Abstract
Rac GTPases are key regulators of cell shape and cytoskeletal organization. While some regulators of Rac activity are known, such as GTPase-activating proteins (GAPs) that repress Rac activity, other Rac regulators remain to be identified. The novel Caenorhabditis elegans WD-repeat protein SWAN-1 was identified in a yeast two-hybrid screen with the LIM domains of the Rac effector UNC-115/abLIM. SWAN-1 was found to also associate physically with Rac GTPases. The swan-1(ok267) loss-of-function mutation suppressed defects caused by the hypomorphic ced-10(n1993) allele and enhanced ectopic lamellipodia and filopodia formation induced by constitutively active Rac in C. elegans neurons. Furthermore, SWAN-1(+) transgenic expression suppressed the effects of overactive Rac, including ectopic lamellipodia and filopodia formation in C. elegans neurons, ectopic lamellipodia formation in cultured mammalian fibroblasts, and cell polarity and actin cytoskeleton defects in yeast. These studies indicate that SWAN-1 is an inhibitor of Rac GTPase function in cellular morphogenesis and cytoskeletal organization. While broadly conserved across species, SWAN-1 family members show no sequence similarity to previously known Rac inhibitors.
CELL and growth cone migration and the establishment and maintenance of cell shape are tightly regulated processes controlled by extracellular cues and intracellular physiology (Tessier-Lavigne and Goodman 1996; Yu and Bargmann 2001; Dickson 2002; Raftopoulou and Hall 2004; Vicente-Manzanares et al. 2005). Rac GTPases of the Rho subfamily are key regulators of cell shape and act in part by controlling the structure and dynamics of the actin cytoskeleton (Hall 1998). Classically, Rac GTPases were identified by their similarity to Rho and were found to induce the formation of veil-like lamellipodial plasma membrane extensions in serum-starved fibroblasts (Hall 1998; Otieno et al. 2005). In Caenorhabditis elegans neurons, Rac activity promotes the formation of both lamellipodia and filopodia, thin finger-like plasma membrane extensions (Struckhoff and Lundquist 2003). Loss-of-function studies in Drosophila and C. elegans demonstrate that Rac GTPases are required for a wide variety of morphogenetic events, including axon pathfinding and cell migration (Dickson 2001; Lundquist 2003). The GTPase regulatory cycle involves active, GTP-bound Racs that hydrolyze GTP to GDP and thus self-inactivate, as the GDP-bound form is inactive. Guanine nucleotide exchange factors (GEFs) mediate the exchange of GDP for GTP and thus favor the GTP-bound active state of Racs. GTPase-activating proteins (GAPs) stimulate the GTPase activity of Racs and thus favor the inactive state (Van Aelst and D'Souza-Schorey 1997). Another class of Rho GTPase negative regulators are the guanine nucleotide dissociation inhibitors (GDIs), which inhibit Rho GTPase association with the plasma membrane where they are active (Van Aelst and D'Souza-Schorey 1997; Michaelson et al. 2001).
In C. elegans, three Rac GTPases (CED-10, MIG-2, and RAC-2) have compensatory, redundant roles in axon pathfinding and in the migration of neurons, embryonic cells during gastrulation, and vulval cells (Lundquist et al. 2001; Kishore and Sundaram 2002; Soto et al. 2002). The actin-binding protein UNC-115/abLIM, which acts with Racs in axon pathfinding, is required for Rac-induced lamellipodia and filopodia in neurons and UNC-115 itself induces the formation of lamellipodia and filopodia in C. elegans neurons and in serum-starved mammalian fibroblasts (Lundquist et al. 1998; Struckhoff and Lundquist 2003; Yang and Lundquist 2005). UNC-115 might be a downstream cytoskeletal effector of Rac GTPases and might control growth cone lamellipodia and filopodia formation in response to Rac signaling during axon pathfinding. The molecular linkage between Racs and UNC-115 in these events is unclear, and molecules that interact physically with UNC-115 remain to be identified. Furthermore, while some Rac inhibitors are known, it will be important to identify all of the molecules that regulate Rac activity to precisely control distinct morphogenetic events.
To identify molecules that act with Rac and UNC-115 in axon pathfinding and cell migration, the LIM domains of UNC-115 were used as bait in a yeast two-hybrid screen of a C. elegans cDNA library. This screen identified a novel, conserved molecule called SWAN-1 (seven-WD-repeat protein of the AN11 family-1), which consists of at least seven WD repeats and a conserved C-terminal tail. In two-hybrid and pull-down assays, SWAN-1 was found to interact both with the UNC-115 LIM domains and with the Racs, suggesting that SWAN-1 is a molecular linker between UNC-115 and the Racs. An array of functional tests in C. elegans, mammalian fibroblasts, and yeast indicate that SWAN-1 represses Rac activity. SWAN-1 displays no similarity to other known Rac negative regulators (e.g., GAPs), indicating that SWAN-1 represents a previously unidentified class of negative regulators of Rac GTPases.
MATERIALS AND METHODS
C. elegans culture and genetics:
C. elegans were cultured using standard techniques (Epstein and Shakes 1995). All experiments were performed at 20°. The swan-1(ok267) mutation was isolated by the C. elegans Gene Knockout Consortium (kindly provided by G. Molder and B. Barstead) and was outcrossed to wild type three times before phenotypic analysis. The homozygosity of swan-1(ok267) in all outcrosses and single and double mutants was confirmed by single-animal polymerase chain reaction (PCR) using primers that amplify a band specific to the ok267 deletion. Germline transformation of C. elegans was performed by standard techniques involving injection of a DNA mixture into the syncytial germline of C. elegans hermaphrodites (Epstein and Shakes 1995). RNA-mediated gene knockdown (RNAi) was performed by feeding nematode strains Escherichia coli expressing double-stranded RNA complementary to either swan-1 or swan-2 (Timmons et al. 2001; Kamath and Ahringer 2003).
Numbers of cell corpses in the region of the pharynx at the mid-threefold stage of embryogenesis (∼750 min postfertilization) were counted using differential interference contrast microscopy. Gonadal distal tip cell migration defects were scored as the percentage of gonad arms that showed defective morphology as previously described (Lundquist et al. 2001). In Table 1, strains harboring a full-length swan-1∷gfp transgene were scored. As a control, nontransgene-bearing siblings from the same brood were also scored and presented in Table 1.
TABLE 1.
swan-1 loss of function suppresses ced-10(n1993) but not mig-2(mu28)
| Genotype | No. of persistent cell corpses/embryoa | % distal tip cell migration defectsb |
|---|---|---|
| + | 0.05 ± 0.3 | 0 |
| n = 50 | n > 200 | |
| swan-1(ok267) | 0.1 ± 0.3 | 3 ± 1 |
| n = 56 | n = 132 | |
| ced-10(n1993) | 14.9 ± 3.8c | 24 ± 4d |
| n = 97 | n = 120 | |
| ced-10(n1993); swan-1(ok267) | 9.6 ± 4.2c | 7 ± 2d |
| n = 89 | n = 113 | |
| ced-10(n1993); swan-1(ok267); Ex[swan-1(+)]e | 15.6 ± 3.4f | 29 ± 3g |
| n = 119 | n = 191 | |
| ced-10(n1993); swan-1(ok267) (non-transgene-bearing sibs)e | 11.7 ± 3.6f | 12 ± 2g |
| n = 115 | n = 174 | |
| mig-2(mu28) | NA | 16 ± 3 |
| n = 145 | ||
| mig-2(mu28); swan-1(ok267) | NA | 17 ± 4 |
| n = 101 |
Persistent corpses in the region of the pharynx were scored in threefold larvae prior to hatching (∼750 min postfertilization) (± standard deviation).
In young adult hermaphrodites, distal tip cell migration was scored as defective if the gonad arm failed to execute its complete migration or if the gonad arm was misguided (± standard error of the proportion).
Distributions are significantly different (t-test, P < 0.001).
Proportions are significantly different (t-test and Fisher's exact analysis, P < 0.001).
Ex[swan-1(+)] is a transgenic array harboring wild-type swan-1. Non-transgene-bearing sibs are the siblings of transgenic animals from the same brood that did not inherit the transgene.
Distributions are significantly different (t-test, P < 0.001).
Proportions are significantly different (t-test and Fisher's exact analysis, P < 0.001).
Significance of differences of means in Table 1 were calculated using the t-test, and significance of differences of proportions in Table 1 and in Figures 4, 5, 7, and 8 were calculated using both the t-test and Fisher's exact analysis.
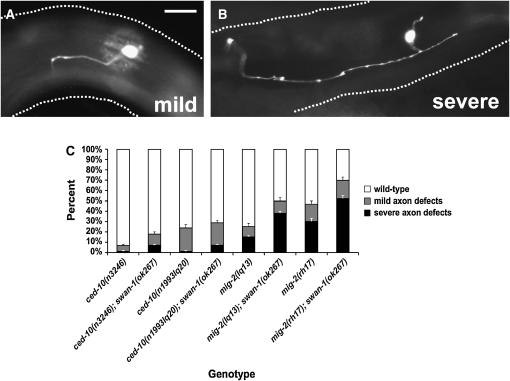
Figure 4.—
SWAN-1 negatively modulates activated Rac. (A and B) Fluorescence micrographs of the PDE neurons of young adult animals with gfp expression driven by the osm-6 promoter in the PDE neuron. Anterior is left and dorsal is up; bar in A, 5 μm. (A) Wild-type PDE morphology. A ciliated dendrite extends dorsally, and a single unbranched axon extends ventrally to the ventral nerve cord (out of focus; dashed line). (B) A PDE neuron with CED-10(G12V) expression. The neuron displayed ectopic protrusive structures that resemble lamellipodia and filopodia. Ectopic axons and axon branches were also observed (data not shown). (C) Loss-of-function swan-1(ok267) enhances the effects of activated CED-10(G12V) and RAC-2(G12V) transgenes. Genotypes analyzed define the x-axis, and the percentages of PDE neurons displaying ectopic lamellipodia/filopodia and ectopic neurites define the y-axis. Error bars represent the standard error of the proportion. (D) Transgenic expression of wild-type swan-1 suppresses transgenic CED-10(G12V) and RAC-2(G12V). The x-axis is compositions of transgenes analyzed. Transgenes included activated rac(G12V) constructs with and without wild-type swan-1 DNA (see materials and methods). Multiple transgenic lines (n) were analyzed for each combination. The y-axis and error bars are as described in C. As determined by a t-test and by Fisher's exact analysis, the effects of swan-1(ok267), swan-1(RNAi), and swan-1(+) expression on CED-10(G12V) and RAC-2(G12V) were significant (P < 0.001).
Figure 5.—
swan-1(ok267) enhances dominant ced-10 and mig-2 alleles. (A and B) Fluorescence micrographs of osm-6∷gfp expression in PDE neurons from ced-10(n3246); swan-1(ok267) animals. Anterior is left and dorsal is up; bar in A, 5 μm. Dashed lines indicate the dorsal and ventral limits of the body of the animal. (A) A mild PDE axon pathfinding defect, where the axon is misguided but reaches the ventral nerve cord (out of focus; dashed line) in the vicinity of the cell body. (B) A severe PDE axon pathfinding defect, where the axon fails to reach the ventral nerve cord. (C) Genotypes analyzed define the x-axis, and the y-axis represents the percentage of PDE neurons in each genotype displaying wild-type morphology (open bars), mild axon pathfinding defects (shaded bars), and severe axon pathfinding defects (solid bars). Open error bars represent the standard error of the proportion of severe axon defects, and solid error bars represent standard error of the proportion of combined mild and severe defects. Differences in the severe category between the activated rac mutation alone and the double with swan-1(ok267) were significant (P < 0.001; t-test and Fisher's exact analysis).
Figure 7.—
SWAN-1 inhibits human Rac1 activity in cultured cells. (A) A serum-starved NIH 3T3 fibroblast transfected with activated human Rac1(Q61L), fixed and stained with rhodamine–phalloidin to visualize the actin cytoskeleton. The arrow indicates a lamellipodial ruffle surrounding the cell. (B) The combinations of transgenes used to transfect serum-starved fibroblasts define the x-axis. EGFP was included in each mix. Rac1 harbored the Q61L mutation, and SWAN-1 was tagged with the Myc epitope. The percentage of transfected cells (as judged by the EGFP expression) displaying lamellipodia define the y-axis. Error bars represent standard error of the proportion. The effects of SWAN-1 expression on Rac1(G12V) activity were significant (P < 0.01) as judged by a t-test and Fisher's exact analysis. (C) A fluorescence micrograph of a serum-starved fibroblast harboring a MYR∷UNC-115∷EGFP transgene. Fluorescence is from MYR∷UNC-115∷EGFP. The arrow indicates a large lamellipodial protrusion with multiple filopodia. D is as described in B. The effects of SWAN-1 expression on MYR∷UNC-115 activity were not significant as judged by a t-test (P = 0.1076) and Fisher's exact analysis. Bar in A, 5 μm.
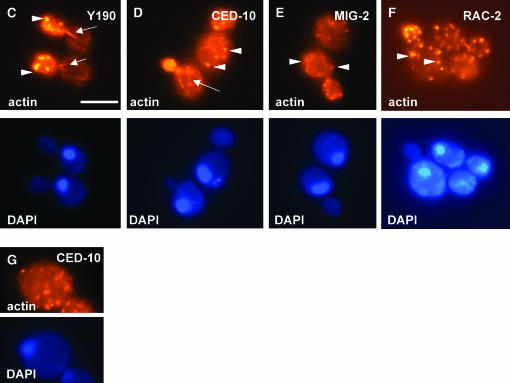
Figure 8.—
SWAN-1 inhibits C. elegans Rac activity in yeast. (A) A growth curve for yeast harboring different combinations of transgenes (x-axis is time point; y-axis is OD600) (see materials and methods). Y190 is the background strain, CED-10 and MIG-2 are fusions of the molecules with the DNA-binding domain of GAL4 in the pAS1CYH two-hybrid vector, and SWAN-1 is a fusion of the molecule to the activation domain of GAL4 in the pACT two-hybrid vector. Error bars represent the standard deviation of two experiments. (B) Rac expression in yeast perturbs polarized cell division. Six hours after dilution to 0.1 OD600, yeast cells harboring different transgenes (x-axis) were fixed and stained with rhodamine–phalloidin and DAPI. On the y-axis is the percentage of cells from each strain in the process of polarized cell budding. Error bars represent standard error of the proportion. (C–G) Fluorescence micrographs of yeast cells from different strains stained with rhodamine–phalloidin to visualize actin (actin) and DAPI to visualize DNA in the nucleus (DAPI). (C) Arrows point to polarized actin cables in the mother cell and arrowheads indicate the actin patches in the daughter cell. (D) The arrow points to a disorganized actin cable in a mother cell. (D–F) Arrowheads indicate actin patches ectopically present in mother cells. (G) A large, unpolarized cell undergoing nonpolarized growth without cell division. Bar in A, 10 μm.
Molecular biology:
All fragments containing coding region that were generated by PCR were sequenced to ensure that no mutations were introduced during the procedure. The sequences of all PCR primers used in this work are available upon request.
Microscopy:
See the figure legends for details about each micrograph. Unless otherwise noted, all images were captured on a Leica DMRE microscope with a 40× Planapo objective and 10× magnifier using a Hamamatsu Orca C4742-94 camera and Openlab software. Images were processed in Photoshop.
UNC-115 LIM domain two-hybrid screen:
Vectors for the two-hybrid system were kindly provided by S. Elledge. The coding region for the UNC-115 LIM domains was amplified by PCR and cloned in frame to the GAL4 DNA-binding domain in the vector pAS1-CYH (the “bait” construct). The yeast strain Y190 was transformed with the bait construct by standard techniques, and autoactivation of the bait construct was tested by growth on 25 mm 3-aminotriazole (3-AT) and X-Gal, which assess expression of HIS5 and LacZ reporter gene expression in Y190. The bait construct showed no autoactivation: it did not allow Y190 yeast to grow on 3-AT (no HIS5 expression), nor did blue colonies result when transgenic yeast were grown on X-Gal (no LacZ expression).
Y190-bait transgenic yeast were transformed with a C. elegans random-primed cDNA library cloned into the pACT vector that harbors the GAL4 activation domain (the library was kindly provided by R. Barstead). Yeast were plated at high density on medium containing 25 mm 3-AT and X-Gal, and colonies that were blue (indicating HIS5 expression and LacZ expression) were selected for further analysis. From an estimated 2 million cDNAs screened, 2 cDNAs representing the swan-1 locus were recovered. These 2 cDNAs did not result in autoactivation when present in Y190 without the bait plasmid, indicating that both plasmids are needed for activation.
The coding regions of the three C. elegans rac genes ced-10, mig-2, and rac-2 and the non-LIM-domain-containing portion of unc-115 were generated by PCR and fused in frame to the GAL4 DNA-binding domain in the pAS1-CYH plasmid. None of these constructs resulted in autoactivation. These constructs were then used in direct two-hybrid tests with the pACT∷SWAN-1 plasmid.
Co-immunoprecipitation experiments:
HeLa cells and HEK293 cells were grown to ∼80% confluency and were transfected using Fugene6 (Roche, Indianapolis). The full-length unc-115 coding region was generated by PCR and cloned in frame to enhanced green fluorescent protein (EGFP) in the pEGFP1 vector (Clontech, Mountain View, CA). The swan-1 coding region was cloned in frame to both the Myc epitope (MYC∷SWAN-1) and the 3xFLAG epitope (FLAG∷SWAN-1) in the pCMV-Myc and pCMV-3xFLAG vectors, respectively (Clontech). MYC∷RAC-2 and FLAG∷RAC-2 plasmids were similarly constructed. In most cases, 16 μg of plasmid were used in single transfections and 8 μg of each plasmid were used in double transfections in 75-cm2 flasks. For RAC-2 plasmids, 8 μg were used because higher concentrations led to cell inviability. After 48 hr of growth after transfection, cells were washed with ice-cold PBS and 1 ml of ice-cold lysis buffer [150 mm NaCl/10 mm Tris (pH 7.4)/1 mm EDTA/1 mm EGTA/0.5% NP-40/0.2 mm PMSF/1% protease inhibitor cocktail; Sigma, St. Louis] was added. Cells were scraped from the bottom of the flasks, incubated for 1 hr at 4° with shaking, and centrifuged for 15 min at 16,000× g in a microcentrifuge at 4°. Supernatants were precleared by incubating with 80 μl of pre-equilibrated protein G Sepharose bead slurry (Amersham Pharmacia Biotech, Piscataway, NJ) for 2 hr at 4°.
An 80-μl bead slurry in 1 ml ice-cold lysis buffer was preloaded with anti-Myc monoclonal antibody (BD Biosciences, Palo Alto, CA) or anti-FLAG M2 monoclonal antibody (Sigma). After 4 hr, the beads were washed four times with lysis buffer and incubated with the precleaned lysates at 4° for 12 hr. Beads were washed in lysis buffer four times and the immune complexes were eluted from the beads by boiling in 40 μl 2× SDS loading buffer with 1 mm DTT for 5 min. The supernatants were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and Western blots using anti-Myc, anti-FLAG, or anti-UNC-115 antibodies (see below) and HRP-labeled secondary antibodies (Kirkegaard and Perry Laboratories, Gaithersburg, MD) with chemiluminescent autoradiography (Pierce, Rockford, IL).
Figure 1, C and D, shows co-immunoprecipitation of UNC-115 and SWAN-1 and RAC-2 and SWAN-1. For each co-immunoprecipitation experiment, the Western blot was also probed with the antibody used for immunoprecipitation to ensure that the immunoprecipitation procedure was robust (Figure 1).
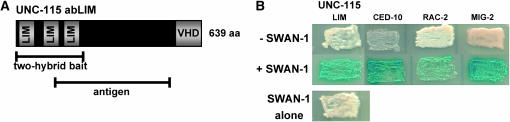
Figure 1.—
SWAN-1 interacts with the UNC-115 LIM domains and with Rac GTPases in a two-hybrid system. (A) A diagram of the 639-residue UNC-115 polypeptide. Indicated are the regions used in the two-hybrid screen and to generate an anti-UNC-115 polyclonal antiserum. (B) Yeast harboring different bait plasmids (across the top) without and with the SWAN-1 prey plasmid (see materials and methods). Yeast were grown on medium containing X-Gal to indicate lacZ activity and a positive interaction (blue). Yeast harboring the bait plasmids or SWAN-1 alone were overgrown to ensure absence of lacZ activity. Images were captured by an Epson flatbed scanner and processed in Photoshop. (C and D) SWAN-1 co-immunoprecipitated with UNC-115 and RAC-2 in HeLa cell lysates. (C) A Western blot using anti-UNC-115 antiserum. Lysates were prepared from cells expressing combinations of MYC∷SWAN-1 and UNC-115∷EGFP. The 100-kDa UNC-115∷EGFP band is indicated, as is the Ig heavy chain of the anti-Myc antibody (50 kDa), which cross-reacts with the secondary antibody. UNC-115∷EGFP was co-immunoprecipitated only when MYC∷SWAN-1 was present. The successful immunoprecipitation of MYC∷SWAN-1 was confirmed by Western blot with an anti-Myc antibody (data not shown). (D) MYC∷RAC-2 co-immunoprecipitated with FLAG∷SWAN-1. Shown are Western blots from two immunoprecipitation experiments. The blots 1 and 3 are the results of a MYC∷RAC-2 immunoprecipitation, and the blots 2 and 4 are the results of a FLAG∷SWAN-1 immunoprecipitation. Across the top the transgenes expressed in cells are indicated for each experiment. The top two blots (1 and 2) show that FLAG∷SWAN-1 co-immunoprecipitated with MYC∷RAC-2 and that MYC∷RAC-2 co-immunoprecipitated with FLAG∷SWAN-1, respectively. The Ig heavy chain from the anti-FLAG antibody (50 kDa), also detected by the secondary antibody, and FLAG-SWAN-1 (45 kDa) were similar in size but could be distinguished (blot 1). The Ig light chain from the anti-Myc antibody (25 kDa) and MYC∷RAC-2 (20 kDa) were more difficult to resolve (blots 2 and 3). In three independent experiments, MYC∷RAC-2 was evident as a thickening of the band below the Ig light chain and by the smaller bands (possible degradation products of MYC∷RAC-2) below 23 kDa (asterisk). The smaller products always correlated with the presence of MYC-RAC-2. Blots 3 and 4 are the controls for immunoprecipitation [MYC∷RAC-2 was immunoprecipitated with anti-Myc antibody (blot 3); and FLAG∷SWAN-1 was immunoprecipitated by anti-FLAG antibody (blot 4)]. FLAG∷SWAN-1 co-immunoprecipitated only in the presence of MYC∷RAC-2; and MYC∷RAC-2 co-immunoprecipitated only in the presence of FLAG∷SWAN-1.
Generation of an UNC-115 antiserum:
The coding region for UNC-115 residues 154–527 was inserted into the pQE9 vector (QIAGEN, Valencia, CA) in frame with the RGS-6-histidine epitope, and fusion protein was produced by bacterial expression. SDS–PAGE gel fragments containing the fusion protein were used as an antigen to immunize two rabbits (Caltag Laboratories, Healdsburg, CA). On Western blots, serum from the immunized rabbits but not preimmune serum recognized a bacterially expressed glutathione-S-transferase (GST) fusion with the same portion of UNC-115 (data not shown) (residues 154–527, produced by inserting the unc-115 fragment into the pGEX4T vector; Amersham Pharmacia Biotech). UNC-115-specific antibodies were purified from the antiserum by their affinity for the GST∷UNC-115 fusion protein using glutathione Sepharose beads (Amersham Pharmacia Biotech). This affinity-purified polyclonal antibody recognized a single band of ∼70 kDa in Western blots of lysates of C. elegans (the estimated size of full-length UNC-115 polypeptide is 72 kDa) (data not shown).
swan-1 expression analysis:
The upstream region of swan-1 excluding the initiator ATG codon (3412–7549 relative to cosmid F53C11) was amplified, inserted into the pPD95.77 GFP expression vector (kindly provided by A. Fire), and used to drive the expression of GFP in transgenic animals. A full-length fusion of the swan-1 coding region to gfp was constructed by amplifying the swan-1 upstream region and entire coding region (3412–9941 relative to cosmid F53C11) from genomic DNA and fusing this fragment in frame to gfp such that the transgene was predicted to encode a full-length SWAN-1 protein with GFP at the C terminus. RNAi of swan-1 resulted in greatly reduced GFP expression from this transgene, suggesting that the transgene encodes a SWAN-1∷GFP molecule (data not shown). swan-1 expression constructs were used at 5 ng/μl in transformation experiments.
SWAN-1 transgenic assays in C. elegans:
For the loss-of-function swan-1(ok267) experiments, previously described constitutively active rac(G12V) transgenes were used (Struckhoff and Lundquist 2003). PDE morphological defects were scored as previously described (Struckhoff and Lundquist 2003): for each genotype, the percentages of PDE neurons exhibiting ectopic neurites and ectopic lamellipodia and filopodia were determined. After construction of each transgene in the swan-1(ok267) background, the transgene was crossed away from swan-1(ok267) and axon defects were scored again to ensure that the transgene caused defects at percentages similar to those before the experiment began. This ensured that the transgenes had not been altered during the course of the experiment and the phenotypic change was due to the presence of swan-1(ok267).
For the swan-1(+) expression experiments, new transgenes were constructed that contained a plasmid harboring the swan-1(+) genomic region (bases 3412–9972 inclusive of cosmid F53C11), a rac(G12V) plasmid, or both. In all cases an osm-6∷gfp plasmid was included as a neuronal marker to visualize the PDE neuron. When constructing transgenic strains, rac(G12V) plasmids were used at 1 ng/μl and other plasmids were used at 5 ng/μl. Multiple transgenic lines were generated for each experiment to ensure uniformity of results, and results from each line were accumulated to derive a percentage of PDE axon defects. In all C. elegans experiments, at least 100 animals were scored for each genotype. Significance of differences in proportions was calculated using the t-test and Fisher's exact analysis.
SWAN-1 transgenic assays in fibroblasts:
Mammalian NIH 3T3 fibroblasts were grown and transformed as previously described (Yang and Lundquist 2005). Briefly, cells grown in six-well plates to ∼50% confluency on polylysine-coated coverslips were transfected by addition of a mixture of the transgene DNA and the Fugene reagent. Cells were transfected with combinations of 1 μg each of the MYC∷SWR-1 construct, a MYR∷UNC-115 construct, and a CFP∷RAC-1(Q61L) construct, which encodes an activated version of human Rac1 fused to cyan fluorescent protein (CFP) (kindly provided by T. Meyer). Cells were cotransfected with egfp to identify transfected cells, and cells transfected with egfp alone were used as a control. After transfection, cells were allowed to grow for 6 hr and deprived of serum (serum-containing medium was replaced by DMEM). Cells were grown for another 12 hr and fixed in 3.7% paraformaldehyde. The fixed cells were treated with 1 μg/ml of rhodamine-labeled phalloidin to visualize the actin cytoskeleton. Cells were mounted in 50% glycerin in PBS for epifluorescence microscopy. For each experiment, at least three transfections were performed and >100 cells were scored to ensure consistency of results.
SWAN-1 transgenic assays in yeast:
All yeast were grown at 30°. For growth curve analysis, Y190 yeast strains harboring different plasmid constructs were grown in selective medium and diluted to ∼0.1 OD600 units in YPD medium. Growth rates were monitored by sampling the OD600 culture densities each hour for 8 hr (see Figure 8). Each strain was tested at least twice, and standard deviations were indicated by error bars in Figure 8. To visualize the yeast actin cytoskeleton with rhodamine-labeled phalloidin, cells were grown in selective medium, diluted to ∼0.1 OD600 units in YPD, and grown to midlog phase (0.3–0.7 OD600) (cells harboring the CED-10 plasmid were assayed after 8 hr). Cells were fixed for 1 hr in 20% formaldehyde in 0.5 m potassium phosphate pH 6.5 and overnight in 4% formaldehyde in 100 mm potassium phosphate pH 6.5. Cells were washed in PBS and permeabilized in 0.2% Triton X-100 in PBS for 15 min, washed three times in PBS, incubated with rhodamine–phalloidin (1 μg/ml) and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (1 μg/ml) in PBS for 1 hr in the dark, washed three times in PBS, and mounted for fluorescence microscopy in PBS with 90% glycerol and 1 mg/ml 1,4-diazabicyclo[2.2.2]octane (DABCO) antifade reagent.
RESULTS
SWAN-1 interacts physically with the UNC-115 LIM domains and with Racs:
UNC-115 is a cytoskeletal linker molecule composed of a C-terminal actin-binding villin headpiece domain (VHD) and three N-terminal LIM domains (Figure 1A) (Lundquist et al. 1998). To identify molecules that bind to the UNC-115 LIM domains, a yeast two-hybrid screen using the three UNC-115 LIM domains as bait was conducted (Figure 1A). From an estimated 2 million C. elegans cDNAs screened, two cDNAs were isolated that encode a novel protein that here is named SWAN-1. The presence of swan-1 two-hybrid fusions with the UNC-115 LIM domain fusion specifically activated both HIS5 expression (data not shown) and lacZ expression (Figure 1B) in the two-hybrid system (see materials and methods). Both swan-1 cDNAs contained the entire swan-1 open reading frame, suggesting that only the full-length SWAN-1 protein is active in the two-hybrid assay. SWAN-1 interacted specifically with the UNC-115 LIM domains, but not with the remainder of the UNC-115 molecule in the two-hybrid system (data not shown).
Because UNC-115 acts genetically downstream of Rac signaling (Struckhoff and Lundquist 2003), it was determined if SWAN-1 could also interact with the three C. elegans Rac molecules in the two-hybrid system. SWAN-1 and each of the three C. elegans Racs, CED-10, MIG-2, and RAC-2, interacted when directly tested in the two-hybrid system (Figure 1B). SWAN-1 interacted with both wild-type RAC-2 (Figure 1B) and constitutively active, GTPase-dead RAC-2(G12V) (data not shown). The LIM domains of UNC-115 did not interact with any of the three Racs in the two-hybrid system, nor did any other region of UNC-115 (data not shown).
To confirm the physical interactions detected in the two-hybrid system, co-immunoprecipitation of SWAN-1 with UNC-115 and RAC-2 was assayed in lysates of HeLa cells in which tagged versions of the molecules were expressed (Figure 1, C and D). EGFP-tagged full-length UNC-115 co-immunoprecipitated with Myc-tagged SWAN-1 (Figure 1C). Experiments with MYC∷RAC-2 and FLAG∷SWAN-1 were consistent with co-immunoprecipitation of the molecules, although MYC∷RAC-2 and FLAG∷SWAN-1 bands were not completely separated from the light and heavy chains of the antibodies used for immunoprecipitation on Western blots (Figure 1D). These experiments were repeated with similar results in HeLa cells and in HEK293 cells (data not shown).
SWAN-1 is a member of a novel, conserved family of seven-WD-repeat proteins:
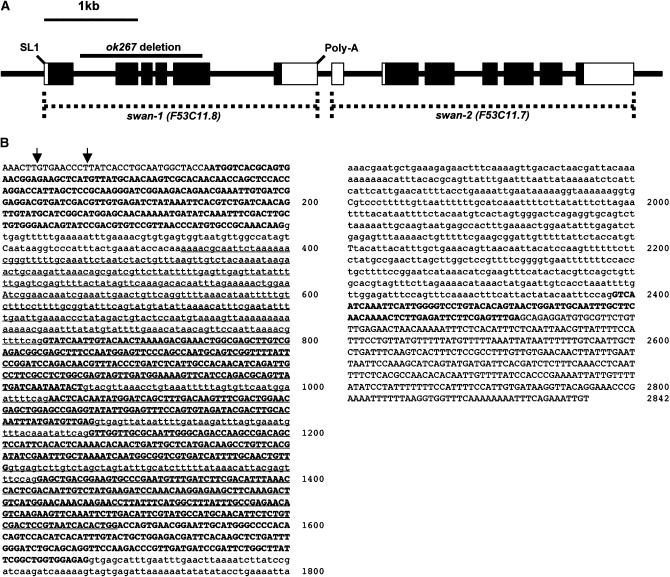
The complete sequences of the two swan-1 cDNAs isolated in the two-hybrid screen were determined, as were the sequences of the independently derived cDNAs yk343g2 and yk326a5 (provided by Y. Kohara) (Figure 2, A and B). The splicing pattern and coding potential for each of the four cDNAs were identical. Reverse-transcription PCR was conducted on the 5′ end of the swan-1 transcript using a 5′ primer complementary to the trans-spliced leader SL1, which revealed that SL1 was spliced onto the 5′-end transcript after position 7464 relative to cosmid F53C11 (Figure 2, A and B). cDNA sequences indicated that the swan-1 transcript was polyadenylated after position 10306 relative to cosmid F53C11 (Figure 2, A and B).
Figure 2.—
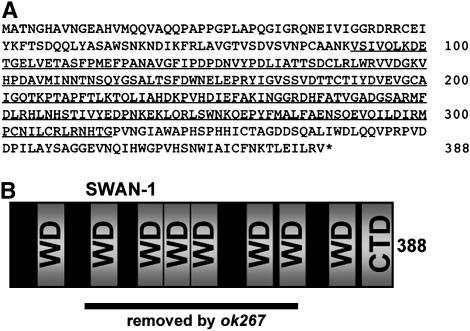
The swan-1 locus. (A) A depiction of the swan-1 and swan-2 loci (5′ is to the left). Boxes represent exons; solid boxes represent coding regions. SL1 indicates where the trans-spliced leader sequence SL1 is attached to the 5′ end of the swan-1 cDNA; and the site of a poly(A) tail on the swan-1 cDNA is shown. The extent of the swan-1(ok267) deletion is indicated. (B) The swan-1 coding region. Uppercase letters represent exons present in three independent cDNAs that were sequenced, and bold letters represent the open reading frame. Lower case indicates intron sequences inferred from the sequence for the F53C11.8 region on WormBase. Underlined sequences represent the extent of the ok267 deletion. Base 1 of the sequence is where an SL1 trans-spliced leader sequence is present in a cDNA generate by 5′ RACE. The two arrows represent the start points of two independent cDNAs isolated in the yeast two-hybrid screen. The sequences of these two cDNAs as well as those of two other independently-derived cDNAs yk326a5 and yk343g2 (kindly provided by Y. Kohara) were determined and were identical throughout the open reading frames of the cDNAs. The end of the sequence is where a poly(A) tail is present in yk1174f05 (data from WormBase).
The swan-1 cDNAs can encode a 388-residue polypeptide that is predicted to contain at least seven WD-repeat elements (Figure 3, A and B) (Neer et al. 1994; Smith et al. 1999). WD repeats are conserved structural elements that form a “β-propeller” structure, with each blade of the propeller corresponding to one WD repeat (Sondek et al. 1996). WD repeats have no known enzymatic activity. Rather, they are thought to form a scaffold for protein–protein interaction. Many proteins with seven WD repeats have been identified, including the β-subunit of G proteins (Smith et al. 1999). However, SWAN-1 is a member of a novel, conserved family of WD-repeat proteins (Figure 3C). The SWAN-1 family is ancient, as obvious SWAN-1 counterparts are found in insects, mammals, protozoa, yeast, and plants (Figure 3C). SWAN-1-family members contain at least seven regions similar to the WD repeat, with a potential eighth region near the C terminus. SWAN-1-family proteins also contain a conserved C-terminal tail that is not found in Gβ or other WD-repeat proteins (Figure 3C). Generally, the first two predicted WD repeats show more variability between species than do the others.
Figure 3.—
The SWAN-1 polypeptide. (A) The amino acid sequence of the 388-residue SWAN-1 polypeptide. Residues whose coding region was removed by the ok267 deletion are underlined. (B) Schematic of the SWAN-1 polypeptide. Regions with similarity to the WD repeat domain are indicated, as is the C-terminal conserved domain (CTD) unique to SWAN-1-family polypeptides. The region of the molecule whose coding region was removed by ok267 is indicated. (C) The SWAN-1 family is conserved in animals, plants, fungi, and protozoa. Predicted amino acid sequences were aligned by ClustalW: Ce, C. elegans SWAN-1 (GenBank accession no. NP_506418) and SWAN-2 (NP_506417); Dm, Drosophila melanogaster (NP_608461); Dr, Danio rerio (zebrafish) (NP_956363); Hs, Homo sapiens (AAH01264); AN11 from Petunia x hybrida (AAC18914); At, Arabidopsis thaliana (NP_172751); Sp, Schizosaccharomyces pombe (CAA21079); Sc, Saccharomyces cerevisiae (NP_015077); Dd, Dictyostelium discoideum (protozoan slime mold) (XP_643620). Residues identical in a majority of the molecules are highlighted in dark shading. Regions similar to the WD repeat (WD1–8) and the conserved C-terminal domain (CTD) are boxed in light shading. Members from the yeast S. cerevisiae and S. pombe include additional amino acid residues, not found in other members, between WD5 and WD6, between WD7 and WD8, and in WD8. The SWAN-1-like molecule from the protozoan D. discoideum contains a run of 38 consecutive serines in the region that corresponds to WD2.
Two SWAN-1 family members in higher plants have known functions. AN11 from petunia is a cytoplasmic molecule involved in Myc and Myb transcription factor activation and anthocyanin pigment biosynthesis in response to light (de Vetten et al. 1997). In Arabidopsis, the SWAN-1-like molecule TTG1 is involved in the Myb-dependent specification of trichome cells on leaves (Walker et al. 1999). However, the Arabidopsis genome also encodes another SWAN-1 family member, AtAN11 (Figure 3C), which is more similar to SWAN-1 than is TTG1. The role of AtAN11 is not known, nor are the roles of SWAN-1 family members in other organisms.
The swan-1 gene corresponds to the gene F53C11.8 in the C. elegans genome. Immediately downstream of the swan-1 gene is the coding region for a gene that encodes another SWAN-1 family member here named SWAN-2 (F53C11.7 in C. elegans genome nomenclature) (Figure 3A). The first exon in a swan-2 cDNA is 114 nucleotides downstream of the site of swan-1 polyadenylation (data not shown). SWAN-2 is 45% identical at the amino acid level to SWAN-1 (Figure 3C). RNA-mediated interference (RNAi) of swan-2 in swan-1(ok267) had no effect on viability, fertility, or gross morphological and behavioral phenotype. Furthermore, neither RNAi of swan-2 nor a swan-2 deletion allele ok964 had an effect on activated CED-10 and RAC-2 in PDE development (data not shown).
swan-1(ok267) suppresses ced-10(n1993):
To determine the role of SWAN-1 in C. elegans, a deletion allele of the gene, called ok267, was obtained from the C. elegans Gene Knockout Consortium (kindly provided by G. Molder and B. Barstead). The swan-1 region from ok267 mutants was amplified by PCR and sequenced to determine the breakpoints of the deletion. The 1189-bp ok267 deletion encompassed bases 7846–9034 relative to cosmid F53C11 (Figure 2, A and B). To ensure that the wild-type swan-1 locus had not been duplicated in the ok267 deletion strain, PCR was performed on swan-1(ok267) genomic DNA using primers that were predicted to amplify a fragment in wild type but not in swan-1(ok267) (one primer was complementary to a region removed by ok267). These primers amplified a fragment from wild type but not from swan-1(ok267) (data not shown).
The ok267 deletion removed the coding region for 223-amino-acid residues of the 388-residue SWAN-1 polypeptide, including all of WD repeats two to six and part of seven (Figure 3, A and B). swan-1(ok267) animals were viable and fertile and displayed no apparent phenotype, including gross morphological or behavioral defects. RNAi directed against swan-1 had no detectable phenotype. Furthermore, no swan-1 transcript was detected by RT–PCR in swan-1(ok267) mutants (data not shown).
Because SWAN-1 interacted physically with UNC-115 and the Racs, double mutants of swan-1(ok267) were built with the rac pathway mutants ced-10 (Lundquist et al. 2001), mig-2(mu28) (Zipkin et al. 1997), unc-73 (Steven et al. 1998), and unc-115 (Lundquist et al. 1998). swan-1(ok267) suppressed cell corpse phagocytosis and gonadal distal tip cell migrations of ced-10(n1993) and enhanced axon defects caused by activated Rac molecules (see below). No effects were observed in swan-1(ok267) double mutants with unc-73 or unc-115 (data not shown).
ced-10 Rac mutations cause defects in phagocytosis of cells undergoing programmed cell death (Reddien and Horvitz 2000) and defects in the migration of the gonadal distal tip cells (Lundquist et al. 2001), resulting in malformed gonads. swan-1(ok267) significantly suppressed the cell corpse engulfment defects and the gonadal distal tip cell (DTC) migration defects of ced-10(n1993) mutants (Table 1). ced-10(n1993) mutants alone displayed 14.9 ± 3.8 persistent cell corpses in the pharyngeal region of late threefold-stage embryos, compared to 9.6 ± 4.2 in swan-1(ok267); ced-10(n1993) double mutants (P < 0.001). Furthermore, 24% of ced-10(n1993) mutants displayed misshapen gonads due to DTC migration errors, whereas swan-1(ok267); ced-10(n1993) double mutants displayed 7% (P < 0.001). swan-1(ok267) alone had little effect on cell corpse engulfment or DTC migration on its own (Table 1). ced-10(n1993) is a hypomorphic allele that retains some ced-10 activity (Reddien and Horvitz 2000; Shakir et al. 2006). That swan-1(ok267) suppressed the defects caused by ced-10(n1993) suggests that the normal role of SWAN-1 might be to repress CED-10 Rac activity [i.e., in the absence of SWAN-1, the remaining activity of ced-10(n1993) is increased]. While swan-1 slightly suppressed the hypomorphic ced-10(n1993) allele, it did not suppress the maternal-effect lethality or gonad defects of the ced-10(n3417) null allele nor did it suppress the lethality or axon pathfinding defects associated with the ced-10(n1993); mig-2(mu28) double mutant (data not shown).
To confirm that the suppression of ced-10(n1993) was due to the swan-1(ok267) deletion, a full-length swan-1∷gfp transgene (predicted to encode a full-length SWAN-1∷GFP fusion protein; see materials and methods) was analyzed for its ability to rescue suppression by swan-1(ok267). The swan-1∷gfp transgene rescued the effects of swan-1(ok267) suppression in ced-10(n1993); swan-1(ok267) mutants (Table 1): transgene-bearing ced-10; swan-1 animals displayed 15.6 ± 3.4 persistent cell corpses and siblings that did not inherit the transgene displayed 11.7 ± 3.6 persistent cell corpses (P < 0.001). Furthermore, suppression of DTC migration defects was rescued (29% vs. 12%, respectively; P < 0.001) (Table 1).
Mutations in the mig-2 Rac gene also cause DTC migration defects (Lundquist et al. 2001). swan-1(ok267) did not suppress the DTC migration defects of mig-2(mu28) (Table 1). As opposed to ced-10(n1993), which retains some CED-10 function, mig-2(mu28) is a null allele that likely causes complete loss of mig-2 function. This might explain why swan-1(ok267) did not suppress mig-2(mu28). Indeed, evidence is presented below that SWAN-1 can regulate MIG-2.
swan-1(ok267) enhances constitutively active Rac in neurons:
To further test the idea that SWAN-1 is a negative Rac regulator, the effect of swan-1 loss of function on Rac activity in development of the PDE neurons was studied. The PDEs are a pair of ciliated sensory neurons that reside in the posterior-lateral region of C. elegans in the postdeirid ganglion (White et al. 1986; Shakir et al. 2006). A single ciliated dendrite extends dorsally from the cell body, and a single axon extends ventrally to the ventral nerve cord, where it branches and runs anteriorly and posteriorly (Figure 4A).
Previous studies indicated that constitutively active Rac molecules, produced by the canonical glycine 12 to valine (G12V) mutation, induce ectopic neurite formation and ectopic lamellipodia and filopodia formation in the PDE neuron in vivo (Figure 4B) (Struckhoff and Lundquist 2003). swan-1(ok267) enhanced the effects of transgenic ced-10(G12V) and rac-2(G12V) (Figure 4C). For example, rac-2(G12V) caused 65% ectopic axon formation and 8% ectopic lamellipod and filopod formation, whereas swan-1(ok267); rac-2(G12V) animals displayed 100 and 74% of the respective defects. While all effects of swan-1(ok267) and swan-1(RNAi) on CED-10(G12V) and RAC-2(G12V) were significant (P < 0.001), the effects of swan-1 on rac-2(G12V) were more pronounced than those on ced-10(G12V). In addition to increasing the penetrance of rac-2(G12V) and ced-10(G12V) defects, swan-1(ok267) also increased the severity of defects. For example, ced-10(G12V) PDEs generally displayed one to two ectopic axons, whereas swan-1(ok267)l ced-10(G12V) PDEs often displayed three or more ectopic axons. Occasionally, so many ectopic axons were present that it became impossible to distinguish which axon was the normal PDE axon. RNAi of swan-1 gave similar but weaker results (Figure 4C), indicating that the effects were due to disruption of swan-1 function. These results demonstrate that swan-1(ok267) enhances the effects of ced-10(G12V) and rac-2(G12V).
The mig-2(rh17) mutant harbors an activating G16 mutation (Zipkin et al. 1997). Neither mig-2(rh17) nor the effects of transgenic expression of MIG-2(G16V) were significantly affected by swan-1(ok267) (Figure 4C).
SWAN-1(+) transgenic expression represses constitutively active Rac:
If SWAN-1 is a negative Rac regulator, overactivity of wild-type SWAN-1 might repress Rac activity. Transgenic expression of wild-type swan-1 suppressed the effects of ced-10(G12V) and rac-2(G12V) (Figure 4D; see materials and methods). For example, rac-2(G12V) trangenes averaged 65% ectopic PDE axons and 6% ectopic lamellipodia and filopodia, whereas rac-2(G12V) transgenes that also contained wild-type swan-1 DNA averaged 16 and 0% defects, respectively. While all effects of SWAN-1(+) expression on CED-10(G12V) and RAC-2(G12V) were significant (P < 0.001), suppression of ced-10(G12V) was weaker than that of rac-2(G12V). swan-1 coexpression had no detectable effect on mig-2(G16V) (Figure 4D).
swan-1(ok267) enhances the neuronal effects of dominant Rac alleles:
In contrast to ced-10(n1993) and mig-2(mu28), the mutations ced-10(n3246), ced-10(n1993lq20), mig-2(rh17), and mig-2(lq13) all cause axon defects on their own, indicating that they are not simple loss-of-function mutations (Shakir et al. 2006). mig-2(rh17) affects the glycine 17 residue of the MIG-2 protein (Zipkin et al. 1997) and is likely to result in constitutively active, GTPase-dead MIG-2 [the equivalent mutation was included in the mig-2(G16V) transgenes]. It is unknown whether ced-10(n3246), ced-10(n1993lq20), or mig-2(lq13) are activating or dominant-negative mutations.
swan-1(ok267) enhanced the axon defects caused by each of these dominant Rac alleles. ced-10(n3246), ced-10(n1993lq20), mig-2(rh17), and mig-2(lq13) caused a variable PDE axon pathfinding phenotype that was categorized as mild (the axon reaches the ventral nerve cord near the normal position but is misguided) or severe (the axon fails to reach the ventral nerve cord near the normal position) (Figure 5, A and B). The percentages of combined mild and severe axon defects of swan-1(ok267) double mutants with ced-10(n3246), mig-2(lq13), and mig-2(rh17) were increased, and the severity of all four increased (P < 0.001; Figure 5C). For example, ced-10(n3246) alone showed 8% axon defects, most of which were of the mild class, and swan-1(ok267); ced-10(n3246) showed 18% defects with 8% of the severe class.
swan-1 is expressed in many cell types including neurons:
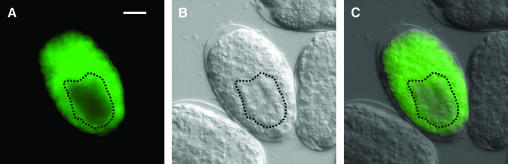
To further understand the role of swan-1 in Rac signaling, the temporal and spatial pattern of swan-1 expression during development was analyzed. Two swan-1 expression transgenes were constructed: a transcriptional reporter consisting of the swan-1 promoter driving green fluorescent protein (gfp) expression and a translation reporter of the full-length swan-1 region fused in frame to gfp such that a full-length SWAN-1 molecule fused to GFP at the C terminus is produced (SWAN-1∷GFP) (see materials and methods). Full-length swan-1∷gfp expression was first detected at ∼100 min postfertilization when embryonic transcription begins. Expression was observed in most cells (Figure 6, A–C), including neuroblasts and neurons, and persisted throughout development into adulthood. The only cells that did not show swan-1∷gfp expression were the intestinal cells and their precursors (Figure 6, A–C). SWAN-1∷GFP accumulated predominantly in the cytoplasm of all cell types analyzed. The transcriptional reporter transgene displayed a temporal and spatial expression pattern identical to that of the full-length fusion transgene (data not shown).
Figure 6.—
swan-1∷gfp is expressed in all cells except the intestine. Micrographs of an embryo, ∼270 min after fertilization, harboring the full-length swan-1∷gfp fusion transgene are shown (see materials and methods). An HCX Planapo 63× objective (1.3 numerical aperture) and a 10× magnifier were used. The dashed line surrounds the intestinal precursor cells. Bar in A, 10 μm. (A) A fluorescence micrograph; (B) a differential interference contrast micrograph; (C) a merged image.
SWAN-1 represses human Rac1 in 3T3 fibroblasts:
The above data indicate that SWAN-1 represses Rac activity in C. elegans neuronal morphogenesis. CED-10 Rac and RAC-2 Rac are very similar to human Rac1 (83% identical at the amino acid level) (Lundquist et al. 2001). Expression of constitutively active Rac1 (the glutamine to leucine mutation at position 61, Q61L) in serum-starved 3T3 fibroblasts led to lamellipodial membrane protrusions along cell edges (Figure 7A). We found that 62% of 3T3 cells expressing Rac1(Q61L) exhibited lamellipodial plasma membrane extensions (Figure 7, A and B) not seen in egfp control-transfected cells. Expression of myc-tagged SWAN-1 alone had little effect on cell morphology (Figure 7B). Fewer cells transfected with both Rac1(Q61L) and SWAN-1 displayed lamellipodial structures (26% compared to 62% for Rac1 alone; P < 0.001) (Figure 7B), suggesting a partial suppression of Rac1(Q61L) activity by SWAN-1.
SWAN-1 might directly repress Rac activity. Alternatively, SWAN-1 might inhibit downstream Rac effectors, such as UNC-115. In C. elegans neurons, no repression of activated UNC-115 by SWAN-1 was detected (data not shown). As previously reported, addition of an N-terminal myristoylation sequence (Myr) from human c-Src targeted UNC-115 to the plasma membrane and caused overactivation of UNC-115, resulting in ectopic lamellipodia and filopodia formation in serum-starved fibroblasts (Figure 7C). In 24% of MYR∷UNC-115-expressing cells, lamellipodial and filopodial membrane protrusions were observed (Figure 7, C and D). Cotransfection of SWAN-1 with MYR∷UNC-115 resulted in 15% of cells exhibiting protrusive morphology, a difference that was not statistically significant (P = 0.1076). While these data indicate that SWAN-1 might slightly repress UNC-115, the effect is not as pronounced as the effect of SWAN-1 on Rac1.
SWAN-1 represses Rac activity in yeast:
In the course of the directed two-hybrid experiments using SWAN-1 and the Racs, it was noted that yeast transformed with CED-10 and MIG-2 (but not RAC-2) were slow-growing and formed small and irregular-shaped colonies. The doubling times of strains harboring CED-10 and MIG-2 grown in liquid culture were increased compared to that of the parent strain Y190 (Figure 8A). CED-10 cells had a doubling time of >10 hr and MIG-2 cells had a doubling time of ∼6 hr, compared to ∼2.5 hr for Y190. Doubling time compared to that of Y190 was not significantly affected by RAC-2 or SWAN-1 expression (data not shown). When cell morphology was analyzed, fewer of the CED-10 and MIG-2 cells were in the process of budding during the log phase of growth compared to the parent Y190 strain (Figure 8B). For example, 85% of Y190 cells were in the process of budding whereas 29% of CED-10 and 45% of MIG-2 cells were budding. SWAN-1 and RAC-2 expression had little effect on percentage of cells undergoing budding (Figure 8B).
CED-10- and MIG-2-expressing cells displayed abnormal morphology, including greatly increased cell size and irregular cell shape (18/21 of CED-10-expressing cells and 24/30 MIG-2-expressing cells). Phalloidin staining revealed that the actin cytoskeleton was disorganized in these cells. In normal budding cells, the actin cytoskeleton is dramatically polarized; the newly forming bud displays actin patches on the cell surface as a result of endocytosis involved in bud growth; and long actin filaments extend from the mother cell to the bud (Adams and Pringle 1984). The actin cytoskeleton of the host strain Y190 displayed this morphology characteristic of asymmetric growth and budding (Figure 8C). In CED-10 and MIG-2 cells undergoing budding, actin patches were often observed both in the bud and in the mother cell (Figure 8, D and E), indicating that cell polarity and asymmetric growth had been disrupted. Furthermore, the long actin filaments were often missing (Figure 8, D and E). When the long filaments were present, they were often disorganized with respect to the bud–mother cell axis (Figure 8D). In the large, irregular, nonbudding cells, actin patches were observed uniformly on the cell surface (Figure 8G), indicating that these cells were undergoing nonpolarized growth without budding. While most RAC-2-expressing cells had normal morphology, some with abnormal polarized growth and actin cytoskeleton disorganization were observed at a low frequency (Figure 8F), which apparently did not significantly interfere with the growth dynamics of RAC-2-expressing yeast. SWAN-1-expressing cells showed no apparent morphology and cytoskeletal defects (data not shown).
The observed defects in budding, polarized growth, and actin disorganization mirrored those seen in selected mutants of the six yeast Rho GTPases (Rho1–5p and Cdc42p) (Dong et al. 2003). Several yeast Rho proteins are required to assemble the long actin filaments, which when absent lead to polarity defects and actin patch disorganization; and Cdc42p is required for the proper orientation of the long actin filaments. While yeast have no true Rac-family GTPases, transgenic expression of CED-10 Rac, RAC-2 Rac, and MIG-2 Rac in yeast might ectopically interfere with the normal roles of the yeast Rho GTPases, resulting in the cytoskeletal and cell polarity defects described above.
The growth defects caused by CED-10, RAC-2, and MIG-2 expression in yeast were reduced when the yeast were cotransformed with SWAN-1. Doubling time decreased in the SWAN-1 cotransformed strains (CED-10-expressing cells went from >10 to 4.5 hr, and MIG-2 cells went from 6 to 3.5 hr), and the percentages of budding cells increased (Figure 8A): 29% of CED-10-expressing cells were budding compared to 64% of CED-10 SWAN-1-expressing cells (P < 0.001). While the SWAN-1 cotransformed cells still showed some cytoskeletal disorganization, it was not as severe as that seen in CED-10 and MIG-2 alone, as indicated by increased percentages of budding cells. These data indicate that SWAN-1 repressed CED-10 and MIG-2 activity in yeast cells.
DISCUSSION
Previous studies indicate that the UNC-115/abLIM actin-binding protein controls lamellipodia and filopodia formation in response to Rac GTPase signaling (Struckhoff and Lundquist 2003; Yang and Lundquist 2005), but the details of this pathway, including other molecules involved, remain unclear. A two-hybrid screen with the UNC-115 LIM domains identified the novel SWAN-1 protein, the C. elegans member of a novel seven- or eight-WD-repeat protein family conserved in yeast, protozoa, plants, and animals. Subsequent directed two-hybrid tests and co-immunoprecipitation experiments revealed that SWAN-1 interacted both with the UNC-115 LIM domains and with each of the three C. elegans Rac-family GTPases CED-10, RAC-2, and MIG-2. SWAN-1 might represent a molecular link between Rac GTPases and UNC-115/abLIM. Functional assays in C. elegans neurons, cultured mammalian 3T3 fibroblasts, and yeast indicate that SWAN-1 is a new class of negative regulator of Rac GTPases.
Expression of activated Rac in C. elegans neurons leads to the formation of ectopic lamellipodia and filopodia that is partially dependent on UNC-115 (Struckhoff and Lundquist 2003). Loss of swan-1 enhanced the effects of CED-10 and RAC-2 activity, and transgenic expression of wild-type swan-1 repressed CED-10 and RAC-2 activity in neurons. In these transgenic assays, no effect of SWAN-1 on MIG-2 activity was detected, despite the fact that SWAN-1 and MIG-2 interacted strongly in the two-hybrid system. swan-1 loss of function did enhance the effects of dominant mig-2 alleles on axon pathfinding, suggesting that SWAN-1 also regulates MIG-2. SWAN-1 also repressed MIG-2 activity in yeast, consistent with the idea that SWAN-1 regulates all three Racs, CED-10, RAC-2, and MIG-2. Expression of C. elegans SWAN-1 also suppressed the lamellipodia formed in 3T3 fibroblasts in response to human Rac1 harboring the activating Q61L mutation, suggesting that the Rac–SWAN-1 interaction has been conserved in the evolutionary divergence of humans and C. elegans.
swan-1 mutation also suppressed the cell corpse engulfment defects and gonadal distal tip cell migration defects of the hypomorphic ced-10(n1993) allele, indicating that SWAN-1 represses CED-10 activity in multiple developmental processes. swan-1 did not suppress the distal tip cell migration defects of mig-2(mu28), which is a predicted null allele of mig-2. ced-10(n1993) retains some ced-10 activity, and this residual activity might be enhanced by loss of swan-1. No residual MIG-2 activity is predicted to be present in the null mig-2(mu28) mutant.
These experiments in C. elegans, mammalian fibroblasts, and yeast support the idea that SWAN-1 inhibits Rac activity. swan-1 mutants have no detectable phenotype on their own, and the effects of swan-1 in rac mutant strains are generally weak albeit consistent across many different assays. Generally, sensitized backgrounds were required to see an effect of swan-1 (e.g., hypomorphic and dominant mutations in rac genes and transgenic expression assays). SWAN-1 inhibited human Rac1 and to a lesser extent MYR∷UNC-115 in fibroblasts. Possibly SWAN-1 is a direct inhibitor of UNC-115. However, no effect of swan-1 on myr∷unc-115 activity in C. elegans neurons was detected, and SWAN-1 inhibited Rac activity in the yeast Saccharomyces cerevisiae, which has no molecule similar to UNC-115 encoded in its genome (Goffeau et al. 1996). These data indicate that SWAN-1 is a direct inhibitor of Rac GTPase activity. The inhibition of MYR∷UNC-115 by SWAN-1 in fibroblasts might be an indirect effect of Rac1 inhibition (i.e., MYR∷UNC-115 might still require Rac activity for full effect). However, it is possible that SWAN-1 is a direct inhibitor of both Rac and MYR∷UNC-115.
The effects of SWAN-1 on Rac activity are weak. The related SWAN-2 molecule showed no detectable functional overlap or similarity to SWAN-1, suggesting that the two molecules have distinct functions. SWAN-1 might be a fine-scale modulator that is involved in precisely tuning the amount of Rac activity. Modest changes in the levels of Rac activity can have distinct physiological outcomes. For example, in C. elegans, reducing the copy number of rac genes can change an attractive semaphorin signal into a repulsive signal (Dalpe et al. 2004). In cultured mammalian fibroblasts, reduction of Rac1 activity by as little as 30% leads to a switch from random peripheral lamellipodia protrusion to directed protrusion and directed cell migration (Pankov et al. 2005). rac genes display redundant functions in C. elegans and Drosophila whereby progressive loss of rac gene activity leads to progressively more severe axon defects (Lundquist et al. 2001; Ng et al. 2002). While there is clear overlap in rac gene function, it is also possible that different levels of rac activity have distinct physiological effects on axon development. SWAN-1 could also be a Rac “fidelity factor” ensuring that background or transient Rac activation is suppressed so that only strong, persistent Rac activity leads to directed protrusion. swan-1(ok267) has no detectable phenotype on its own, but it is possible that a subtle phenotype was not revealed in our endpoint phenotypic analysis (e.g., the growth cones of swan-1 animals might behave differently from those of wild type, but they might eventually reach their targets).
GTPase-activating proteins and guanine nucleotide dissociation inhibitors are a well-characterized family of Rac negative regulators. GAPs inhibit small GTPases by activating the GTPase activity of the molecules, favoring the inactive, GDP-bound state, and GDIs prevent GTPase interaction with the plasma membrane (Van Aelst and D'Souza-Schorey 1997; Michaelson et al. 2001). SWAN-1 does not resemble any known GAPs or GDIs, and, as expected, SWAN-1 does not act as a GAP on human Rac1 (I. Blasutig and A. Pawson, personal communication). SWAN-1 interacted physically with both wild-type and G12V-mutant-activated Rac molecules, and swan-1 interacted genetically with a variety of rac dominant mutations, including the activating G12V and Q61L mutations and the dominant mutations ced-10(n3246), ced-10(n1993lq20), and mig-2(lq13), which affect Rac by unknown mechanisms. SWAN-1 might bind to Racs and lock them in an inactive conformation, SWAN-1 might block sites of interaction with Rac effectors regardless of the GTP-bound state of Rac, or SWAN-1 might regulate Rac interaction with the plasma membrane.
UNC-115/abLIM and Racs act in the same pathway to promote protrusive activity. SWAN-1 was isolated by its ability to interact with UNC-115 LIM domains, and SWAN-1 is a negative regulator of Rac GTPases in the pathway. UNC-115/abLIM might act as a scaffold for a complex that includes both activators and inhibitors of protrusive activity. SWAN-1 might be recruited with the Rac effector UNC-115/abLIM to Racs to precisely control levels of Rac activity or to prevent spurious, transient Rac activity so that directed protrusion can occur.
Acknowledgments
We thank E. Struckhoff for technical assistance; R. Barstead, G. Molder, and the C. elegans Gene Knockout Consortium for C. elegans strains; R. Barstead for providing the two-hybrid cDNA library; T. Meyer for providing the Rac1(Q61L) clone; K. Neufeld and T. C. Gamblin for reagents and technical assistance; and the Caenorhabditis Genetics Center, sponsored by the National Center for Research Resources, for providing C. elegans strains. This work was supported by National Institutes of Health (NIH) grant NS40945 and National Science Foundation grant IBN93192 to E.A.L. and NIH grant P20 RR016475 from the INBRE Program of the National Center for Research Resources.
References
- Adams, A. E., and J. R. Pringle, 1984. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J. Cell Biol. 98: 934–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpe, G., L. W. Zhang, H. Zheng and J. G. Culotti, 2004. Conversion of cell movement responses to Semaphorin-1 and Plexin-1 from attraction to repulsion by lowered levels of specific RAC GTPases in C. elegans. Development 131: 2073–2088. [DOI] [PubMed] [Google Scholar]
- de Vetten, N., F. Quattrocchio, J. Mol and R. Koes, 1997. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants and animals. Genes Dev. 11: 1422–1434. [DOI] [PubMed] [Google Scholar]
- Dickson, B. J., 2001. Rho GTPases in growth cone guidance. Curr. Opin. Neurobiol. 11: 103–110. [DOI] [PubMed] [Google Scholar]
- Dickson, B. J., 2002. Molecular mechanisms of axon guidance. Science 298: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Dong, Y., D. Pruyne and A. Bretscher, 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell Biol. 161: 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, H. F., and D. C. Shakes, 1995. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press, New York.
- Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon et al., 1996. Life with 6000 genes. Science 274: 546, 563–567. [DOI] [PubMed] [Google Scholar]
- Hall, A., 1998. Rho GTPases and the actin cytoskeleton. Science 279: 509–514. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., and J. Ahringer, 2003. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30: 313–321. [DOI] [PubMed] [Google Scholar]
- Kishore, R. S., and M. V. Sundaram, 2002. ced-10 Rac and mig-2 function redundantly and act with unc-73 trio to control the orientation of vulval cell divisions and migrations in Caenorhabditis elegans. Dev. Biol. 241: 339–348. [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., 2003. Rac proteins and the control of axon development. Curr. Opin. Neurobiol. 13: 384–390. [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., R. K. Herman, J. E. Shaw and C. I. Bargmann, 1998. UNC-115, a conserved protein with predicted LIM and actin-binding domains, mediates axon guidance in C. elegans. Neuron 21: 385–392. [DOI] [PubMed] [Google Scholar]
- Lundquist, E. A., P. W. Reddien, E. Hartwieg, H. R. Horvitz and C. I. Bargmann, 2001. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128: 4475–4488. [DOI] [PubMed] [Google Scholar]
- Michaelson, D., J. Silletti, G. Murphy, P. D'Eustachio, M. Rush et al., 2001. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 152: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer, E. J., C. J. Schmidt, R. Nambudripad and T. F. Smith, 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371: 297–300. [DOI] [PubMed] [Google Scholar]
- Ng, J., T. Nardine, M. Harms, J. Tzu, A. Goldstein et al., 2002. Rac GTPases control axon growth, guidance and branching. Nature 416: 442–447. [DOI] [PubMed] [Google Scholar]
- Otieno, C. J., J. Bastiaansen, A. M. Ramos and M. F. Rothschild, 2005. Mapping and association studies of diabetes related genes in the pig. Anim. Genet. 36: 36–42. [DOI] [PubMed] [Google Scholar]
- Pankov, R., Y. Endo, S. Even-Ram, M. Araki, K. Clark et al., 2005. A Rac switch regulates random versus directionally persistent cell migration. J. Cell Biol. 170: 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftopoulou, M., and A. Hall, 2004. Cell migration: Rho GTPases lead the way. Dev. Biol. 265: 23–32. [DOI] [PubMed] [Google Scholar]
- Reddien, P. W., and H. R. Horvitz, 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2: 131–136. [DOI] [PubMed] [Google Scholar]
- Shakir, M. A., J. S. Gill and E. A. Lundquist, 2006. Interactions of UNC-34 Enabled with Rac GTPases and the NIK kinase MIG-15 in Caenorhabditis elegans axon pathfinding and neuronal migration. Genetics 172: 893–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, T. F., C. Gaitatzes, K. Saxena and E. J. Neer, 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185. [DOI] [PubMed] [Google Scholar]
- Sondek, J., A. Bohm, D. G. Lambright, H. E. Hamm and P. B. Sigler, 1996. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature 379: 369–374. [DOI] [PubMed] [Google Scholar]
- Soto, M. C., H. Qadota, K. Kasuya, M. Inoue, D. Tsuboi et al., 2002. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 16: 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven, R., T. J. Kubiseski, H. Zheng, S. Kulkarni, J. Mancillas et al., 1998. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92: 785–795. [DOI] [PubMed] [Google Scholar]
- Struckhoff, E. C., and E. A. Lundquist, 2003. The actin-binding protein UNC-115 is an effector of Rac signaling during axon pathfinding in C. elegans. Development 130: 693–704. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne, M., and C. S. Goodman, 1996. The molecular biology of axon guidance. Science 274: 1123–1133. [DOI] [PubMed] [Google Scholar]
- Timmons, L., D. L. Court and A. Fire, 2001. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263: 103–112. [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and C. D'Souza-Schorey, 1997. Rho GTPases and signaling networks. Genes Dev. 11: 2295–2322. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares, M., D. J. Webb and A. R. Horwitz, 2005. Cell migration at a glance. J. Cell Sci. 118: 4917–4919. [DOI] [PubMed] [Google Scholar]
- Walker, A. R., P. A. Davison, A. C. Bolognesi-Winfield, C. M. James, N. Srinivasan et al., 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J. G., E. Southgate, J. N. Thomson and S. Brenner, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Yang, Y., and E. A. Lundquist, 2005. The actin-binding protein UNC-115/abLIM controls formation of lamellipodia and filopodia and neuronal morphogenesis in Caenorhabditis elegans. Mol. Cell. Biol. 25: 5158–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T. W., and C. I. Bargmann, 2001. Dynamic regulation of axon guidance. Nat. Neurosci. 4(Suppl.): 1169–1176. [DOI] [PubMed] [Google Scholar]
- Zipkin, I. D., R. M. Kindt and C. J. Kenyon, 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90: 883–894. [DOI] [PubMed] [Google Scholar]