, Zn2+, Ca2+, Cd2+, or Fe. We found that ilr3-1 roots were clearly less sensitive to exogenous Mn2+ than wild type (Figure 4C) but had responses more similar to wild type to Co2+, Zn2+, Ca2+, Cd2+, and Fe (Figure 4C; data not shown). Because the metal response assays were conducted on medium lacking sucrose, this analysis also revealed that ilr3-1 had a somewhat short root on medium lacking sucrose (Figure 4C), but normal root elongation on sucrose-supplemented medium (Figure 4, A and B).
Ectopic ilr3-1 expression in wild type recapitulates ilr3-1 mutant phenotypes:
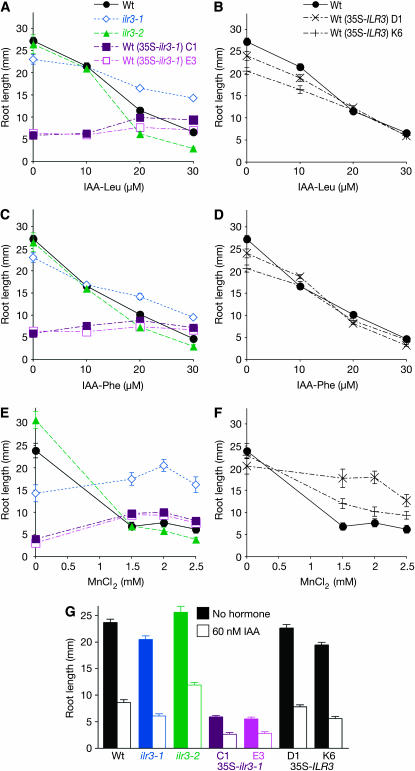
To confirm that the dominant lesion we identified in ilr3-1 was responsible for the ilr3-1 mutant phenotypes, we subcloned ilr3-1 and ILR3 cDNAs behind the cauliflower mosaic virus 35S promoter in the 35SpBARN plant transformation vector (LeClere and Bartel 2001). We transformed these constructs into wild-type plants and assayed homozygous lines derived from the transformants for root elongation on unsupplemented medium and medium containing IAA, IAA–Leu, IAA–Phe, or MnCl2.
Whereas we observed ilr3-1 root elongation defects only on medium lacking sucrose (Figure 4), we found that ectopic expression of the ilr3-1 cDNA in wild type reduced root elongation approximately fivefold on medium both with (Figure 5, A, C, and G) and without (Figure 5E) sucrose. Interestingly, the roots of these 35S–ilr3-1 plants were completely resistant to the inhibitory effects of IAA–Leu and IAA–Phe at concentrations that resulted in a fivefold reduction in wild-type root length (Figure 5, A and C). Similarly, exogenous MnCl2 did not inhibit 35S–ilr3-1 roots, but rather slightly promoted elongation (Figure 5E). To test whether this apparent insensitivity reflected a general inability to further reduce root elongation, we tested the response of 35S–ilr3-1 roots to IAA. We found that they responded by further reducing elongation (Figure 5G), consistent with the wild-type response of the ilr3-1 mutant to free IAA (Figures 4B and 5G). Resistance to IAA–Leu, IAA–Phe, and MnCl2 resulting from ectopic expression of the ilr3-1 cDNA in wild-type plants indicates that the lesion we identified in ilr3-1 is responsible for the phenotypes observed in the dominant ilr3-1 mutant. In contrast to 35S–ilr3-1 roots, roots of 35S–ILR3 plants more closely resembled wild type on medium with or without IAA–amino acid conjugates (Figure 5, B and D), indicating that ILR3 is not normally limiting for conjugate resistance. In contrast, ILR3 expression is limiting for MnCl2 resistance, as plants overexpressing wild-type ILR3 were resistant to MnCl2 (Figure 5F).
Figure 5.—
Recapitulating ilr3-1 mutant phenotypes by overexpressing ilr3-1 in wild type. Col-0 (Wt), ilr3-1, ilr3-2, and two independent lines each for Wt (35S–ILR3) and Wt (35S–ilr3-1) were grown on unsupplemented medium or medium containing the indicated concentrations of IAA–Leu (A and B), IAA–Phe (C and D), MnCl2 (E and F), or IAA (G). Medium in A–D and G contained 0.5% sucrose; that in E and F lacked sucrose. Root lengths were measured after 8 days of growth in yellow-filtered light (n ≥ 10). Error bars represent standard errors of the means.
To confirm that the overexpression constructs were having the expected effects on ILR3 transcript levels, we used quantitative real-time reverse transcriptase PCR (qRT–PCR) with gene-specific oligonucleotides and probes. We found that the 35S–ILR3 seedlings accumulated ∼19- to 24-fold more ILR3 transcript than wild type, whereas levels were increased only ∼3-fold in 35S–ilr3-1 seedlings (Figure 6A). This result suggests that the ilr3-1 mRNA is less stable than the ILR3 mRNA or that seedlings ectopically expressing high levels of ilr3-1 are compromised and not recovered following transformation. Indeed, even the modest overexpression of ilr3-1 that we observed was accompanied by a dramatic short-root phenotype that we did not observe in the ilr3-1 mutant or in 35S–ILR3 lines (Figure 5, A, C, and E), and we recovered fewer viable transformants using the 35S–ilr3-1 construct than the 35S–ILR3 construct (see materials and methods).
Figure 6.—
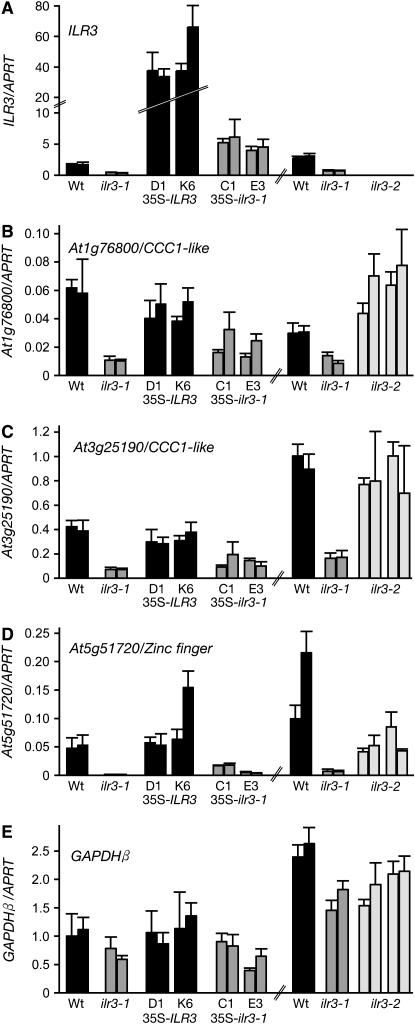
Specific transcripts misregulated in ilr3 mutants. ILR3 (A) and the indicated putative ILR3 target mRNAs (B–D) were quantified in total RNA prepared from 7-day-old seedlings from Col-0 (Wt), ilr3-1, two independent lines each for Wt (35S–ILR3) and Wt (35S–ilr3-1), and progeny of two ilr3-2 plants using quantitative real-time RT–PCR with gene-specific oligonucleotides and probes. GAPDHβ is a nontarget control message (E). mRNA levels are displayed relative to the level of an APRT control mRNA (Woodward and Bartel 2005a) in the sample, and bars represent mean values for three PCR replicates for two independent reverse transcription reactions on biological replicates. Error bars show the standard deviations of the means for three PCR replicates of a single reverse transcription reaction.
Loss of ILR3 increases auxin conjugate responsiveness:
To examine consequences of reduced ILR3 function, we identified a second ilr3 allele from the Salk Institute Genomic Analysis Laboratory sequence-indexed collection of insertion mutants (Alonso et al. 2003). This mutant, designated ilr3-2 (SALK_004997), has a T-DNA inserted 190 bp into the first ILR3 intron (Figure 1C). This insertion could allow translation of the first exon, truncating the protein prior to the bHLH or any other conserved region (Figure 2A). Using qRT–PCR with ILR3-specific oligonucleotides, we did not detect intact ILR3 transcripts in homozygous ilr3-2 (Figure 6A). Thus, ilr3-2 is likely to confer a null phenotype, although we have not explored the possibility that ilr3-2 encodes a partially functional or neomorphic protein fragment. We analyzed ilr3-2 root elongation on compounds to which the ilr3-1 mutant is resistant and found that ilr3-2 roots are slightly more sensitive than wild type to exogenous IAA–Leu and IAA–Phe (Figure 5, A and C). In contrast, the ilr3-2 mutant had nearly wild-type root length on MnCl2 (Figure 5E). Like ilr3-1, ilr3-2 displayed wild-type sensitivity to free IAA (Figure 5G).
We examined root growth of heterozygous ilr3-2/ILR3 seedlings on medium containing IAA–Leu and found that ilr3-2 is recessive (data not shown). Because the ilr3-2 loss-of-function allele increases sensitivity to auxin conjugates, we conclude that ILR3 normally reduces responsiveness to IAA–Leu and IAA–Phe. This result implies that the dominant ilr3-1 mutant confers a gain of function rather than causing a dominant-negative effect, as it displays IAA-conjugate resistance phenotypes opposite to those of the ilr3-2 loss-of-function allele.
Genes with altered expression in ilr3-1 seedlings:
Because ILR3 encodes an apparent transcription factor, we sought to determine if the auxin conjugate resistance of ilr3-1 was accompanied by reduced expression of genes known to be necessary for conjugate responsiveness. We used qRT–PCR with gene-specific oligonucleotides and probes to assay expression of ILR2 and the auxin conjugate hydrolase genes ILR1 and IAR3 in RNA prepared from 7-day-old ilr3-1 and wild-type seedlings grown on unsupplemented medium. We found nearly wild-type mRNA levels in ilr3-1 (Table 1), suggesting that ILR3 does not regulate expression of these IAA-conjugate sensitivity genes.
TABLE 1.
Gene expression in 7-day-old ilr3-1 mutant and wild-type seedlings
| Microarray
|
qRT–PCR: | ||||
|---|---|---|---|---|---|
| Signala
|
Fold changeb: | Fold changec: | |||
| Transcript | Encoded protein | ilr3-1 | Wt | ilr3-1/Wt | ilr3-1/Wt |
| At5g54680 | ILR3 bHLH–leucine zipper transcription factor | 168 | 2122 | −12.6* | −4.4 ± 0.7 |
| At5g51720 | Expressed protein (zinc-finger domain) | 94 | 1226 | −13.0* | −37.4 ± 4.8 |
| At1g76800 | Putative metal transporter (Ccc1p-like) | 56 | 267 | −4.4* | −5.7 ± 0.2 |
| At3g25190 | Putative metal transporter (Ccc1p-like) | 662 | 2050 | −2.9* | −5.7 ± 0.3 |
| At1g21140 | Putative metal transporter (Ccc1p-like) | 189 | 525 | −2.7* | −4.1 ± 0.5 |
| At3g12900 | Oxidoreductase-like | 284 | 946 | −3.2* | 1.0 ± 0.5 |
| At3g02875 | ILR1 IAA–Leu hydrolase | 107 | 145 | −1.3* | −1.3 ± 0.3 |
| At1g51760 | IAR3 IAA–Ala hydrolase | ND | ND | ND | −1.4 ± 0.2 |
| At3g18485 | ILR2 novel protein | ND | ND | ND | −1.1 ± 0.04 |
| At1g68100 | IAR1 ZIP-like metal transporter | 793 | 769 | −1.0 | ND |
| At1g24180 | IAR4 pyruvate dehydrogenase E1α | 1737 | 1813 | 1.1 | ND |
Mean signal from three biological replicates. ND, not determined. (Probes for IAR3 and ILR2 are not present on the Affymetrix ATH1 chips.)
Asterisks indicate fold changes that are significant (P < 0.05) according to two-tailed t-tests assuming unequal variance.
Error represents the standard deviation of the fold change values from three PCR replicates of each of two biological replicates.
To identify potential ILR3 targets, we compared global transcript levels in ilr3-1 and wild-type seedlings using Affymetrix ATH1 Arabidopsis “whole-genome” microarray chips, which monitor expression of ∼22,000 genes. We analyzed three biological replicates of RNA prepared from 7-day-old wild-type and ilr3-1 seedlings (Figure 7; supplemental Table 2 at http://www.genetics.org/supplemental/). This analysis revealed two transcripts potentially downregulated >10-fold in ilr3-1: ILR3 itself and At5g51720, which encodes an apparent C2H2 zinc-finger protein (Bateman et al. 2002). In addition, four transcripts appeared to be downregulated between 2.5- and 4-fold in ilr3-1, and these encode an oxidoreductase homolog (At3g12900) and three Ccc1p-like putative metal transporters (Fu et al. 1994; Lapinskas et al. 1996; Li et al. 2001). The Arabidopsis genome contains six CCC1-like genes (Figure 8A); the three CCC1-like genes misregulated in ilr3-1 may be the more highly expressed members of the family (Figure 8B). In addition to identifying potential ILR3 targets, the microarray analysis confirmed that ILR1 mRNA levels were similar to wild type in ilr3-1 and revealed that IAR1 and IAR4, additional genes required for conjugate responsiveness (Lasswell et al. 2000; LeClere et al. 2004), had unchanged transcript levels in ilr3-1 (Table 1).
Figure 8.—
Ccc1p-like membrane proteins in plants. (A) Ccc1p-like Arabidopsis and rice (Os) proteins were aligned as in the Figure 2 legend. Names of proteins with ILR3-regulated accumulation of the encoding mRNAs are shown in black, and others are in gray. Identical residues in at least four proteins are shaded in black; chemically similar residues are shaded in gray. Solid lines over the alignment indicate predicted transmembrane domains in At1g21140; lines below the alignment indicate predicted transmembrane domains in At2g01770. (B) CCC1-like genes are expressed in various tissues. Bars represent average transcript level from Genevestigator (Zimmermann et al. 2004) compiled microarray data plus standard errors. Genes depicted in solid bars are ILR3-regulated; those shown in shaded bars are not. Expression data are shown for entire seedlings (S; n = 320 chips), roots (R; n = 187), rosettes (L; n = 576), inflorescences (I; n = 139), and cell suspension (C; n = 42) of wild-type Col-0. At3g43630 is not shown because the ATH1 chip lacks probes specific for this transcript.
We sought to confirm the microarray results using qRT–PCR with gene-specific oligonucleotides and probes for each of the six mRNAs that were most dramatically affected in ilr3-1. In each case except At3g12900, we found mRNA levels reproducibly lower in multiple RNA preparations of ilr3-1 seedlings compared to wild type (Figure 6, B–D, Table 1, and data not shown). In addition to ilr3-1, the confirmed misregulated transcripts in ilr3-1 seedlings encode three uncharacterized Ccc1p-like putative metal transporters and the uncharacterized zinc-finger protein.
To determine whether ectopic expression of the ilr3-1 cDNA, which recapitulated the root response phenotypes of ilr3-1 (Figure 5), also conferred gene expression changes observed in the ilr3-1 mutant, we measured levels of the putative ILR3-regulated mRNAs in the ilr3-1 and ILR3 overexpression lines. We found that two CCC1 transporter-like transcripts and the At5g51720 zinc-finger transcript accumulated to lower levels in 35S–ilr3-1 lines (Figure 6, B–D), confirming that the ilr3-1 lesion is responsible for the decreased expression of these putative target mRNAs. In contrast, we did not detect altered levels of messages misregulated in ilr3-1 plants when wild-type ILR3 was overexpressed (Figure 6, B–D), consistent with our finding that seedlings overexpressing ILR3 respond like wild type to IAA conjugates (Figure 5, B and D).
In the T-DNA insertion ilr3-2 allele, the transcripts misregulated in ilr3-1 were not dramatically affected. Expression of the CCC1-like gene At3g25190 resembled wild type in ilr3-2 seedlings (Figure 6C). At1g76800, another CCC1-like gene, had slightly higher (approximately twofold) expression in ilr3-2 compared to wild type (Figure 6B). At5g51720, encoding the zinc-finger domain protein, had slightly reduced (approximately twofold) expression in ilr3-2 seedlings (Figure 6D).
Altered ion homeostasis in ilr3 mutants:
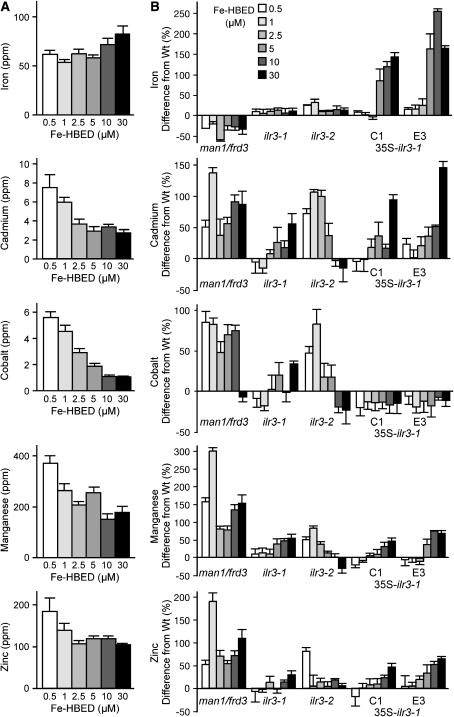
To determine whether the observed CCC1-like transcript level changes were accompanied by metal ion level alterations in ilr3 mutants, we used inductively coupled plasma–mass spectrometry (ICP–MS) to quantify metal levels. Initial experiments suggested that supplemental Fe differentially altered the elemental profile of ilr3-1 plants, so we examined the effects of Fe nutrition in wild type, ilr3-1, ilr3-2 and wild-type plants transformed with 35S–ilr3-1 by treating plants with a range of supplemental Fe concentrations from 0.5 to 30 μm. As a control, we included the man1/frd3-3 mutant (Delhaize 1996; Rogers and Guerinot 2002), which has constitutively low shoot Fe accompanied by elevated levels of other ions (Delhaize 1996; Rogers and Guerinot 2002; Lahner et al. 2003).
We found that supplemental Fe had a modest (1.3-fold) effect on Fe accumulation in wild-type leaves and that incremental increases in Fe in the fertilization solution resulted in decreased levels of leaf Cd, Co, Mn, and Zn (Figure 9A), consistent with the known Fe scavenging response of Arabidopsis (Yi and Guerinot 1996; Vert et al. 2002). man1/frd3-3 leaves accumulated less Fe and more Cd, Mn, and Zn than wild type at all supplemental Fe levels (Figure 9B; supplemental Figure 1 at http://www.genetics.org/supplemental/). Strikingly, we found that supplemental Fe resulted in ∼3-fold increases in Fe accumulation in 35S–ilr3-1 leaves (Figure 9B; supplemental Figure 1). In addition, the reduction of other metals in response to Fe fertilization was attenuated in 35S–ilr3-1 lines and the ilr3-1 dominant mutant. For example, Cd2+ levels declined 2.8-fold in Fe-treated wild type, but only 1.7-fold in ilr3-1 and 1.4-fold in 35S–ilr3-1 (Figure 9; supplemental Figure 1). We observed similar attenuation of the Fe-responsive decreases in Zn2+ and Mn2+ levels in ilr3-1 and 35S–ilr3-1 leaves (Figure 9; supplemental Figure 1). Conversely, the Fe response of the loss-of-function ilr3-2 allele displayed a heightened amplitude compared to wild type, showing increased levels of Cd, Co, Mn, and Zn in low-Fe conditions, accompanied by slightly reduced levels of some of these elements in Fe-replete conditions. Together, these changes approximately doubled the amplitude of Fe-responsive Cd, Co, and Mn diminution in the ilr3-2 mutant (Figure 9; supplemental Figure 1). We conclude that ILR3 plays a role in metal homeostasis in response to Fe nutrition, perhaps by regulating transcript levels of certain Ccc1p-like putative metal transporters.
Figure 9.—
Altered ionomic responses to iron in ilr3 mutants. Col-0 (Wt), man1/frd3-3, ilr3-1, ilr3-2, and two independent lines of Wt (35S–ilr3-1) were grown in short-day conditions and watered twice weekly with the indicated concentrations of Fe–HBED. Leaf samples from 5-week-old plants were analyzed for As2+, B+, Ca2+, Cd2+, Co2+, Cu2+, Fe3+, K+, Li+, Mg2+, Mn2+, Mo2+, Na+, Ni2+, Pb2+, PO43−, Se2+, and Zn2+ using ICP–MS. Ion profiles of the lines were compared, and levels of selected ions that responded differently to iron in wild type and the mutants are shown. (A) Bars depict median parts per million (ppm) of the indicated metals in wild-type plants (n = 12); error bars represent half of the interquartile range. (B) Median percentage of change from the wild-type value of the indicated mutants at each iron concentration. Error bars represent half of the combined interquartile range.
IAA–Leu sensitivity of ilr3 mutants depends on iron nutrition:
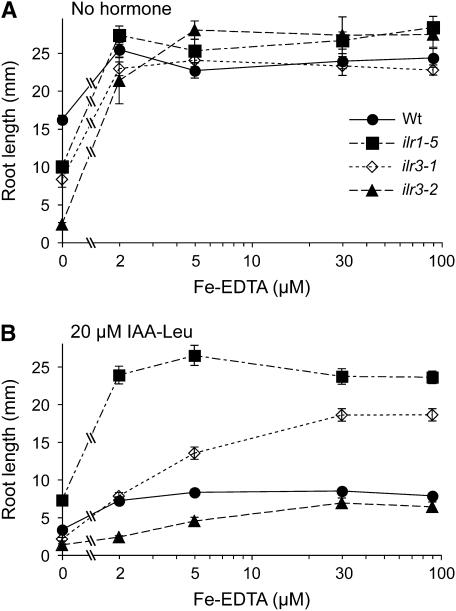
Because we noted alterations in ilr3 mutant metal homeostasis in response to Fe nutrition (Figure 9), we tested whether the aberrant ilr3 IAA–Leu response phenotypes depended on Fe nutrition. Our normal growth medium (Haughn and Somerville 1986) supplies Fe in the form of 50 μm ferric EDTA, and we found that Fe–EDTA levels between 2 and 90 μm supported normal root growth in both wild-type and ilr3 mutants (Figure 10A). Moreover, the sensitivity of wild type and the resistance of the ilr1-5 IAA–Leu hydrolase mutant to root growth inhibition by 20 μm IAA–Leu did not appreciably vary between 2 and 90 μm Fe (Figure 10B). In marked contrast to ilr1-5, the dominant ilr3-1 mutant required high Fe for maximal IAA–Leu resistance, displaying a wild-type IAA–Leu response at 2 μm Fe, intermediate resistance at 5 μm, and substantial IAA–Leu resistance at 30 and 90 μm Fe. Conversely, the recessive ilr3-2 IAA–Leu supersensitive mutant was most IAA–Leu responsive at low Fe levels, with IAA–Leu sensitivity approaching wild type at higher Fe levels. The Fe dependence of IAA–Leu response alterations in both gain- and loss-of-function ilr3 mutants is consistent with the possibility that the conjugate responsiveness changes in ilr3 mutants are secondary to alterations in metal homeostasis.
Figure 10.—
Dependence of ilr3 IAA–Leu phenotypes on the metal environment. (A) Root lengths of wild-type Col-0, the amidohydrolase mutant ilr1-5, the gain-of-function ilr3-1 allele, and the loss-of-function ilr3-2 allele grown for 8 days at 22° with continuous illumination under yellow filters on media containing 0.5% sucrose and various concentrations of Fe–EDTA. (B) The plant types from A were grown on media containing 0.5% sucrose and various concentrations of Fe–EDTA with 20 μm IAA–Leu. Points represent means plus or minus standard errors of the means (n ≥ 6). ilr3-1 roots were significantly longer than wild type on 20 μm IAA–Leu supplemented with 5, 30, and 90 μm Fe–EDTA, and ilr3-2 roots were significantly shorter than wild type on 20 μm IAA–Leu supplemented with 2 and 5 μm Fe–EDTA (P ≤ 0.001 in two-tailed t-tests assuming unequal variance).
DISCUSSION
ILR3, the founding member of the Arabidopsis bHLH–leucine zipper protein family:
The gene defective in ilr3 encodes a basic helix-loop-helix transcription factor. Heim et al. (2003) divided the Arabidopsis bHLH proteins into 12 groups, and ILR3/bHLH105 is the first of 13 group IV members to be functionally characterized. The Arabidopsis bHLH–ZIP proteins, in which the bHLH domain is followed directly by a leucine zipper motif, constitute two subgroups (ILR3/bHLH105, bHLH115, bHLH034, and bHLH104 and bHLH047, bHLH011, and bHLH121) within group IV. Other members of the ILR3 subgroup are between 63% (bHLH115) and 40% (bHLH034 and bHLH104) identical to ILR3 (Figure 2A), whereas other Arabidopsis bHLH–leucine zipper proteins are between 28% (bHLH121) and 21% (bHLH011 and bHLH047) identical to ILR3 (Figure 2B).
Although there are only seven bHLH proteins with canonical leucine zippers in Arabidopsis (Buck and Atchley 2003; Heim et al. 2003; Toledo-Ortiz et al. 2003), bHLH–ZIP proteins are common in animals. Phylogenetic analysis suggests that juxtaposition of bHLH and leucine zipper domains occurred independently in the plant and animal lineages (Buck and Atchley 2003). ILR3 family members are widely conserved in flowering plants, with likely orthologs in rice, poplar, tomato, soybean, Medicago, and grape (Figure 2B and data not shown). The presence of monocot homologs in both bHLH–ZIP subgroups suggests the possibility of conserved, distinct functions for each group. Further, ILR3 homologs are present in pine, suggesting an ancient function for the bHLH–ZIP proteins in seed plants (Figure 2B).
ILR3 is expressed in many tissues during Arabidopsis development (Figure 2C). In particular, we noted ILR3 promoter-driven GUS expression in root tips, root and shoot vasculature, anthers, siliques, hydathodes, and stipules (Figure 3), suggesting that ILR3 functions at multiple developmental stages. These tissues include areas in which the IAA-conjugate hydrolase genes are expressed (Rampey et al. 2004), consistent with the possibility that ILR3 regulates hydrolase activity. As hydrolase transcript levels are nearly wild type in ilr3-1, this regulation is likely indirect (see below).
ilr3 mutants:
The dominant ilr3-1 allele (Figure 1A) confers decreased sensitivity to certain IAA–amino acid conjugates (Figures 4A, 5, A and C, and 10B) and to Mn2+ (Figures 4C and 5E). The recessive ilr3-2 allele contains a T-DNA inserted in the first intron (Figure 1C) and lacks intact ILR3 mRNA (Figure 6A). ilr3-2 seedlings have increased sensitivity to IAA–amino acid conjugates (Figures 5, A and C, and 10B), suggesting that the dominant ilr3-1 lesion confers a gain of function. The C-terminal domain missing in ilr3-1 does not contain recognizable motifs, but is conserved in other members of the ILR3 bHLH subgroup (Heim et al. 2003), including those in rice (Figure 2A). Other bHLH proteins have similarly positioned transcriptional activation (Franks and Crews 1994; Gerber et al. 1997; Ema et al. 1999) or repression domains (Sato et al. 1994; Fisher et al. 1996; Fujitani et al. 1999). As the bHLH and leucine zipper domains remain intact in ilr3-1, it is tempting to speculate that the ilr3-1 protein still dimerizes and binds DNA via the intact bHLH and leucine zipper domains, but the missing C-terminal domain prevents proper modulation of gene expression. The dominant nature of the ilr3-1 mutation could result from altered activity or stability of ilr3-1 homo- or heterodimers.
Although expressing ilr3-1 from the 35S promoter recapitulated certain aspects of the ilr3-1 phenotype, such as conjugate resistance and gene expression changes, other 35S–ilr3-1 phenotypes were more severe. The striking root elongation defects (Figure 5) and dramatic Fe accumulation (Figure 9B) observed in 35S–ilr3-1 plants may result from increased ilr3-1 protein levels relative to the ilr3-1 mutant. Alternatively, 35S–ilr3-1 phenotypic severity could be enhanced by ectopic ilr3-1 expression in cells where ILR3 is normally not expressed. Either of these scenarios could cause ilr3-1 to interact with nontarget cis elements or to dimerize with normally unavailable partners, resulting in neomorphic phenotypes. We attempted to explore interactions between ILR3 and other Arabidopsis proteins, but both ILR3 and ilr3-1 proteins activate transcription in the yeast two-hybrid assay (data not shown), precluding use of this method to identify potential ILR3 dimerization partners.
Seedling phenotypes on exogenous IAA–amino acid conjugates and on exogenous metals do not correlate perfectly with ILR3 status. Although ilr3-1 and wild-type plants expressing 35S–ilr3-1 are resistant to IAA–Leu, IAA–Phe, and exogenous Mn, wild-type plants expressing 35S–ILR3 are resistant to Mn while remaining sensitive to IAA conjugates (Figure 5). Thus, resistances to IAA–amino acid conjugates and Mn are separable, and the latter appears more affected by changes in ILR3 level than does IAA-conjugate responsiveness. These results are consistent with our hypothesis that the primary function of ILR3 is in regulating metal homeostasis, which secondarily influences conjugate hydrolysis (see below). Future studies of ion homeostasis in 35S–ILR3 plants may allow dissection of which metals or what threshold metal levels are most relevant to IAA-conjugate sensitivity.
Transcripts misregulated in ilr3-1:
Analysis of whole-genome microarrays comparing gene expression in wild-type and ilr3-1 seedlings identified several genes potentially misregulated in ilr3-1 (Figure 7). Using quantitative real-time RT–PCR, we confirmed that five of these genes are misregulated in ilr3-1 (Table 1, Figure 6). At5g51720 is downregulated >10-fold in ilr3-1 (Table 1, Figure 6D) and encodes a 108-amino-acid protein with an apparent C2H2 zinc-finger domain (Pfam; Bateman et al. 2002), suggesting a role in DNA-binding or protein–protein interactions. Although apparent At5g51720 homologs are present in other plants, including pine, wheat, rice, poplar, cotton, and soybean (data not shown), the functions of these proteins have not been reported.
Three CCC1-like genes also are downregulated in ilr3-1 (Table 1, Figures 6 and 7). Yeast CCC1 has been isolated in several metal homeostasis screens (Fu et al. 1994; Lapinskas et al. 1996; Li et al. 2001). Ccc1p has been localized to vacuolar (Li et al. 2001) and Golgi (Lapinskas et al. 1996) membranes, and CCC1 overexpression results in vacuolar Fe and Mn accumulation (Li et al. 2001), suggesting that Ccc1p transports Fe2+ and Mn2+ from the cytoplasm to intracellular stores.
Like yeast Ccc1p, the six Arabidopsis Ccc1p-like proteins contain five predicted transmembrane domains (Figure 8A). Plant homologs of Ccc1p have not been functionally characterized, although a soybean CCC1 family member is annotated as a nodulin (Delauney et al. 1990). At2g01770 is 33% identical to yeast Ccc1p and shares an extended region between the second and third transmembrane domains; expression of this gene is not detectably misregulated in ilr3-1. Another group of five proteins are 21–24% identical to At2g01770 and yeast Ccc1p, 58–86% identical to one another, and lack the extended loop present in yeast Ccc1p and At2g01770 (Figure 8A). The three CCC1-like genes with reduced expression in ilr3-1 fall in this latter, more divergent group. The similarity between these potential ILR3 targets and the yeast Ccc1p Fe2+ and Mn2+ transporter suggests that ILR3 might modulate metal homeostasis by changing transporter levels.
As predicted by the altered Mn response and misregulation of CCC1-like genes, ion homeostasis is disrupted in ilr3 mutants (Figure 9; supplemental Figure 1). In the presence of low environmental Fe, the plant Fe scavenging response increases uptake and translocation of not only Fe, but also Mn, Cd, Co, and Zn (Delhaize 1996; Yi and Guerinot 1996; Vert et al. 2002). All of these Fe-coregulated metals are misregulated in ilr3 mutants. In particular, the amplitude of the reduced accumulation of Mn, Cd, Co, and Zn that follows Fe supplementation is dampened in the ilr3-1 gain-of-function mutant and increased in the ilr3-2 loss-of-function mutant, suggesting that ILR3 is involved in coordinating homeostasis of Fe and coregulated metals.
It remains to be determined which, if any, potential target genes identified here are directly regulated by ILR3 or which, if any, of these genes are involved in ilr3 phenotypes. ILR3 is in the subfamily of bHLH proteins expected to recognize both E- and G-boxes (Toledo-Ortiz et al. 2003), and we found that each misregulated gene contains two to six E-boxes but no G-boxes within 1 kb of the initiator ATG (data not shown), indicating that the identified genes could be direct ILR3 targets. One possibility is that ILR3 directly regulates At5g51720 expression and that At5g51720 influences CCC1-like gene expression. As At5g51720 lacks close relatives in the Arabidopsis genome, it would be interesting to determine if plants defective in At5g51720 have altered responses to IAA conjugates or metals. However, no insertional mutants disrupting At5g51720 are currently available in public collections (http://signal.salk.edu/cgi-bin/tdnaexpress).
Transcript levels of putative ILR3 targets were only subtly affected in the ilr3-2 T-DNA insertional mutant compared to the dominant ilr3-1 mutant (Figure 6). It is possible that an ILR3 homolog, such as bHLH115 (Figure 2), partially compensates for loss of ILR3 and that future analyses of mutants defective in both of these putative transcription factors will reveal more dramatic transcript alterations in loss-of-function alleles.
Models for ILR3 function:
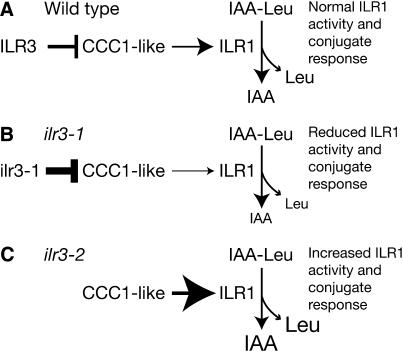
ILR3 is an apparent transcription factor important for IAA-conjugate responsiveness. The similar conjugate resistance profiles of ilr3-1, ilr1 (Figure 4A), and ilr2 (Magidin et al. 2003) initially suggested that ILR1 or ILR2 might be ILR3 targets. However, ILR1 and ILR2 transcript levels were unaltered in ilr3-1 mutant seedlings. Similarly, transcript levels of the IAR1, IAR3, and IAR4 genes, which affect conjugate sensitivity (Davies et al. 1999; Lasswell et al. 2000; LeClere et al. 2004), were unaltered in ilr3-1 (Table 1). These data suggested that ILR3 might regulate ILR1 activity rather than message levels. Microarray analysis revealed that ILR3 may directly or indirectly target genes involved in metal homeostasis, supporting a model in which perturbed metal homeostasis affects ILR1 activity (Figure 11). In support of this hypothesis, ilr3-1 defects in IAA–Leu response and ion homeostasis are most apparent at high Fe, whereas ilr3-2 defects are most apparent at low Fe (Figures 9 and 10). The ILR1 amidohydrolase requires Mn or Co for activity and is predicted to be localized to the ER lumen (Bartel and Fink 1995; LeClere et al. 2002). In yeast, Ccc1p is suggested to transport Fe2+ and Mn2+ ions from the cytosol into the vacuole (Li et al. 2001). If the Ccc1p-like proteins misregulated in ilr3-1 are also metal transporters, we speculate that reduced CCC1-like transcript levels in ilr3-1 might limit metal cofactor availability in the compartment in which ILR1 resides, thereby reducing ILR1 conjugate hydrolase activity and conjugate responsiveness (Figure 11B). Moreover, the increased IAA-conjugate sensitivity of ilr3-2 seedlings suggests increased ILR1 hydrolase activity and conjugate responses, perhaps because CCC1-like expression is freed from ILR3 repression (Figure 11C). As either Mn or Co can serve as ILR1 cofactors (Bartel and Fink 1995; LeClere et al. 2002), the dependence of ilr3-1 and ilr3-2 mutant phenotypes on environmental Fe (Figure 10) could reflect Fe nutrition effects on Mn or Co levels (Figure 9) or localization. Further studies on the Arabidopsis CCC1-like genes are needed to test this model; analyses of CCC1-like mutant phenotypes and Ccc1p-like transport activity may be particularly informative.
Figure 11.—
Hypothetical model for ILR3 regulation of metal homeostasis and IAA-conjugate metabolism. (A) In wild-type plants, ILR3 may repress expression of the Ccc1p-like metal transporters, which helps maintain proper ion homeostasis and normal ILR1 hydrolase activity and conjugate responses. (B) CCC1-like gene expression is reduced in the ilr3-1 mutant, altering ion homeostasis, which reduces ILR1 activity, resulting in reduced conjugate responses. (C) Plants lacking ILR3 (ilr3-2) and close homologs may insufficiently repress CCC1-like expression, causing increased ILR1 activity and increased conjugate responses.
The conjugate response profiles of the ilr3-1 (Figure 4A) and ilr2 mutants (Magidin et al. 2003) resemble that of the ilr1 hydrolase mutant (Bartel and Fink 1995) more than that of the iar3 hydrolase mutant (Davies et al. 1999). As ILR1 does not appear to be a transcriptional target of ILR3, this result implies that ILR1 and IAR3 reside in different subcellular compartments or that ILR1 is particularly sensitive to the local metal environment. Our attempts to directly determine the subcellular localization of the hydrolases by expressing GFP-tagged proteins from native promoters have been unsuccessful, perhaps because of the low basal expression of these genes (Rampey et al. 2004).
ILR3 is the second Arabidopsis bHLH transcription factor implicated in metal transport regulation. Expression of the FIT1 bHLH transcription factor gene is upregulated in roots of Fe-deficient plants (Colangelo and Guerinot 2004). fit1 mutants are inviable without Fe supplementation, and many transcripts normally induced during Fe starvation are no longer induced in the fit1 mutant, indicating that FIT1 is a positive regulator of Fe-responsive genes (Colangelo and Guerinot 2004). The ZIP metal transporter IRT1 (Eide et al. 1996) is undetectable in fit1, although IRT1 transcripts remain present. FIT1 therefore may regulate expression of a gene that affects IRT1 turnover (Colangelo and Guerinot 2004). It appears that the FIT1 and ILR3 bHLH transcription factors have different targets, as none of the 72 transcripts misregulated in fit1 (Colangelo and Guerinot 2004) were also misregulated more than twofold in ilr3-1 (data not shown). [The only FIT1-regulated gene that initially appeared to be misregulated in the ilr3-1 microarrays was the putative oxidoreductase transcript (At3g12900), but we were unable to verify this transcript as misregulated in ilr3-1 using qRT–PCR.] It is intriguing that the FIT1 bHLH protein (Figure 2B) regulates a ZIP transporter and the ILR3 bHLH protein regulates a process (IAA-conjugate sensitivity) influenced by the ZIP-like IAR1 protein (Lasswell et al. 2000). It will be interesting to determine whether further parallels exist between FIT1 and ILR3 functions.
The identification of three genes (ILR3, ILR2, and IAR1) implicated in metal homeostasis from auxin-conjugate sensitivity screens suggests that these screens are very sensitive to metal perturbations. Only single alleles of ilr3 and ilr2 (Magidin et al. 2003) have been recovered from the screens, suggesting that additional components important for both metal homeostasis and auxin metabolism await discovery. These components might include other members of the bHLH–leucine zipper-encoding family of which ILR3 is the founding member.
Acknowledgments
We thank Diana Dugas for GAPDHβ and APRT qRT–PCR primer and probe design and advice on qRT–PCR analysis; Sara Leibovich for ilr3-1 phenotypic analysis and ilr3-2 isolation; Thomas McKnight for microarray experiments; Mary Lou Guerinot for the man1/frd3-3 mutant; and Naxhiely Martinez, Melanie Monroe-Augustus, Dereth Phillips, Jeanne Rasbery, and Lucia Strader for critical comments on the manuscript. The Salk Institute Genomic Analysis Laboratory generated the sequenced-indexed T-DNA insertion mutant (ilr3-2), and the Arabidopsis Biological Resource Center at Ohio State University provided ilr3-2 seeds. This research was supported by the Robert A. Welch Foundation (C-1309 to B.B.) and the National Science Foundation Arabidopsis 2010 program (to D.E.S.). R.A.R. and A.W.W were recipients of Houston Livestock Show and Rodeo scholarships.
References
- Alonso, J. M., A. N. Stepanova, T. J. Leisse, C. J. Kim, H. Chen et al., 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al., 1999. Current Protocols in Molecular Biology. Greene Publishing Associates/Wiley-Interscience, New York.
- Bailey, P. C., C. Martin, G. Toledo-Ortiz, P. H. Quail, E. Huq et al., 2003. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., and G. R. Fink, 1994. Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91: 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B., and G. R. Fink, 1995. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268: 1745–1748. [DOI] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C. J., and J. R. Ecker, 1994. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144. [DOI] [PubMed] [Google Scholar]
- Bialek, K., and J. D. Cohen, 1992. Amide-linked indoleacetic acid conjugates may control levels of indoleacetic acid in germinating seedlings of Phaseolus vulgaris. Plant Physiol. 100: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, M. J., and W. R. Atchley, 2003. Phylogenetic analysis of plant basic helix-loop-helix proteins. J. Mol. Evol. 56: 742–750. [DOI] [PubMed] [Google Scholar]
- Clough, S. J., and A. F. Bent, 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colangelo, E. P., and M. L. Guerinot, 2004. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, R. T., D. H. Goetz, J. Lasswell, M. N. Anderson and B. Bartel, 1999. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delauney, A. J., C. I. Cheon, P. J. Snyder and D. P. Verma, 1990. A nodule-specific sequence encoding a methionine-rich polypeptide, nodulin-21. Plant Mol. Biol. 14: 449–451. [DOI] [PubMed] [Google Scholar]
- Delhaize, E., 1996. A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 111: 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, D. J., M. Broderius, J. Fett and M. L. Guerinot, 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi et al., 1999. Molecular mechanisms of transcriptional activation by HLF and HIFα in response to hypoxia: their stabilization and redox-induced interaction with CBP/p300. EMBO J. 18: 1905–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, A. L., S. Ohsako and M. Caudy, 1996. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16: 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, R. G., and S. T. Crews, 1994. Transcriptional activation domains of the single-minded bHLH protein are required for CNS midline cell development. Mech. Dev. 45: 269–277. [DOI] [PubMed] [Google Scholar]
- Fu, D., T. Beeler and T. Dunn, 1994. Sequence, mapping and disruption of CCC1, a gene that cross-complements the Ca(2+)-sensitive phenotype of csg1 mutants. Yeast 10: 515–521. [DOI] [PubMed] [Google Scholar]
- Fujitani, Y., Y. Kajimoto, T. Yasuda, T. Matsuoka, H. Kaneto et al., 1999. Identification of a portable repression domain and an E1A-responsive activation domain in Pax4: a possible role of Pax4 as a transcriptional repressor in the pancreas. Mol. Cell. Biol. 19: 8281–8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber, A. N., T. R. Klesert, D. A. Bergstrom and S. J. Tapscott, 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11: 436–450. [DOI] [PubMed] [Google Scholar]
- Haughn, G. W., and C. Somerville, 1986. Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204: 430–434. [Google Scholar]
- Heim, M. A., M. Jakoby, M. Werber, C. Martin, B. Weisshaar et al., 2003. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747. [DOI] [PubMed] [Google Scholar]
- Huang, L., C. P. Kirschke, Y. Zhang and Y. Y. Yu, 2005. The ZIP7 gene (Slc39a7) encodes a zinc transporter involved in zinc homeostasis of the Golgi apparatus. J. Biol. Chem. 280: 15456–15463. [DOI] [PubMed] [Google Scholar]
- Jefferson, R. A., T. A. Kavanagh and M. W. Bevan, 1987. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., J. S dchell and G. P. Rédei, 1992. T-DNA transformation and insertion mutagenesis, pp. 224–273 in Methods in Arabidopsis Research, edited by C. Koncz, N.-H. Chua and J. Schell. World Scientific, Singapore.
- Konieczny, A., and F. M. Ausubel, 1993. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4: 403–410. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, M., and G. Sandberg, 2001. Quantitative analysis of indole-3-acetic acid metabolites in Arabidopsis. Plant Physiol. 127: 1845–1853. [PMC free article] [PubMed] [Google Scholar]
- Lahner, B., J. Gong, M. Mahmoudian, E. L. Smith, K. B. Abid et al., 2003. Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21: 1215–1221. [DOI] [PubMed] [Google Scholar]
- Lapinskas, P. J., S. J. Lin and V. C. Culotta, 1996. The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol. Microbiol. 21: 519–528. [DOI] [PubMed] [Google Scholar]
- Lasswell, J., L. E. Rogg, D. C. Nelson, C. Rongey and B. Bartel, 2000. Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12: 2395–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last, R. L., and G. R. Fink, 1988. Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310. [DOI] [PubMed] [Google Scholar]
- LeClere, S., and B. Bartel, 2001. A library of Arabidopsis 35S-cDNA lines for identifying novel mutants. Plant Mol. Biol. 46: 695–703. [DOI] [PubMed] [Google Scholar]
- LeClere, S., R. A. Rampey and B. Bartel, 2004. IAR4, a gene required for auxin conjugate sensitivity in Arabidopsis, encodes a pyruvate dehydrogenase E1α homolog. Plant Physiol. 135: 989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClere, S., R. Tellez, R. A. Rampey, S. P. T. Matsuda and B. Bartel, 2002. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277: 20446–20452. [DOI] [PubMed] [Google Scholar]
- Li, L., O. S. Chen, D. M. Ward and J. Kaplan, 2001. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276: 29515–29519. [DOI] [PubMed] [Google Scholar]
- Magidin, M., J. K. Pittman, K. D. Hirschi and B. Bartel, 2003. ILR2, a novel gene regulating IAA conjugate sensitivity and metal transport in Arabidopsis thaliana. Plant J. 35: 523–534. [DOI] [PubMed] [Google Scholar]
- Michaels, S. D., and R. M. Amasino, 1998. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 14: 381–385. [DOI] [PubMed] [Google Scholar]
- Neff, M. M., J. D. Neff, J. Chory and A. E. Pepper, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14: 387–392. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., J. Cho, D. Lee, F. Liang, I. Holt et al., 2001. The TIGR gene indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey, R. A., S. LeClere, M. Kowalczyk, K. Ljung, G. Sandberg et al., 2004. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135: 978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, E. E., and M. L. Guerinot, 2002. FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Y., E. Asamizu, D. Shibata, Y. Nakamura, T. Kaneko et al., 2001. Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res. 8: 153–161. [DOI] [PubMed] [Google Scholar]
- Sato, R., J. Yang, X. Wang, M. J. Evans, Y. K. Ho et al., 1994. Assignment of the membrane attachment, DNA binding, and transcriptional activation domains of sterol regulatory element-binding protein-1 (SREBP-1). J. Biol. Chem. 269: 17267–17273. [PubMed] [Google Scholar]
- Stasinopoulos, T. C., and R. P. Hangarter, 1990. Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2001. PAUP*. Phylogenetic Analysis Using Parsimony (and Other Methods). Sinauer Associates, Sunderland, MA.
- Tam, Y. Y., E. Epstein and J. Normanly, 2000. Characterization of auxin conjugates in Arabidopsis. Low steady-state levels of indole-3-acetyl-aspartate, indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol. 123: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko, E., E. Rojo, J. León and J. J. Sánchez-Serrano, 1997. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 115: 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., E. Huq and P. H. Quail, 2003. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G., N. Grotz, F. Dedaldechamp, F. Gaymard, M. L. Guerinot et al., 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz, A., S. Park, J. P. Slovin, J. Ludwig-Müller, Y. S. Momonoki et al., 2002. A gene encoding a protein modified by the phytohormone indoleacetic acid. Proc. Natl. Acad. Sci. USA 99: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A. W., and B. Bartel, 2005. a The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol. Biol. Cell 16: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward, A. W., and B. Bartel, 2005. b Auxin: regulation, action, and interaction. Ann. Bot. 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Y., and M. L. Guerinot, 1996. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844. [DOI] [PubMed] [Google Scholar]
- Zimmermann, P., M. Hirsch-Hoffmann, L. Hennig and W. Gruissem, 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]