Abstract
The major repeat sequence (MRS) is known to play a role in karyotypic variation in Candida albicans. The MRS affects karyotypic variation by expanding and contracting internal repeats, by altering the frequency of chromosome loss, and by serving as a hotspot for chromosome translocation. We proposed that the effects of the MRS on translocation could be better understood by examination of the effect of the MRS on a similar event, mitotic recombination between two chromosome homologs. We examined the frequency of mitotic recombination across an MRS of average size (∼50 kb) as well as the rate of recombination in a 325-kb stretch of DNA adjacent to the MRS. Our results indicate that mitotic recombination frequencies across the MRS were not enhanced compared to the frequencies measured across the 325-kb region adjacent to the MRS. Mitotic recombination events were found to occur throughout the 325-kb region analyzed as well as within the MRS itself. This analysis of mitotic recombination frequencies across a large portion of chromosome 5 is the first large-scale analysis of mitotic recombination done in C. albicans and indicates that mitotic recombination frequencies are similar to the rates found in Saccharomyces cerevisiae.
CANDIDA albicans is an interesting organism because despite its obligatory diploid genome and ability to mate and generate tetraploids (Hull et al. 2000; Magee and Magee 2000; Bennett and Johnson 2003; Legrand et al. 2004), it has not been shown to undergo any type of meiotic recombination or sporulation (reviewed in Johnson 2003). The apparent lack of a complete sexual cycle excludes this mechanism of generating diversity within the genome of C. albicans, and analyses of the karyotypes and genomes of clinical strains have found widespread variation such as loss of heterozygosity, chromosome length polymorphisms, changes in DNA fingerprint patterns, and chromosome translocation (Merz et al. 1988; Asakura et al. 1991; Pfaller et al. 1994; Diaz-Guerra et al. 1997; Stephan et al. 2002). The presence of this extensive heterozygosity further supports the lack of meiosis, since homologs that cannot pair precisely prevent meiotic segregation. Genomic variation has been associated with drug resistance phenotypes and altered cell physiology (Iwaguchi et al. 2001; Jain et al. 2001; Rustchenko et al. 1997) and has also been found to occur during the course of infection (Lockhart et al. 1996), suggesting that variant genotypes are selected for in vivo. Therefore, there must be alternative mechanisms in addition to random mutation and meiotic recombination that allow for the significant diversity and adaptability seen in the growth of C. albicans in the host.
Two potential mechanisms that may provide for rapid evolutionary change in the host are mitotic recombination and genome rearrangement. Typically, determination of mitotic recombination rates in diploid sexual organisms, such as Saccharomyces cerevisiae, can be done by generating haploid spores and directly identifying recombinant haplotypes. Unfortunately, the identification of recombinant haplotypes and the determination of a more general mitotic recombination frequency are made more difficult by the lack of a complete sexual cycle in C. albicans. An estimate of the basal mitotic recombination rate in C. albicans has been made previously by studying the frequency of the recombination event required for URA3 loopout in the commonly used Ura-blaster cassette. In vitro experiments using the Ura-blaster demonstrated mitotic recombination rates of ∼3 × 10−6/kb/generation (Enloe et al. 2000). However, the genomewide applicability of this estimate is questionable since the loopout of the URA3 gene may represent a special case.
The major repeat sequence (MRS) is an intermediate repeat sequence that is found on all but one C. albicans chromosome. Its structure is complex: one end is a 6-kb sequence called RB2, the central part has multiple repeats of a 2-kb sequence called RPS, and centromere proximal is a different 8-kb sequence called HOK (Chibana et al. 1994; Chindamporn et al. 1998). Earlier studies showed that the size of the MRS on a given homolog of chromosome 5 in C. albicans was inversely correlated with the frequency of appearance of that homolog among cells that acquired the ability to grow on sorbose as a sole carbon source (Lephart et al. 2005). The MRS also has a well-documented history of association with genome rearrangements and translocations in numerous clinical isolates (Chu et al. 1993; Lockhart et al. 1995; Chibana et al. 2000; Iwaguchi et al. 2000, 2001). Chromosome rearrangements have also been induced at a high frequency in the laboratory by introduction of foreign DNA into the MRS (Iwaguchi et al. 2004). However, spontaneous translocation is an infrequent event in C. albicans in the laboratory and has rarely been observed. The lack of a selection for the products of translocation between heterologous chromosomes makes it hard to determine a translocation frequency or to study what may affect its occurrence both in vitro and in vivo.
However, since translocation in C. albicans is likely to be a mitotic recombination event, it should be possible to examine that process and the effect that the MRS may have on it by assaying recombination between two chromosome homologs. The ability to generate strains that are monosomic for chromosome 5 in C. albicans (Janbon et al. 1998) makes it possible to identify recombinant chromosomes by the selectable markers that they can obtain only via specific recombination events with their homologs. In this article we describe such an assay and show that the rate of mitotic recombination in C. albicans is comparable to that in S. cerevisiae and is not enhanced across the MRS of chromosome 5.
MATERIALS AND METHODS
Mitotic recombination tester strain creation:
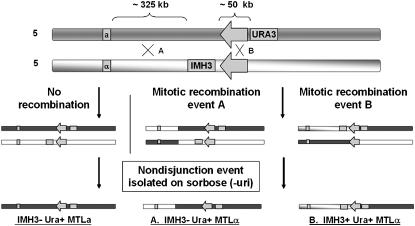
Strain CAI-4 was used to create the mitotic recombination tester (MRT)Ø strain. Strain MRTØ was constructed with an IMH3 gene inserted near the HOK of the chromosome 5 MRS on the MTLα homolog and a URA3 gene inserted near the RB2 of the chromosome 5 MRS on the MTLa homolog.
A 2.7-kb DNA fragment containing the IMH3 gene was cloned into a PCR product amplified from sequence located 1.8 kb from the HOK on the MTL side of the chromosome 5 MRS. This fragment (now including the IMH3 gene) was used to transform strain CAI-4, selecting for mycophenolic acid resistance, and verification of the proper integration was conducted by PCR. Construction of MRTØ was completed by the integration of a 1.6-kb DNA fragment containing the URA3 gene at a site 2 kb from the RB2 of the chromosome 5 MRS on the MTLa homolog. The integrating DNA was created by PCR using oligonucleotides URA3nRB2 F (5′-CTGTTATTAAAGTGTTATGAGTGTTTTAGCTTTCGGCGAAATCCGCAAGAACCGAGGTCTAGAAGGACCACCTTTGATTG-3′) and URA3nRB2 R (5′-CCACTAGCAATTAATTAATACGCATACTCATTTATCACTGTCGGAAATATGGTCCTAATGCTGGTAAAGATTCGAACGAC-3′), 80-mers that contained ∼60 bp of homology to unique sequences 1.6 kb from the RB2 of the chromosome 5 MRS and ∼20 bp of URA3 sequence to drive a PCR reaction across the URA3 gene. These oligos were used in a PCR reaction with the URA3 ORF as template to create the URA3nRB2 cassette. The URA3nRB2 cassette was transformed into the above IMH3+ CAI-4 strain and URA+ transformants were selected on minimal medium and verified by PCR. IMH3+/URA3+ strains were grown on sorbose medium to select for the loss of either chromosome 5 homolog to verify independent segregation of the markers to opposite chromosome 5 homologs. Figure 1 (top) shows the configuration of the two chromosome 5 homologs of the strain.
Figure 1.—
Construction and utility of strain MRTØ. This strain was constructed from strain CAI-4 and contains the IMH3 gene adjacent to the chromosome 5 MRS HOK subunit on the chromosome 5α homolog and the URA3 gene adjacent to the RB2 subunit of the MRS on the 5a homolog. Recombination events that occur between the MTL locus and the IMH3 gene (event A) and recombination events that occur in the MRS between the IMH3 gene and the URA3 gene (event B) are detectable in strains that have become homozygous for chromosome 5 after growth on min-sorbose medium. Recombination events are identified by plating to min-sorbose medium, selecting for sorbose-utilizing (SOU+) colonies that retain the URA3 gene, and identifying MTLα URA+ recombinant strains by PCR. Of these MTLα SOU+ recombinants, those that also contain the IMH3 gene by PCR must have undergone a recombination event in the MRS rather than in the region between the IMH3 gene and the MTL locus.
Mitotic recombination assay:
Cells were picked from a stock plate of MRTØ and used to start a 2-ml liquid YPD culture that was incubated at 30° overnight. Fifty-microliter aliquots of a 10−4 dilution of this overnight culture were used to start 20 independent 2-ml liquid YPD cultures. The secondary cultures were grown at 30° for ∼24 hr or ∼16 generations. Appropriate dilutions of the secondary cultures were made for each type of medium the cells were to be plated on. A 10−4 dilution of each secondary culture was made and 50 μl were plated to 3 YPD plates per culture for a viable cell count. A 1:200 dilution of each secondary culture was made and 100 μl were plated to one sorbose plus uri plate per culture for determination of the sorbose utilization frequency for each culture. A 10−2 dilution of each secondary culture was made and 100 μl were plated to one sorbose plate per culture to assay recombination across the control MTL–MRS interval and to assay recombination across the MRS.
C. albicans grew on YPD plates for 2 days at 30° and individual colonies were counted to determine the viable cell count of the secondary cultures and to establish how many cells were plated to the sorbose media for each culture. All sorbose plates were grown for at least 7 days at 30°. Colonies on sorbose plus uri plates were counted to determine the sorbose utilization frequency for each culture. Colonies on sorbose plates were picked and patched to a YPD plate. These patch plates were grown for 2 days at 30°. Cells were then picked from the patches for use in colony PCR of the MTL and/or IMH3 loci.
Colony PCR:
A 20-μl pipetman tip was used to pick a small amount of cells from a colony and resuspend them in 20 μl of colony PCR incubation solution (1.2 m sorbitol, 100 mm sodium phosphate pH 7.4, and 2.5 mg/ml yeast lytic enzyme; ICN Biochemicals, Aurora, OH). This solution was incubated at room temperature for 15 min and 1 μl was used as template for the PCR reaction. Twenty-microliter PCR reactions used dNTPs (Fisher Scientific, Pittsburgh) at 200 mm, 0.4 units of Taq (Sigma, St. Louis), 1× PCR buffer, and MgCl2 at 3.75 mm. Thermocycler (PTC-200; MJ Research, Waltham, MA) conditions for PCR were as follows: 93° for 5 min; 29 cycles of 93° for 45 sec, 68° ramped down to 55° at 0.2°/sec, 55° for 30 sec, and 68° for 1 min; 68° for 10 min; and hold at 4°.
Fluctuation analysis:
Two statistical methods of fluctuation analysis were used to analyze the mitotic recombination data. The Lea–Coulson method of the median is required to compensate for jackpot events that tend to skew the mean of data that fit a Poisson curve. The Lea–Coulson method of the median (Rosche and Foster 2000) was used to estimate recombinant frequency generated across the MTL–MRS interval in the MRTØ experiments but could not be used to estimate recombinant frequency across the MRS due to the paucity of recombinants detected across that interval. In its place, the p0 method, as described in (Rosche and Foster 2000), can be used to estimate the mean of a set of data that fit a Poisson distribution by determining the negative natural log of the ratio of cultures that had no recombinants to total cultures. Therefore, while the p0 method could be used to estimate the recombination frequency across the MRS interval, as most cultures produced no recombinants, it could not be used to estimate the rate of recombination across the MTL–MRS interval, as most cultures in those experiments produced recombinants.
SNP sequencing of mitotic recombinants:
Genomic DNA was used as template for PCR, utilizing the SNP markers 1922/2344, PDE1, 1969/2162, and 1341/2493, previously designed in Forche et al. (2005). The PCR products were purified and quantified and then placed into 96-well plates for sequencing. Sequencing reactions were run by the DNA Sequencing and Analysis Facility of the BioMedical Genomics Center at the University of Minnesota.
RESULTS
The MRTØ strain was constructed from strain CAI-4 and contains the IMH3 gene adjacent to the chromosome 5 MRS HOK subunit on the MTLα homolog and the URA3 gene adjacent to the RB2 subunit of the chromosome 5 MRS on the MTLa homolog (Figure 1). This strain was designed to assay recombination events that occurred between the MTL locus and the IMH3 gene (325 kb) as well as across the MRS, between the IMH3 gene and the URA3 gene (50 kb). To identify recombination events, the strain was plated to sorbose medium lacking uridine. Since growth on sorbose selects strains that have lost one homolog of chromosome 5, only those that have retained the portion of the MTLa homolog containing the URA3 gene are expected to grow on this medium. These include both cells that have retained the entire 5a homolog and those mitotic recombinants that have retained a URA3-containing portion of the chromosome. In the latter case, since the cells will be transiently monosomic, the eventual disomic (euploid) product will be homozygous for the recombinant chromosome 5. The occurrence of a recombination event between the MTL locus and the URA3 gene will result in some Sou+ colonies that are both URA3 and MTLα. Among these MTLα SOU+ recombinants will be some that have undergone a recombination event in the region between the IMH3 gene and the MTL locus and some that have recombined in the MRS; these last ones will contain the IMH3 gene from the MTLα homolog.
To measure recombination, a set of 20 secondary cultures was inoculated with ∼103 cells each from a primary culture of MRTØ and was grown for 24 hr (∼16 generations). At the end of that time, samples of each culture were plated on sorbose medium and grown for 1 week at 30°. The colonies that grew were assayed for MTL configuration and presence of the IMH3 gene by PCR. Cells that were MTLα were assumed to have undergone a recombination event between URA3 and MTL. Those that were also IMH3 positive were assumed to have undergone a recombination event between URA3 and IMH3 (i.e., across the MRS); IMH3-negative recombinants must have crossed over between MTL and IMH3.
To determine the true recombination rate we used a variant of the Luria–Delbruck equation, the Lea–Coulson method of the median to obtain the rate of recombination events per cell between the MTL locus and the IMH3 gene for each experiment. This number was then divided by the size of the interval in kilobases (325 kb MTL–MRS), the number of generations grown (16 generations), and the sorbose frequency (measured specifically for each experiment, average 7.8 × 10−4) to obtain the recombination frequency per kilobase per generation. Table 1 shows that the range of values for the four separate experiments was quite narrow. The average frequency found across the 325-kb MTL–IMH3 region was 2.82 × 10−6/kb/generation.
TABLE 1.
Recombination frequency in MRTØ
| Experiment | Cultures with event A recombinants/total cultures | Total event A recombinants/cells plated | Recombination frequency A |
|---|---|---|---|
| 1 | 12/20 | 38/1.2 × 106 | 2.90 × 10−6/kb/generation |
| 2 | 20/20 | 70/3.0 × 106 | 2.90 × 10−6/kb/generation |
| 3 | 10/15 | 30/1.35 × 106 | 2.79 × 10−6/kb/generation |
| 4 | 10/10 | 28/1.1 × 106 | 2.70 × 10−6/kb/generation |
| Experiment | Cultures with event B recombinants/total cultures | Total event B recombinants/cells plated | Recombination frequency B |
| 1 | 0/20 | 0/1.2 × 106 | 0 |
| 2 | 1/20 | 1/3.0 × 106 | 6.25 × 10−7/kb/generation |
| 3 | 3/15 | 3/1.35 × 106 | 1.44 × 10−6/kb/generation |
| 4 | 1/10 | 1/1.1 × 106 | 1.25 × 10−6/kb/generation |
The number of recombinants that occurred across the MRS was too low to produce statistically significant results when analyzed by the Lea–Coulson method. The p0 method, a less accurate method of obtaining a rate of recombination, was used to estimate the rate of recombination events per cell between the URA3 gene and the IMH3 gene (across the MRS) for each experiment. This rate was divided by the size of the interval in kilobases (50-kb MRS), the number of generations grown (16 generations), and the sorbose frequency to obtain the recombination frequency per kilobase per generation. This resulted in a rate across the MRS that was at least twofold lower than the rate per kilobase obtained across the 325-kb region between the MRS and the MTL locus (Table 1). This result indicates that the MRS is not highly recombinogenic under these conditions, despite its size and repetitive nature.
The level of mitotic recombination measured between MTL and IMH3 might be due to a mitotic recombination “hotspot,” leading to a heightened recombination rate. If this were true, most of the recombinants would be found in a small region. This possibility was tested by analyzing the SNP profile of the recombinants. Forche et al. (2004) have constructed single-nucleotide polymorphism (SNP) maps for the two chromosome 5 homologs, as well as determined the haplotypes. We utilized four of the SNPs identified by Forche et al. in our analysis; three of these are between the MTL locus and the IMH3 position near the MRS in strain MRTØ and one is located on 5I outside of the analyzed region as a control. All SNPs should be homozygous in our selected recombinants, since they have passed through a monosomic stage. On the basis of the SNP profile of each, we could determine the region in which the recombination occurred.
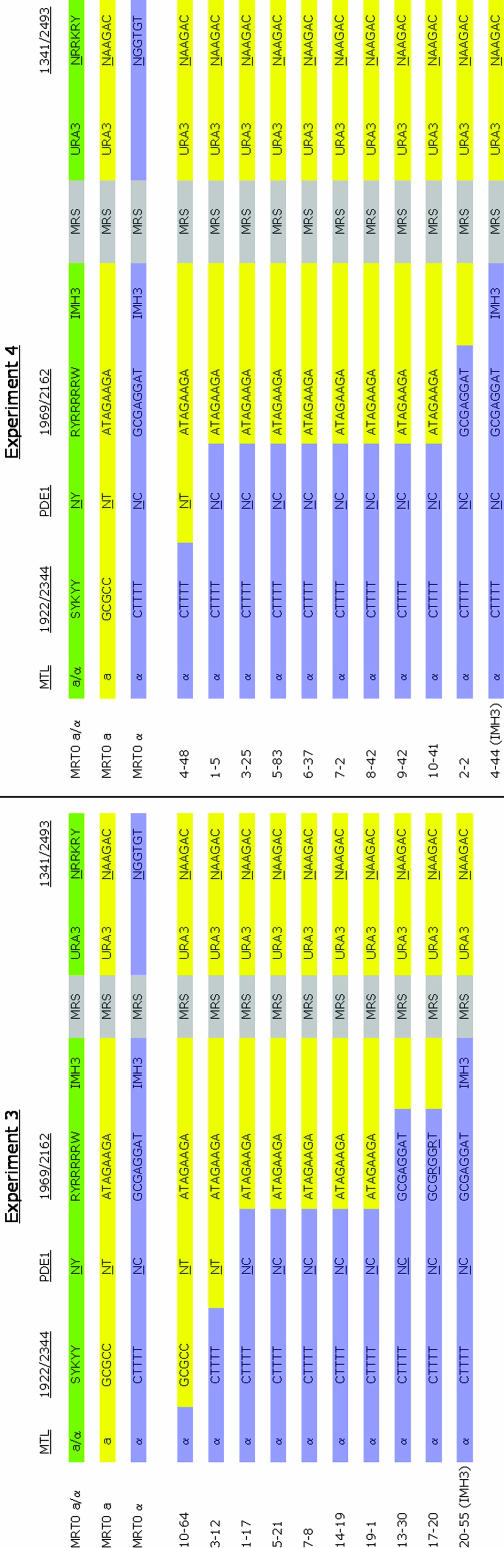
One recombinant from each secondary culture from experiments 3 and 4 was analyzed for its SNP profile, on the basis of the sequence of PCR products generated by primers designed to amplify the SNP markers (Figure 2). At least one of each of the recombinants identifiable by these SNP markers was found and the collective results of the recombinant genotyping from both experiments are found in Figure 3. While the majority of the recombination events did occur in the PDE1–1969/2162 interval, this is not altogether surprising as at 180 kb it is the largest interval analyzed and is >2.5 times the size of the next largest interval of 70 kb (1922/2344–PDE1). In fact, when taken together, there were 13 recombination events identified in the 180-kb PDE1–1969/2162 interval and 8 identified in the remaining intervals that total 195 kb (and include the MRS). Therefore, these results indicate that the recombination frequency determined in the MRTØ experiments was the result of crossover events that spanned the entire interval analyzed and was not entirely due to a single recombination hotspot in this strain.
Figure 2.—
SNP sequencing of MRTØ recombinants from experiments 3 and 4. A color-coded map of the chromosome 5 genotype of the MRTØ recombinants analyzed from experiments 3 and 4 is shown. Light blue corresponds to the MTLα/α genotype while the yellow shading refers to the MTLa/a genotype. The heterozygous genotype of an MTLa/α strain is shown at the top in green. The recombinant strains identified have genetic information solely from the MTLα genotype to the left of the site of recombination and only MTLa genetic content to the right, indicating a crossover event.
Figure 3.—
MRTØ recombinant genotype analysis. Scale representation of the locations of the SNP and selective markers on chromosome 5 is shown. The numbers of strains identified in experiments 3 and 4 that underwent a recombination event between the indicated adjacent markers on chromosome 5 are shown.
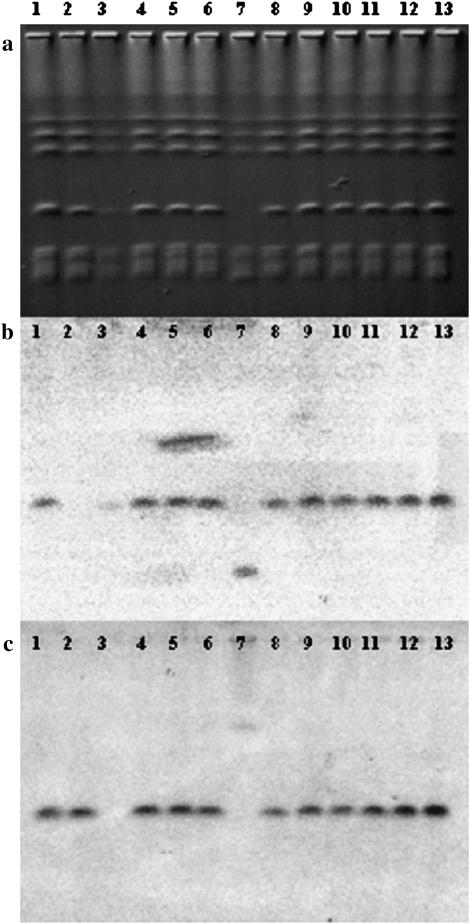
To characterize the recombination process further, we analyzed the karyotype of at least one recombinant per culture from MRTØ experiments 3 and 4 using CHEF pulsed-field gel electrophoresis, transferred the karyotypes to a membrane, and probed with MTLα1 and URA3. The resulting Southern blots, shown in Figure 4, b and c, verified that these markers, which were used to determine the interval analyzed, were associated with the chromosome 5 band in every case but one, providing more evidence that a recombination event between the two chromosome 5 homologs and not some other sort of genomic rearrangement had occurred.
Figure 4.—
Karyotype and Southern analysis of MRTØ recombinants . CHEF gel of a representative sample of MRTØ recombinant strains (a), probed by Southern blot with either MTLα1 (b) or URA3 (c), is shown. Lane 1 shows strain MRTØ MTLa/α; lane 2, MRTØ MTLa; lane 3, MRTØ MTLα; lane 4, MRTØ recombinant (rec) 1-17; lane 5, MRTØ rec 3-12; lane 6, MRTØ rec 10-64; lane 7, MRTØ rec 17-20; lane 8, MRTØ rec 20-55; lane 9, MRTØ rec 1-5; lane 10, MRTØ rec 2-2; lane 11, MRTØ rec 7-2; lane 12, MRTØ rec 8-42; lane 13, MRTØ rec 4-44. Lanes 4–8 are from experiment 3 and lanes 9–13 are from experiment 4.
All of the karyotypes analyzed were indistinguishable from the parent except for one case in which a translocation involving the remaining chromosome 5 homolog had occurred (Figure 4a, lane 7). The translocation has led to the MTLα locus migrating at the position of chromosome 7 and URA3 migrating in a band the size of chromosome 2. To our knowledge this is one of the first identifications of a spontaneously occurring in vitro translocation event. The remaining recombinants had no karyotypic variants, once more supporting the conclusion that most of the events we measured were simple mitotic recombinations.
DISCUSSION
Previous research has shown that the MRS is a preferred site for translocations between heterologous chromosomes and that these translocations can alter the phenotype of the resultant cell (Chu et al. 1993; Chibana et al. 2000). The control of the translocation process is not understood, and most translocations seem to occur either during growth in the host or when induced by chromosome damage in vitro (Iwaguchi et al. 2004). And since heterologous translocations do not alter the genetic composition of the affected cell (just its genomic arrangement), they are difficult to detect except by analysis of the karyotypes of all potential translocatants. These considerations make the measurement of the frequency of translocation very difficult. However, heterologous translocation events in C. albicans probably occur via mitotic recombination events, and recombination events between homologous chromosomes likely occur via a similar mechanism. Hence, study of mitotic recombination should provide insight into the translocation process. But a difficulty in measuring the frequency of mitotic recombination is the obligate diploid nature of C. albicans; it is impossible to generate haploid progeny of the recombinants. In addition, the presence of two nearly identical homologs makes it hard to select for the occurrence of recombinant chromosomes or to identify their haplotypes when they arise.
Growth of C. albicans on sorbose requires the loss of one homolog of chromosome 5; when the selective pressure is removed, the remaining homolog becomes disomic and homozygous (Janbon et al. 1998, 1999). This system allowed us to construct a strain that would allow for the large-scale screening of chromosome 5 recombinant haplotypes. The sort of experimental system we have devised, a strain that can yield a recombination product carrying selectable recessive markers on chromosome 5 and then render it homozygous by growth on sorbose, should be applicable in a variety of other experiments to examine recombination in C. albicans. However, this assay is limited by the inability to select for monosomy of chromosome homologs other than chromosome 5, thus limiting the applicability of this assay to the analysis of recombination on other chromosomes. In addition, the insertion of heterozygous DNA, in the form of our selectable markers (URA3 and IMH3), onto chromosome homologs may also have an effect on the mitotic recombination rates measured near those sites of insertion. Interestingly, the proposed inhibition of recombination measured across the MRS may have been due to the insertion of heterozygous DNA so near the MRS and may actually serve as a preliminary model for translocation between heterologous chromosomes.
The results reported here demonstrate that mitotic recombination in C. albicans is a common event; in the course of our experiments almost 1 in 100 cells underwent a recombination event between chromosome 5 homologs over a 325-kb interval between the MTL and MRS on chromosome 5. This translates to an average mitotic recombination rate of 2.82 ± 0.08 × 10−6 events/kb/generation, which is very close to the reported value for the recombination event required for URA3 loopout from the UAU cassette (∼3 × 10−6/division) (Enloe et al. 2000). Interestingly, spontaneous mitotic recombination rates in S. cerevisiae have also been measured at about ∼2 × 10−6 to 1 × 10−5/kb/generation, depending on the gene (Montelone et al. 1988). Thus, while both the Enloe estimation of recombination frequency at the URA3 locus and the frequency reported in this article on chromosome 5 may represent special cases corresponding to their specific intervals, the general agreement of the rates found by Enloe and this article to rates found in S. cerevisiae makes a strong argument of their relevance as an estimate of mitotic recombination in the genome of C. albicans.
Despite these similarities in the mitotic recombination frequencies previously measured, the recombination rate generated by this assay may not be directly comparable to those of other studies due to the inability of this assay to detect all recombinants in the experimental population and to the unknown effects that exposure to sorbose may have on recombination frequency. The sorbose utilization frequency was typically 7.8 × 10−4 sorbose-utilizing cells per cell plated (i.e., of 1300 cells that are plated to sorbose media, only one acquired the ability to grow) and was included in the determination of the final recombination frequency. Therefore, if only one recombinant colony was detected on a sorbose recombination assay plate, it was deduced that 1300 recombinants originally existed in the total cells that were plated. However, it is possible that the carbon source starvation that occurs upon plating to sorbose may artificially enhance mitotic recombination rates prior to the loss of one chromosome 5 homolog.
Mitotic recombination rates in most eukaryotes are much lower than those seen in meiosis and this is thought to be mainly due to a lack of a structured pairing of homologs in mitosis and the lack of the formation of chiasmata. The MRS may have evolved the function of providing a site for mitotic recombination resulting in chromosome translocation and an increase in the amount of genomic diversity in an organism that lacks a meiotic cycle. Although our experiments did not show an enhanced recombination rate at the MRS on chromosome 5 under standard in vitro growth conditions, by serving as a major source of homology throughout the genome, the MRSs clearly facilitate translocation between heterologous chromosomes. Additionally, the MRS may have a dual role in producing genomic variability as the facilitation of heterologous translocation allows for the process of nondisjunction to create further genomic diversity in a strain. The translocation process produces two reciprocal recombinant chromosomes that can undergo the same nondisjunction process that other chromosomes are vulnerable to. The loss of a recombinant chromosome would introduce a partial hemizygosity for two separate chromosomes and thus potentially a new phenotype unattainable by the loss of a nonrecombinant homolog. Interestingly, the size of an MRS has also been shown to affect the frequency of chromosome nondisjunction (Lephart et al. 2005); thus this repeated sequence may be involved in the regulation of both translocation and nondisjunction.
The MRS remains a tantalizing structure within the genome of C. albicans; it has interesting connections to essential functions and chromosome dynamics and yet seems to be dispensable to chromosome 3 (naturally) and chromosome 5 (deleted in Lephart et al. 2005). We have now shown that the MRS on chromosome 5 does not enhance mitotic recombination under standard in vitro growth conditions and rather may slightly inhibit mitotic recombination. Perhaps this result is not surprising though, as the majority of translocated strains have been recovered from clinical specimens, possibly indicating a role for stress in the enhancement of the recombination frequency near the MRS or in general. Nevertheless, the MRS has been shown to serve as an important vehicle for the generation of genomic diversity and ultimately phenotypic diversity in C. albicans and deserves to be understood in greater detail.
Acknowledgments
We are grateful to Anja Forche for providing the SNP marker primers and sequences and for technical assistance. We are grateful to Bebe Magee and the members of the Magee lab and Judith Berman, David Kirkpatrick, and Dana Davis for helpful discussions. This work was supported by National Institutes of Health grants AI16567 and AI46351 and by contract AI05406 awarded to P.T.M. from the National Institute of Allergy and Infectious Diseases. P.R.L. was supported by training grant T32 GM08347 from the National Institute of General Medical Sciences. Sequence data for Candida albicans were obtained from the Stanford Genome Technology Center website at http://www.sequence.stanford.edu/group/ Candida. Sequencing of C. albicans was accomplished with the support of the National Institute of Dental Research and the Burroughs Wellcome Fund.
References
- Asakura, K., S. Iwaguchi, M. Homma, T. Sukai, K. Higashide et al., 1991. Electrophoretic karyotypes of clinically isolated yeasts of Candida albicans and C. glabrata. J. Gen. Microbiol. 137: 2531–2538. [DOI] [PubMed] [Google Scholar]
- Bennett, R. J., and A. D. Johnson, 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22: 2505–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana, H., S. Iwaguchi, M. Homma, A. Chindamporn, Y. Nakagawa et al., 1994. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. J. Bacteriol. 176: 3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibana, H., J. L. Beckerman and P. T. Magee, 2000. Fine-resolution physical mapping of genomic diversity in Candida albicans. Genome Res. 10: 1865–1877. [DOI] [PubMed] [Google Scholar]
- Chindamporn, A., Y. Nakagawa, I. Mizuguchi, H. Chibana, M. Doi et al., 1998. Repetitive sequences (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology 144: 849–857. [DOI] [PubMed] [Google Scholar]
- Chu, W. S., B. B. Magee and P. T. Magee, 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J. Bacteriol. 175: 6637–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Guerra, T. M., J. V. Martinez-Suarez, F. Laguna and J. L. Rodriguez-Tudela, 1997. Comparison of four molecular typing methods for evaluating genetic diversity among Candida albicans isolates from human immunodeficiency virus-positive patients with oral candidiasis. J. Clin. Microbiol. 35: 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe, B., A. Diamond and A. P. Mitchell, 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182: 5730–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., P. T. Magee, B. B. Magee and G. May, 2004. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3: 705–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche, A., G. May and P. T. Magee, 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell 4: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, C. M., R. M. Raisner and A. D. Johnson, 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289: 307–310. [DOI] [PubMed] [Google Scholar]
- Iwaguchi, S., M. Suzuki, N. Sakai, Y. Nakagawa, P. T. Magee et al., 2004. Chromosome translocation induced by the insertion of the URA blaster into the major repeat sequence (MRS) in Candida albicans. Yeast 21: 619–634. [DOI] [PubMed] [Google Scholar]
- Iwaguchi, S. I., T. Kanbe, T. Tohne, P. T. Magee and T. Suzuki, 2000. High-frequency occurrence of chromosome translocation in a mutant strain of Candida albicans by a suppressor mutation of ploidy shift. Yeast 16: 411–422. [DOI] [PubMed] [Google Scholar]
- Iwaguchi, S. I., M. Sato, B. B. Magee, P. T. Magee, K. Makimura et al., 2001. Extensive chromosome translocation in a clinical isolate showing the distinctive carbohydrate assimilation profile from a candidiasis patient. Yeast 18: 1035–1046. [DOI] [PubMed] [Google Scholar]
- Jain, P., Z. K. Khan, E. Bhattacharya and S. A. Ranade, 2001. Variation in random amplified polymorphic DNA (RAPD) profiles specific to fluconazole-resistant and -sensitive strains of Candida albicans. Diagn. Microbiol. Infect. Dis. 41: 113–119. [DOI] [PubMed] [Google Scholar]
- Janbon, G., F. Sherman and E. Rustchenko, 1998. Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95: 5150–5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbon, G., F. Sherman and E. Rustchenko, 1999. Appearance and properties of L-sorbose-utilizing mutants of Candida albicans obtained on a selective plate. Genetics 153: 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A., 2003. The biology of mating in Candida albicans. Nat. Rev. Microbiol. 1: 106–116. [DOI] [PubMed] [Google Scholar]
- Legrand, M., P. Lephart, A. Forche, F. M. Mueller, T. Walsh et al., 2004. Homozygosity at the MTL locus in clinical strains of Candida albicans: karyotypic rearrangements and tetraploid formation. Mol. Microbiol. 52: 1451–1462. [DOI] [PubMed] [Google Scholar]
- Lephart, P. R., H. Chibana and P. T. Magee, 2005. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot. Cell 4: 733–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schroppel, T. Srikantha et al., 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart, S. R., B. D. Reed, C. L. Pierson and D. R. Soll, 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J. Clin. Microbiol. 34: 767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee, B. B., and P. T. Magee, 2000. Induction of mating in Candida albicans by construction of MTLa and MTLalpha strains. Science 289: 310–313. [DOI] [PubMed] [Google Scholar]
- Merz, W. G., C. Connelly and P. Hieter, 1988. Variation of electrophoretic karyotypes among clinical isolates of Candida albicans. J. Clin. Microbiol. 26: 842–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelone, B. A., M. F. Hoekstra and R. E. Malone, 1988. Spontaneous mitotic recombination in yeast: the hyper-recombinational rem1 mutations are alleles of the RAD3 gene. Genetics 119: 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller, M. A., J. Rhine-Chalberg, S. W. Redding, J. Smith, G. Farinacci et al., 1994. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J. Clin. Microbiol. 32: 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche, W. A., and P. L. Foster, 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko, E. P., D. H. Howard and F. Sherman, 1997. Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology 143: 1765–1778. [DOI] [PubMed] [Google Scholar]
- Stephan, F., M. S. Bah, C. Desterke, S. Rezaiguia-Delclaux, F. Foulet et al., 2002. Molecular diversity and routes of colonization of Candida albicans in a surgical intensive care unit, as studied using microsatellite markers. Clin. Infect. Dis. 35: 1477–1483. [DOI] [PubMed] [Google Scholar]