Abstract
Substitution mapping was used to refine the localization of blood pressure (BP) quantitative trait loci (QTL) within the congenic region of S.R-Edn3 rats located at the q terminus of rat chromosome 3 (RNO3). An F2(S × S.R-Edn3) population (n = 173) was screened to identify rats having crossovers within the congenic region of RNO3 and six congenic substrains were developed that carry shorter segments of R-rat-derived RNO3. Five of the six congenic substrains had significantly lower BP compared to the parental S rat. The lack of BP lowering effect demonstrated by the S.R(ET3 × 5) substrain and the BP lowering effect retained by the S.R(ET3 × 2) substrain together define the RNO3 BP QTL-containing region as ∼4.64 Mb. Two nonoverlapping substrains, S.R(ET3 × 1) and S.R(ET3 × 6), had significantly lower BP compared to the S strain, indicating the presence of two distinct BP QTL in the RNO3 q terminus. The RNO3 q terminus was fine mapped with newly developed polymorphic markers to characterize the extent of the congenic regions. The two RNO3 BP QTL regions were thus defined as within intervals of 0.05–1.12 and 0.72–1.25 Mb, respectively. Also important was our difficulty in fine mapping and marker placement in this portion of the rat genome (and thus candidate gene identification) using the available genomic data, including the rat genome sequence.
HYPERTENSION, or high blood pressure (BP), is a disorder predisposing humans and animals to cardiovascular diseases, including stroke, heart failure, and renal failure. BP is a quantitative trait showing continuous variation from low to high values in outbred populations of both humans and animals. In some humans (and animals), increased dietary NaCl intake leads to increased BP, a phenomenon referred to as “salt sensitivity” (Pickering 1990). In others, increased dietary NaCl intake does not lead to higher BP (“salt resistance”). The inbred Dahl salt-sensitive (SS/Jr) rat is the most widely studied genetic model of salt-sensitive hypertension. Initially, Dahl salt-sensitive and Dahl salt-resistant rat strains were derived from outbred Sprague–Dawley stock by recurrent selection of rats exhibiting extremely high or low BP values, respectively, in the context of a massive dietary NaCl intake (Dahl et al. 1962). Later, inbred Dahl salt-hypertension-sensitive and salt-hypertension-resistant (SR/Jr) rat strains were developed by Rapp and colleagues (Rapp and Dene 1985). In SS/Jr (hereafter, S) rats, supplemental dietary NaCl increases BP, while supplemental dietary NaCl has little or no effect on the BP of SR/Jr (hereafter, R) rats (Rapp and Dene 1985). Segregating populations and congenic strains bred from these inbred strains have been used to screen for (and confirm) chromosomal locations responsible for heritable strain differences in BP, i.e., BP quantitative trait loci (QTL) (reviewed by Rapp 2000; Garrett and Joe 2006).
Multiple BP QTL on rat chromosome 3 (RNO3) were identified using linkage analysis in several different segregating rat populations. We previously used interval mapping to identify two putative BP QTL, separated by >90 cM on RNO3 in an F1(S × R) × S population given an excessive dietary NaCl intake (Cicila et al. 1999). A congenic rat strain, S.R-Edn3, was constructed by introgressing the more distal RNO3 BP QTL into the S strain. S.R-Edn3 rats had significantly lower BP compared to S rats, confirming the existence of a BP QTL in the RNO3 q terminus (Cicila et al. 1999). In the above studies, S-rat RNO3 BP QTL allele(s) were associated with higher BP. Our objective for this study was to use substitution mapping to further delimit this RNO3 BP QTL-containing region, narrowing the interval used to identify candidate genes for future study. Another RNO3 q-terminal BP QTL was also identified using a salt-loaded F2(S × BN) population, although in contrast with our results, overdominance was observed at this BP QTL (Kato et al. 1999).
Linkage analysis in segregating rat populations bred with Dahl S rats have identified RNO3 BP QTL located more proximally to those described above. They were identified in the following segregating populations: F1(S × R) × S (Cicila et al. 1999), F2(S × BN) (Stoll et al. 2000, 2001; Moreno et al. 2003), F2(S × Lew) (Garrett et al. 1998), and F2(S × SHR) (Garrett et al. 2000; Siegel et al. 2003). In these studies, with the exception of those using F2(S × SHR) rats, where noted, S-rat RNO3 BP QTL allele(s) were associated with higher BP. In the studies using populations bred from two hypertensive strains, S-rat RNO3 BP QTL allele(s) were associated with lower BP (Garrett et al. 2000; Siegel et al. 2003).
The broad chromosomal region covered by the LOD plots for the F2(S × Lew) (Garrett et al. 1998) and the more proximal F1(S × R) × S (Cicila et al. 1999) RNO3 BP QTL suggests that multiple genes underlie these QTL and that different subsets of genes may affect BP in different segregating populations. Indeed, Palijan et al. (2003) used overlapping RNO3 congenic strains, bred by introgressing Lewis-rat intervals into an S-rat background, to confirm the presence of at least two BP QTL, with opposing effects on BP, in this more proximal RNO3 region. A congenic strain incorporating both BP-QTL-containing intervals found that Lewis-rat alleles associated with decreased BP were epistatic to Lewis-rat alleles associated with lower BP (Palijan et al. 2003). Herrera et al. (2001) also identified epistasis involving an RNO3 locus associated with increased BP in an F2(S × R) population, although no independent RNO3 BP QTL was observed.
While genome scans using segregating rodent populations have been effective in determining rough chromosomal location for QTL, their imprecision has presented difficulties in further studying the gene(s) responsible for these observed effects (Darvasi et al. 1993). Congenic strains establish definitive limits for QTL-containing regions, which, in turn, can be narrowed by selecting congenic substrains retaining the QTL while carrying progressively smaller amounts of donor chromosome (Rapp and Deng 1995). This strategy has been effective in confirming BP QTL locations in rat models (reviewed by Rapp 2000; Cicila et al. 2001; Joe and Garrett 2005) and has identified two genes in rat models of complex traits responsible, in part, for QTL: 11β-hydroxylase (Cyp11b1) in the Dahl BP model (Cicila et al. 2001) and neutrophil cytosolic factor 1 (Ncf1) in both an experimentally induced arthritis model (Olofsson et al. 2003) and an experimental autoimmune encephalomyelitis model (Becanovic et al. 2006).
The present study utilized substitution mapping of the S.R-Edn3 congenic strain to demonstrate the presence of two distinct BP QTL located at the RNO3 q terminus, whereas the initial linkage analysis suggested the presence of single BP QTL (Cicila et al. 1999). We also describe fine mapping of these two q-terminal RNO3 BP QTL-containing intervals, each ∼1 Mb in size, as well as potential candidate genes located within them.
MATERIALS AND METHODS
Inbred rat strains and genetic crosses:
Inbred Dahl SS/Jr and SR/Jr rat strains were developed (Rapp and Dene 1985) from outbred stock originally obtained from Dahl (Dahl et al. 1962). S and R rats used to make the genetic crosses and develop congenic strains were from the colony at the University of Toledo College of Medicine. Two backcross F1(S × R) × S populations (n = 150 rats) were used to construct a detailed linkage map of RNO3. The breeding and phenotyping of these populations were previously described in detail (Cicila et al. 1997, 1999).
Development of S.R-Edn3 congenic substrains:
Congenic substrains were developed by crossing the previously described S.R-Edn3 congenic strain (Cicila et al. 1999) with S rats to yield F1(S × S.R-Edn3) rats heterozygous for the introgressed region of R-rat-derived RNO3. F1(S × S.R-Edn3) rats were intercrossed to obtain a population of F2(S × S.R-Edn3) (n = 173) rats, which was screened to identify rats having crossovers within this region of introgressed chromosome. Rats containing seven different classes of recombinant chromosomes were identified and used to develop six different congenic substrains carrying smaller portions of R-rat RNO3, as follows. Genomic DNA was isolated from the tails of progeny rats and genotyped for the D3Wox1, D3Mgh1, and D3Mco7 microsatellite markers, located in the congenic region of S.R-Edn3. F2(S × S.R-Edn3) rats carrying recombinant chromosomes were then genotyped using additional polymorphic markers located within the congenic region of S.R-Edn3.
Rats carrying the appropriate recombinant chromosomes were crossed with S rats to duplicate the recombinant chromosome, and the resulting heterozygous progeny were intercrossed. Intercross progeny were genotyped to identify those carrying two copies of the same recombinant chromosome. Rats homozygous for a recombinant chromosomal segment were crossed to fix the recombinant chromosome and establish six congenic substrains: S.R(D3Mco36–D3Mco46), S.R(D3Mco36–D3Got166), S.R(D3Mco36–D3Got159), S.R(D3Mco78–D3Got130), S.R(D3Mco24–D3Got130), and S.R(D3Mco39–D3Got130). These six congenic strains are referred to as S.R(ET3 × 1), S.R(ET3 × 2), S.R(ET3 × 3), S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 7), respectively.
Phenotyping:
BP and heart weight (HW) were measured for male rats of each congenic substrain and a group of age- and weight-matched control S rats. Each set of congenic substrain rats (n = 20) and control S rats (n = 20) was bred, housed, and studied concomitantly to minimize environmental effects. Rats were weaned at 30 days of age and placed on a low-salt diet (0.4% NaCl, Harlan Teklad diet TD7034). Four animals (two of each strain) were randomly assigned to each cage. At 40–42 days of age rats were fed 2% NaCl diet (Harlan Teklad diet TD94217) for 24 days and their BP was measured (see below).
Systolic BP was measured using the tail-cuff microphonic method (Buñag and Butterfield 1982) on conscious restrained rats warmed to 28°. Operators were unaware of the identity of the rats during these measurements. The BP of each rat was measured for 4 consecutive days. BP values for each day were the mean of three to four consistent readings. The final BP value used was the mean of the four daily BP values. Rats were killed by pentobarbital overdose, and body and heart weights measured.
Kidney weight measurement began following the first reports of kidney weight QTL in S-rat populations (Stoll et al. 2001), after the start of phenotypic testing of the congenic substrains. Kidney (decapsulated) weights were measured for S.R(ET3 × 1), S.R(ET3 × 2), S.R(ET3 × 6), and S.R(ET3 × 7) congenic substrain rats, along with concomitantly raised control S rats. Kidney weights were measured only in the second groups of S.R(ET3 × 1), S.R(ET3 × 2), and S.R(ET3 × 7) congenic substrain rats phenotyped.
Genotyping:
DNA for genotyping rats during congenic substrain development was extracted from tail biopsy material using the QIAamp tissue DNA kit (QIAGEN, Valencia, CA). PCR amplification and gel electrophoresis were performed as previously described (Cicila et al. 1997). Additional microsatellite markers were placed on the RNO3 linkage map to further define the extent of R-rat chromosome introgressed into the congenic strain and substrains. Markers were selected from the following sources: (1) Massachusetts Institute of Technology (Cambridge, MA) (http://www.genome.wi.mit.edu), (2) Wellcome Trust Centre for Human Genomics (Oxford, UK) (http://www.well.ox.ac.uk), (3) Otsuka GEN Research Institute [Otsuka Pharmaceutical, Tokushima, Japan (ratmap.ims-u.tokyo.ac.jp/)], (4) Rat Genome Database (http://rgd.mcw.edu/), and (5) University of Toledo College of Medicine (http://www.meduohio.edu/depts/physiology/research/rat/marker.html). Markers were placed by genotyping the combined F1(S × R) × S backcross population and the F2(S × S.R-Edn3) intercross populations described above. Linkage maps were developed using the Map Manager QT (Manly and Olson 1999) program obtained from Kenneth F. Manly (State University of New York, Buffalo). Potential errors in typing, i.e., loci involved in double-recombination events, were retyped to confirm or correct the results.
For this study, additional microsatellite markers were developed, using genomic sequence information from mouse and rat databases to further characterize the QTL-containing regions located at the RNO3 q terminus. Fine mapping of the RNO3 q terminus was achieved by designing 47 additional PCR-primer sets that distinguished S-rat from R-rat alleles (supplemental Table 2 at http://www.genetics.org/supplemental/). Forty-four of these additional primer sets amplify novel microsatellites and 3 primer sets (D3Mco40, D3Mco79, and D3Mco80) were developed to better amplify repetitive sequences contained in the D3Got166, D3Mco3, and D3Mco4 microsatellites, respectively. Primer sets to identify microsatellites distinguishing S-rat from R-rat alleles were designed, using the Oligo-Lite 6.21 program (Molecular Biology Insights, Cascade, CO).
Microsatellites (D3Mco34–39 and D3Mco41–43) were first identified using the BLAST program to identify rat BAC clone sequences from the Baylor College of Medicine database (Rat Genome Project, http://hgsc.bcm.tmc.edu/projects/rat/) that contained portions of the orthologous region of mouse chromosome. Later, microsatellites (D3Mco40, D3Mco44–80) were developed from rat genomic sequences extracted from the rat genome sequence database at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=rat&chr=3). The six congenic substrains described above were genotyped for all newly developed microsatellite markers.
Comparative mapping:
Sixty-one genetic markers on RNO3 polymorphic between S and R rats (47 newly developed and 14 established microsatellite markers) were placed on mouse and rat genomic sequence maps. Blocks of sequence (500–800 bp) centered on the repetitive sequence of each microsatellite were compared with the rat (Build 3) and mouse (Build 35.1) genomic sequence databases (NCBI) using the BLAST program.
QTL located on RNO3 (and orthologous mouse chromosome 2 and human chromosome 20 intervals) were identified using the Rat Genome Database (http://rgd.mcw.edu/objectSearch/qtlQuery.jsp) and Mouse Genome Informatics (http://www.informatics.jax.org/) sites, as well as by review of the literature. If flanking markers or LOD-support intervals were not available, QTL-containing regions were assumed to extend 15 cM from the location of the LOD-plot peak. QTL involving the heart, kidney, fat/lipid metabolism, and glucose sensitivity/resistance were considered as potentially blood pressure related.
Statistical analysis:
Phenotypic differences were assessed by Student's t-test using the StatView 5.1 (Abacus Concepts, Mountain View, CA) computer program, with P < 0.05 the criterion for significance. Where significant differences in final body weight were observed between a congenic substrain and concomitantly raised control S rats, the strong linear relationships between (1) HW and body weight (BW) and (2) kidney weight (KW) and BW allowed regression analysis to remove its influence from the measurement of these organ weights.
RESULTS
Characterizing q-terminal RNO3 BP QTL by substitution mapping:
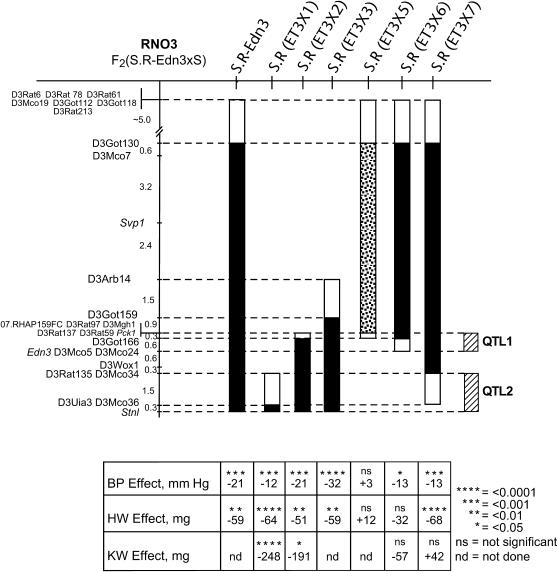
The introgressed portion of R-rat RNO3 in the original S.R-Edn3 (Cicila et al. 1999) congenic strain contained low-BP QTL allele(s) and low-HW QTL allele(s), as well as R-rat alleles for endothelin 3 (Edn3), phosphoenolpyruvate carboxykinase 1 (Pck1), guanine nucleotide-binding protein, α-stimulating complex locus (Gnas), and other genes. Substitution mapping using newly developed congenic substrains further delimited the introgressed region of R-rat chromosome containing the gene(s) responsible for these QTL. Six novel congenic substrains were developed, three extending from the distal end of the S.R-Edn3 congenic region [S.R(ET3 × 1), S.R(ET3 × 2), and S.R(ET3 × 3)] and three extending from the proximal end of the S.R-Edn3 congenic region [S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 7)] (Figure 1). Male rats from these six congenic strains were characterized for BP, HW, and KW along with concomitantly raised S rats (Table 1).
Figure 1.—
Genetic maps of RNO3 showing locations of congenic segments. A linkage map of selected markers on rat chromosome 3 (RNO3) using an F2(S.R-Edn3 × S) population of 173 rats is shown, with distances between loci expressed in centimorgans. The known extent of R-rat-derived RNO3 carried by each congenic strain and substrain is designated by the solid portion of the bars to the right of the linkage map, with open portions of the bars designating intervals containing the recombinant endpoints of R-rat-derived chromosome. The portion of donor strain chromosome carried by the initial congenic strain, S.R-Edn3, is shown by the leftmost bar, with the portions carried by the congenic substrains [S.R(ET3 × 1), S.R(ET3 × 2), S.R.(ET3 × 3), S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 7)] shown by adjacent bars. The bars are solid if the congenic substrain had a significantly reduced BP compared to the parental S strain and stippled if no BP effect was observed. Hatched bars indicate the QTL1 and QTL2 intervals (see discussion). The bottom part indicates the deviation of blood pressure (BP, in millimeters Hg), heart weight (HW, in milligrams), and kidney weight (KW, in milligrams) of the congenic strain and its substrains compared to those of concomitantly raised control S rats. A significant negative deviation indicates that an R-rat QTL allele was present in the congenic region. ns, no significant difference for the quantitative trait in the congenic substrain, compared to S rats. nd, not determined. BP and HW values for S.R-Edn3 are from Cicila et al. (1999).
TABLE 1.
Comparison of chromosome 3 congenic substrains with the S strain
| Strain | N | Blood pressure (mm Hg) | Heart weight (mg) | Body weight (g) | Kidney weight (mg) |
|---|---|---|---|---|---|
| S.R(ET3 × 1) | 40 | 201.3 ± 2.2 | 1070 ± 10 | 313.7 ± 2.6 | 2833 ± 37 |
| S | 40 | 213.0 ± 2.5 | 1134 ± 9 | 318.6 ± 2.6 | 3081 ± 36 |
| Differencea | −11.7 | −64 | −4.9 | −248 | |
| P-value | 0.0009 | <0.0001 | 0.20 | <0.0001 | |
| S.R(ET3 × 2) | 40 | 197.5 ± 2.5 | 1121 ± 15 | 303.9 ± 2.0 | 2839 ± 54 |
| S | 40 | 218.3 ± 2.6 | 1172 ± 17 | 298.6 ± 2.0 | 3030 ± 60 |
| Difference | −20.8 | −51 | +5.3 | −191 | |
| P-value | 0.0001 | 0.001 | 0.05 | 0.02 | |
| S.R(ET3 × 3) | 20 | 178.6 ± 4.0 | 1150 ± 11 | 300.5 ± 2.0 | ND |
| S | 20 | 210.6 ± 3.4 | 1209 ± 14 | 288.1 ± 2.8 | |
| Difference | −32.0 | −59 | +12.4 | ||
| P-value | <0.0001 | 0.002 | <0.0001 | ||
| S.R(ET3 × 5) | 20 | 209.1 ± 4.8 | 1281 ± 15 | 296.3 ± 3.0 | ND |
| S | 20 | 206.6 ± 2.4 | 1269 ± 15 | 301.8 ± 2.4 | |
| Difference | +2.5 | +12 | −5.5 | ||
| P-value | 0.65 | 0.58 | 0.16 | ||
| S.R(ET3 × 6) | 19 | 190.9 ± 3.6 | 1087 ± 13 | 307.7 ± 1.9 | 2842 ± 51 |
| S | 20 | 203.6 ± 4.2 | 1119 ± 16 | 304.7 ± 2.1 | 2899 ± 51 |
| Difference | −12.7 | −32 | +3.0 | −57 | |
| P-value | 0.03 | 0.12 | 0.29 | 0.44 | |
| S.R(ET3 × 7) | 39 | 191.8 ± 2.6 | 1082 ± 11 | 319.1 ± 1.8 | 3079 ± 48 |
| S | 41 | 204.7 ± 2.1 | 1150 ± 10 | 326.2 ± 2.1 | 3037 ± 30 |
| Difference | −12.9 | −68 | −7.1 | +42 | |
| P-value | 0.0002 | <0.0001 | 0.01 | 0.46 |
Blood pressure, heart weight, body weight, and kidney weight values for the S and congenic substrain rats are presented as the mean ± standard error of the mean. Congenic substrains are defined in Figure 1. Male S and congenic substrain rats were maintained on a low-salt diet until 40 days of age and were then fed a 2% NaCl diet for 28 days. Substrains S.R(ET3 × 1), S.R(ET3 × 2), and S.R(ET3 × 7) were tested twice. Statistical significance was analyzed using a t-test. ND, not determined.
Difference values equal congenic-rat value minus S-rat value. Comparisons were made between concomitantly raised and phenotyped sets of S and RNO3 congenic rats.
S.R(ET3 × 1), S.R(ET3 × 2), S.R(ET3 × 3), S.R(ET3 × 6), and S.R(ET3 × 7) rats showed significantly lower BP, compared to concomitantly raised S rats, with only S.R(ET3 × 5) rats showing no significant difference in BP (Table 1). S.R(ET3 × 1), S.R(ET3 × 2), S.R(ET3 × 3), and S.R(ET3 × 7) rats also showed significantly lower HW compared to concomitantly studied S rats, consistent with the lower BP observed in these congenic substrains, with only S.R(ET3 × 5) and S.R(ET3 × 6) rats showing no significant difference in HW. S.R(ET3 × 1) and S.R(ET3 × 2) rats had significantly lower KW compared to S rats, while S.R(ET3 × 6) and S.R(ET3 × 7) showed no significant difference in KW (Table 1). The KWs of S.R(ET3 × 3) and S.R(ET3 × 5) rats were not measured. The same pattern of congenic strain differences in KW was observed whether total kidney weight or the left and the right kidney weights were measured separately (data not shown).
Significant differences in BW were observed only in comparisons of S.R(ET3 × 3) and S.R(ET3 × 7) rats with concomitantly raised S rats. Significant differences in BW-adjusted HW were observed in S.R(ET3 × 3) and S.R(ET3 × 7) rats, compared with concomitantly raised S rats (data not shown), confirming the presence of low-HW QTL alleles in these congenic substrains. Significant BW-adjusted KW differences were observed in the S.R(ET3 × 1) and S.R(ET3 × 2) substrains compared to concomitantly raised S rats (data not shown), confirming the presence of low-KW QTL alleles in the S.R(ET3 × 1) and S.R(ET3 × 2) congenic substrains. No difference in the BW-adjusted KW was observed between male S.R(ET3 × 7) and S rats (data not shown).
Mapping the RNO3 q terminus:
Three different approaches were employed: radiation hybrid mapping, physical mapping, and comparative mapping. Both agreements and conflicts were identified while integrating our substitution mapping data with that obtained with these three mapping approaches. Radiation hybrid mapping was used to characterize the RNO3 q terminus because of the many rearrangements of contigs that occurred during the early builds of the rat genomic sequence. We first mapped rat microsatellite markers known to be within our congenic region, as well as primer sets designed from genes located in the orthologous region of mouse chromosome 2 (MMU2, supplemental Figure 1 at http://www.genetics.org/supplemental/). Six of the newly developed RNO3 markers (see below) were also placed on this radiation hybrid map (supplemental Figure 1).
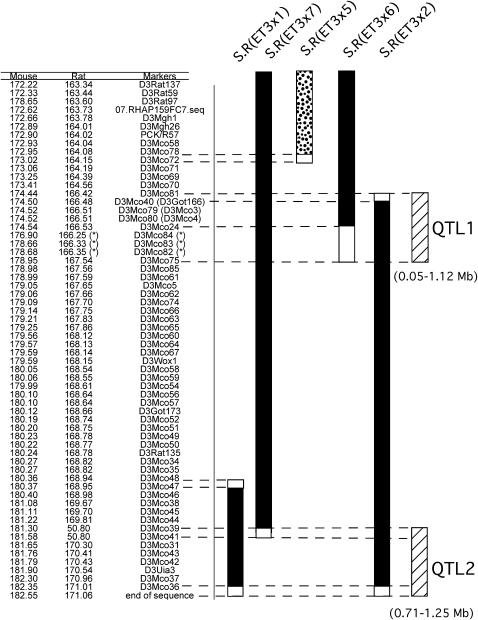
To further characterize the RNO3 q terminus 44 novel microsatellite markers polymorphic between S and R rats were developed to more precisely define the regions containing the crossovers in the congenic substrains and thus better define the limits of the congenic regions. Simple-sequence repeats were identified from sequences of BACs and RNO3 genomic contigs corresponding to the distal ends of S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 7), the RNO3 q terminus [i.e., distal ends of S.R(ET3 × 1) and S.R(ET3 × 2)], and the proximal ends of S.R(ET3 × 1) and S.R(ET3 × 2). The locations of theses novel RNO3 q-terminal microsatellites in the rat and mouse genomes were determined by BLAST using the genomic sequence databases at the NCBI site and the six RNO3 congenic substrains were genotyped for each of the novel polymorphic markers, as well as 11 other previously described markers polymorphic between Dahl S and R rats (Figure 2). RNO3 marker ordering in Figure 2 was primarily based upon the available RNO3 genomic sequence, but also utilized information from the MMU2 genomic sequence, the R-rat RNO3 content within the six RNO3 congenic substrains, and an F2(S.R-Edn3 × S) population (data not shown).
Figure 2.—
Locations of polymorphic microsatellite markers and their genotypes in congenic substrains in the RNO3 q terminus. The locations of sequences in both the mouse and rat genomes are described, which surround the simple-sequence repeats used to design RNO3 genetic markers polymorphic between Dahl S and R rats. Sequence locations were determined using the BLAST computer program using mouse and rat genome sequence data on the NCBI web site and are given in megabases. Loci marked with asterisks (*) did not map to the RNO3 locations predicted from the genomic sequence map (as shown). The reasons for these discrepancies are discussed in detail in results. The known extents of R-rat-derived RNO3 carried by five of the congenic substrains [S.R(ET3 × 1), S.R(ET3 × 7), S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 2)] are designated by the solid portions of the bars to the right, with open portions of the bars designating intervals containing the recombinant endpoints of R-rat-derived chromosome. The bars are solid if the congenic substrain had a significantly reduced BP compared to the parental S strain and stippled if no BP effect was observed. Note that only the distal portions of the congenic regions of S.R(ET3 × 5), S.R(ET3 × 6), and S.R(ET3 × 7) are shown. Hatched bars indicate the QTL1 and QTL2 intervals (see discussion).
Considerable agreement was found between the RNO3 and MMU2 q-terminal regions with respect to both microsatellite marker order and spacing, especially over the most distal ∼3.5 Mb (D3Mco75–D3Mco36) of RNO3 (Figure 2), as well as in two more proximal intervals (D3Mco24–D3Mco40 and D3Mco70–D3Rat137).
The exceptions within the first interval, D3Mco39 and D3Mco41, were derived from BAC sequences. The placement of these two loci in RNO3 is strongly supported by the allelic content of the congenic substrains, with S.R(ET3 × 1), S.R(ET3 × 2), and S.R(ET3 × 3) carrying R-rat alleles for these loci and S.R(ET3 × 5) and S.R(ET3 × 6) carrying S-rat alleles. These two loci define the distal end of the S.R(ET3 × 7) congenic region, with this congenic substrain carrying an R-rat allele for D3Mco39 and an S-rat allele for D3Mco41 (Figure 2). Placement of D3Mco39 and D3Mco41 at the RNO3 q terminus by radiation hybrid mapping (supplemental Figure 1 at http://www.genetics.org/supplemental/) and in orthologous positions on MMU2 by in silico mapping provides further support for their placement between D3Mco44 and D3Mco31 at the RNO3 q terminus (Figure 2).
Two additional problematic intervals were identified in the RNO3 q-terminus map. The first results from three microsatellite markers, D3Mco82, D3Mco83, and D3Mco84 (indicated with asterisks in Figure 2), expected to map between D3Mco70 and D3Mco81 on the basis of the location of the genomic sequences from which they were designed. However, a linkage map developed from a large F2[S.R(ET3 × 2) × S] population placed these three loci between D3Mco24 and D3Mco75 (data not shown). This placement was also confirmed by S.R(ET3 × 1), S.R(ET3 × 5), and S.R(ET3 × 6) carrying S-rat alleles and S.R(ET3 × 2), S.R(ET3 × 3), and S.R(ET3 × 7) carrying R-rat alleles for these three loci. The placement of D3Mco82, D3Mco83, and D3Mco84 markers on RNO3 shown in Figure 2 was also supported by the location of orthologous sequences in the mouse genome.
The second interval lies between D3Mco40 and D3Mco70. Two microsatellite markers, D3Mco76 and D3Mco77, designed from sequences expected to be located between D3Mco70 and D3Mco3, distinguished S-rat and R-rat alleles. However, because none of the congenic substrains carried R-rat alleles for these markers, these markers cannot be located at the RNO3 q terminus within the S.R(Edn3) congenic region. Collectively, these results indicate serious problems with the draft rat genomic sequence in this interval, which, in part, are resolvable on the basis of biological evidence of recombinations occurring in the RNO3 congenic substrains we report here.
DISCUSSION
The RNO3 q terminus contains two distinct BP QTL:
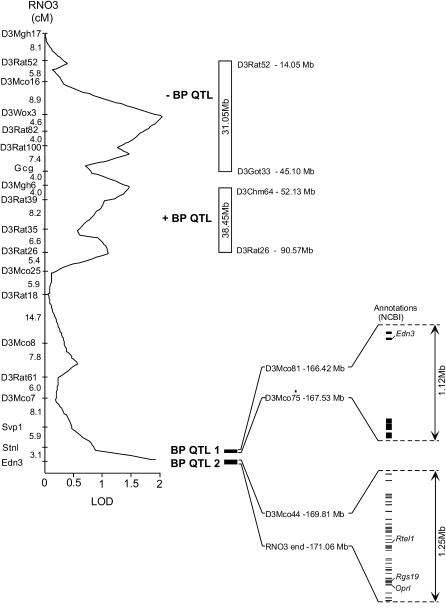
Our previous study with an F1(S × R) backcross population identified at least two distinct BP QTL (Cicila et al. 1999), although only the q-terminal BP QTL was confirmed using congenic substrains. Palijan and co-workers developed congenic strains to identify BP QTL of contrasting effects (Palijan et al. 2003) in more proximal RNO3 intervals located near the second RNO3 BP QTL identified in the F1(S × R) backcross population (Figure 3). Compared to the large chromosomal intervals (31.05 and 38.45 Mb) that contain these more proximal RNO3 BP QTL (Palijan et al. 2003), the q-terminal RNO3 BP QTL reported in this study represent a high degree of resolution by fine mapping, i.e., close to 1-Mb intervals for both QTL1 and QTL2 (Figure 3). None of the other RNO3 BP QTL (reviewed by Garrett and Joe 2006) have been confirmed by the development of congenic strains.
Figure 3.—
Blood pressure QTL locations on rat chromosome 3 characterized in Dahl rats using congenic substrains. The size and location of the RNO3 BP QTL-containing regions found in the Dahl rat model, characterized using congenic strains and substrains, are shown in the context of a previous linkage analysis of BP in an F1(S × R) × S backcross population (Cicila et al. 1999) with a display of annotated sequences (NCBI) contained in the q-terminal portion of RNO3. A LOD plot for BP on RNO3 in an F1(S × R) × S population (Cicila et al. 1999) is shown on the left. To the right of the BP LOD plot are bars showing the location and extent of the four RNO3 BP QTL identified in segregating populations bred using Dahl S rats and a normotensive strain. The intervals containing the two q-terminal RNO3 BP QTL (QTL1 and QTL2) described in this study are shown below and are indicated by solid bars. For both of the q-terminal RNO3 BP QTL, R-rat alleles were associated with decreased BP. Unfilled bars indicate the locations of two, more proximal, RNO3 BP QTL. These more proximal RNO3 BP QTL were first identified using an F2(LEW × S) population (Garrett et al. 1998) and later characterized using a set of overlapping congenic strains (Palijan et al. 2003). This analysis indicated that the proximal portion of RNO3 also contained two distinct BP QTL-containing intervals, one carrying Lewis-rat alleles associated with decreased BP (−BP QTL) and the other carrying Lewis-rat alleles associated with increased BP (+BP QTL).
The presence of at least two distinct BP QTL at the RNO3 q terminus was demonstrated by two substrains with nonoverlapping congenic regions, S.R(ET3 × 1) and S.R(ET3 × 6) (Figures 1 and 2), having significantly lower BP compared to concomitantly raised S rats (Table 1). The decreased KW observed in S.R(ET3 × 1), but not S.R(ET3 × 6) or S.R(ET3 × 7) rats, compared to the parental S strain (Table 1) corroborates the distinctiveness of these two BP QTL. Despite the low LOD scores obtained in our previous linkage studies (Cicila et al. 1994, 1999) and the limitation of tail-cuff methods to detect small BP differences (reviewed by Kurtz et al. 2005) we were able to confirm BP QTL at the RNO3 q terminus. This could be particularly important with regard to the S.R(ET3 × 6) congenic strain, where a small difference (13 mm Hg) in BP was observed, but significant differences in HW and KW were not observed.
No significant differences in BP or HW were observed between S.R(ET3 × 5) and S rats, indicating that the R-rat RNO3 alleles introgressed into this congenic substrain have no net effect on these quantitative traits. Thus, it is unlikely that BP QTL are located within the distal portion (>8.0 cM) of R-rat RNO3 introgressed into the initial congenic strain, S.R-Edn3 (Cicila et al. 1999). This would also be true for the same proximal portion of the congenic regions of S.R(ET3 × 6) and S.R(ET3 × 7) substrains that were derived from S.R-Edn3. The congenic regions of substrains S.R(ET3 × 2) and S.R(ET3 × 3) contain R-rat-derived alleles that result in substantially greater BP effects (defined by the difference in BP for concomitantly raised S rats and congenic substrains) compared to those of substrains S.R(ET3 × 1) and S.R(ET3 × 6) (−20.8 and −32.0 mm Hg compared to −11.7 and −12.8 mm Hg, respectively). The simplest interpretation of the above data is that substrains S.R(ET3 × 2) and S.R(ET3 × 3) contain both sets of low-BP QTL alleles, which are carried separately by S.R(ET3 × 1) and S.R(ET3 × 6). The smaller of these two substrains, S.R(ET3 × 2), contains 4.54–4.64 Mb of R-rat RNO3 and has significantly lower BP, HW, and KW compared to the parental, S strain (Table 1, Figure 1) and defines the limits of the QTL-containing region.
The relatively small size of the introgressed region of the S.R(ET3 × 1) congenic substrain (2.04–2.11 Mb) and its effects on BP, HW, and KW (Table 1) suggest that the gene(s) responsible for the more distal of the two q-terminal RNO3 BP QTL act independently. The same cannot be said about the more proximal of the q-terminal RNO3 BP QTL. This RNO3 BP QTL is found only in the S.R(ET3 × 6) substrain, which carries a much larger congenic region, or the S.R(ET3 × 2) and S.R(ET3 × 3) substrains whose congenic regions also incorporate the more distal of the q-terminal RNO3 BP QTL. This suggests that QTL1 might not exert its effects independently and thus epistatic interactions between QTL1 and other loci at the RNO3 q terminus cannot be ruled out. Epistatic interactions responsible for BP effects have previously been observed for BP QTL located on the same chromosomes, with interdependent effects observed for two closely linked RNO5 BP QTL (Garrett and Rapp 2002) and a closely linked RNO3 QTL able to suppress the BP-lowering effects of another on RNO3 (Palijan et al. 2003).
Defining the limits of the two RNO3 BP QTL-containing intervals:
Fine mapping the RNO3 q terminus with 44 newly designed polymorphic microsatellite markers more precisely defines the limits of these two BP QTL-containing regions. The more proximal RNO3 q-terminal BP QTL-containing interval (QTL1) is defined as a 0.05- to 1.12-Mb overlap of the S.R(ET3 × 6) and S.R(ET3 × 2) congenic regions (Figure 3). The QTL1 interval is delimited by D3Mco81 and D3Mco75 markers and does not overlap with that of the S.R(ET3 × 5) congenic interval, which lacks low-BP QTL allele(s). Comparative mapping indicates that the interval orthologous to QTL1 in MMU2 is of similar size, 0.04–2.46 Mb (an interval delimited by sequences similar to D3Mco84 and D3Mco81). This analysis excludes Pck1 and Gnas1, but not Edn3, from consideration as candidate genes for the more proximal RNO3 q-terminal BP QTL.
The end of the RNO3 q-arm and that portion of the S.R(ET3 × 1) congenic region not overlapping with the S.R(ET3 × 7) congenic region delimit the more distal RNO3 BP QTL-containing interval (QTL2) (Figure 3). QTL2 is delimited by D3Mco44 and the end of the RNO3 sequence, defining an interval of 0.71–1.25 Mb. Comparative mapping of this chromosomal interval on the orthologous portion of MMU2 is of similar size, 0.87–1.25 Mb (an interval delimited by a sequence similar to D3Mco39 and the end of the MMU2 sequence). The rationale for this is that S.R(ET3 × 7), whose congenic region overlaps that of S.R(ET3 × 1), does not carry low-kidney-weight QTL allele(s), which both S.R(ET3 × 1) and S.R(ET3 × 2) carry. The magnitude of the observed BP effect of the R-rat alleles carried by the S.R(ET3 × 7) substrain makes it unlikely that it harbors low-BP alleles from both QTL.
S.R(ET3 × 2) and S.R(ET3 × 3) congenic regions contain both the QTL1 and the QTL2 intervals, with both substrains having significantly lower BP and HW compared to S rats. The observed BP differences suggest that the S.R(ET3 × 2) and S.R(ET3 × 3) substrains carry low-BP alleles from both RNO3 q-terminal BP QTL that exert additive effects. Association of a KW difference with only one BP QTL suggests that the alleles responsible for the different BP QTL act in different tissues and/or pathways.
Genes (and putative genes) within the QTL1 and QTL2 intervals:
Genes and putative genes predicted to encode proteins were identified in the QTL1 and QTL2 intervals using annotations from the NCBI and Ensembl (http://www.ensembl.org/Rattus_norvegicus/index.html) rat genome sites. The QTL1 interval contains six sequences predicted to be protein encoding with NCBI annotations and eight sequences with Ensembl annotations. Endothelin 3 (Edn3), among the few genes within this interval, is the most interesting candidate. Edn3 is a member of the endothelin gene family that encodes the most potent known endogenous vasoactive peptides and, as such, has long been considered as a candidate gene for BP (Lüscher et al. 1993; Cicila et al. 1994; Marteau et al. 2005). However, it is possible that other genes identified (e.g., cadherin 26) or presently unknown within this interval might also be responsible or contribute to the effects of RNO3 BP QTL1.
The QTL2 interval contains 42 sequences with NCBI annotations predicted to be protein encoding, including 9 unique to NCBI. The QTL2 interval also contains 35 sequences with Ensembl annotations predicted to be protein encoding, including 2 unique to Ensembl. Four noncoding RNAs were annotated by Ensembl, including rno-mir-124a-3, which is similar to miRNAs identified in mouse (Dostie et al. 2006) and human (Lagos-Quintana et al. 2002), with the human miRNA mapping to HSA20.
The QTL2 interval contains genes involved in regulating transcription (SRY-box-containing gene 18, Sox18; basic helix-loop-helix transcriptional regulator β4, Bhlhb4; myelin transcription factor 1, Myt1; glucocorticoid modulatory element-binding protein 2, Gmeb2; and similar to PPARα-interacting complex 285-kDa protein), signal transduction (stathmin-like 3, Stmn3; src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites, Srms; protein tyrosine kinase 6, Ptk6; opioid receptor-like 1, Oprl; and regulator of G protein signaling 19, Rgs19), vesicle transport (cysteine string protein, Dnajc5), protein methylation [protein-l-isoaspartate (d-aspartate) O-methyltransferase domain containing 2, Pcmtd2], telomere length (regulator of telomere length 1, Rtel1), and the extracellular matrix (α1-type XX collagen, Col20a1). While the above genes can be considered as potential candidates for BP QTL2, three deserve further discussion: Oprl, Rgs19, and Rtel1.
Oprl mediates concentration-dependent vasorelaxation responses to nociceptin in rat arterial rings (Hugghins et al. 2000) and has been proposed to be involved in the pathophysiology of arterial hypertension (reviewed by Malinowska et al. 2002). Rgs19 is located adjacent to (and opposite of) Oprl and shares a common promoter (Ito et al. 2000). Rgs19 has been shown to regulate the expression of many G-protein-coupled receptors, including opioid receptors, with in vitro experiments indicating that Rgs19 negatively regulates Oprl preferentially compared to other opioid receptors (Xie et al. 2005). Rtel1 is discussed in the Comparative QTL mapping section, below. Again, it is possible that other, novel genes may be responsible for, or contribute to, RNO3 BP QTL2.
Comparative QTL mapping:
BP QTL:
No BP QTL were identified on regions orthologous to the RNO3 q terminus in the mouse (MMU2) or human (HSA20) from searches of the Rat Genome Database and Mouse Genome Informatics sites. However, two studies associated alleles for the Gs protein α-subunit (Gsα) gene (GNAS1) with BP differences in humans (Jia et al. 1999; Abe et al. 2002), although another study did not find linkage (Kato et al. 1997). Gnas1 is located proximal to the congenic region of S.R(ET3 × 2) and not within QTL2, but within the congenic region of S.R(ET3 × 3). Previous studies found evidence of decreased Gsα and altered β-adrenoreceptor GS coupling in vascular smooth muscle cell membranes of Milan hypertensive rats, compared to Milan normotensive rats (Clark et al. 1993, 1994).
Related quantitative traits:
QTL for several other quantitative traits related to BP have also been mapped to the RNO3 q terminus in rat genetic models. These include QTL for hyperglycemia following a partial pancreatectomy (Ogino et al. 1999), renal blood flow (Stoll et al. 2001), renal vascular resistance (Moreno et al. 2003), and insulin-dependent diabetes susceptibility (Martin et al. 1999). These traits are potential intermediate phenotypes that may contribute to BP regulation.
No BP QTL have been found in the human genome region orthologous to the RNO3 q terminus, the HSA20 q terminus. However, QTL for serum triglyceride (Li et al. 2005) and cholesterol levels (Imperatore et al. 2000), as well as body fat and fasting insulin levels (Lembertas et al. 1997), have been identified in orthologous regions of the human genome.
While no BP QTL have been identified in the region of the mouse genome orthologous to the RNO3 q terminus, the MMU2 q terminus, QTL for non-insulin-dependent diabetes (Komatsu et al. 2002) and obesity (Mehrabian et al. 1998; Diament et al. 2004; Estrada-Smith et al. 2004; Jerez-Timaure et al. 2004, 2005; Rocha et al. 2004) have been described. Again, these traits are potential intermediate phenotypes that may contribute to BP regulation.
A telomere length QTL also mapped to this MMU2 interval (Zhu et al. 1998) and a candidate gene, regulator of telomere length (Rtel1), was identified within this interval (Ding et al. 2004). Inactivation of Rtel1 by homologous recombination results in embryonic lethality, as well as telomere shortening in undifferentiated embryonic stem cells (Ding et al. 2004). Telomere dysfunction has been implicated in the pathogenesis of age-related cardiovascular diseases (Serrano and Andrés 2004), including hypertension both in animal models (Hamet et al. 2001; Cao et al. 2002) and in humans (Jeanclos et al. 2000; Benetos et al. 2001).
Overall, comparative QTL mapping suggests that the two q-terminal RNO3 BP QTL reported here represent novel genetic determinants of BP that remain undetected in other species. It remains to be determined whether the functional effects of the two q-terminal RNO3 BP QTL are exerted through the related phenotypes reported by the overlapping QTL in rats, mice, or humans.
Another important conclusion from this study is that for certain regions of the rat genome, particularly the termini of chromosomes, the quality of the available rat genomic data (including the rat genomic sequence) made fine mapping and marker placement difficult and complicated our efforts to characterize QTL-containing intervals and identify causal genes.
Acknowledgments
We acknowledge the technical assistance of Phyllis K. Farms, Shane Yerga-Woolwine, Larisa N. Cicila, and Thomas Mohlman. We thank Andrew McSweeny for critical reading of this manuscript. We thank Siwon Lee for assistance with graphic design. This work was supported by grants to G. T. Cicila and S. J. Lee from the National Institutes of Health (HL62338 and HL68994). G. T. Cicila is an Established Investigator of the American Heart Association.
References
- Abe, M., J. Nakura, Y. Yamamoto, J. J. Jin, Z. Wu et al., 2002. Association of GNAS1 gene variant with hypertension depending on smoking status. Hypertension 40: 261–265. [DOI] [PubMed] [Google Scholar]
- Becanovic, K., M. Jagodic, J. R. Sheng, I. Dahlman, F. Aboul-Enein et al., 2006. Advanced intercross line mapping of Eae5 reveals Ncf-1 and CLDN4 as candidate genes for experimental autoimmune encephalomyelitis. J. Immunol. 175: 6055–6064. [DOI] [PubMed] [Google Scholar]
- Benetos, A., K. Okuda, M. Lajemi, M. Kimura, F. Thomas et al., 2001. Telomere length as an indicator of biological aging: the gender effect and relation with pulse pressure and pulse wave velocity. Hypertension 37: 381. [DOI] [PubMed] [Google Scholar]
- Buñag, R., and J. Butterfield, 1982. Tail cuff blood pressure measurement without external preheating in awake rats. Hypertension 4: 898–903. [DOI] [PubMed] [Google Scholar]
- Cao, Y., H. Li, F.-T. Mu, O. Ebisui, J. W. Funder et al., 2002. Telomerase activation causes vascular smooth muscle cell proliferation in genetic hypertension. FASEB J. 16: 96–98. [DOI] [PubMed] [Google Scholar]
- Cicila, G. T., J. P. Rapp, K. D. Bloch, T. W. Kurtz, M. Pravenec et al., 1994. Cosegregation of the endothelin-3 locus with blood pressure and relative heart weight in inbred Dahl rats. J. Hypertens. 12: 643–650. [PubMed] [Google Scholar]
- Cicila, G. T., O. I. Dukhanina, T. W. Kurtz, R. Walder, M. R. Garrett et al., 1997. Blood pressure and survival of a chromosome 7 congenic strain derived from Dahl rats. Mamm. Genome 8: 896–902. [DOI] [PubMed] [Google Scholar]
- Cicila, G. T., C. Choi, H. Dene, S. J. Lee and J. P. Rapp, 1999. Two blood pressure/cardiac mass quantitative trait loci are present on chromosome 3 of Dahl salt-sensitive and salt-resistant rats. Mamm. Genome 10: 112–116. [DOI] [PubMed] [Google Scholar]
- Cicila, G. T., M. R. Garrett, S. J. Lee, J. Liu, H. Dene et al., 2001. High resolution mapping of the blood pressure QTL on chromosome 7 using Dahl rat congenic strains. Genomics 72: 51–60. [DOI] [PubMed] [Google Scholar]
- Clark, C. J., G. Milligan and J. M. C. Connell, 1993. Guanine nucleotide regulatory protein alterations in the Milan hypertensive rat strain. J. Hypertens. 11: 1161–1169. [PubMed] [Google Scholar]
- Clark, C. J., G. Milligan and J. M. C. Connell, 1994. Guanine nucleotide regulatory protein alterations in young Milan hypertensive strain rats. Biochim. Biophys. Acta 1225: 149–157. [DOI] [PubMed] [Google Scholar]
- Dahl, L. K., M. Heine and L. Tassinari, 1962. Effects of chronic excess salt ingestion: evidence that genetic factors play an important role in the susceptibility to experimental hypertension. J. Exp. Med. 115: 1173–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvasi, A., A. Weinreb, V. Minke, J. I. Weller and M. Soller, 1993. Detecting marker-QTL linkage and estimating QTL gene effect and map location using a saturated genetic map. Genetics 134: 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diament, A. L., P. Farahani, S. Chiu, J. Fisler and C. H. Warden, 2004. A novel mouse chromosome 2 congenic strain with obesity phenotypes. Mamm. Genome 15: 452–459. [DOI] [PubMed] [Google Scholar]
- Ding, H., M. Schertzer, X. Wu, M. Gertsenstein, S. Selig et al., 2004. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117: 873–886. [DOI] [PubMed] [Google Scholar]
- Dostie, J., Z. Mourelatos, M. Yang, A. Sharma and G. Dreyfyss, 2006. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA 9: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Smith, D., L. W. Castellani, H. Wong, P. Z. Wen, A. Chui et al., 2004. Dissection of multigenic obesity traits in congenic mouse strains. Mamm. Genome 15: 14–22. [DOI] [PubMed] [Google Scholar]
- Garrett, M. P., H. Dene, R. Walder, Q. Zhang, G. T. Cicila et al., 1998. Genomic scan and congenic strains for blood pressure QTL using Dahl salt-sensitive rats. Genome Res. 8: 711–723. [DOI] [PubMed] [Google Scholar]
- Garrett, M. R., and B. Joe, 2006. Genetic analysis of inherited hypertension in the rat, pp. 177–200 in Handbook of Hypertension, edited by A. Dominiczak. Elsevier Science, The Netherlands (in press).
- Garrett, M. R., and J. P. Rapp, 2002. Two closely linked interactive blood pressure QTL on rat chromosome 5 defined using Dahl rats. Physiol. Genomics 8: 81–86. [DOI] [PubMed] [Google Scholar]
- Garrett, M. R., Y. Saad, H. Dene and J. P. Rapp, 2000. Blood pressure QTL that differentiate Dahl salt-sensitive and spontaneously hypertensive rats. Physiol. Genomics 3: 33–38. [DOI] [PubMed] [Google Scholar]
- Hamet, P., N. Thorin-Trescases, P. Moreau, P. Dumas, B. S. Tea et al., 2001. Excess growth and apoptosis: Is hypertension a case of accelerated aging of cardiovascular cells? Hypertension 37: 760–766. [DOI] [PubMed] [Google Scholar]
- Herrera, V. L., L. V. Lopez and N. Ruiz-Opazo, 2001. α1 Na,K-ATPase and Na,K,2Cl-cotransporter/D3mit3 loci interact to increase susceptibility to salt-sensitive hypertension in Dahl SHSD rats. Mol. Med. 7: 125–134. [PMC free article] [PubMed] [Google Scholar]
- Hugghins, S. Y., H. C. Champion, G. Cheng, P. J. Kadowitz and J. R. Jeter, 2000. Vasorelaxant responses to endomorphins, nociceptin, albuterol, and adrenomedullin in isolated rat aorta. Life Sci. 67: 471–476. [DOI] [PubMed] [Google Scholar]
- Imperatore, G., W. C. Knowler, D. J. Pettitt, S. Kobes, J. H. Fuller et al., 2000. A locus influencing total serum cholesterol on chromosome 19: results from an autosomal genomic scan of serum lipid concentrations in Pima Indians. Arterioscler. Thromb. Vasc. Biol. 20: 2651–2656. [DOI] [PubMed] [Google Scholar]
- Ito, E., G. X. Xie, K. Maruyama and P. Pierce Palmer, 2000. A core-promoter region functions bi-directionally for human opioid-receptor-like gene ORL1 and its 5′-adjacent gene GAIP. J. Mol. Biol. 304: 259–270. [DOI] [PubMed] [Google Scholar]
- Jeanclos, E., N. J. Schork, K. O. Kyvik, M. Kimura, J. H. Skurnick et al., 2000. Telomere length inversely correlates with pulse pressure and is highly familial. Hypertension 36: 195–200. [DOI] [PubMed] [Google Scholar]
- Jerez-Timaure, N. C., F. Kearney, E. B. Simpson, E. J. Eisen and D. Pomp, 2004. Characterization of QTL with major effects on fatness and growth on mouse chromosome 2. Obes. Res. 12: 1408–1420. [DOI] [PubMed] [Google Scholar]
- Jerez-Timaure, N. C., E. J. Eisen and D. Pomp, 2005. Fine mapping of a QTL region with large effects on growth and fatness on mouse chromosome 2. Physiol. Genomics 21: 411–422. [DOI] [PubMed] [Google Scholar]
- Jia, H., A. D. Hingorani, P. Sharma, R. Hopper, C. Dickerson et al., 1999. Association of the GSa gene with essential hypertension and response to β-blockade. Hypertension 34: 8–14. [DOI] [PubMed] [Google Scholar]
- Joe, B., and M. R. Garrett, 2005. Substitution mapping: using congenic strains to detect genes controlling blood pressure, pp. 41–58 in Contemporary Cardiology: Cardiovascular Genomics, edited by M. K. Raizada, J. F. R. Paton, S. Kasparov and M. J. E. Katovich. Humana Press, Totowa, NJ.
- Kato, N., C. Julier, F. Soubrier, X. Jeunemaitre, P. Corvol et al., 1997. Search for susceptibility loci to hypertension in a region of human chromosome 20q, the homologous region to rat chromosome 3. Am. J. Hypertens. 10(4 Pt. 2): 33A. [Google Scholar]
- Kato, N., G. Hyne, M. Bihoreau, D. Gauguier, G. Lathrop et al., 1999. Complete genome searches for quantitative trait loci controlling blood pressure and related traits in four segregating populations derived from Dahl hypertensive rats. Mamm. Genome 10: 259–265. [DOI] [PubMed] [Google Scholar]
- Komatsu, S., H. Kiyosawa, A. Yoshiki, Y. Okazaki, M. Yoshino et al., 2002. Identification of seven loci for static glucokinesis and dynamic glucokinesis in mice. Mamm. Genome 13: 293–298. [DOI] [PubMed] [Google Scholar]
- Kurtz, T. W., K. A. Griffin, A. K. Bidani, R. L. Davisson and J. E. Hall, 2005. Recommendations for blood pressure measurement in humans and experimental animals: part 2: blood pressure measurement in experimental animals: a statement for professionals from the Subcommittee of Professional and Public Education of the American Heart Association Council on High Blood Pressure Research. Hypertension 45: 299–310. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., R. Rauhut, A. Yalcin, J. Meyer, W. Lendeckel et al., 2002. Identification of tissue-specific microRNAs from the mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lembertas, A. Y., L. Perusse, Y. C. Chagnon, J. S. Fisler, C. H. Warden et al., 1997. Identification of an obesity quantitative trait locus on mouse chromosome 2 and evidence of linkage to body fat and insulin on the human homologous region 20q. J. Clin. Invest. 100: 1240–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-D., C. Dong, D. Li, C. Garrigan and R. A. Price, 2005. A genome scan for serum triglyceride in obese nuclear families. J. Lipid Res. 46: 432–438. [DOI] [PubMed] [Google Scholar]
- Lüscher, T. F., B.-G. Seo and F. R. Bühler, 1993. Potential role of endothelin in hypertension: controversy on endothelin in hypertension. Hypertension 21: 752–757. [DOI] [PubMed] [Google Scholar]
- Malinowska, B., G. Godlewski and E. Schlicker, 2002. Function of nociceptin and opiod OP4 receptors in the regulation of the cardiovascular system. J. Physiol. Pharmacol. 53: 301–324. [PubMed] [Google Scholar]
- Manly, K., and J. Olson, 1999. Overview of QTL mapping software and introduction to Map Manager QT. Mamm. Genome 10: 327–334. [DOI] [PubMed] [Google Scholar]
- Marteau, J.-B., M. Zaiou, G. Siest and S. Visvikis-Siest, 2005. Genetic determinants of blood pressure regulation. J. Hypertens. 23: 2127–2143. [DOI] [PubMed] [Google Scholar]
- Martin, A. M., E. P. Blankenhorn, M. N. Maxson, M. Zhao, J. Leif et al., 1999. Non-major histocompatibility complex-linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP-BB/Wor rat to the WF rat. Diabetes 48: 50–58. [DOI] [PubMed] [Google Scholar]
- Mehrabian, M., P. Z. Wen, J. Fisler, R. C. Davis and A. J. Lusis, 1998. Genetic loci controlling body fat, lipoprotein metabolism, and insulin levels in a multifactorial mouse model. J. Clin. Invest. 101: 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno, C., P. Dumas, M. L. Kaldunski, P. J. Tonellato, A. S. Greene et al., 2003. Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol. Genomics 15: 243–257. [DOI] [PubMed] [Google Scholar]
- Ogino, T., D. H. Moralejo, M. Zhu, K. Toide, S. Wei et al., 1999. Identification of possible quantitative trait loci responsible for hyperglycaemia after 70% pancreatectomy using a spontaneously diabetogenic rat. Genet. Res. 73: 29–36. [DOI] [PubMed] [Google Scholar]
- Olofsson, P., J. Holmberg, J. Tordsson, S. Lu, B. Akerstrom et al., 2003. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet. 33: 25–32. [DOI] [PubMed] [Google Scholar]
- Palijan, A., J. Dutil and A. Y. Deng, 2003. Quantitative trait loci with opposing blood pressure effects demonstrating epistasis on Dahl rat chromosome 3. Physiol. Genomics 15: 1–8. [DOI] [PubMed] [Google Scholar]
- Pickering, T. G., 1990. Nutritional influences on the pathophysiology of hypertension, pp. 3–16 in Nutritional Factors in Hypertension, edited by H. Langford, B. Levine and L. Ellenbogen. Alan R. Liss, New York.
- Rapp, J. P., 2000. Genetic analysis of inherited hypertension in the rat. Physiol. Rev. 8: 135–172. [DOI] [PubMed] [Google Scholar]
- Rapp, J. P., and H. Dene, 1985. Development and characteristics of inbred strains of Dahl salt-sensitive and salt-resistant rats. Hypertension 7: 340–349. [PubMed] [Google Scholar]
- Rapp, J. P., and A. Y. Deng, 1995. Detection and positional cloning of blood pressure quantitative trait loci: Is it possible? Identifying the genes for genetic hypertension. Hypertension 25: 1121–1128. [DOI] [PubMed] [Google Scholar]
- Rocha, J. L., E. J. Eisen, L. D. Van Vleck and D. Omp, 2004. A large-sample QTL study in mice: II. Body composition. Mamm. Genome 15: 100–113. [DOI] [PubMed] [Google Scholar]
- Serrano, A. L., and V. Andrés, 2004. Telomeres and cardiovascular disease: Does size matter? Circ. Res. 94: 575–584. [DOI] [PubMed] [Google Scholar]
- Siegel, A.-K., M. Planert, S. Rademacher, A. P. Mehr, P. Kossmehl et al., 2003. Genetic loci contribute to the progression of vascular and cardiac hypertrophy in salt-sensitive spontaneous hypertension. Arterioscler. Thromb. Vasc. Biol. 23: 1211–1217. [DOI] [PubMed] [Google Scholar]
- Stoll, M., A. E. Kwitek-Black, A. W. Cowley, Jr., E. I. Harris, S. B. Harrap et al., 2000. New target regions for human hypertension via comparative genomics. Genome Res. 10: 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll, M., A. W. Cowley, Jr., P. J. Tonellato, A. S. Greene, M. L. Kaldunski et al., 2001. A genomic-systems biology map for cardiovascular function. Science 294: 1723–1726. [DOI] [PubMed] [Google Scholar]
- Xie, G.-X., Y. Yanagisawa, E. Ito, K. Maruyama, X. Han et al., 2005. N-terminally truncated variant of the mouse GAIP/RGS19 lacks selectivity of full-length GAIP/RGS19 in regulating ORL1 receptor signaling. J. Mol. Biol. 353: 1081–1092. [DOI] [PubMed] [Google Scholar]
- Zhu, L., K. S. Hathcock, P. Hande, P. M. Lansdorp, M. F. Seldin et al., 1998. Telomere length regulation in mice is linked to a novel chromosome locus. Proc. Natl. Acad. Sci. USA 95: 8648–8653. [DOI] [PMC free article] [PubMed] [Google Scholar]