Abstract
To identify novel functions for the Cdc34/SCF ubiquitination complex, we analyzed genomewide transcriptional profiles of cdc53-1 and cdc34-2 Saccharomyces cerevisiae mutants. This analysis revealed altered expression for several gene families, including genes involved in the regulation of cell wall organization and biosynthesis. This led us to uncover a role for the Cdc34/SCF complex in the regulation of cell wall integrity. In support of this, cdc53-1 and cdc34-2 mutants exhibit phenotypes characteristic of cell wall integrity mutants, such as SDS sensitivity and temperature-sensitive suppression by osmotic stabilizers. Examination of these mutants revealed defects in their induction of Slt2 phosphorylation, indicating defects in Pkc1-Slt2 MAPK signaling. Consistent with this, synthetic genetic interactions were observed between the genes encoding the Cdc34/SCF complex and key components of the Pck1-Slt2 MAPK pathway. Further analysis revealed that Cdc34/SCF mutants have reduced levels of active Rho1, suggesting that these defects stem from the deregulated activity of the Rho1 GTPase. Altering the activity of Rho1 via manipulation of the Rho1-GAPs LRG1 or SAC7 affected Cdc34/SCF mutant growth. Strikingly, however, deletion of LRG1 rescued the growth defects associated with Cdc34/SCF mutants, whereas deletion of SAC7 enhanced these defects. Given the differential roles that these GAPs play in the regulation of Rho1, these observations indicate the importance of coordinating Cdc34/SCF activity with specific Rho1 functions.
SEVERAL signaling networks exist within eukaryotic cells to mediate the complex series of events regulating cellular proliferation. A major impetus in promoting cell proliferation is the coordinated expression of distinct families of genes encoding factors that promote the transition between distinct cell cycle phases. Many of these gene families are under the control of signaling pathways that respond to extracellular signals such as nutrient availability or stress. In this way, eukaryotic cells have developed an elegant method of responding to varying environmental conditions. One of the many means by which gene expression can be modulated is through ubiquitination of transcriptional regulatory proteins. This involves the covalent ligation of ubiquitin (Ub) onto protein substrates through an enzymatic cascade consisting of a Ub-activating enzyme, a Ub-conjugating enzyme, and a Ub ligase. Ub attachment or the assembly of poly(Ub) chains onto a protein may destine it for a variety of fates, commonly targeting that protein for degradation by the 26S proteasome. The relationship between Ub and transcription can be observed at many different levels, such as in the regulation of signal transduction effectors and the function of transcription factors (Muratani and Tansey 2003).
The Cdc34 Ub-conjugating enzyme together with the Skp1/Cdc53/Rbx1/F-box (SCF) Ub-ligase functions as a key regulator of several transcriptional events. Studies in Saccharomyces cerevisiae have defined several essential roles for this complex. One of the best-studied roles for this complex in transcription is the regulation of the Gcn4 transcription factor, which functions to regulate genes responsible for the biosynthesis of amino acids, vitamins, and purines in response to amino acid starvation (Braus 1991; Natarajan et al. 2001). Under favorable growth conditions, the Cdc34/SCF complex ubiquitinates Gcn4, targeting it for proteasomal degradation. In this manner, Gcn4 protein levels are kept low, thereby sustaining low-level expression of its target genes. However, under conditions of amino acid starvation, Gcn4 protein levels are stabilized, allowing for activation of its target genes (Hinnebusch 1997; Irniger and Braus 2003). Interestingly, under these conditions, Cdc34/SCF-mediated ubiquitination of Gcn4 is also required for the transcriptional activation of these genes, indicating that Ub also plays a direct role in transcriptional activation (Lipford et al. 2005).
Many other similar examples of Cdc34/SCF-mediated control of transcriptional regulation also exist, including the regulation of the transcriptional factors Met4 (Kaiser et al. 2000; Rouillon et al. 2000) and Tec1 (Bao et al. 2004), the transcriptional repressor Mth1 (Spielewoy et al. 2004), as well as several components of signaling pathways that affect gene expression, such as the mitogen-activated protein kinase (MAPK) Ste7 (Wang et al. 2003b). The widespread importance of the Cdc34/SCF complex in the regulation of transcriptional events prompted us to examine its transcriptional roles on a global nature. Using DNA microarrays to examine the transcriptional variance in cdc53-1 and cdc34-2 mutants, we identified a novel role for the Cdc34/SCF complex in the regulation of cell wall integrity.
In S. cerevisiae, cell wall integrity is maintained by an integrated network of signaling pathways that function to regulate cell wall metabolism and actin reorganization during the cell cycle and in response to stress. These pathways initially converge on, and then diverge from, the GTPase Rho1 (Gustin et al. 1998; Levin 2005). In response to cell wall stress, plasma membrane sensors stimulate a process leading to an increase in the activity of Rho1. Several distinct roles have been identified for Rho1, including (1) activating the protein kinase Pkc1 and subsequently the Slt2 MAPK pathway (Nonaka et al. 1995; Martin et al. 2000); (2) participating as a regulatory component of the 1,3-β-glucan synthase enzyme, which is essential for cell wall biosynthesis (Drgonova et al. 1996; Qadota et al. 1996); (3) mediating actin polymerization via the regulation of the formin proteins Bni1 and Bnr1 (Imamura et al. 1997); (4) activating the transcription factor Skn7 (Alberts et al. 1998); and (5) regulating vesicle transport (Guo et al. 2001). Several regulators of Rho1 activity exist to direct its differential roles, including guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). The two major GEFs that positively regulate Rho1 function in response to cell wall signals are Rom1 and Rom2 (Ozaki et al. 1996; Bickle et al. 1998). Negative regulation of Rho1 is achieved by several different GAPs, including Sac7 and Lrg1, each of which act to regulate distinct functions of Rho1. For example, Sac7 has been implicated as a regulator of actin polarization (Dunn and Shortle 1990; Schmidt et al. 1997) whereas Lrg1 has been implicated as a negative regulator of 1,3-β-glucan synthase activity (Watanabe et al. 2001; Fitch et al. 2004).
In this study we show that yeast strains possessing mutations in the Cdc34/SCF complex exhibit cell wall integrity defects. Our analysis suggests that these defects stem from the misregulation of Rho1 activity, implicating a role for the Cdc34/SCF complex in its regulation. Consistent with this, we observe that signaling via the Slt2 MAPK pathway, which lies downstream of Rho1 activation, is compromised in Cdc34/SCF mutants. Furthermore, we observe strong genetic interactions between genes encoding components of the Cdc34/SCF complex and the Slt2 pathway. Together, these results suggest that the Cdc34/SCF complex plays a key role in regulating yeast cell integrity.
MATERIALS AND METHODS
Yeast strains:
All yeast strains used were isogenic to K699 (Table 1). Strains harboring sic1∷kanr, slt2∷kanr, bck2∷kanr, sac7∷kanr, or lrg1∷kanr deletions were generated by homologous recombination using PCR products. Each PCR product was synthesized from genomic DNA derived from the appropriate kanr deletion strain (Open Biosystems) and included the kanr gene flanked by ∼100–500 bp of DNA sequence from the upstream and downstream flanking regions of the target gene. Each strain was subsequently transformed with the appropriate PCR product and plated onto YPD medium containing 200 μg/ml G418 to select for integrants. For slt2∷kanr and bck2∷kanr strains, selection was carried out on medium containing both G418 and 1 m sorbitol. The resulting knockout strains were then confirmed by PCR and functional analysis.
TABLE 1.
Yeast strains used in this study
| Yeast strains | Genotype | Source/reference |
|---|---|---|
| K699 (W303) | MATaade2-101 his3-11,151eu2-3,112 trpl-1 ura3-1 canl-100 | Willems (1996) |
| MTY740 | W303 MATacdc53-1 | Willems (1996) |
| MTY670 | W303 MATacdc34-2 | Willems (1996) |
| BVY036 | W303 MATaIrgl∷kanr | This study |
| BVY037 | W303 MATacdc34-2 Irg 1∷kanr | This study |
| BVY038 | W303 MATacdc53-1 Irg 1∷kanr | This study |
| BVY039 | W303 MATasIt2∷kanr | This study |
| BVY040 | W303 MATacdc34-2 sIt2∷kanr | This study |
| BVY041 | W303 MATacdc53-1 sIt2∷kanr | This study |
| BVY086 | W303 MATasicl∷kanr | This study |
| BVY087 | W303 MATacdc34-2 sicl∷kanr | This study |
| BVY088 | W303 MATacdc53-1 sicl∷kanr | This study |
| BVY121 | W303 MATasac7∷kanr | This study |
| BVY122 | W303 MATacdc34-2 sac7∷kanr | This study |
| BVY123 | W303 MATacdc53-1 sac7∷kanr | This study |
| BVY130 | W303 MATabckl∷kanr | This study |
| BVY131 | W303 MATacdc34-2 bckl∷kanr | This study |
| BVY132 | W303 MATacdc53-1 bckl∷kanr | This study |
Growth media:
Yeast strains were cultured in rich liquid medium (1% yeast extract, 2% bacto-peptone) containing 2% dextrose (YPD). Exceptions included the slt2∷kanr and bck2∷kanr strains, which were grown in YPD medium supplemented with 1 m sorbitol. Strains transformed with the YCp111.GAL (YCp), YCp111.GAL-Lrg1 (YCp-Lrg1), or YCp111.GAL-Sac7 (YCp-Sac7) plasmids were cultured in liquid SD medium (0.67% yeast nitrogen base lacking amino acids, supplemented with the required amino acids) containing 2% raffinose (SD-R). Solid YPD, SD-D (2% dextrose), and SD-G (2% galactose) media were prepared as above except that 2% bacto-agar was added to each. For the cell integrity plating experiments, YPD solid medium was supplemented with 1 m sorbitol, 0.5 m NaCl, or sodium dodecyl sulfate (SDS) to a final concentration of either 0.005% or 0.0075%.
Microarray analysis:
Wild-type, cdc53-1, and cdc34-2 cells were grown at 30° in YPD to an OD600 of 0.8. Three separate cultures of each strain were generated for subsequent microarray analysis. Cells were collected by centrifugation and washed twice with ice-cold water and total RNA was immediately extracted from the cells by the hot-acid phenol method (Kohrer and Domdey 1991). Messenger RNA (mRNA) was then isolated using the Easy-mRNA kit (QIAGEN, Chatsworth, CA), and cDNA was synthesized from the mRNA by reverse transcription, and biotin-labeled cRNA was constructed and purified using the procedures described by Affymetrix. UV spectroscopy was used to quantitate the RNA.
Biotin-labeled cRNA was hybridized to Affymetrix yeast S98 whole-genome oligonucleotide microarray chips according to the manufacturer's procedures. The chips were processed using the Affymetrix Fluidics Station 400 and the arrays were imaged using the Affymetrix GeneArray Scanner (570 nm, 3-μm pixel resolution). Each array was scanned twice and the images were averaged. The acquired images were then analyzed using the default parameters in Affymetrix's MicroArray Suite 5.0 (MAS 5.0). Subsequently the data was analyzed using Micro DB, Data Mining Tool 3.0 (DMT 3.0) and GeneSpring 6.2.
Statistical analysis (MAS 5.0) revealed that 84 and 85% of the genome in the cdc53-1 and cdc34-2 cells, respectively, was significantly present. Approximately 14% of the genes were absent in all the samples analyzed. The present genes from the triplicate cdc53-1 and cdc34-2 samples were compared in every various combination to those from the triplicate wild-type samples (for example, nine different fold changes were calculated for the cdc53-1 strain by comparing the three cdc53-1 samples to the three different wild-type samples), and from this an average fold gene expression was generated.
Plating experiments:
For all plating experiments, a single colony of each strain examined was used to inoculate YPD liquid medium. Each culture was incubated at 30° and grown to midlog phase (OD600 = ∼0.5–1.0) prior to plating. A dilution series of each culture was subsequently prepared, and 105, 104, 103, and 102 cells were spotted onto four separate plates of the appropriate solid culture medium. The plates were then incubated at 30°, 33°, 35°, and 37° for 3 days prior to documentation. All plating experiments were carried out in duplicate using two separate colonies and were confirmed by at least one repetition of the experiment.
For LRG1 overexpression the wild-type, cdc34-2, and cdc53-1 strains were transformed either with an empty control plasmid (YCp111.GAL/YCp) or with the same plasmid carrying the LRG1 coding sequence under control of the GAL1 promoter (YCp111.GAL-Lrg1/YCpLrg1). Plating experiments using each transformed strain followed the same procedure as described above except that single colonies were initially used to inoculate SD-R liquid medium. A dilution series of each culture was subsequently prepared and the appropriate volume of each spotted onto SD-D or SD-G solid medium lacking leucine. Plates were then incubated at 30° for 3 days prior to documentation.
YCp111.GAL was constructed using a fragment of the pESC(Trp) plasmid (Invitrogen, San Diego) that contains a multiple cloning sequence, a transcriptional terminator, and the GAL1 and GAL10 promoters. This fragment was generated by PCR using oligonucleotides that introduced a 5′ PstI and a 3′ MfeI restriction site. These sites were used to ligate this fragment into the PstI and EcoRI restriction sites of the YCplac111 plasmid. The LRG1 coding region was PCR amplified from genomic DNA and inserted into the EcoRI and SstI sites of the YCp111.GAL multiple cloning sequence to generate YCp111.GAL-Lrg1.
Flow cytometry:
To assess the cell cycle position of various cultures, the DNA content of the cells was determined. An aliquot of each culture was taken, and the cells were collected by centrifugation. Cells were then fixed in 70% ethanol followed by overnight incubation at 4°. Fixed cells were subsequently stained with propidium iodide and analyzed by flow cytometry using a FACScan instrument (Becton Dickinson, San Jose, CA), as described previously (Epstein and Cross 1992).
Slt2 phosphorylation assays:
Cultures were grown in YPD liquid medium at 30° to an OD600 of ∼0.5 after which they were shifted to 37° or into YPD liquid medium supplemented with 2 mm caffeine. Aliquots of each culture were taken prior to the shift as well as at 15-, 30-, and 60-min time points after the shift for immunoblot analysis. The level of Slt2 or phosphorylated Slt2 present at each time point was determined by immunoblot analysis. To this end, a 1.5-ml aliquot was taken at each time point, the cells were collected by centrifugation, and the cell pellet was immediately frozen in liquid nitrogen. Cells were subsequently lysed by resuspending the pellet in SDS load buffer (500 mm Tris–HCl, pH 6.8, 20% glycerol, 10% SDS, 0.1% bromophenol blue, 100 mm DTT) followed by boiling for 5 min. The volume of SDS load buffer used for each aliquot was normalized on the basis of the OD600 of each culture at each time point. Cells lysates were clarified by centrifugation and then separated by electrophoresis through a 10% SDS–polyacrylamide gel followed by transfer to a polyvinylidene difluoride membrane. To detect dually phosphorylated Slt2, the membranes were probed with an antiphospho-p44/42 MAPK (Thr202/Tyr204) primary antibody (Cell Signaling Technology and New England Biolabs, Beverly, MA) diluted 1:1000 in TBS-T (50mm Tris–HCl, pH 7, 150 mm NaCl, 0.1% Tween-20) with 2% w/v skim milk powder overnight at 4°. Membranes were then washed with TBS-T and probed with a HRP-conjugated anti-rabbit (Cell Signaling Technology and New England Biolabs) secondary antibody at 1:1000 dilution in TBS-T for 2 hr at room temperature. Total Slt2 was detected by stripping and reprobing the membranes with an anti-Mpk1 primary antibody (Santa Cruz Biotechnology) at 1:100 dilution in TBS-T followed by a HRP-conjugated anti-mouse antibody (Santa Cruz Biotechnology) at a dilution of 1:1000 in TBS-T using the same conditions as described for detection of phosphorylated Slt2.
Rho1 GTP assays:
Wild-type, cdc34-2, and cdc53-1 strains were transformed either with an empty control plasmid (YCp111.GAL) or with the same plasmid carrying the RHO1 coding sequence followed by a HA epitope tag under control of the GAL1 promoter (YCp-Rho1-HA). These cells were grown to midlog phase at 30° in SD-R and then galactose was added to a final concentration of 0.2% and the cells were grown for an additional 2 hr to induce a moderate level of Rho1-HA expression. Cells were then lysed in GPLB buffer (20 mm Tris–HCl, pH 7.5, 150 mm NaCl, 5 mm MgCl2, 0.5% NP40, 5 mm glycerophosphate, 5 mm NaF, 1 mm DTT, 1 mm PMSF) and the extracts were incubated either with GST–rhotekin-binding domain bound to glutathione agarose beads (GST–RBD) (Cytoskeleton) or with GST bound to glutathione agarose beads for 1 hr at 4°. The beads were then washed five times with GPLB buffer, and SDS load buffer was added. The samples were then analyzed by immunoblotting with a mouse anti-HA antibody (12CA5, Roche).
Microscopy:
To examine cell morphology, cells from an exponentially growing culture were initially fixed by the addition of 4% formaldehyde and subsequently incubated at room temperature for 10 min. Cells were then collected by centrifugation, resuspended in phosphate buffered saline (PBS) containing 4% formaldehyde, and incubated at room temperature for 1 hr. The samples were washed twice using PBS, briefly sonicated at the lowest setting to disperse cell clumps, and then spotted onto a microscope slide for analysis. Cells were visualized by DIC microscopy using a Zeiss Axioskop 2 microscope and documented using a Spot digital camera and Spot software 3.0.4 (Diagnostic Instruments).
RESULTS
The transcriptional profiles of cdc53-1 and cdc34-2 mutants reflect Cdc34/SCF function:
In an attempt to uncover novel functions for the Cdc34/SCF ubiquitination complex, we analyzed the transcriptional profiles of the cdc53-1 and cdc34-2 temperature-sensitive mutants using DNA microarray analysis. To mitigate the potentially confounding effects of heat-stress-related transcriptional changes, we performed our analysis at the permissive temperature of 30°. We reasoned that at this temperature the mutants would grow efficiently with minimal defects, yet may still exhibit sufficient transcriptional variation to delineate novel roles. Transcription profiles were generated from three independent, asynchronous, midlog phase cultures of cdc53-1 and cdc34-2 mutants and compared to that of wild-type cells (see supplemental Table 1 at http://www.genetics.org/supplemental/ for the complete data set).
Relative to wild-type cells, the cdc53-1 and cdc34-2 mutants exhibited increased gene expression (twofold or higher) for 175 and 146 genes, respectively, and a decreased expression (twofold or more) for 19 and 17 genes, respectively. Comparison of the transcription profiles for the two mutants showed a high correlation (r2 = 0.534) with an overlap of 92 genes showing an induction and 9 genes showing a reduction of twofold or more. Overall, 229 different genes were induced and 27 genes were repressed for expression when the two gene sets were combined.
The similar effects that the cdc53-1 and cdc34-2 mutations had on global transcription indicated that Cdc53 and Cdc34 affect the expression of a common set of genes, an observation consistent with the known roles for the Cdc34/SCF complex in transcriptional regulation. Analysis of the microarray data revealed enrichment for several groups of genes, many of which are regulated by Cdc34/SCF targets, including (1) the transcriptional activator Gcn4, which regulates the expression of genes involved in the metabolism and biosynthesis of small molecules including amino acids, vitamins, cofactors, and purine bases; (2) the transcriptional activator Met4, which regulates the expression of genes involved in sulfur metabolism and biosynthesis of the sulfur containing the amino acids methionine and cysteine; (3) the transcription factors Tec1 and Ste12 and the signaling effectors Ste7 and Ste3, which regulate various aspects of the mating signaling pathway; and (4) Mth1, which downregulates the expression of genes involved in glucose metabolism and transport.
Taken together, the microarray analysis confirms that the data set is of high confidence as known functions for the Cdc34/SCF complex were identified. Therefore, we demonstrate that examining Cdc34/SCF function via the analysis of the cdc53-1 and cdc34-2 transcriptional profiles at a temperature permissive for growth is a valid approach.
Novel functions for the cdc53-1 and cdc34-2 mutants are revealed by the microarray analysis, including an induced expression of cell-wall-related genes:
In addition to genes known to be regulated by the Cdc34/SCF complex, our microarray analysis identified changes in expression of several gene clusters that had not been previously linked to Cdc34/SCF function. Most notably, these included genes involved in sporulation, heavy metal ion homeostasis, and cell wall organization and biosynthesis (Tables 2 and 3).
TABLE 2.
Selected groups of genes induced in the cdc53-1 and cdc34-2 mutants
| Biological process (based on gene ontology) | P-value | Genes | Genes in input cluster/genes in category | Fold enrichmenta | Cdc34/SCF target(s) regulating this process |
|---|---|---|---|---|---|
| Amino acid metabolism (GO:0006520) | 1.011e-10 | GDH3, HIS4, LYS20, ARO3, MET32, ARO10, SER3, STR3, ARG4, ARO9, LYS1, ARG3, MET3, CPA2, MET14, MET1, ECM40, MET2, LYS9, ARG1, HIS3, CPA1, ICL2, MET16 | 24/140 | 4.8 | Gcn4, Met4 |
| Sulfur utilization (GO:0006791) | 5.288e-08 | MET10, MET3, ECM17, MET14, MET1, MET16 | 6/8 | 20.9 | Met4 |
| Vitamin metabolism (GO:0006766) | 2.857e-05 | TKL2, RIB5, BNA1, BNA2, TH111, BNA5, SNO1, SNZ1, SNZ2, THI21 | 10/57 | 4.9 | Gcn4 |
| Sodium transport (GO:0006814) | 0.003759 | ENA5, ENA2 | 2/3 | 18.6 | Gcn4 |
| Cell–cell adhesion (GO:0016337) | 0.00734 | AGA2, MUC1 | 2/4 | 13.9 | Gcn4 |
| Sporulation (GO:0030435) | 0.007634 | UBC5, SPO71, SPS1, PRB1, SHC1, SGA1, SPS19, SPO21 | 8/80 | 2.8 | ? |
| Purine base metabolism (GO:0006144) | 0.01 | ADE1, MTD1, ADE2 | 3/13 | 6.4 | Gcn4 |
| Cell wall organization and biogenesis (GO:0007047) | 0.03079 | ECM13, YDL222C, TIR1, GSC2, ECM17, PIR3, KTR2, ECM40, PKH2 | 9/122 | 2.1 | ? |
| Heavy metal ion homeostasis (GO:0030006) | 0.03227 | PCA1, SIT1, ISU2, 1SU1 | 4/34 | 3.3 | ? |
| Signal transduction of mating signal (GO:0007330) | 0.03769 | AFR1, MFA1, STE3 | 3/21 | 4.0 | Tec1, Ste7 |
Table was generated by analyzing the combined list of genes induced in the cdc53-1 and cdc34-2 data sets using Funspec (Robinson et al. 2002).
Fold enrichment is the percentage of category genes induced in the mutant/percentage of category genes found in the genome.
TABLE 3.
Selected groups of genes repressed in the cdc53-1 and cdc34-2 mutants
| Biological process (based on gene ontology) | P-value | Genes | Genes in input cluster/genes genes in category | Fold enrichmenta | Cdc34/SCF target(s) regulating this process |
|---|---|---|---|---|---|
| Ammonium transporter (GO:0015696) | 6.341e-08 | MEP1, MEP2, MEP3 | 3/3 | 232 | ? |
| Hexose transport (GO:0008645) | 0.003633 | HTX1, HTX4 | 2/22 | 21 | Mthl |
Table was generated by analyzing the combined list of genes induced in the cdc53-1 and cdc34-2 data sets using Funspec (Robinson et al. 2002).
Fold enrichment is the percentage of category genes induced in the mutant/percentage of category genes found in the genome.
Considering that the majority of the genes (at least 70%) for which expression changes by twofold or more in cdc53-1 and cdc34-2 mutants can be explained by the misregulation of well-characterized Cdc34/SCF targets, there exists a clear enrichment for genes involved in cell wall organization and biogenesis among those left currently unexplained. At least nine genes with gene ontology (GO) annotations related to cell wall synthesis were upregulated by twofold or greater in our microarray analysis (Table 2). Furthermore, a comparison of our data set with studies of cells that have undergone cell wall damage show a significant overlap in clusters of induced genes, which tend to be enriched for regulators of cell wall biosynthesis (Garcia et al. 2004; Lesage et al. 2004; supplemental Figure 1 at http://www.genetics.org/supplemental/). Of the genes induced by twofold or more, the transcription of at least three (FKS2/GSK2, PIR3, and KTR2) are critical under conditions that compromise the cell wall (Boorsma et al. 2004; Garcia et al. 2004). In particular, FKS2, the gene encoding the catalytic subunit of the 1,3-β-glucan synthase enzyme, was upregulated. This enzyme is a key player in the generation of cell wall material and genes encoding this complex are known to be induced following cell wall insult and in mutants with chronic cell wall injuries. In addition to these genes, other genes that likely affect the yeast cell wall were also upregulated. In particular, there was a large enrichment for genes encoding known glycosyl–phosphatidylinositol-anchored cell wall proteins (21.6-fold enrichment). Apart from providing structural support, these mannoproteins are incorporated into the yeast cell wall to accommodate changes to environmental conditions as well as to mediate cell wall reorganization during the cell cycle (Mouyna et al. 2000). More generally, the induction of several of these cell-wall-specific genes parallels that observed in response to cellular stress, such as osmotic and temperature stress (Boorsma et al. 2004), as well as cell wall damage that may occur as a result of reduced 1,3-β-glucan incorporated into the cell wall (Terashima et al. 2000). Since our analysis was performed at 30° under rich growth conditions, the alteration of expression of these cell-wall-specific genes suggests a role for the Cdc34/SCF complex in maintaining cell wall integrity distinct from that of its previously characterized functions.
The cdc53-1 and cdc34-2 mutants exhibit cell wall integrity defects:
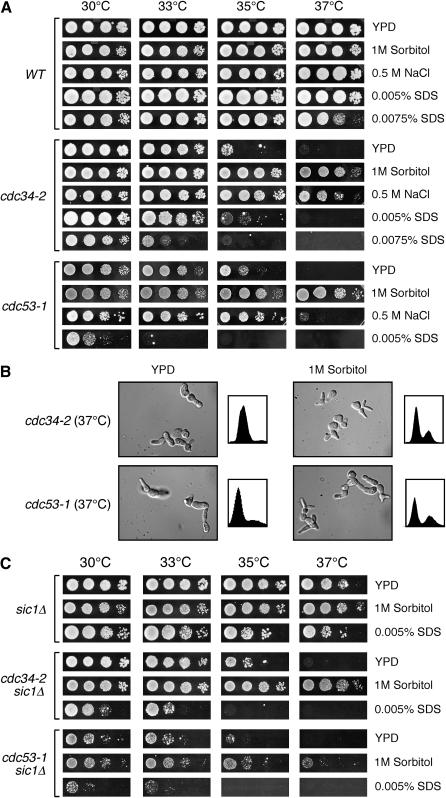
The increased transcription of cell-wall-related genes within the cdc53-1 and cdc34-2 mutants may be a consequence of cell wall defects associated with these mutants. If so, we anticipated that these mutants would manifest phenotypes representative of cell wall integrity defects, such as (1) temperature sensitivity; (2) suppression of the temperature sensitivity by the presence of high concentrations of extracellular solutes, such as sorbitol or NaCl; and (3) sensitivity to low levels of the anionic detergent SDS (Levin and Bartlett-Heubusch 1992; Paravicini et al. 1992; Martin et al. 1996). We found that the temperature sensitivity of both cdc53-1 and cdc34-2 mutants was markedly suppressed by the presence of 1 m sorbitol or 0.5 m NaCl in the growth media (Figure 1A). While these mutants grew at elevated temperature under these growth conditions, they still displayed an elongated multi-budded phenotype and an accumulation of cells in the G1 phase of the cell cycle, indicating that their cell cycle defects were not fully suppressed (Figure 1B). This suggested that the primary reason for suppression of the temperature-sensitive growth defects in these mutants resulted from cell wall stabilization rather than from stabilization of the Cdc34 or Cdc53 heat-labile proteins in the presence of osmotic stabilizers. Further evidence for cell wall defects was observed when the mutants were grown on medium containing low levels of SDS. When grown on medium containing 0.005% SDS, the nonpermissive temperature of the cdc53-1 mutant was drastically reduced (Figure 1A). The cdc34-2 strain also showed SDS sensitivity; however, a slightly higher SDS concentration of 0.0075% was required to observe a strong phenotype. On the basis of these observations, the cdc53-1 and cdc34-2 mutants exhibit phenotypes consistent with cell wall integrity defects suggestive of a role for the Cdc34/SCF complex in the maintenance of the yeast cell wall.
Figure 1.—
The cdc53-1 and cdc34-2 mutants display cell integrity defects. (A) The cdc53-1 and cdc34-2 mutants display phenotypes consistent with cell integrity defects. Wild-type (WT), cdc53-1, and cdc34-2 cells were grown on YPD or YPD containing 1 m sorbitol, 0.5 m NaCl, 0.0050% SDS, or 0.0075% SDS. The growth of the cells incubated at 30°, 33°, 35°, and 37° is shown. (B) cdc53-1 and cdc34-2 mutants grown in the presence of 1 m sorbitol display morphological and cell cycle defects. Cultures of cdc53-1 and cdc34-2 cells were grown in YPD media or YPD media containing 1 m sorbitol at 30° until midlog phase. These cells were shifted to 37° and samples were taken from each temperature and examined for morphology by microscopy and for DNA content as shown. (C) Deletion of SIC1 does not suppress the cdc34-2 and cdc53-1 cell integrity defects. sic1Δ, cdc34-2 sic1Δ, and cdc53-1 sic1Δ cells were grown on YPD and YPD containing 1 m sorbitol or 0.0050% SDS. The growth of the cells incubated at 30°, 33°, 35°, and 37° is shown.
A possible cause for the cdc34-2 and cdc53-1 cell integrity phenotypes might stem from Sic1 stabilization. Failure to degrade Sic1 in Cdc34/SCF mutants causes hyperpolarized growth that leads to an elongated, multi-budded phenotype (Schwob et al. 1994; Verma et al. 1997). It is possible that this aberrant morphology perturbs the cell wall sufficiently to cause the observed cell integrity phenotypes. If true, it would be expected that deletion of SIC1 in the cdc34-2 and cdc53-1 backgrounds, which is known to suppress their morphological defects, would also suppress the cell integrity defects associated with these strains. However, rather than suppressing the cdc53-1 and cdc34-2 cell integrity defects, the deletion of SIC1 either had no effect or exacerbated the SDS sensitivity of these mutants, respectively (Figure 1C). In fact, deletion of SIC1 in the cdc53-1 mutant showed very strong synthetic growth defects under all conditions tested. Moreover, when the double mutants were grown in the presence of 1 m sorbitol, a partial suppression of the growth defects was observed (Figure 1C). Therefore, these results indicate that the cell wall integrity defects associated with the cdc53-1 and cdc34-2 mutants occur independently of hyperpolarized growth caused by Sic1 accumulation, and rather reveal the importance Sic1 in mitigating these defects.
The cdc53-1 and cdc34-2 mutants are defective in the induction of Slt2 phosphorylation:
Our data are consistent with the notion that the cdc53-1 and cdc34-2 mutations cause cell wall defects, likely leading to the increased transcription of cell-wall-related genes. These cell wall defects do not appear to result simply from the aberrant morphology associated with these mutants, and therefore it is possible that the Cdc34/SCF complex plays a direct role in cell wall maintenance that is compromised in the cdc53-1 and cdc34-2 mutants. To uncover such a function, we considered the effect that these mutations might have on signaling through cellular signaling pathways and in particular on the cell-integrity-regulating Slt2 MAPK pathway.
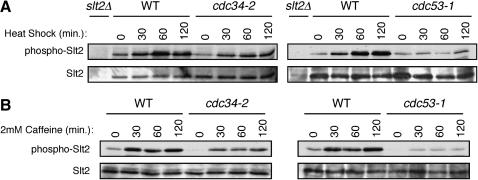
The Slt2 signaling pathway is central to the maintenance of yeast cell wall integrity and consists of a cascade of phosphorylation events that leads to the phosphorylation of the Slt2 kinase in response to cell-wall-related stress (Levin 2005). Activation of Slt2 ultimately leads to the expression of genes required for cell wall biosynthesis and cell cycle progression (Igual et al. 1996; Madden et al. 1997; Jung et al. 2002). Misregulation of this pathway often leads to cell integrity phenotypes such as those observed in the cdc53-1 and cdc34-2 mutants (Levin and Bartlett-Heubusch 1992; Paravicini et al. 1992; Martin et al. 1996). We therefore monitored the induction of Slt2 phosphorylation in the wild-type, cdc53-1, and cdc34-2 cells following different conditions that are known to induce the phosphorylation of Slt2, such as heat shock or treatment with caffeine (Martin et al. 2000). Wild-type cells showed a strong induction of Slt2 phosphorylation in response to heat shock (Figure 2A) or caffeine treatment (Figure 2B) as previously described (Martin et al. 2000). The cdc34-2 strain also showed an induction of Slt2 phosphorylation, but to a slightly lesser extent than that seen for the wild-type cells (Figure 2, A and B). By comparison, almost no induction of Slt2 phosphorylation was observed in the cdc53-1 mutant after any of the conditions tested (Figure 2, A and B). Therefore, these observations indicate that the cdc53-1 mutation, and to a lesser extent the cdc34-2 mutation, leads to a defect in the induction of Slt2 phosphorylation. Furthermore, the degree to which the cdc53-1 and cdc34-2 mutations exhibit SDS sensitivity correlates with the extent to which Slt2 phosphorylation is compromised in each mutant. Together, these observations indicate that the cell integrity phenotype and the strength of this phenotype relate to the relative degree to which Slt2 signaling is defective in each mutant.
Figure 2.—
The cdc34-2 and cdc53-1 mutants display defects in Slt2 phosphorylation. (A) Wild-type (WT), cdc34-2, and cdc53-1 cells were grown in YPD medium to midlog phase at 30° and then shifted to 37°. Samples were taken before (0 min) or at the indicated time points (15, 30, and 60 min) following heat shock. As a negative control, a sample from the slt2Δ strain after 60 min of growth at 37° was analyzed. (B) WT, cdc34-2, and cdc53-1 cells were grown in YPD medium to midlog phase at 30° and then shifted in YPD containing 2 mm caffeine at 30°. Samples were taken before (0 min) or at the indicated time points (15, 30, and 60 min) following caffeine treatment. The samples were analyzed for phosphorylated Slt2 and total Slt2 protein levels by immunoblotting.
The Slt2 MAPK pathway is required for maintaining cell wall integrity in cdc53-1 and cdc34-2 mutants:
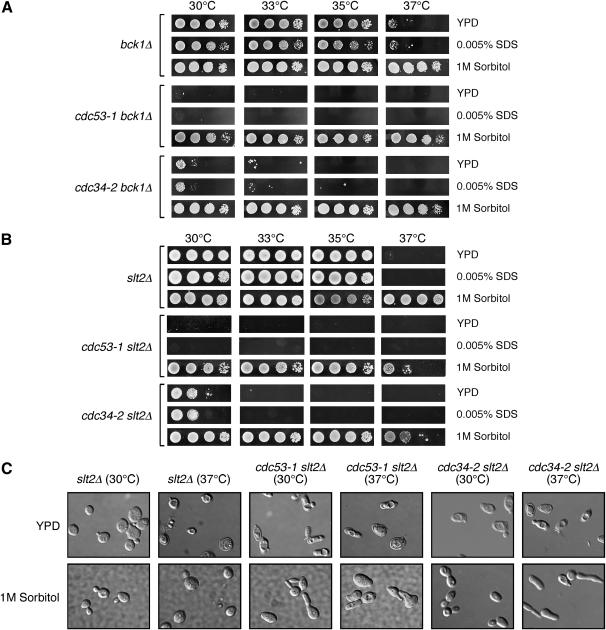
We reasoned that, if Slt2 misregulation contributes directly to the cell integrity defects observed in the cdc53-1 and cdc34-2 mutants, then eliminating Slt2 signaling in these mutants should enhance their growth defects. Deletion of SLT2 or its upstream activating kinase BCK1 is known to block signaling via this MAPK pathway, resulting in temperature-sensitive growth defects that are suppressed by growth on media containing 1 m sorbitol (Figure 3, A and B; Lee et al. 1993). Microscopic inspection of the slt2Δ cells revealed that these cells undergo cell lysis at 37°, and this was suppressed by growth in 1 m sorbitol as previously reported (Figure 3C; Madden et al. 1997). Severe synthetic growth defects were observed at all temperatures tested when either SLT2 or BCK1 were deleted in the cdc34-2 or the cdc53-1 mutants, and these defects were also suppressed by growth on 1 m sorbitol media (Figure 3, A and B). Like slt2Δ cells, microscopic inspection of cdc34-2 slt2Δ and cdc53-1 slt2Δ cells revealed a cell lysis phenotype that could be suppressed by the presence of 1 m sorbitol (Figure 3C). Interestingly, the SLT2 or BCK1 deletion strains did not display any significantly increased sensitivity to SDS as compared to growth on YPD media (Figures 3, A and B). The observed synthetic phenotype indicates that an intact cell integrity pathway is required to maintain cdc53-1 and cdc34-2 mutant viability and further indicates that at least some of the defects that occur in these mutants are independent of this signaling pathway. In particular, since the slt2Δ and bck1Δ cells do not display SDS sensitivity, it appears that this phenotype in the cdc53-1 and cdc34-2 mutants occurs as a result of other defects. Taken together, these observations suggest that the Cdc34/SCF complex mediates some aspect of cell wall integrity that is independent of, but affects, Slt2 signaling.
Figure 3.—
Abrogation of the Slt2 signaling pathway in cdc53-1 and cdc34-2 mutants results in severe synthetic lysis defects. (A) Deletion of BCK2 in cdc34-2 and cdc53-1 mutants results in synthetic growth defects. bck2Δ, cdc34-2 bck2Δ, and cdc53-1 bck2Δ cells were grown on YPD and YPD containing 1 m sorbitol or 0.005% SDS at the indicated temperatures. (B) Deletion of SLT2 in cdc34-2 and cdc53-1 mutants results in synthetic growth defects. slt2Δ, cdc34-2 slt2Δ, and cdc53-1 slt2Δ cells were grown on YPD plates and YPD plates containing 1 m sorbitol or 0.0050% SDS at the indicated temperatures. (C) Deletion of SLT2 results in cell lysis defects. slt2Δ, cdc34-2 slt2Δ, and cdc53-1 slt2Δ cells were grown in YPD containing 1 m sorbitol at 30° to midlog phase and then shifted to 37°, or transferred into YPD medium without sorbitol and grown at 30° or 37° for 6 hr. Samples from each were analyzed by microscopy.
Cdc34/SCF mutants possess defects in distinct Rho1 function(s):
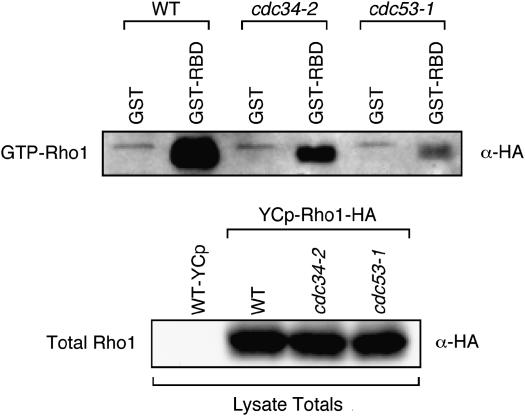
One potential link between cell wall integrity and the phenotypes observed in the cdc53-1 and cdc34-2 mutants is the Rho1 GTP-binding protein. Rho1 is a central mediator of many aspects of cell wall integrity, and its misregulation severely impairs cell growth (Levin 2005). Interestingly, low levels of SDS treatment have been shown to deregulate Rho1 activity (Bickle et al. 1998), which has the potential to affect the phosphorylation of Slt2. This suggested the possibility that the cell wall integrity defects in the cdc53-1 and cdc34-2 mutants might result from alterations in Rho1 activity. To test this, we examined the levels of GTP-bound Rho1 in these mutants. We transformed a RHO1-HA-expressing plasmid into wild-type, cdc53-1, and cdc34-2 cells and measured the ability of Rho1-HA in these cells to bind to the rhotekin-binding domain (RBD), a domain that tightly interacts with GTP-bound Rho1 (Ren et al. 1999; Garcia et al. 2006). Using GST–RBD coupled to beads, we precipitated GTP-bound Rho1 from wild-type, cdc53-1, and cdc34-2 cell extracts and found that the levels of GTP-bound Rho1 in cdc53-1 and cdc34-2 mutants were considerably less than that in wild-type cells (Figure 4). This interaction was specific since GST beads alone did not precipitate any Rho1-HA and was not due to variation in Rho1-HA protein levels as the total Rho1-HA expressed in these cells was similar. This indicates that the active state of Rho1 is reduced in these Cdc34/SCF mutants. Interestingly, the amounts of GTP-bound Rho1 in each mutant correlated with their defects in phospho-Slt2 induction and the extent of their cell integrity phenotypes, suggesting that the cell integrity defects observed in these cells are a result of deregulated Rho1 activity.
Figure 4.—
cdc34-2 and cdc53-1 mutants have reduced levels of GTP-bound Rho1. WT, cdc34-2, and cdc53-1 cells were transformed with empty YCP.GAL or YCP.GAL-Rho1-HA and then grown in the presence of galactose to induce the expression of HA-Rho1. Extracts were prepared from these cells, which expressed approximately equal amounts of HA-tagged Rho1 (bottom), and incubated with either GST–RBD glutathione beads to pull down GTP-bound Rho1 or GST glutathione beads as a control. The beads were washed and then analyzed for their retention of GTP-Rho1-HA by immunoblotting with an anti-HA antibody (top).
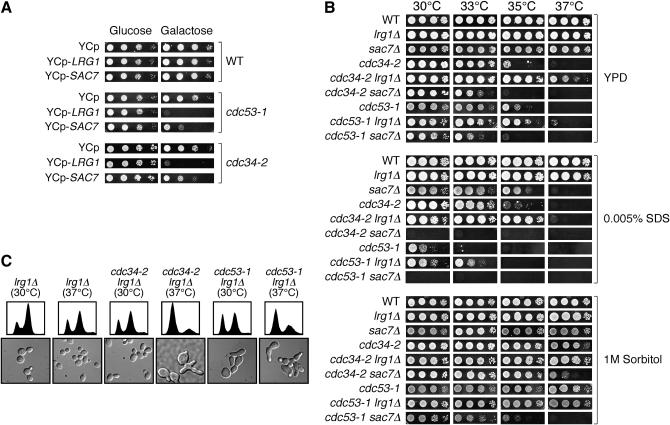
To try to gain further insight into how Rho1's function might be affected, we tested whether we could alter the phenotypes of cdc53-1 and cdc34-2 mutants by modulating the activity of Rho1. We initially pursued this by overexpressing RHO1 in these mutants, but found that overexpression of RHO1 alone had no phenotypic effects on their growth (data not shown). We also tested the expression of a hyperactive allele of Rho1 (Q58H) in wild-type, cdc53-1, and cdc34-2 mutants, but found that the expression of this gene was extremely toxic to the mutants (data not shown) as well as to the wild-type cells as previously reported (Madaule et al. 1987). Therefore, we chose to affect Rho1 function indirectly by manipulating the expression of Rho1-specific regulators. In particular, we placed the genes encoding the Rho1-specific GAPs Lrg1 and Sac7 under control of a galactose (GAL)-inducible promoter. These GAPs induce Rho1's GTPase activity and so function to reduce GTP-bound Rho1 levels, effectively acting as negative regulators. Galactose induction of LRG1 or SAC7 expression had no effect on the growth of wild-type cells when compared to growth on glucose media (GAL-repressing conditions) at 30° (Figure 5A). In contrast, the galactose induction of either LRG1 or SAC7 expression in the cdc53-1 and cdc34-2 mutants was toxic (Figure 5A), particularly in the case of LRG1 expression. Given the role of these GAPs, their overexpression likely further reduces the GTP-bound Rho1 levels in these mutants, indicating that proper regulation of Rho1 activity is essential in Cdc34/SCF mutants.
Figure 5.—
CDC34 and CDC53 display genetic interactions with the Rho1-GAPs LRG1 and SAC7. (A) Overexpression of LRG1 or SAC7 is toxic to cdc53-1 and cdc34-2 mutants. Wild-type (WT), cdc34-2, and cdc53-1 cells carrying an empty control plasmid (YCP) or with plasmids expressing LRG1 or SAC7 from the GAL1 promoter (YCP-LRG1 and YCP-SAC7, respectively) were grown on medium containing glucose (left) or galactose (right) at 30° for 3 days. (B) Deletion of LRG1 suppresses the growth defects associated with cdc53-1 and cdc34-2 mutants, whereas the deletion of SAC7 enhances these defects. cdc53-1 and cdc34-2 mutants having either LRG1 or SAC7 deleted were grown on YPD, YPD containing 1 m sorbitol, or YPD containing 0.0050% SDS at the indicated temperatures. (C) cdc53-1 lrg1Δ and cdc34-2 lrg1Δ mutants display cell cycle and morphological defects. cdc53-1 and cdc34-2 cells having LRG1 deleted were grown in YPD at 30° to midlog phase and shifted to 37°. Samples from both temperatures were examined for morphology by microscopy and for DNA content as shown.
To investigate the consequences of increasing Rho1 activity, and to try to delineate which function(s) of Rho1 might be compromised in the Cdc34/SCF mutants, we examined the effects of deleting SAC7 or LRG1 in cdc53-1 and cdc34-2 mutants. Both of these GAPS operate to reduce the levels of active GTP-bound Rho1; however, each of these proteins has been shown to regulate distinct aspects of Rho1 function. Thus, deletion of LRG1 or SAC7 would lead to a stimulation of Rho1 activity, but could also bias Rho1 activity toward a certain functional pathway. Both GAPs may affect signaling leading to Slt2 phosphorylation; however, deletion of LRG1 would negatively affect Rho1 function pertaining to its 1,3-β-glucan synthase activity (Watanabe et al. 2001). In contrast, deletion of SAC7 would affect Rho1 function pertaining to its actin cytoskeleton remodeling activity (Dunn and Shortle 1990; Schmidt et al. 2002). Therefore, deletion of the genes encoding these GAPs in the context of Cdc34/SCF mutants could reveal interactions between the function(s) of the Cdc34/SCF complex and Rho1-specific roles.
We observed that the deletion of SAC7 in wild-type cells resulted in SDS sensitivity at high temperatures, but had no obvious growth defects on YPD (Figure 5B). Deletion of SAC7 in the cdc53-1 or cdc34-2 mutants had a slight effect on cell growth on YPD media and also resulted in extreme sensitivity to SDS at all temperatures tested. Interestingly, the defects associated with the cdc34-2 sac7Δ mutant could be efficiently suppressed by the presence of 1 m sorbitol, whereas this was not the case with the cdc53-1 sac7Δ mutant. Together, these results establish a genetic relationship between Sac7 and the Cdc34/SCF complex and suggest that a particular function of Rho1 that is regulated by Sac7 may be defective in the cdc53-1 and cdc34-2 mutants.
Deletion of LRG1 had a much different effect. No growth differences were observed between lrg1Δ and wild-type cells under the conditions that we tested, but when LRG1 was deleted in the cdc53-1 and cdc34-2 mutants, we observed a suppression of their growth defects on YPD in both the presence and the absence of SDS (Figure 5B). The cell cycle defects associated with these mutants were not fully suppressed since these cells still displayed an elongated multi-budded morphology and had an accumulation of 1 n DNA content (Figure 5C). Therefore, deletion of LRG1 likely does not affect the function of the Cdc34/SCF complex in Sic1 degradation, but rather must suppress other defects within the cdc53-1 and cdc34-2 mutants that could alleviate the defects in these cells sufficiently to allow for growth. Deletion of LRG1 leads to increased activity via the Pkc1-Slt2 pathway (Lorberg et al. 2001), even when combined with the cdc53-1 and cdc34-2 mutants (data not shown). This increased pathway activity may contribute to the effects observed; however, deletion of SAC7 also leads to similar increases in Pkc1-Slt2 pathway activity (Martin et al. 2000; Schmidt et al. 2002; data not shown). Therefore, activation of this pathway alone is not capable of explaining the suppressive effects observed from the deletion of LRG1. A likely defect suppressed by deletion of LRG1 may relate to its role in 1,3-β-glucan synthesis, as Lrg1 functions primarily as a negative regulator of this Rho1 function. Although a definitive cause for the suppression by LRG1 deletion has not been delineated, these results support the notion that the Cdc34/SCF complex functions to mediate cell integrity by regulating the activity of Rho1.
DISCUSSION
Our global expression analysis of cdc53-1 and cdc34-2 mutants revealed that the Cdc34/SCF complex regulates multiple aspects of cellular growth. In some cases the observed effect directly correlated with a known role for the Cdc34/SCF complex in transcriptional regulation. These included pathways that regulate methionine and general amino acid biosynthesis. In other cases, however, subsets of genes not previously known to be regulated by the Cdc34/SCF complex were found to be upregulated within cdc53-1 and cdc34-2 mutants, including groups that regulate sporulation, heavy metal ion homeostasis, and cell wall organization and biosynthesis.
The observed induction of sporulation-specific genes was interesting since our analysis was done in haploid cells proliferating in rich medium, conditions that normally preclude the induction of these genes. With the exception of PRB1, all of the sporulation-specific genes identified are normally induced in the middle and late phases of meiotic differentiation (Chu et al. 1998). Consistent with this finding, most of these genes possess middle sporulation elements within their promoters and are induced by the meiosis-specific transcription factor Ndt80 (Chu et al. 1998). These observations implicate the Cdc34/SCF complex in the repression of these sporulation-specific genes in haploid cells. The induction of genes regulating heavy metal ion homeostasis were also interesting, given recent studies suggesting a role for Cdc34 in the resistance of yeast to methylmercury (Furuchi et al. 2002).
The link between the Cdc34/SCF complex and cell wall integrity was further substantiated by our observations that (1) these mutants display growth phenotypes consistent with cell integrity defects; (2) genetic interactions occur between CDC34 or CDC53 and genes composing the Slt2 MAPK pathway, a critical regulator of cell wall integrity; (3) cdc53-1 and cdc34-2 mutants display defects in the regulation of Slt2 phosphorylation, a key event in coordinating cell wall biosynthesis and cell cycle progression; (4) genetic interactions occur between CDC34 or CDC53 and genes encoding regulators of Rho1, a major player in cell wall synthesis and remodeling; and (5) active GTP-bound Rho1 levels are reduced in cdc53-1 and cdc34-2 mutants.
The connection between Cdc34/SCF function and cell integrity was generated in large part from observations that cdc34-2 or cdc53-1 mutants exhibit sensitivity to low concentrations of SDS. This treatment imposes stress on the cell wall, thereby lowering the permissive temperature at which these mutants proliferate. These data, combined with the observation that high concentrations of the osmotic stabilizers sorbitol or NaCl raise the nonpermissive temperature for their proliferation, suggest that mutations in the Cdc34/SCF complex result in cell wall defects. A trivial explanation for these defects may be the inability of Cdc34/SCF mutants to degrade the Cdk inhibitor Sic1 at elevated temperatures. Increased Sic1 levels lead to a prolonged G1 phase of the cell cycle that in turn causes the formation of single or multiple elongated buds, which could cause sufficient stress to produce the observed cell integrity defects. However, we believe this possibility is unlikely, given that (1) cdc34-2 sic1Δ and cdc53-1 sic1Δ cells, which do not display elongated multiple buds and proceed through the G1-S transition of the cell cycle, remain SDS sensitive and their growth is partially suppressed by the presence of 1 m sorbitol; (2) suppression of growth by high concentrations of sorbitol does not alleviate the elongated multi-budded phenotype associated with cdc34 and cdc53 mutants, indicating that sorbitol functions to osmotically stabilize cell wall defects rather than to stabilize the mutant proteins within the cell; (3) SDS treatment reduces the restrictive temperature of these mutants to temperatures at which Sic1 is efficiently degraded; and (4) there is an increased expression of cell-wall-related genes even at a temperature at which Sic1 is efficiently degraded. Of particular interest is the increased expression of FKS2, which comprises a component of the cell wall synthesis machinery and whose expression is normally increased in response to cell wall stress. Together, these data indicate a novel function for the Cdc34/SCF complex in the regulation of cell wall integrity that is distinct from its cell cycle function related to Sic1 degradation. Nevertheless, the importance of Sic1 regulation in maintaining cell growth does become apparent when SIC1 is deleted, as cdc34-2 sic1Δ and cdc53-1 sic1Δ cells have greater defects, emphasizing the importance of a proper coordination between cell cycle progression and cell wall synthesis.
It is logical that a relationship exists between factors that control cell integrity and cell division, given the extensive cell wall remodeling and synthesis that occurs throughout the cell cycle. This increased level of cell wall metabolism coincides with the formation of a nascent bud and subsequent polarized bud growth. Under these conditions, the synthesis of proteins engaged in the assembly of cell wall components is induced and these proteins, along with required materials, are directed toward the emerging bud in a controlled fashion (Cabib et al. 1998). This involves a restructuring of the actin cytoskeleton and includes the concerted action of many pathways. Several cell cycle regulators (e.g., Sic1, Cln2, and Cdc6) as well as effectors of actin cytoskeleton remodeling (e.g., Gic2) have been identified as Cdc34/SCF substrates. Therefore, a strategy linking these processes by a common regulator, such as the Cdc34/SCF complex, likely exists.
Several results suggest that cell integrity defects seen in the cdc53-1 and cdc34-2 mutants arise from the misregulation of Rho1 function. This conclusion is supported by the observations that the levels of activated GTP-bound Rho1 are reduced in these mutants and that genetic interactions exist between genes encoding the Cdc34/SCF complex and the Rho1-GAPs Lrg1 and Sac7. Inhibition of Rho1 activity via overexpression of these negative regulators is severely toxic to the cdc53-1 and cdc34-2 mutants, demonstrating the importance of properly controlled Rho1 activity in these cells. Interestingly, these GAPs affect the Cdc34/SCF mutants to different degrees, with overexpression of LRG1 causing a stronger growth defect as compared to SAC7. Although both GAPs negatively affect Rho1 activity, they have been shown to affect distinct roles for Rho1. For example, Lrg1 has been observed to regulate the 1,3-β-glucan synthesis activity of Rho1, whereas Sac7 predominately regulates the actin organization and MAPK-regulating functions of Rho1. This suggests that the decrease in active GTP-Rho1 levels that is observed in the Cdc34/SCF mutants likely exists in the pool of Rho1 that is regulated by Lrg1, possibly functioning to regulate 1,3-β-glucan synthesis. This possibility is supported by the elevated expression of FKS2, a gene encoding a component of the 1,3-β-glucan synthesis enzyme, in the cdc34-2 and cdc53-1 mutants. Furthermore, deletion of LRG1 suppresses both growth and cell integrity defects associated with cdc53-1 and cdc34-2 mutants. This suppressive effect is specific to cell integrity defects, given that at elevated temperatures cdc34-2 lrg1Δ and cdc53-1 lrg1Δ retain an elongated multi-budded morphology associated with a defect in Sic1 degradation.
Our observations indicate that the balance and specificity of Rho1 activity is crucial in Cdc34/SCF mutants. This is evident when SAC7 is deleted, which likely shifts the active pool of GTP-Rho1 from its role in 1,3-β-glucan synthesis toward its roles in actin polymerization. This results in severe cell integrity defects, particularly with respect to SDS sensitivity. It has been previously reported that disturbing the cell wall with low amounts of SDS results in an increased GDP/GTP exchange activity of Rho1 (Bickle et al. 1998), a similar effect to the deletion of SAC7. As such, deletion of SAC7 combined with the presence of SDS likely leads to increased Rho1 activity in a certain function that the cdc53-1 and cdc34-2 mutants are not capable of coping with, therefore resulting in the severe defects observed. Sac7 has been shown to have an important role in actin polymerization and Pkc1-Slt2 pathway activation, suggesting that the proper regulation of these activities is essential for maintaining cell viability in the cdc53-1 and cdc34-2 mutants.
The analysis of signaling via the Slt2 pathway confirms that defects in Slt2 phosphorylation do exist in these mutants and suggests that the cell wall integrity defects observed in these cells are, in part, a consequence of defective signaling through this pathway. The misregulation of Slt2 phosphorylation is far more pronounced in the cdc53-1 mutant than in the cdc34-2 mutant, correlating with the extreme sensitivity that the cdc53-1 mutant displays toward SDS and to its more reduced levels of GTP-bound Rho1. The importance of Slt2 pathway activity in these mutants is highlighted by the fact that the deletion of BCK1 or SLT2 results in severe lysis defects. These defects are likely due to cell wall disruption, as they can be suppressed by the presence of 1 m sorbitol. This indicates that the expression of downstream target genes of the Slt2 pathway is important for maintaining the integrity of the cell wall in these mutants and is consistent with the enrichment of cell wall biogenesis genes observed in our microarray profiling. However, the defects observed in the Cdc34/SCF mutants are not solely due to the misregulation of the Slt2 pathway, since other cell integrity defects are observed, such as SDS sensitivity. A substantial overlap (see supplemental Figure 1 at http://www.genetics.org/supplemental/) of gene regulation in cdc53-1 and cdc34-2 mutants as compared to hyperactive rho1 and pkc1 mutants lends further support to the idea that the misregulation of Slt2 signaling may occur as a consequence of upstream defects at the level of Rho1 (Roberts et al. 2000). In fact several clusters of similarly regulated genes are evident between these mutants, including a cluster of genes enriched for mediators of cell wall organization and biogenesis (P = 5.66e-09). Interestingly, similar clusters are also observed when the data sets from the cdc53-1 and cdc34-2 mutants are compared with those from cells that have undergone cell wall damage either from mutations in key cell wall regulators (Lagorce et al. 2003) or from cell-wall-disrupting reagents (Garcia et al. 2004) (see supplemental Figure 1 at http://www.genetics.org/supplemental/). This correlation suggests that a large number of genes involved in the regulation of cell wall organization and biosynthesis may be affected in the cdc53-1 and cdc34-2 mutants (more than only the nine genes that met our cutoff of being induced by twofold or greater), further supporting our observations that the Cdc34/SCF complex mediates aspects of yeast cell wall integrity.
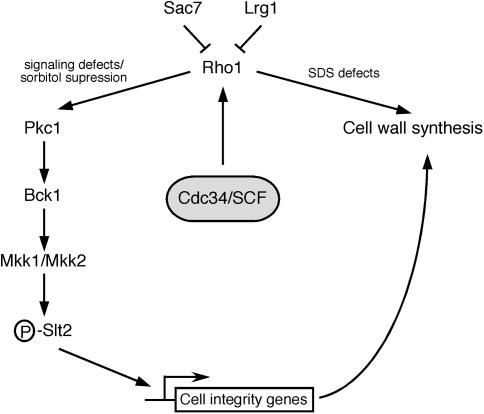
As the Cdc34/SCF complex functions to ubiquitinate proteins, its cell integrity function likely reflects this activity. Candidate targets include negative regulators of cell wall synthesis or regulators Rho1 activity. An appealing possibility is that the direct regulation of the Rho1-GAPs (Figure 6), particularly Lrg1, is influenced by Cdc34/SCF-dependent ubiquitination and subsequent degradation. This would provide a logical explanation of our data since Rho1-GAPs have the potential to downregulate both the MAPK cascade and cell wall synthesis. Alternatively, Rho1 itself may be targeted for ubiquitination by the Cdc34/SCF complex in a context-dependent manner. Supporting this possibility, Rho1 has been identified as a ubiquitinated protein in yeast (Peng et al. 2003). Furthermore, its mammalian homolog, RhoA, is ubiquitinated and targeted for degradation at distinct subcellular locations (Wang et al. 2003a).
Figure 6.—
Model for the role of the Cdc34/SCF complex in the maintenance of cell wall integrity. Depicted is our model for the role of the Cdc34/SCF complex in the regulation of cell wall integrity. In this model, the Cdc34/SCF complex regulates Rho1 function, thereby affecting several pathways, including the Slt2 signaling pathway as well as 1,3-β-glucan synthesis. This would result in transcriptional variation of genes required for cell wall biogenesis and organization. Several potential Cdc34/SCF targets exist, which could include Rho1 or negative regulators of Rho1 such as the Rho1-GAPs.
Taken together, these observations identify a novel role for the Cdc34/SCF ubiquitination complex in the mediation of yeast cell integrity. Future studies not only should identify specific cell integrity targets, but also should provide insight into the extent to which the cell integrity functions of Cdc34/SCF overlap or are coordinated with its role in cell cycle regulation. Furthermore, signaling from the yeast cell wall and the role of the cell integrity MAPK pathway in mediating cell proliferation show similarities with pathways involved in signaling from the extracellular matrix of mammalian cells. Thus, it will be of interest to determine if mammalian homologs of the yeast Cdc34/SCF complex regulate signaling from the extracellular matrix in a similar fashion.
Acknowledgments
This work was made possible by grants from the National Cancer Institute of Canada. D.S. is supported by grants from the Alberta Heritage Foundation for Medical Research and the Canadian Institutes for Health Research.
References
- Alberts, A. S., N. Bouquin, L. H. Johnston and R. Treisman, 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 273: 8616–8622. [DOI] [PubMed] [Google Scholar]
- Bao, M. Z., M. A. Schwartz, G. T. Cantin, J. R. Yates, III and H. D. Madhani, 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell 119: 991–1000. [DOI] [PubMed] [Google Scholar]
- Bickle, M., P. A. Delley, A. Schmidt and M. N. Hall, 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17: 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma, A., H. de Nobel, B. ter Riet, B. Bargmann, S. Brul et al., 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21: 413–427. [DOI] [PubMed] [Google Scholar]
- Braus, G. H., 1991. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol. Rev. 55: 349–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., J. Drgonova and T. Drgon, 1998. Role of small G proteins in yeast cell polarization and wall biosynthesis. Annu. Rev. Biochem. 67: 307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein et al., 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. [DOI] [PubMed] [Google Scholar]
- Drgonova, J., T. Drgon, K. Tanaka, R. Kollar, G. C. Chen et al., 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272: 277–279. [DOI] [PubMed] [Google Scholar]
- Dunn, T. M., and D. Shortle, 1990. Null alleles of SAC7 suppress temperature-sensitive actin mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 2308–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, C. B., and F. R. Cross, 1992. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes Dev. 6: 1695–1706. [DOI] [PubMed] [Google Scholar]
- Fitch, P. G., A. E. Gammie, D. J. Lee, V. B. de Candal and M. D. Rose, 2004. Lrg1p is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics 168: 733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuchi, T., G. W. Hwang and A. Naganuma, 2002. Overexpression of the ubiquitin-conjugating enzyme Cdc34 confers resistance to methylmercury in Saccharomyces cerevisiae. Mol. Pharmacol. 61: 738–741. [DOI] [PubMed] [Google Scholar]
- Garcia, R., C. Bermejo, C. Grau, R. Perez, J. M. Rodriguez-Pena et al., 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279: 15183–15195. [DOI] [PubMed] [Google Scholar]
- Garcia, P., V. Tajadura, I. Garcia and Y. Sanchez, 2006. Rgf1p is a specific Rho1-GEF that coordinates cell polarization with cell wall biogenesis in fission yeast. Mol. Biol. Cell 17: 1620–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, W., F. Tamanoi and P. Novick, 2001. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol. 3: 353–360. [DOI] [PubMed] [Google Scholar]
- Gustin, M. C., J. Albertyn, M. Alexander and K. Davenport, 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62: 1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch, A. G., 1997. Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272: 21661–21664. [DOI] [PubMed] [Google Scholar]
- Igual, J. C., A. L. Johnson and L. H. Johnston, 1996. Coordinated regulation of gene expression by the cell cycle transcription factor Swi4 and the protein kinase C MAP kinase pathway for yeast cell integrity. EMBO J. 15: 5001–5013. [PMC free article] [PubMed] [Google Scholar]
- Imamura, H., K. Tanaka, T. Hihara, M. Umikawa, T. Kamei et al., 1997. Bni1p and Bnr1p: downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 16: 2745–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger, S., and G. H. Braus, 2003. Controlling transcription by destruction: the regulation of yeast Gcn4p stability. Curr. Genet. 44: 8–18. [DOI] [PubMed] [Google Scholar]
- Jung, U. S., A. K. Sobering, M. J. Romeo and D. E. Levin, 2002. Regulation of the yeast Rlm1 transcription factor by the Mpk1 cell wall integrity MAP kinase. Mol. Microbiol. 46: 781–789. [DOI] [PubMed] [Google Scholar]
- Kaiser, P., K. Flick, C. Wittenberg and S. I. Reed, 2000. Regulation of transcription by ubiquitination without proteolysis: Cdc34/SCF(Met30)-mediated inactivation of the transcription factor Met4. Cell 102: 303–314. [DOI] [PubMed] [Google Scholar]
- Kohrer, K., and H. Domdey, 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194: 398–405. [DOI] [PubMed] [Google Scholar]
- Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken et al., 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278: 20345–20357. [DOI] [PubMed] [Google Scholar]
- Lee, K. S., K. Irie, Y. Gotoh, Y. Watanabe, H. Araki et al., 1993. A yeast mitogen-activated protein kinase homolog (Mpk1p) mediates signalling by protein kinase C. Mol. Cell. Biol. 13: 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage, G., A. M. Sdicu, P. Menard, J. Shapiro, S. Hussein et al., 2004. Analysis of β-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167: 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69: 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., and E. Bartlett-Heubusch, 1992. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J. Cell Biol. 116: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford, J. R., G. T. Smith, Y. Chi and R. J. Deshaies, 2005. A putative stimulatory role for activator turnover in gene expression. Nature 438: 113–116. [DOI] [PubMed] [Google Scholar]
- Lorberg, A., H. P. Schmitz, J. J. Jacoby and J. J. Heinisch, 2001. Lrg1p functions as a putative GTPase-activating protein in the Pkc1p-mediated cell integrity pathway in Saccharomyces cerevisiae. Mol. Genet. Genomics 266: 514–526. [DOI] [PubMed] [Google Scholar]
- Madaule, P., R. Axel and A. M. Myers, 1987. Characterization of two members of the rho gene family from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 84: 779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden, K., Y. J. Sheu, K. Baetz, B. Andrews and M. Snyder, 1997. SBF cell cycle regulator as a target of the yeast PKC-MAP kinase pathway. Science 275: 1781–1784. [DOI] [PubMed] [Google Scholar]
- Martin, H., M. C. Castellanos, R. Cenamor, M. Sanchez, M. Molina et al., 1996. Molecular and functional characterization of a mutant allele of the mitogen-activated protein-kinase gene SLT2(MPK1) rescued from yeast autolytic mutants. Curr. Genet. 29: 516–522. [DOI] [PubMed] [Google Scholar]
- Martin, H., J. M. Rodriguez-Pachon, C. Ruiz, C. Nombela and M. Molina, 2000. Regulatory mechanisms for modulation of signaling through the cell integrity Slt2-mediated pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275: 1511–1519. [DOI] [PubMed] [Google Scholar]
- Mouyna, I., T. Fontaine, M. Vai, M. Monod, W. A. Fonzi et al., 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275: 14882–14889. [DOI] [PubMed] [Google Scholar]
- Muratani, M., and W. P. Tansey, 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4: 192–201. [DOI] [PubMed] [Google Scholar]
- Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts et al., 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21: 4347–4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka, H., K. Tanaka, H. Hirano, T. Fujiwara, H. Kohno et al., 1995. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 14: 5931–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki, K., K. Tanaka, H. Imamura, T. Hihara, T. Kameyama et al., 1996. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15: 2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Paravicini, G., M. Cooper, L. Friedli, D. J. Smith, J. L. Carpentier et al., 1992. The osmotic integrity of the yeast cell requires a functional PKC1 gene product. Mol. Cell. Biol. 12: 4896–4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., D. Schwartz, J. E. Elias, C. C. Thoreen, D. Cheng et al., 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926. [DOI] [PubMed] [Google Scholar]
- Qadota, H., C. P. Python, S. B. Inoue, M. Arisawa, Y. Anraku et al., 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272: 279–281. [DOI] [PubMed] [Google Scholar]
- Ren, X. D., W. B. Kiosses and M. A. Schwartz, 1999. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 18: 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer et al., 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287: 873–880. [DOI] [PubMed] [Google Scholar]
- Robinson, M. D., J. Grigull, N. Mohammad and T. R. Hughes, 2002. FunSpec: a web-based cluster interpreter for yeast. BMC Bioinformatics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon, A., R. Barbey, E. E. Patton, M. Tyers and D. Thomas, 2000. Feedback-regulated degradation of the transcriptional activator Met4 is triggered by the SCF(Met30)complex. EMBO J. 19: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., M. Bickle, T. Beck and M. N. Hall, 1997. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88: 531–542. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., T. Schmelzle and M. N. Hall, 2002. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 45: 1433–1441. [DOI] [PubMed] [Google Scholar]
- Schwob, E., T. Bohm, M. D. Mendenhall and K. Nasmyth, 1994. The B-type cyclin kinase inhibitor p40SIC1 controls the G1 to S transition in S. cerevisiae. Cell 79: 233–244. [DOI] [PubMed] [Google Scholar]
- Spielewoy, N., K. Flick, T. I. Kalashnikova, J. R. Walker and C. Wittenberg, 2004. Regulation and recognition of SCFGrr1 targets in the glucose and amino acid signaling pathways. Mol. Cell. Biol. 24: 8994–9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima, H., N. Yabuki, M. Arisawa, K. Hamada and K. Kitada, 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264: 64–74. [DOI] [PubMed] [Google Scholar]
- Verma, R., R. S. Annan, M. J. Huddleston, S. A. Carr, G. Reynard et al., 1997. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278: 455–460. [DOI] [PubMed] [Google Scholar]
- Wang, H. R., Y. Zhang, B. Ozdamar, A. A. Ogunjimi, E. Alexandrova et al., 2003. a Regulation of cell polarity and protrusion formation by targeting RhoA for degradation. Science 302: 1775–1779. [DOI] [PubMed] [Google Scholar]
- Wang, Y., Q. Ge, D. Houston, J. Thorner, B. Errede et al., 2003. b Regulation of Ste7 ubiquitination by Ste11 phosphorylation and the Skp1-Cullin-F-box complex. J. Biol. Chem. 278: 22284–22289. [DOI] [PubMed] [Google Scholar]
- Watanabe, D., M. Abe and Y. Ohya, 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1,3-beta-glucan synthesis. Yeast 18: 943–951. [DOI] [PubMed] [Google Scholar]