Abstract
Maize B-A translocations result from reciprocal interchanges between a supernumerary B chromosome and an arm of an essential A chromosome. Because of the frequent nondisjunction of the B centromere at the second pollen mitosis, B-A translocations have been used to locate genes to chromosome arms and to study the dosage effects of specific A segments. Compound B-A translocations (B-A-A translocations) are created by bringing together a simple B-A translocation with an A-A translocation in which breakpoints in the A-A and B-A translocations are in the same arm. Recombination in the region of shared homology of these A chromosome segments creates a B-A-A translocation. Success in creating and testing for a new B-A-A translocation requires that the B-A translocation be proximal to the A-A translocation and that the A-A translocation be proximal to the tester locus. The breakpoints of most of the A-A translocations have been cytologically defined by earlier investigators. Previous investigators have produced 16 B-A-A translocations and one B-A-A-A translocation, which collectively define 35 A chromosome breakpoints. We have enlarged this group by creating 64 new B-A-A translocations. We present a summary of the total of 81 B-A-A translocations showing their distribution among the chromosome arms and the 163 cytologically defined chromosome segments delimited by them. We also illustrate the method of construction of these B-A-A stocks and their uses.
THERE are many useful genetic and cytogenetic resources. Two of these cytogenetic resources are particularly notable in maize. The first is a large set of reciprocal translocation stocks. These involve interchanges between nonhomologous pairs of the standard (A) chromosomes and are termed A-A translocations. There are ∼870 different A-A translocation stocks that are available from the Maize Genetics Cooperation Stock Center in Urbana, Illinois. Their chromosome breakpoints were cytologically characterized by Longley (1961) and the complete list of the available stocks and their breakpoints are listed in the Maize Genetics and Genomics Database website (http://www.maizegdb.org). The second resource is the set of simple B-A translocation stocks. These stocks each consist of an interchange between a B chromosome and an A chromosome (Roman 1947; Beckett 1991, 1993). Because centromeres of B chromosomes frequently undergo nondisjuntion at the second pollen mitosis, B-A chromosomes are useful for several purposes, including the determination of the chromosome arm location of a gene of interest (Beckett 1991, 1993) and dosage analyses (Birchler 1980, 1981, 1993a; Birchler et al. 2001).
When breakpoints of B-A and A-A translocations are in the same A chromosome arm, a recombination event can yield a B-A-A compound translocation (Rakha and Robertson 1970).
The purpose of this article is to report on the construction of 64 new B-A-A translocations that, together with the 17 previously constructed stocks, has expanded the number of these useful cytogenetic stocks to a total of 81 compound B-A-A translocations. We describe the procedures and the A-A translocation stocks and simple B-A translocation stocks that were brought together to produce the rare recombinants comprising the new B-A-A translocations. In addition, we provide our observations on the behavior of these stocks and how to most efficiently identify, propagate, and utilize the B-A-A stocks. We have undertaken this project because the B-A-A stocks can be employed to analyze cytologically defined chromosome segments and to relate genes and other DNA sequences located in these segments to their locations on the maize genetic and DNA physical maps. Such analyses will aid in integrating the maize cytological map with these other maps to create a unified maize genetic map. In addition, by expanding the number and detailed coverage of these cytogenetic tools, the creation of new B-A-A stocks will make possible a finer level of dosage analysis of cytologically defined chromosome segments and a more precisely defined location of a gene of interest to a small chromosome region.
MATERIALS AND METHODS
The 18 simple B-A translocation stocks used in this study are listed in Table 1. There are a large number of simple B-A translocations; these 18 were chosen because they compose a basic set of simple B-A translocations carrying the greatest extent of the A chromosome arm involved in the interchange (Beckett 1991, 1993). The breakpoints are described as the proportional distance from the centromere to the end of the normal chromosome arm where the exchange is located. For example, TB-1Sb has a breakpoint at 1S.05, which means the chromosome 1 centromere retains only 5% of its original short arm, the remainder being replaced by a portion of the B chromosome. The remaining 95% of 1S is attached to the B chromosome. The breakpoints of the B chromosomes are not presented.
TABLE 1.
Simple B-A translocation stocks
| B-A stock | Breakpoint in A chromosome | Tester for propagating B-A |
|---|---|---|
| TB-1Sb | 1S.05 | dek1, sh-1322A, vp5 |
| TB-1La | 1L.20 | bz2 |
| TB-2Sa | 2S.50 | r1 |
| TB-3Sb | 3S.50 | cl1 |
| TB-3La | 3L.10 | a1 |
| TB-4Sa | 4S.25 | cp2, bt2, su1 |
| TB-4Lf | 4L.15 | c2 |
| TB-5Sc | 5S.30 | a2 |
| TB-5La | 5L.10 | pr |
| TB-6Sa | 6S.50 | dek28, wr*-N1389A |
| TB-6Lc | 6L.11 | y1, de-1400 |
| TB-7Sc | 7S.? | vp9, o2 |
| TB-7Lb | 7L.30 | o5 |
| TB-8Lc | 8L.24 | pro1 |
| TB-9Sd | 9S.08 | c1, sh1, bz1 |
| TB-9Lc | 9L.10 | dek13, dek30 |
| TB-10Sc | 10S.30 | y9 |
| TB-10L19 | 10L.cent | r1 |
The A-A translocation stocks crossed onto the B-A stocks, to create the B-A/A-A stocks that were used to successfully screen for recombinants (new B-A-A translocations), are listed in Table 2. For each stock, the breakpoints for both chromosomes involved with the translocation are shown.
TABLE 2.
A-A chromosomal translocation stocks used to create new B-A-A translocations
| A-A stock | Chromosome arm breakpoints |
|---|---|
| T1-4 (064-20) | 1S.23, 4L.19 |
| T1-6 (055-10) | 1S.29, 61.48 |
| T1-9 (7535) | 1S.33, 9S.27 |
| T1-6 (7352) | 1S.40, 6L.60 |
| T1-4 (8602) | 1S.41, 4L.81 |
| T1-6 (7097) | 1S.46, 6L.62 |
| T1-3 (8995) | 1S.49, 3L.06 |
| T1-4 (4308) | 1S.65, 4L.58 |
| T1-3 (5597) | 1S.77, 3L.48 |
| T1-10 g | 1S.80, 10L.21 |
| T1-4 (002-19) | 1S.87, 4L.42 |
| T1-4 h | 1S.94, 4L.52 |
| T1-7 (4891) | 1L.12, 7L.69 |
| T1-10 a | 1L.29, 10L.33 |
| T1-9 (4997-6) | 1L.37, 9S.28 |
| T1-5 (0707-12) | 1L.39, 5S.71 |
| T1-6 (070-1) | 1L.40, 6L.58 |
| T1-10 c | 1L.43, 10L.74 |
| T1-5 (7212) | 1L.44, 5S.28 |
| T1-10 d | 1L.50, 10L.68 |
| T1-8 b | 1L.59, 8L.82 |
| T1-5 (8041) | 1L.80, 5S.10 |
| T1-10 (001-3) | 1L.86, 10L.48 |
| T2-3 (5304) | 2S.62, 3L.29 |
| T2-3 e | 2S.76, 3L.48 |
| T2-5 (015-3) | 2L.16, 5S.69 |
| T2-9 (062-11) | 2L.21, 9S.53 |
| T3-9 (5643) | 3S.55, 9L.64 |
| T3-5 g | 3L.01, 5S.73 |
| T3-10 c | 3L.22, 10L.30 |
| T3-9 g | 3L.40, 9L.14 |
| T3-6 (8672) | 3L.47, 6L.87 |
| T3-4 (6534) | 3L.48, 4L.89 |
| T3-9 b | 3L.48, 9L.53 |
| T3-10 (036-15) | 3L.48, 10L.64 |
| T3-6 (7162) | 3L.52, 6L.53 |
| T3-9 (4727) | 3L.54, 9L.42 |
| T3-5 b | 3L.61, 5L.57 |
| T3-5 (7043) | 3L.63, 5L.61 |
| T3-7 (5471) | 3L.64, 7L.58 |
| T3-7 d | 3L.64, 7L.81 |
| T4-9 (6222) | 4L.03, 9S.68 |
| T4-9 (6504) | 4L.09, 9S.83 |
| T4-9 (4373) | 4L.29, 9L.39 |
| T4-6 (8764) | 4L.32, 6L.90 |
| T4-7 (4483) | 4L.39, 7L.61 |
| T4-5 (006-7) | 4L.43, 5S.25 |
| T4-5 f | 4L.50, 5L.80 |
| T4-10 (6587) | 4L.55, 10L.51 |
| T4-6 (8927) | 4L.70, 6L.18 |
| T5-6 (6671) | 5S.49, 6L.35 |
| T5-9 (4790) | 5L.34, 9L.45 |
| T5-6 (4666) | 5L.35, 6L.86 |
| T5-10 (006-11) | 5L.49, 10L.52 |
| T5-10 (7142) | 5L.73, 10L.17 |
| T7-9 (6482) | 7L.01, 9S.97 |
| T7-9 (7074) | 7L.03, 9S.80 |
| T7-9 a | 7L.63, 9S.07 |
| T9-10 (8630) | 9S.28, 10L.37 |
| T9-10 (059-10) | 9S.31, 10L.53 |
| T9-10 (3688) | 9S.49, 10L.02 |
The designations for the A-A stocks and their chromosome arm breakpoints are from Longley (1961). See the Maize Genetics and Genomics Database website (http://www.maizegdb.org) for a complete listing with corrections.
Construction of B-A-A translocations requires two steps (Rakha and Robertson 1970; Birchler 1993b). First, a particular chromosome arm of interest was identified. For 18 of the 20 chromosome arms of maize there is a simple B-A translocation; the exceptions are for the long arm of chromosome 2 (2L) and the short arm of chromosome eight (8S). For creating each new B-A-A translocation stock, a simple B-A stock was crossed with pollen from a stock containing an A-A reciprocal translocation where one of the chromosome arms involved in the reciprocal interchange is the same A chromosome arm as the chromosome arm involved in the simple B-A chromosome. The other chromosome arm involved in the reciprocal interchange is the A chromosome region of interest, which will become the distal-most segment of the new B-A-A translocation. It is necessary that the B-A translocation be proximal to the A-A translocation and that the A-A translocation be proximal to the locus to the marker of the tester stock.
Second, the progeny of the cross of the B-A stock by the A-A stock were grown and plants that were B-A/A-A heterozygotes were identified by pollen semisterility; these heterozygotes contained one copy of the B-A chromosome and one copy of the two interchange chromosomes involved in the A-A translocation, as well as an A-B chromosome and a normal chromosome of each of the chromosomes involved in the A-A interchange.
Crosses from these plants were made onto appropriate tester stocks. The tester stock carried recessive kernel alleles of genes borne on the chromosome arm homologous to the one that will be recombined with the B-A chromosome to constitute the distal-most segment of the new B-A-A translocation. For those chromosome arms bearing an aleurone color factor, a recessive color allele was the first choice for a tester. (See the supplemental data at http://www.genetics.org/supplemental/ for a detailed description of the identification and selection of suitable pollen parent plants and the need for the presence of the A-B chromosome and for a detailed consideration of kernel tester stocks best suited for screening for new B-A-A translocations.)
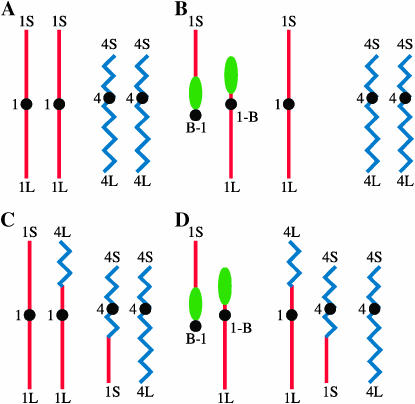
The chromosome constitutions with regard to chromosome 1 and chromosome 4 are shown for a normal maize plant lacking any translocation, a plant heterozygous for a simple B-A translocation involving chromosome arm 1S (TB-1Sb), a plant heterozygous for a reciprocal translocation between chromosome arms 1S and 4L[(1-4 (002-19)], and a plant heterozygous for both the TB-1Sb and the T1-4 (002-19) translocation in Figure 1, A–D, respectively.
Figure 1.—
Chromosome constitution of plants with normal and translocated chromosomes: (A) A plant with a pair of normal chromosome 1s and a pair of normal chromosome 4s. (B) A plant heterozygous for a B-A translocation. The B-A is the TB-1Sb, the A-B is the 1-B, and a normal chromosome 1 and a pair of normal chromosome 4's are also present. (C) A plant heterozygous for an A-A translocation between chromosomes arms 1S and 4L. This translocation is named T1-4 (002-19) 1S.87 4S.42. A normal chromosome 1 and a normal chromosome 4 are also present. (D) A plant heterozygous for the B-A translocation TB-1Sb and for the T1-4 (002-19) translocation. The B-A (TB-1Sb), the A-B (1-B) chromosome, the 1-4 chromosome, the 4-1 chromosome, and a normal chromosome 4 are present. The normal chromosomes other than chromosome 1 and chromosome 4 are not shown in Figures 1–4.
A crossover event can yield a new B-A-A translocation:
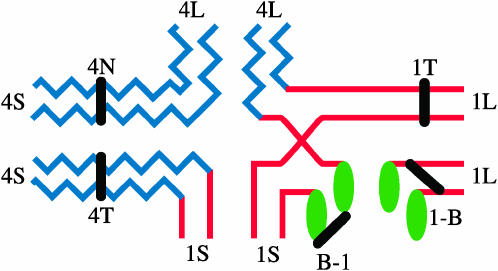
A new B-A-A translocation chromosome can result from a crossover event during the first meiotic division of meiocytes in plants that are B-A/A-A heterozygotes, as shown in the example in Figure 1D. The chromosomes in this example undergo homologous pairing and synapsis and can be seen at the pachytene stage in the configuration in Figure 2. The homologous chromosomes are shown in their duplicated form and a single crossover event is shown involving an interchange between the 1S regions of the B-A (TB-1Sb) and one of the A-A [T1-4 (002-19)] chromosomes.
Figure 2.—
Diagram of the pairing configuration of the chromosomes in Figure 1D. The chromosomes are at the pachytene stage of meiosis I and a crossover is shown in the 1S region of the B-A chromosome and the homologous 1S region of the 1-4 chromosome. Centromeres are indicated by black bars and are labeled according to their chromosome (4N, normal chromosome 4; 4T, translocation 4-1; 1T, translocation 1-4; B-1 and 1-B). One of the two products of this crossover event will be a new B-A-A chromosome while the other product is a reconstituted normal chromosome 1.
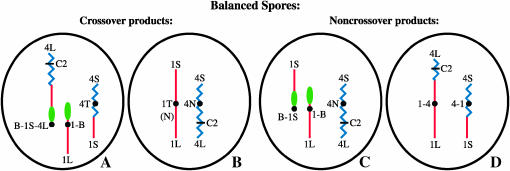
The results of the completion of the meiotic sequence in a microsporocyte are shown for the four genetically balanced haploid microspores diagrammed in Figure 3. Other meiotic segregation products that are genetically unbalanced and result in ∼50% pollen abortion are formed, but these are not shown. Four types of microspores are shown in Figure 3: Figure 3A shows the balanced microspore containing the new B-A-A (B-1-4) recombinant chromosome, the A-B (1-B) chromosome, and the complementary A-A (4-1) chromosome. These three chromosomes together contain all regions of chromosome 1 and the B-A-A and the A-A together contain all regions of chromosome 4. Figure 3B contains the balanced reciprocal recombination product, a normal chromosome 1, and a normal chromosome 4. The noncrossover balanced microspore products contain the B-A, the A-B and a normal chromosome 4 (Figure 3C), and the 1-4 and 4-1 translocation chromosomes (Figure 3D).
Figure 3.—
Four genetically balanced haploid microspores are produced by the meiotic divisions of the doubly heterozygous microsporocyte shown in Figure 2. The genetically unbalanced microspores that also are produced in equal frequency are not shown but they result in ∼50% pollen abortion. Note that the crossover product microspores contain (A) the new B-1-4 chromosome, the 1-B chromosome, and a 4-1 translocated chromosome or (B) a normal chromosome 1 that was reconstituted by the crossover event and is labeled 1T(N) and a normal chromosome 4. The noncrossover product microspores contain (C) the simple B-1, the 1-B, and a normal chromosome 4 or (D) a 1-4 chromosome and a 4-1 chromosome.
Detecting a new B-A-A translocation:
It is apparent from Figure 3 that one of the four functional pollen grains that will develop from the four microspores shown in Figure 3 will contain the new B-A-A chromosome and can potentially be recovered in a screening test when crossed onto an appropriate female tester stock. A relatively efficient procedure for screening to detect new B-A-A translocations is to cross the B-A/A-A stock as a pollen parent onto a tester stock carrying a recessive allele for a kernel trait. The new B-A-A chromosome can be recovered in the embryo of a kernel displaying a recessive endosperm phenotype. Returning to our example, a pollen grain developing from the microspore shown in Figure 3A will include a generative nucleus containing the B-1-4, the 1-B, and the 4-1 chromosome. When the generative nucleus divides to produce two sperm nuclei, the B-A-A chromatids frequently undergo nondisjunction, resulting in a hyperploid sperm containing two B-1-4 chromosomes, a 1-B chromosome, and the 4-1 chromosome, while the other sperm becomes a hypoploid gamete lacking the B-1-4 chromosome but containing the 1-B and the 4-1 chromosome.
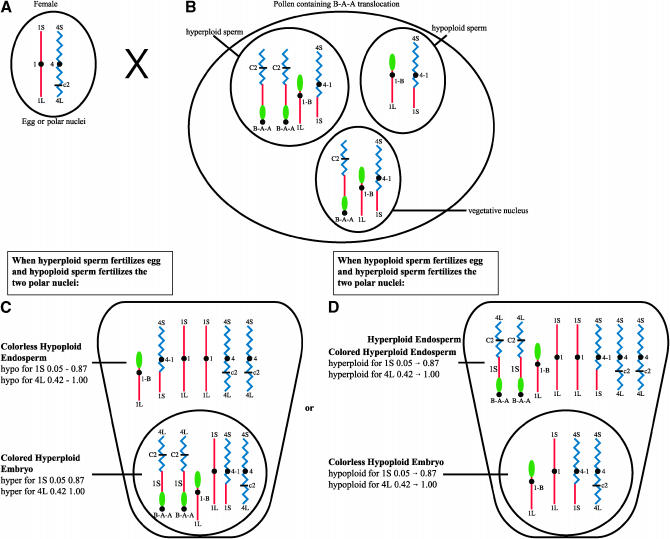
In the example described above, the “colorless” tester stock bears the recessive c2 allele located distal to the breakpoint at 0.42 on 4L while the 4L chromosome segment comprising the distal portion of the B-1-4 chromosome carries the dominant C2 allele, which is required for anthocyanin pigmentation of the endosperm and embryo. When this tester stock (Figure 4A) is crossed by pollen of the constitution described above (containing a hyperploid sperm and a hypoploid sperm) as shown in Figure 4B, then double fertilization in the embryo sacs of the tester results in two classes of kernels: (1) those with colorless hypoploid (c2 c2_) endosperm and colored hyperploid (C2 C2 c2) embryos (Figure 4C) and (2) those displaying a reciprocal coloration, with colored hyperploid (C2 C2 c2 c2) endosperm and colorless hypoploid (c2_) embryos (Figure 4D). (The kernel constitution resulting from double fertilization of an embryo sac of this same tester stock by sperm from pollen with a normal chromosome constitution is shown in supplemental Figure 1 at http://www.genetics.org/supplemental/.)
Figure 4.—
A tester plant homozygous for the recessive aleurone color factor c2 will produce embryo sacs containing egg cells and polar nuclei that each have a haploid normal chromosome set and carry the recessive c2 allele on chromosome arm 4L (A). When double fertilization of the egg and polar nuclei is accomplished by sperm of a pollen grain containing a B-A-A and the corresponding A-B (as shown in Figure 3A) and nondisjunction has occurred with the B-A-A chromosome (B), two kinds of nonconcordant kernels can result. Fertilization of the egg by the hyperploid sperm carrying the B-A-A (bearing the dominant C2 allele) results in a colored embryo, while fertilization of the two polar nuclei by the hypoploid sperm (lacking any c2 allele) results in a colorless endosperm (C). When the reciprocal event occurs and the hypoploid sperm fertilizes the egg and the hyperploid sperm fertilizes the two polar nuclei, a colorless embryo and a colored endosperm are the result (D).
Kernels with colorless endosperm and colored embryos contain in their embryos the B-A-A translocation, as well as the A-B chromosome and the A-A chromosome. These are the kernels of primary interest because they can be used to propagate the B-A-A translocation. This type of kernel is shown in Figure 4C. Confirmation that these kernels actually possess the desired B-A-A is obtained by repeating the testcross in the next generation. The recovery of numerous kernels with colorless endosperm and colored embryos confirms the identity and propagation of the new B-A-A translocation.
RESULTS
Frequency of identifying new B-A-A translocations:
The scope of the crossing efforts required to identify new B-A-A translocations is illustrated by the results of crossing pollen of 59 different B-A/A-A combinations onto appropriate tester stocks. A total of 230 ears were recovered from these crosses and a positive result (subsequently confirmed by genetic testing) was obtained with 15 ears representing nine different new B-A-A combinations. For five of these B-A-A translocations, a single ear bearing one to four rare recombinant kernels was obtained, while the other four combinations were recovered with two or three ears bearing one to five recombinant kernels. The total number of recombinant kernels among the 15 ears was 32, resulting in an average of ∼2.1 recombinant kernels borne on each of these ears. All nine of the positive events resulted from crosses onto aleurone color tester stocks.
New and previously created B-A-A chromosome translocation stocks:
Prior to the onset of this study, a total of 17 B-A-A translocation stocks were created. The first eight B-A-A translocations were created by Rakha and Robertson (1970) and addressed the need for B-A stocks that would uncover portions of the short arm and the long arm of chromosome two and the long arm of chromosome four. Subsequently, nine additional B-A-A stocks were created. These 17 stocks and their descriptions and citations of origins are listed in Table 3. Here we report on the characteristics of the 64 new B-A-A translocation stocks produced in this study. The 17 previously reported B-A-A stocks subdivided chromosome arms 1S, 1L, 2S, 2L, 3L, 4L, 5L, 7L, and 9S into two (7L) to eight regions (3L). The 64 additional B-A-A stocks described here further subdivide these arms and also subdivide the additional chromosome arms of 3S, 5S, 6L, 8L, 9L, and 10L. Together, these 81 B-A-A translocations subdivide 15 of the 20 chromosome arms (except 4S, 6S, 7S, 8S, and 10S); and the number of subdivided regions ranges as high as 22 regions on chromosome arm 3L.
TABLE 3.
New and previously created B-A-A translocation stocks
| No. in Figure 5 | Designation | A-A chromosome breakpoints | Uncovered chromosome regions | Tester for distal A region |
|---|---|---|---|---|
| 1 | TB-1Sb-4L064-20 | 1S.23, 4L.19 | 1S.05-.23; 4L.19-1.00 | c2 |
| 2 | TB-6Lc-1S055-10 | 1S.29, 6L.48 | 6L.11-.48; 1S.29-1.00 | 1S deks |
| 3 | TB-1Sb-9S7535 | 1S.33, 9S.27 | 1S.05-.33; 9S.27-1.00 | c1 |
| 4 | TB-6Lc-1S7352 | 1S.40, 6L.60 | 6L.11-.60; 1S.40-1.00 | 1S deks |
| 5 | TB-4Lf-1S8602 | 1S.41, 4L.81 | 4L.15-.81; 1S.41-1.00 | 1S deks |
| 6 | TB-6Lc-1S7097 | 1S.46, 6L.62 | 6L.11-.62; 1S.46-1.00 | 1S deks |
| 7 | TB-1Sb-3L8995 | 1S.49, 3L.06 | 1S.05-.49; 3L.06-1.00 | a1 |
| 8 | TB-1Sb-2L4464a | 1S.53, 2L.28 | 1S.05-.53; 2L.28-1.00 | w3 |
| 9 | TB-1Sb-2L4464-4Lfb | 1S.53, 2L.28, 4L.12 | 1S.05-53; 2L.28-75; 4L.12-1.00 | c2 |
| 10 | TB-4Lf-1S4308 | 1S.65, 4L.58 | 4L.15-.58; 1S.65-1.00 | 1S deks |
| 11 | TB-1Sb-2Lca | 1S.77, 2L.33 | 1S.05-.77; 2L.33-1.00 | w3 |
| 12 | TB-1Sb-3L5597 | 1S.77, 3L.48 | 1S.05-.77; 3L.48-1.00 | a1 |
| 13 | TB-1Sb-10Lg | 1S.80, 10L.21 | 1S.05-.80; 10L.21-1.00 | r1 |
| 14 | TB-1Sb-4L002-19 | 1S.87, 4L.42 | 1S.05-.87; 4L.42-1.00 | c2 |
| 15 | TB-1Sb-4Lh | 1S.94, 4L.52 | 1S.05-.94; 4L.52-1.00 | c2 |
| 16 | TB-7Lb-1L4891 | 1L.12, 7L.69 | 7L.30-.69; 1L.12-1.00 | bz2 |
| 17 | TB-10L19-1La | 1L.29, 10L.33 | 10L cent-.33; 1L.29-1.00 | bz2 |
| 18 | TB-9Sd-1L4997-6 | 1L.37, 9S.28 | 9S.08-.28; 1L.37-1.00 | bz2 |
| 19 | TB-1La-3L4759-3c | 1L.39, 3L.20 | 1L.2-.39; 3L.20-1.00 | a1 |
| 20 | TB-5Sc-1L0707-12 | 1L.39, 5S.71 | 5S.30-.71; 1L.39-1.00 | bz2 |
| 21 | TB-6Lc-1L070-1 | 1L.40, 6L.58 | 6L.11-.58; 1L.40-1.00 | bz2 |
| 22 | TB-10L19-1Lc | 1L.43, 10L.74 | 10L cent-.74; 1L.43-1.00 | bz2 |
| 23 | TB-1La-5S7212 | 1L.44, 5S.28 | 1L.20-.44; 5S.28-1.00 | a2 |
| 24 | TB-1La-4L4692a | 1L.46, 4L.15 | 1L.2-.46; 4l.15-1.00 | c2 |
| 25 | TB-10L19-1Ld | 1L.50, 10L.68 | 10L cent.-.68; 1L.50-1.00 | bz2 |
| 26 | TB-1La-3Lec | 1L.58, 3L.45 | 1L.2-.58; 3L.45-1.00 | a1 |
| 27 | TB-8Lc-1Lb | 1L.59, 8L.82 | 8L.24-.82; 1L.59-1.00 | bz2 |
| 28 | TB-1La-3L5267c | 1L.72, 3L.73 | 1L.2-.72; 3L.73-1.00 | a1 |
| 29 | TB-1La-5S8041e | 1L.80, 5S.10 | 1L.2-.80; 5S.10-1.00 | a2 |
| 30 | TB-1La-5S8041 | 1L.80, 5S.10 | 1L.2-.80; 5S.10-1.00 | a2 |
| 31 | TB-1La-10L001-3 | 1L.86, 10L.48 | 1L.2-.86; 10L.48-1.00 | r1 |
| 32 | TB-1La-3L5242c | 1L.90, 3L.65 | 1L.2-.90; 3L.65-1.00 | a1 |
| 33 | TB-3La-2S6270a | 2S.46, 3L.60 | 3L.10-.60; 2S.46-1.00 | al1 |
| 34 | TB-2Sa-3L5304 | 2S.62, 3L.29 | 2S.5-.62; 3L.29-1.00 | a1 |
| 35 | TB-2Sa-3Le | 2S.76, 3L.48 | 2S.5-.76; 3L.48-1.00 | a1 |
| 36 | TB-5Sc-2L015-3 | 2L.16, 5S.69 | 5S.3-.69; 2L.16-1.00 | w3 |
| 37 | TB-9Sd-2L062-11 | 2L.21, 9S.53 | 9S.08-.53; 2L.21-1.00 | w3 |
| 38 | TB-3La-2L7285a | 2L.26, 3L.39 | 3L.1-.39; 2L.26-1.00 | w3 |
| 39* | TB-1Sb-2L4464a | 2L.28, 1S.53 | 1S.05-.53; 2L.28-1.00 | w3 |
| 40* | TB-1Sb-2L4464-4Lfb | 2L.28, 1S.53 4L.12 | 1S.05-.53; 2L.28-.75; 4L.12-1.00 | c2 |
| 41* | TB-1Sb-2Lca | 2L.33, 1S.77 | 1S.05-.77; 2L.33-1.00 | w3 |
| 42 | TB-9Lc-3S5643 | 3S.55, 9L.64 | 9L.10-.64; 3S.55-1.00 | 3S deks |
| 43 | TB-5Sc-3Lg | 3L.01, 5S.73 | 5S.3-.73; 3L.01-1.00 | a1 |
| 44* | TB-1Sb-3L8995 | 3L.06, 1S.49 | 1S.05-.49; 3L.06-1.00 | a1 |
| 45 | TB-5La-3L5521d | 3L.17, 5L.48 | 5L.1-.48; 3L.17-1.00 | a1 |
| 46* | TB-1La-3L4759-3c | 3L.20, 1L.39 | 1L.2-.39; 3L.20-1.00 | a1 |
| 47 | TB-10L19-3Lc | 3L.22, 10L.30 | 10L cent-.30; 3L.22-1.00 | a1 |
| 48* | TB-2Sa-3L5304 | 3L.29, 2S.62 | 2S.5-.62; 3L.29-1.00 | a1 |
| 49* | TB-3La-2L7285a | 3L.39, 2L.26 | 3L.1-.39; 2L.26-1.00 | w3 |
| 50 | TB-3La-9Lg | 3L.40, 9L.14 | 3L.10-.40; 9L.14-1.00 | 9L deks |
| 51* | TB-1La-3Lec | 3L.45, 1L.58 | 1L.2-.58; 3L.45-1.00 | a1 |
| 52 | TB-6Lc-3L8672 | 3L.47, 6L.87 | 6L.11-.87; 3L.47-1.00 | a1 |
| 53* | TB-1Sb-3L5597 | 3L.48, 1S.77 | 1S.05-.77; 3L.48-1.00 | a1 |
| 54* | TB-2Sa-3Le | 3L.48, 2S.76 | 2S.5-.76; 3L.48-1.00 | a1 |
| 55 | TB-4Lf-3L6534 | 3L.48, 4L.89 | 4L.15-.89; 3L.48-1.00 | a1 |
| 56 | TB-3La-9Lb | 3L.48, 9L.53 | 3L.10-.48; 9L.53-1.00 | 9L deks |
| 57 | TB-9Lc-3Lb | 3L.48, 9L.53 | 9L.10-.53; 3L.48-1.00 | a1 |
| 58 | TB-3La-10L036-15 | 3L.48, 10L.64 | 3L.10-.48; 10L.64-1.00 | r1 |
| 59 | TB-10L19-3L036-15 | 3L.48, 10L.64 | 10L cent-.64; 3L.48-1.00 | a1 |
| 60 | TB-6Lc-3L7162 | 3L.52, 6L.53 | 6L.11-.53; 3L.52-1.00 | a1 |
| 61 | TB-3La-9L4727 | 3L.54, 9L.42 | 3L.10-.54; 9L.42-1.00 | 9L deks |
| 62* | TB-3La-2S6270a | 3L.60, 2S.46 | 3L.10-.60; 2S.46-1.00 | al1 |
| 63 | TB-5La-3Lbd | 3L.61, 5L.57 | 5L.1-.57; 3L.61-1.00 | a1 |
| 64 | TB-5La-3Lb | 3L.61, 5L.57 | 5L.1-.57; 3L.61-1.00 | a1 |
| 65 | TB-5La-3L7043d | 3L.63, 5L.61 | 5L.1-.61; 3L.63-1.00 | a1 |
| 66 | TB-5La-3L7043 | 3L.63, 5L.61 | 5L.1-.61; 3L.63-1.00 | a1 |
| 67 | TB-7Lb-3L5471 | 3L.64, 7L.58 | 7L.3-.58; 3L.64-1.00 | a1 |
| 68 | TB-7Lb-3Ld | 3L.64, 7L.81 | 7L.3-.81; 3L.64-1.00 | a1 |
| 69* | TB-1La-3L5242c | 3L.65, 1L.90 | 1L.2-.90; 3L.65-1.00 | a1 |
| 70 | TB-3La-6L054-12 | 3L.72, 6L.75 | 3L.1-.72; 6L.75-1.00 | 6L deks |
| 71* | TB-1La-3L5267c | 3L.73, 1L.72 | 1L.2-.72; 3L.73-1.00 | a1 |
| 72 | TB-9Sb-4L6222a | 4L.03, 9S.68 | 9S.4-.68; 4L.03-1.00 | c2 |
| 73 | TB-9Sd-4L6222 | 4L.03, 9S.68 | 9S.08-.68; 4L.03-1.00 | c2 |
| 74 | TB-7Lb-4L4698a | 4L.08, 7L.74 | 7L.3-.74; 4L.08-1.00 | c2 |
| 75 | TB-9Sb-4L6504a | 4L.09, 9S.83 | 9S.4-.83; 4L.09-1.00 | c2 |
| 76 | TB-9Sd-4L6504 | 4L.09, 9S.83 | 9S.08-.83; 4L.09-1.00 | c2 |
| 77* | TB-1Sb-2L4464-4Lfb | 4L.12, 1S.53, 2L.28 | 1S.05-.53; 2L.28-.75; 4L.12-1.00 | c2 |
| 78* | TB-1La-4L4692a | 4L.15, 1L.46 | 1L.2-.46; 4L.15-1.00 | c2 |
| 79* | TB-1Sb-4L064-20 | 4L.19, 1S.23 | 1S.05-.23; 4L.19-1.00 | c2 |
| 80 | TB-9Lc-4L4373 | 4L.29, 9L.39 | 9L.1-.39; 4L.29-1.00 | c2 |
| 81 | TB-6Lc-4L8764 | 4L.32, 6L.90 | 6L.11-.90; 4L.32-1.00 | c2 |
| 82 | TB-7Lb-4L4483 | 4L.39, 7L.61 | 7L.3-.61; 4L.39-1.00 | c2 |
| 83* | TB-1Sb-4L002-19 | 4L.42, 1S.87 | 1S.05-.87; 4L.42-1.00 | c2 |
| 84 | TB-4Lf-5S006-7 | 4L.43, 5S.25 | 4L.15-.43, 5S.25-1.00 | a2 |
| 85 | TB-5La-4Lf | 4L.50, 5L.80 | 5L.1-.80; 4L.50-1.00 | c2 |
| 86* | TB-1Sb-4Lh | 4L.52, 1S.94 | 1S.05-.94; 4L.52-1.00 | c2 |
| 87 | TB-4Lf-10L6587 | 4L.55, 10L.51 | 4L.15-.55; 10L.51-1.00 | r1 |
| 88* | TB-4Lf-1S4308 | 4L.58, 1S.65 | 4L.15-.58; 1S.65-1.00 | 1S deks |
| 89 | TB-4Lf-6L8927 | 4L.70, 6L.18 | 4L.15-.70; 6L.18-1.00 | 6L deks |
| 90* | TB-4Lf-1S8620 | 4L.81, 1S.41 | 4L.15-.81; 1S.41-1.00 | 1S deks |
| 91* | TB-4Lf-3L6534 | 4L.89, 3L.48 | 4L.15-.89; 3L.48-1.00 | a1 |
| 92* | TB-1La-5S8041e | 5S.10, 1L.80 | 1L.2-.80; 5S.10-1.00 | a2 |
| 93* | TB-1La-5S8041 | 5S.10, 1L.80 | 1L.2-.80; 5S.10-1.00 | a2 |
| 94* | TB-4Lf-5S006-7 | 5S.25, 4L.43 | 4L.15-.43; 5S.25-1.00 | a2 |
| 95* | TB-1La-5S7212 | 5S.28, 1L.44 | 1L.20-.44; 5S.28-1.00 | a2 |
| 96 | TB-6Lc-5S6671 | 5S.49, 6L.35 | 6L.11-.35; 5S.49-1.00 | a2 |
| 97* | TB-5Sc-2L015-3 | 5S.69, 2L.16 | 5S.3-.69; 2L.16-1.00 | w3 |
| 98* | TB-5Sc-1L0707-12 | 5S.71, 1L.39 | 5S.30-.71; 1L.39-1.00 | bz2 |
| 99* | TB-5Sc-3Lg | 5S.73, 3L.01 | 5S.3-.73; 3L.01-1.00 | a1 |
| 100 | TB-9Lc-5L4790 | 5L.34, 9L.45 | 9L.1-.45; 5L.34-1.00 | pr |
| 101 | TB-6Lc-5L4666 | 5L.35, 6L.86 | 6L.11-.86; 5L.35-1.00 | pr |
| 102* | TB-5La-3L5521d | 5L.48, 3L.17 | 5L.1-.48; 3L.17-1.00 | a1 |
| 103 | TB-5La-10L006-11 | 5L.49, 10L.52 | 5L.1-.49, 10L.52-1.00 | r1 |
| 104* | TB-5La-3Lbd | 5L.57, 3L.61 | 5L.1-.57, 3L.61-1.00 | a1 |
| 105* | TB-5La-3Lb | 5L.57, 3L.61 | 5L.1-.57, 3L.61-1.00 | a1 |
| 106* | TB-5La-3L7043d | 5L.61, 3L.63 | 5L.1-.61, 3L.63-1.00 | a1 |
| 107* | TB-5La-3L7043 | 5L.61, 3L.63 | 5L.1-.61, 3L.63-1.00 | a1 |
| 108 | TB-5La-10L7142 | 5L.73, 10L.17 | 5L.1-.73; 10L.17-1.00 | r1 |
| 109* | TB-5La-4Lf | 5L.80, 4L.50 | 5L.1-.80; 4L.50-1.00 | c2 |
| 110* | TB-4Lf-6L8927 | 6L.18, 4L.70 | 4L.15-.70; 6L.18-1.00 | 6L deks |
| 111* | TB-6Lc-5S6671 | 6L.35, 5S.49 | 6L.11-.35, 5S.49-1.00 | a2 |
| 112* | TB-6Lc-1S055-10 | 6L.48, 1S.29 | 6L.11-.48; 1S.29-1.00 | 1S deks |
| 113* | TB-6Lc-3L7162 | 6L.53, 3L.52 | 6L.11-.53, 3L.52-1.00 | a1 |
| 114* | TB-6Lc-1L070-1 | 6L.58, 1L.40 | 6L.11-.58; 1L.40-1.00 | bz2 |
| 115* | TB-6Lc-1S7352 | 6L.60, 1S.40 | 6L.11-.60; 1S.40-1.00 | 1S deks |
| 116* | TB-6Lc-1S7097 | 6L.62, 1S.46 | 6L.11-.62; 1S.46-1.00 | 1S deks |
| 117* | TB-3La-6L054-12 | 6L.75, 3L.72 | 3L.1-.72; 6L.75-1.00 | 6L deks |
| 118* | TB-6Lc-5L4666 | 6L.86, 5L.35 | 6L.11-.86; 5L.35-1.00 | pr |
| 119* | TB-6Lc-3L8672 | 6L.87, 3L.47 | 6L.11-.87; 3L.47-1.00 | a1 |
| 120* | TB-6Lc-4L8764 | 6L.90, 4L.32 | 6L.11-.90; 4L.32-1.00 | c2 |
| 121 | TB-9Sd-7L6482 | 7L.01, 9S.97 | 9S.08-.97; 7L.01-1.00 | 7L deks |
| 122 | TB-9Sd-7L7074 | 7L.03, 9S.80 | 9S.08-.80; 7L.03-1.00 | 7L deks |
| 123* | TB-7Lb-3L5471 | 7L.58, 3L.64 | 7L.3-.58; 3L.64-1.00 | a1 |
| 124* | TB-7Lb-4L4483 | 7L.61, 4L.39 | 7L.3-.61; 4L.39-1.00 | c2 |
| 125 | TB-7Lb-9Sa | 7L.63, 9S.07 | 7L.3-.63; 9S.07-1.00 | c1 |
| 126* | TB-7Lb-1L4891 | 7L.69, 1L.12 | 7L.30-.69; 1L.12-1.00 | bz2 |
| 127* | TB-7Lb-4L4698a | 7L.74, 4L.08 | 7L.3-.74; 4L.08-1.00 | c2 |
| 128* | TB-7Lb-3Ld | 7L.81, 3L.64 | 7L.3-.81; 3L.64-1.00 | a1 |
| 129* | TB-8Lc-1Lb | 8L.82, 1L.59 | 8L.24-.82; 1L.59-1.00 | bz2 |
| 130* | TB-7Lb-9Sa | 9S.07, 7L.63 | 7L.3-.63; 9S.07-1.00 | c1 |
| 131* | TB-1Sb-9S7535 | 9S.27, 1S.33 | 1S.05-.33; 9S.27-1.00 | c1 |
| 132* | TB-9Sd-1L4997-6 | 9S.28, 1L.37 | 9S.08-.28; 1L.37-1.00 | bz2 |
| 133 | TB-10L19-9S8630 | 9S.28, 10L.37 | 10L cent-.37; 9S.28-1.00 | c1 |
| 134 | TB-10L19-9S059-10 | 9S.31, 10L.53 | 10L cent.-.53; 9S.31-1.00 | c1 |
| 135 | TB-9Sd-10L3688 | 9S.49, 10L.02 | 9S.08-.49; 10L.02-1.00 | r1 |
| 136* | TB-9Sd-2L062-11 | 9S.53, 2L.21 | 9S.08-.53; 2L.21-1.00 | w3 |
| 137* | TB-9Sd-4L6222 | 9S.68, 4L.03 | 9S.08-.68; 4L.03-1.00 | c2 |
| 138* | TB-9Sb-4L6222a | 9S.68, 4L.03 | 9S.4-.68; 4L.03-1.00 | c2 |
| 139* | TB-9Sd-7L7074 | 9S.80, 7L.03 | 9S.08-.80; 7L.03-1.00 | 7L deks |
| 140* | TB-9Sb-4L6504a | 9S.83, 4L.09 | 9S.4-.83; 4L.09-1.00 | c2 |
| 141* | TB-9Sd-4L6504 | 9S.83, 4L.09 | 9S.08-.83; 4L.09-1.00 | c2 |
| 142* | TB-9Sd-7L6482 | 9S.97, 7L.01 | 9S.08-.97; 7L.01-1.00 | 7L deks |
| 143* | TB-3La-9Lg | 9L.14, 3L.40 | 3L.10-.40; 9L.14-1.00 | 9L deks |
| 144* | TB-9Lc-4L4373 | 9L.39, 4L.29 | 9L.1-.39; 4L.29-1.00 | c2 |
| 145* | TB-3La-9L4727 | 9L.42, 3L.54 | 3L.10-.54; 9L.42-1.00 | 9L deks |
| 146* | TB-9Lc-5L4790 | 9L.45, 5L.34 | 9L.1-.45; 5L.34-1.00 | pr |
| 147* | TB-3La-9Lb | 9L.53, 3L.48 | 3L.10-.48; 9L.53-1.00 | 9L deks |
| 148* | TB-9Lc-3Lb | 9L.53, 3L.48 | 9L.10-.53; 3L.48-1.00 | a1 |
| 149* | TB-9Lc-3S5643 | 9L.64, 3S.55 | 9L.10-.64; 3S.55-1.00 | 3S deks |
| 150* | TB-9Sd-10L3688 | 10L.02, 9S.49 | 9S.08-.49; 10L.02-1.00 | r1 |
| 151* | TB-5La-10L7142 | 10L.17, 5L.73 | 5L.1-73; 10L.17-1.00 | r1 |
| 152* | TB-1Sb-10Lg | 10L.21, 1S.80 | 1S.05-.80; 10L.21-1.00 | r1 |
| 153* | TB-10L19-3Lc | 10L.30, 3L.22 | 10L cent-.30; 3L.22-1.00 | a1 |
| 154* | TB-10L19-1La | 10L.33, 1L.29 | 10L cent-.33; 1L.29-1.00 | bz2 |
| 155* | TB-10L19-9S8630 | 10L.37, 9S.28 | 10L cent-.37; 9S.28-1.00 | c1 |
| 156* | TB-1La-10L001-3 | 10L.48, 1L.86 | 1L.2-.86; 10L.48-1.00 | r1 |
| 157* | TB-4Lf-10L6587 | 10L.51, 4L.55 | 4L.15-.55; 10L.51-1.00 | r1 |
| 158* | TB-5La-10L006-11 | 10L.52, 5L.49 | 5L.1-.49; 10L.52-1.00 | r1 |
| 159* | TB-10L19-9S059-10 | 10L.53, 9S.31 | 10L cent.-.53; 9S.31-1.00 | c1 |
| 160* | TB-3La-10L036-15 | 10L.64, 3L.48 | 3L.10-.48; 10L.64-1.00 | r1 |
| 161* | TB-10L19-3L036-15 | 10L.64, 3L.48 | 10L cent-.64; 3L.48-1.00 | a1 |
| 162* | TB-10L19-1Ld | 10L.68, 1L.50 | 10L cent.-.68; 1L.50-1.00 | bz2 |
| 163* | TB-10L19-1Lc | 10L.74, 1L.43 | 10L cent-.71; 1L.43-1.00 | bz2 |
See Figure 5 for the distribution of the A-A chromosome breakpoints on the chromosomes. The numbering of the B-A-A translocations in this table corresponds to the numbering of the chromosome arm breakpoints in Figure 5. For each B-A-A translocation stock, there are two breakpoints in Figure 5 that correspond to the two numbered entries in this table for that B-A-A translocation. The second of the two entries is marked by an asterisk (*) following its number. New B-A-A translocations are designated with roman letters and previously created B-A-A translocations are designated with italics.
Shadley and Weber (1984).
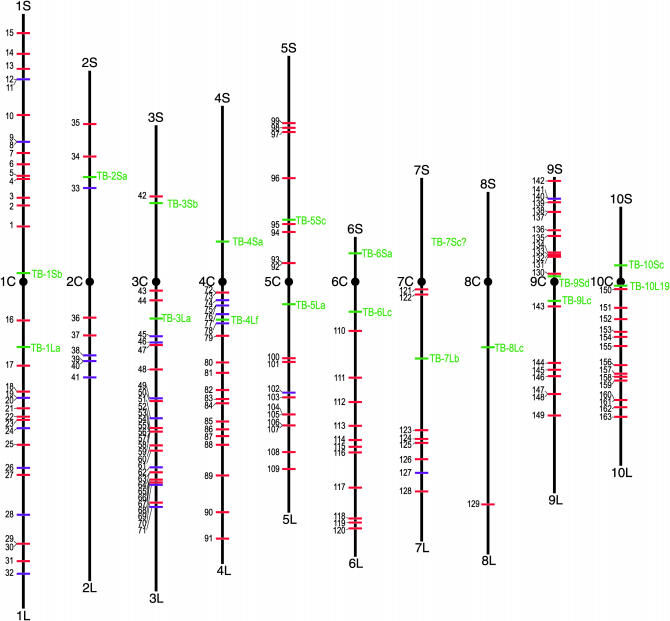
The distribution of the breakpoints of the B-A-A translocations among the chromosome arms is presented in Figure 5. The numbering of the chromosome arm breakpoints in Figure 5 corresponds to the numbering of the B-A-A translocations listed in Table 3. For each B-A-A translocation stock, there are two breakpoints in Figure 5 and these correspond to the two entries in Table 3 for that B-A-A translocation. For example, entry number 14 in Table 3 describes TB-1Sb-4L002-19, and this translocation appears again in Table 3 as number 83*, with the asterisk indicating that this is the second entry for this B-A-A stock. This B-A-A stock is entered into Table 3 in two locations to facilitate searching for B-A-A stocks that subdivide either chromosome arm 1S or 4L. The B-A-A stock can be used to produce hyperploid and hypoploid dosages for the chromosome arm regions of 1S.05-.87 and 4L.42-1.00. The B-A-A stocks are listed sequentially in Table 3 with breakpoints proceeding distally from those closest to the centromere outward toward the distal end of the chromosome arm. In Figure 5, the chromosome arm breakpoints are shown in a parallel fashion. For example, in the case of the new B-A-A TB-1Sb-4L002-19, a breakpoint is shown in the distal region of chromosome arm 1S corresponding to a site at 87% of the cytological distance outward from the centromere on 1S. This breakpoint is number 14 in Figure 5, which corresponds to entry number 14 in Table 3. In addition, for TB-1Sb-4L002-19, a breakpoint is shown near the midregion of chromosome arm 4L, corresponding to a site at 42% of the cytological distance outward from the centromere of 4L. This breakpoint is number 83* in Figure 5, which corresponds to entry number 83* in Table 3.
Figure 5.—
Cytological map of maize showing the chromosome arm breakpoints of the A-A translocations used to create the B-A-A translocations. The chromosomes are shown as vertical lines in proportion to each other and the arms are numbered at their ends. Centromeres are shown as solid circles at their approximate positions. Short horizontal lines indicate the cytological position of breakpoints described for each of the translocations used in this project. The numbers shown to the left of the horizontal lines marking the breakpoints on the chromosome arms correspond to the entries for each of the B-A-A translocations listed and described in Table 3. Also shown are the breakpoint sites for the A chromosome arm of the simple B-A translocations, which are marked by green lines and labeled TB-1Sb, etc. The breakpoints of the old B-A-A translocations are marked by purple lines and the breakpoints of the new B-A-A translocations are marked by red lines. This cytological map is based on that of Neuffer et al. (1997).
Extending the range of chromosome arms uncovered:
An examination of Figure 5 and Table 3 reveals that, prior to this study, the simple B-A translocations indicated in Figure 5 provided the greatest extent of the uncovering of gene loci for most of the chromosome arms. The three exceptions to this pattern are found on chromosome arms 2S, 2L, and 4L. Whereas TB-2Sa uncovers 2S from 0.50 to the distal end of the arm, the B-A-A translocation TB-3La-2S6270 has been available and uncovers 2S from 0.46 distally. For chromosome arm 2L, there is no simple B-A available but the B-A-A translocation TB-3La-2L7285 that uncovers 2L from 0.26 distally has been available. Whereas TB-4Lf uncovers 4L from 0.15 to 0.20 to the distal end of the arm, the B-A-A translocation TB-9Sb-4L6222 that uncovers 4L from 0.03 distally has been available.
The new B-A-A translocations described here extend the amount of chromosome arm that can be uncovered for chromosome arms 1L, 2L, 3L, 5S, and 7L. The new B-A-A translocation TB-7Lb-1L4891 uncovers 1L from 0.12 distally; the new B-A-A translocation TB-5Sc-2L015-3 uncovers 2L from 0.16 distally; the new B-A-A translocation TB-5Sc-3Lg uncovers 3L from 0.01 distally; the new B-A-A translocation TB-1La-5S8041 uncovers 5S from 0.10 distally (this is a recreation of this B-A-A translocation; see entries 29 and 30 in Table 3); and the new B-A-A translocation TB-9Sd-7L6482 uncovers 7L from 0.01 distally.
Additionally, the new B-A-A translocations include TB-9Sd-4L6222. This B-A-A translocation differs from TB-9Sb-4L6222 inasmuch as the simple B-A used to create this new stock was TB-9Sd, which uncovers 9S from 0.08 distally. Consequently, whereas both TB-9Sb-4L6222 and TB-9Sd-4L6222 uncover chromosome arm 4L from 0.03 distally, the former B-A-A uncovers 9S from 0.4 distally to 0.68 while the latter B-A-A uncovers 9S from 0.08 distally to 0.68. These two B-A-A translocations can be readily distinguished by crossing onto a wx1 tester because the waxy1 locus is distal to 0.08 but proximal to 0.4 on 9S.
Three previously created B-A-A translocations, all involving TB-5La and chromosome arm 3L, merit consideration. These three B-A-A stocks (TB-5La-3L5521 5L.48 3L.17, TB-5La-3Lb 5L.57 3L.61, and TB-5La-3L7043 5L.61 3L.63) were created by Shadley and Weber (1984) and are listed as entries 102*, 104*, and 106* in Table 3. These three B-A-A stocks may be available from the Maize Genetics Cooperation Stock Center but their pedigree identity is uncertain inasmuch as the listing for each of these stocks states that the exact identity of the stock is unknown and that it could be any one of these three B-A-A stocks. We have created new versions of two of these stocks, TB-5La-3Lb and TB-5L-3L 7043, and we plan to deposit these along with all of the other new B-A-A translocation stocks with the stock center.
DISCUSSION
When used in combinations, B-A-A translocations can localize a gene locus or DNA sequence to a specific cytologically defined chromosome segment (Beckett 1973). An example of this is the cytogenetic localization of the alcohol dehydrogenase 1 (Adh1) locus in maize to a segment of chromosome arm 1L (Birchler 1980). Four B-A-A translocations were synthesized and all involved TB-lLa and four different 1L-3L A-A translocations. A fifth B-A-A translocation (TB-lLa-5S8041) was obtained from Donald Robertson. These five B-A-A translocations subdivided chromosome arm lL into five regions (0.20–0.39, 0.20–0.58, 0.20–0.72, 0.20–0.80, and 0.20–0.90). When these B-A-A stocks were crossed onto the Adh1 tester stock, only the B-A-A uncovering lL from 0.20 distally to 0.90 uncovered the Adh1 locus. These results led to the conclusion that the Adh1 locus lies within the cytological segment between 0.80 and 0.90 on chromosome arm 1L.
This approach to gene localization could be applied to locating individual recessive mutations that are members of a series of mutations with similar phenotypes and are located on the same chromosome arm. For example, an analysis of 51 embryo-specific (emb) maize mutants using the basic set of simple B-A translocations identified the putative chromosome arm location of 25 of these mutations (Clark and Sheridan 1991). Among these 25, 8 mutations were uncovered by crosses using TB-1Sb as the pollen parent. A total of 11 B-A-A translocations are now available for segmental analysis of chromosome arm 1S and we are utilizing them to determine the cytologically defined arm segments containing the emb mutant loci.
Maize endosperm development is strongly influenced by the dosage of certain of the chromosome arms inherited through the pollen. When a chromosome region that contributes to endosperm formation is absent from the sperm, then a reduced kernel size, “the small kernel effect,” results. The chromosome arms that most strongly exhibit this effect are 1S, 1L, 4S, 5S, 6L, 7S, 7L, and 10L (Birchler and Hart 1987; Birchler 1993c). Simple B-A translocations have been used to examine the dosage effect of individual chromosome arms and Birchler and Hart (1987) observed that the small kernel effect of chromosome arm 10L was enhanced by the presence of an extra copy of chromosome arm 4S. The B-A-A translocations described in this article provide for an expanded range of investigations of the dosage effect of chromosome segments not only on endosperm development, but also on plant height (Beckett 1991) in maize aneuploids (Lee et al. 1996a,b) and nuclear genomes (Guo and Birchler 1994; Auger et al. 2001; Birchler and Auger 2004).
Analysis of maize gametophyte mutants can readily identify gene loci that are essential for either male or female gametophyte development (Vollbrecht and Hake 1995). But the identification of mutations affecting both male and female gametophyte development poses the challenge of how to obtain transmission of the mutant gametophyte lethal allele through either the pollen or the embryo sac to propagate the mutation. A promising approach is to use tertiary trisomic stocks to provide the normal allele to cover the mutant allele during gametophyte development. Tertiary trisomic stocks can be obtained from B-A translocation stocks (Auger and Birchler 2002) for many of the maize chromosome arms. The numerous B-A-A translocations described here can be used to generate tertiary trisomic stocks that will increase the extent of coverage of a gametophyte lethal allele for chromosome arms 3L, 4L, and 5S and provide the opportunity to delimit the location of the lethal mutation to a cytologically defined segment for many of the chromosome arms.
Acknowledgments
We thank The Maize Genetics Cooperation Stock Center at the University of Illinois, Urbana, Illinois, for providing translocation stocks and other stocks and for agreeing to maintain the new B-A-A translocations stocks and to make them available to the scientific community. We thank Jack Beckett for his assistance and advice during the early stages of this study, Don Robertson and David Weber for permission to cite their Maize News Letter articles, and Holly LaFerriere, Elizabeth Gruman, Joel Schwartz-Moretti, and Jonathan Labonte for their technical assistance. This research was supported by a North Dakota Experimental Program to Stimulate Competitive Research seed money award and by a National Science Foundation Plant Genome Research Program grant 0321565 (to W.F.S.).
References
- Auger, D. L., and J. A. Birchler, 2002. Maize tertiary trisomic stocks derived from B-A translocations. J. Hered. 93: 42–47. [DOI] [PubMed] [Google Scholar]
- Auger, D. L., K. J. Newton and J. A. Birchler, 2001. Nuclear gene dosage effects upon the expression of maize mitochondrial genes. Genetics 157: 1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, J. B., 1973. Segmental analysis of maize chromosomes by means of compound derivatives of B-A translocations. Genetics 74: s18–s19. [Google Scholar]
- Beckett, J. B., 1991. Cytogenetic, genetic and breeding applications of B-A translocations in maize, pp. 491–527 in Chromosome Engineering in Plants, edited by T. Tsuchia and P. K. Gupta. Elsevier, Amsterdam.
- Beckett, J. B., 1993. Locating recessive genes to chromosome arm with B-A translocations, pp. 315–327 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag. New York.
- Birchler, J. A., 1980. The cytogenetic localization of the alcohol dehydrogenase-1 locus in maize. Genetics 94: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., 1981. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics 97: 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., 1993. a Dosage analysis using B-A translocations, pp. 328–329 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Birchler, J. A., 1993. b Construction of compound B-A translocations, pp. 332–334 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Birchler, J. A., 1993. c Dosage analysis of maize endosperm development. Annu. Rev. Genet. 27: 181–204. [DOI] [PubMed] [Google Scholar]
- Birchler, J. A., and D. L. Auger, 2004. Biological consequences of dosage dependent gene regulation in multicellular eukaryotes, pp. 89–105 in The Biology of Genetic Dominance, edited by R. A. Veitia. Landes Bioscience, Georgetown, TX.
- Birchler, J. A., and J. A. Hart, 1987. Interaction of endosperm size factors in maize. Genetics 117: 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler, J. A., U. Bhadra, M. Pal Bhadra and D. L. Auger, 2001. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes and quantitative traits. Dev. Biol. 234: 275–288. [DOI] [PubMed] [Google Scholar]
- Clark, J. K., and W. F. Sheridan, 1991. Isolation and characterization of 51 embryo-specific mutations of maize. Plant Cell 3: 935–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, M., and J. S. Birchler, 1994. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science 262: 1999–2002. [DOI] [PubMed] [Google Scholar]
- Lee, E. A., E. Coe and L. Darrah, 1996. a Genetic variation in dosage effects in maize aneuploids. Genome 39: 711–721. [DOI] [PubMed] [Google Scholar]
- Lee, E. A., L. Darrah and E. Coe, 1996. b Dosage effects on morphological and quantitative traits in maize aneuploids. Genome 39: 898–908. [DOI] [PubMed] [Google Scholar]
- Longley, A. E., 1961. Breakage points for four corn translocation series and other corn chromosome aberrations. USDA-ARS Crop Research Bulletin no. 34-16.
- Neuffer, M. G., E. H. Coe and S. R. Wessler, 1997. Mutants of Maize. Cold Spring Harbor Laboratory Press, Plainview, NY.
- Rakha, F. A., and D. S. Robertson, 1970. A new technique for the production of A-B translocations and their use in genetic analysis. Genetics 65: 223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, D. S., 1975. a A new compound A-B translocation TB-5S, 1L (8041). Maize Genet. Coop. Newsl. 49: 79–81. [Google Scholar]
- Robertson, D. S., 1975. b An A-B translocation with segments from three different A chromosomes. Maize Genet. Coop. Newsl. 49: 81–82. [Google Scholar]
- Roman, H., 1947. Mitotic nondisjunction in the case of interchanges involving the B-type chromosome in maize. Genetics 32: 391–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadley, J. D., and D. F. Weber, 1984. Placement of sporophyte mutants within the long arm of chromosome 5. Maize Genet. Coop. Newsl. 58: 160–161. [Google Scholar]
- Vollbrecht, E., and S. Hake, 1995. Deficiency analysis of female gametogenesis in maize. Dev. Genet. 16: 44–63. [Google Scholar]