Abstract
The nuclear pore complex (NPC) is embedded in the nuclear envelope where it mediates transport between the cytoplasm and nucleus and helps to organize nuclear architecture. We previously isolated sonB1, a mutation encoding a single amino acid substitution within the Aspergillus nidulans SONBnNup98 NPC protein (nucleoporin). Here we demonstrate that this mutation causes marked DNA damage sensitivity at 42°. Although SONBnNup98 has roles in the G2 transition, we demonstrate that the G2 DNA damage checkpoint is functional in the sonB1 mutant at 42°. The MRN complex is composed of MRE11, RAD50, and NBS1 and functions in checkpoint signaling, DNA repair, and telomere maintenance. At 42° we find that the DNA damage response defect of sonB1 mutants causes synthetic lethality when combined with mutations in scaANBS1, the A. nidulans homolog of NBS1. We provide evidence that this synthetic lethality is independent of MRN cell cycle checkpoint functions or MREAMRE11-mediated DNA repair functions. We also demonstrate that the single A. nidulans histone H2A gene contains the C-terminal SQE motif of histone H2AX isoforms and that this motif is required for the DNA damage response. We propose that the sonB1 nucleoporin mutation causes a defect in a novel part of the DNA damage response.
THE nuclear pore complex (NPC) is an evolutionarily conserved structure made up of multiple copies of ∼30 different NPC proteins (nucleoporins) embedded in the nuclear envelope (for review see Hetzer et al. 2005; Tran and Wente 2006). The NPC restricts diffusion of proteins and nucleic acids between the nucleus and cytoplasm and facilitates active nucleocytoplasmic transport through the nuclear envelope. Other roles for the NPC are only just beginning to be understood. For example, in Saccharomyces cerevisiae the NPC has been demonstrated to play roles in tethering telomeres to the nuclear periphery, which helps to facilitate transcriptional silencing of subtelomeric genes (Galy et al. 2000; Feuerbach et al. 2002; Therizols et al. 2006). Somewhat paradoxically, certain nucleoporins have been demonstrated to preferentially associate with transcriptionally active genes (Ishii et al. 2002; Casolari et al. 2004, 2005; Dilworth et al. 2005; Menon et al. 2005; Schmid et al. 2006). Interestingly, budding yeast nucleoporin null alleles that display sensitivity to DNA-damaging agents have been identified (Galy et al. 2000; Bennett et al. 2001; Chang et al. 2002; Loeillet et al. 2005; Therizols et al. 2006). Although the mechanism leading to DNA damage sensitivity of these nucleoporin nulls is currently not known, it is likely that NPC function is required for the normal DNA damage response.
In response to DNA damage, cells both activate DNA repair pathways and enforce checkpoints to arrest cell cycle progression until DNA has been repaired (for review see Zhou and Elledge 2000; McGowan and Russell 2004). In the presence of DNA damage, the G2 DNA damage checkpoint prevents mitotic entry via tyrosine phosphorylation of the cyclin-dependent kinase Cdc2 (Cdk1 or NIMXCdc2 in Aspergillus nidulans). Tyrosine-phosphorylated Cdc2 must be dephosphorylated for mitosis to occur and mutation of tyrosine to nonphosphorylatable phenylalanine (cdc2F mutants) results in premature mitotic entry in the presence of DNA damage (Ye et al. 1997; for review see Zhou and Elledge 2000). The evolutionarily conserved phosphatidyl inositol 3-kinase-like kinases (PIKK) ATR and ATM function to signal both repair and checkpoint pathways in response to DNA damage. The central importance of ATR and ATM is underscored by human diseases such as ataxia telangiectasia, which result from mutation of these genes. More recently the MRE11, RAD50, NBS1 (MRN) complex has been demonstrated to function early in the DNA damage response together with the ATM and ATR kinases, and mutations in MRN genes have been linked to the diseases Nijmegen breakage syndrome (NBS) and ataxia telangiectasia-like disorder (Uziel et al. 2003; Difilippantonio et al. 2005; Lee and Paull 2005; Stiff et al. 2005; You et al. 2005; Jazayeri et al. 2006; for review see Abraham and Tibbetts 2005; Stavridi and Halazonetis 2005; Zhang et al. 2006). The MRN complex has roles in cell cycle checkpoint signaling as well as in DNA repair and telomere maintenance (for review see D'Amours and Jackson 2002; Zhang et al. 2006). Mre11p has DNA nuclease, strand dissociation, and strand annealing activities, while RAD50 has similarity to structural maintenance of chromosome proteins and is thought to form a dimer that bridges DNA strands at a double-strand break (for review see D'Amours and Jackson 2002; Stavridi and Halazonetis 2005; Zhang et al. 2006). The precise function of NBS1 is less clear although it contains a forkhead-associated (FHA) and breast cancer C terminus (BRCT) domain, suggesting that it binds phosphorylated proteins (Becker et al. 2006; for review see D'Amours and Jackson 2002; Stavridi and Halazonetis 2005; Zhang et al. 2006). Indeed, NBS1 has been demonstrated to bind the γ-H2AX phosphoserine epitope, which is phosphorylated early in the DNA damage response by the ATM/ATR kinases in nucleosomes surrounding DNA damage (Downs et al. 2000; Kobayashi et al. 2002; Celeste et al. 2003; Nakamura et al. 2004; Unal et al. 2004; for review see Vidanes et al. 2005).
A. nidulans has long been utilized as a model genetic system and the cell cycle and DNA damage response in this organism is well characterized (for review see Osmani and Ye 1996; Goldman et al. 2002; Goldman and Kafer 2004; Osmani and Mirabito 2004). Temperature-sensitive mutants of the A. nidulans nimA kinase reversibly arrest in G2 at the nonpermissive temperature of 42° even though the Cdc2/cyclinB kinase is fully activated (Osmani et al. 1987). This is likely because the Cdc2/cyclinB kinase is cytoplasmic at a nimA1 G2 arrest and cannot enter the nucleus (Wu et al. 1998). We have previously isolated mutations in two nucleoporins, SONAGle2 and SONBnNup98, which suppress a nimA1 G2 arrest and allow entry into mitosis (Wu et al. 1998; De Souza et al. 2003). Both SONAGle2 and SONBnNup98 disperse from the NPC during the partial disassembly of the NPC in A. nidulans (De Souza et al. 2004). It is likely that these NPC mutants suppress the nimA1 G2 arrest by allowing sufficient Cdc2/cyclinB and tubulin into the nucleus to allow mitotic entry (Wu et al. 1998; De Souza et al. 2004). Here we show that the sonB1 mutation displays a high degree of sensitivity to DNA-damaging agents at 42° but that this DNA damage sensitivity is independent of the G2 DNA damage checkpoint. Epistasis analysis indicates that SONBnNup98 functions on a different pathway of the DNA damage response from those involving UVSCRad51, UVSHRad18, γ-H2AX phosphorylation, and the G2 DNA damage checkpoint. The defect in sonB1 mutants that causes DNA damage sensitivity also results in synthetic lethality at 42° when combined with mutations in scaANBS1, which encodes the A. nidulans homolog of NBS1 (Bruschi et al. 2001; Semighini et al. 2003). Similar synthetic lethality was not observed between sonB1 and mreAMRE11 mutants, suggesting that sonB1 synthetic lethality with scaANBS1 mutants is independent of the DNA repair activities of the MRN complex. We provide evidence that the synthetic lethality between sonB1 and scaANBS1 mutants is also independent of the cell cycle checkpoint functions of SCAANBS1. Our data suggest that the SONBnNup98 nucleoporin may have a novel role in the DNA damage response.
MATERIALS AND METHODS
General techniques:
Media and general techniques for A. nidulans culture, transformation, and DAPI staining for chromosome mitotic index were as previously described (Osmani et al. 1987, 1991, 1994; Oakley and Osmani 1993; Ye et al. 1995; Wu et al. 1998). Western analysis was carried out preparing lysates in sample buffer containing 6 m urea as described previously (De Souza et al. 2000). The phospho-Cdc2 (Tyr15) antibody was purchased from Cell Signaling Technology.
DNA damage sensitivity assays:
Quiescent conidiospores and germlings were tested for sensitivity to UV irradiation as previously described (Ye et al. 1997) using a microprocessor-controlled UV crosslinker (FBUVXL-1000; Fischer Biotech; 254 nm). Hydroxyurea, 1,2,7,8 diepoxyoctane (DEO), methyl methanesulfonate (MMS), and camptothecin were all purchased from Sigma (St. Louis) and added to media at the appropriate concentrations immediately prior to pouring plates. All plates were prewarmed to 32° or 42° as appropriate prior to inoculation. Entry into mitosis after MMS treatment of either conidiospores or germlings arrested at the G2 nimA5 arrest point was as described previously (Ye et al. 1997).
Plasmid constructs:
Serine 129 of the A. nidulans histone H2A gene (May and Morris 1987) in plasmid pRG3-H2A-H2B (De Souza et al. 2003) was mutated to alanine using the Quickchange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to generate plasmid pRG3 H2A S129A-H2B. Introduction of the appropriate mutation was confirmed by sequencing.
A. nidulans strains:
Genotypes of strains used in this study are listed in supplemental Table 1 at http://www.genetics.org/supplemental/. Although the mreAMRE11 disruption strain (TMRE) was previously reported as sterile (Semighini et al. 2003), we were able obtain viable progeny in sexual crosses. The H2A S129A mutant was constructed by a two-step gene replacement (Ye et al. 1996). GR5 (pyrG89, wA3, pyroA4) and CDS40 (pyrG89, wA2, pyroA4, sonB1) were transformed with plasmid pRG3 H2A S129A-H2B and single-site integration at the histone H2A/H2B locus confirmed by Southern blot analysis and PCR using primers external to the region of duplication (Yang et al. 2004). Plasmid loss was selected for with 5-fluoroorotic acid (Osmani et al. 1994) and evictants maintaining the H2A S129A mutation were selected for by screening for DNA damage sensitivity. Introduction of the H2A S129A mutation was confirmed by PCR amplification and sequencing of the histone H2A locus. Double-mutant strains generated between nucleoporin mutants and DNA damage response mutants were confirmed by crossing strains back to a wild-type strain to recover the single-mutant phenotypes. Strains IM69, MKF11, SCA299-16, and TMRE were kind gifts from Gustavo Goldman (Universidade de São Paulo, São Paulo, Brazil).
RESULTS
The sonB1 NPC mutant is highly DNA damage sensitive at 42°:
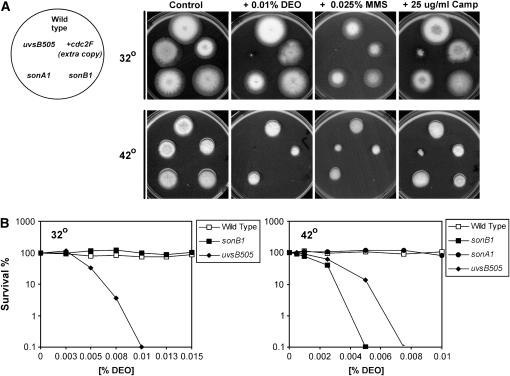
The nimA1 temperature-sensitive mutation causes cells to arrest in G2 of the cell cycle at the restrictive temperature of 42°. Intriguingly, we previously isolated single point mutations in two essential NPC proteins, SONAGle2 and SONBnNup98, which suppress the nimA1 G2 arrest and allow mitotic entry at 42° (Wu et al. 1998; De Souza et al. 2003). This suggests that the sonA1 and sonB1 NPC mutants are defective in some aspect of G2 regulation at 42°. As loss of G2 checkpoint functions over mitotic entry can cause DNA damage sensitivity, we tested the ability of sonA1 and sonB1 mutants to form a colony in the presence of DNA-damaging agents. Wild-type and the G2 checkpoint-deficient uvsB505ATR and cdc2F mutant strains were used as controls (Ye et al. 1997; De Souza et al. 1999). Strikingly, the sonB1 mutant displayed marked sensitivity to the DNA alkylating agents MMS and DEO at 42° but behaved similarly to wild type at 32° (Figure 1A). Similar results were obtained using survival assays that indicated that the sonB1 mutant was more sensitive than the uvsB505ATR mutant to DEO at 42o (Figure 1B). Moreover, this DNA damage sensitivity was specific to the sonB1 mutant as the sonA1 nucleoporin mutant remained viable at 42° in the presence of DEO or MMS (Figure 1, A and B). Importantly, the DNA damage sensitivity of sonB1 mutants at 42° was not due to general cellular stress as sonB1 mutants were not sensitive to nocodazole, or camptothecin at 42° (Figure 1A; data not shown). Thus the sonB1 mutation causes a defect in the DNA damage response specifically at the temperature at which it suppresses the nimA1 G2 arrest. It is particularly intriguing that a single amino acid substitution within an essential NPC protein (De Souza et al. 2003) would cause sensitivity to DNA-damaging agents.
Figure 1.—
The sonB1 mutant is highly sensitive to DNA-damaging agents at 42°. (A) Colony formation of wild-type (R153), uvsB505ATR (CDS314), cdc2F (FRY24), sonA1 (CDS365), and sonB1 (CDS364) strains under the indicated conditions. Plates were incubated for 3 days. Note that the sectoring of the cdc2F mutant at 32° in the presence of DEO or camptothecin is due to loss of the cdc2F allele by plasmid eviction, leaving the wild-type allele. (B) Viability of the indicated strains (250 conidiospores spread/plate, two plates/strain) at 32° and 42° in the presence of different concentrations of the DNA alkylating agent DEO. Viability was assessed after 3 days incubation.
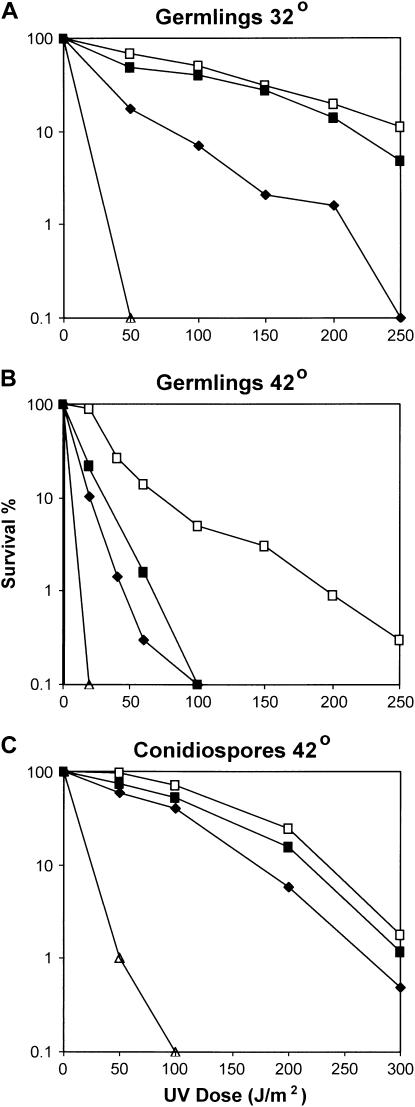
In A. nidulans, mutants with defective G2 checkpoint functions are sensitive to DNA damage only if damage is elicited after cells have entered the cell cycle but not if damage is elicited to quiescent conidiospores (Ye et al. 1997; De Souza et al. 1999). This is because after DNA damage has occurred, quiescent conidiospores take several hours to enter the cell cycle, allowing time for DNA repair before DNA replication and mitotic entry. We therefore compared the UV irradiation sensitivity of sonB1 germlings and quiescent conidiospores at 42°. The cdc2AF mutant, which is defective in the G2 checkpoint over mitotic entry (Ye et al. 1997), was also tested as a control. The sonB1 mutant behaved similarly to the cdc2AF mutant with germlings, but not conidiospores, displaying sensitivity to UV irradiation at 42° (Figure 2). In contrast, both germlings and conidiospores of the uvsH304Rad18 DNA-repair-deficient mutant (Kafer and Mayor 1986; Yoon et al. 1995) were sensitive to UV irradiation elicited at 42°. Notably, the UV irradiation sensitivity of sonB1 germlings was restricted to 42° (Figure 2), similar to the case for sonB1 sensitivity to MMS and DEO (Figure 1). These data are consistent with the sonB1 mutant potentially having a defective G2 DNA damage checkpoint at 42°, the temperature at which this mutation suppresses the nimA1 G2 arrest.
Figure 2.—
Differential UV sensitivity of sonB1 quiescent conidiospores and germlings at 42°. Conidiospores (250/plate, two plates/strain) of (□) wild type (GR5), (♦) cdc2AF (FRY20-1), (▪) sonB1 (CDS40), and (▵) uvsH304Rad18 (A329) were spread onto plates and either were allowed to germinate at (A) 32° or (B) 42° for 6 hr prior to UV irradiation or (C) were immediately UV irradiated. After irradiation, plates were incubated at the indicated temperatures for 2 days to allow colony formation. The percentage survival after UV irradiation is expressed as the percentage of colonies produced in the absence of treatment. This experiment was performed twice with similar results.
In sonB1 mutants, Cdc2 undergoes tyrosine phosphorylation in response to DNA damage:
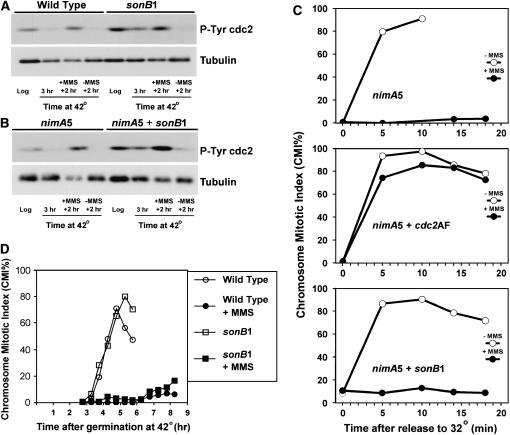
Similar to humans, the G2 DNA damage checkpoint prevents mitotic entry through a pathway leading to tyrosine phosphorylation of Cdc2 in A. nidulans (Ye et al. 1997). We therefore examined the ability of sonB1 mutants to tyrosine phosphorylate Cdc2 in response to DNA damage at 42°. Asynchronous wild-type or sonB1 log-phase cultures were shifted to 42° for 3 hr and then either treated or not treated with MMS for an additional 2 hr at 42°. Lysates were prepared at each stage and immunoblotted with an antibody specific for tyrosine-phosphorylated Cdc2. Cdc2 tyrosine phosphorylation increased similarly in either wild-type or the sonB1 mutant in response to DNA damage (Figure 3A). To ensure that cells were in G2 prior to the addition of MMS, we also performed this experiment in strains that contained the nimA5 temperature-sensitive mutation, which arrests cells in G2 at the restrictive temperature of 42° (Osmani et al. 1987). At nimA5 arrest, Cdc2 was not tyrosine phosphorylated in the absence of DNA damage, but became tyrosine phosphorylated following the addition of MMS (Figure 3B), consistent with our previous results (Ye et al. 1997). As shown in Figure 3B, sonB1 mutants were able to tyrosine phosphorylate Cdc2 in response to DNA damage elicited at the G2 nimA5 arrest point. This provides strong evidence that the G2 DNA damage checkpoint pathway leading to Cdc2 tyrosine phosphorylation is functional in sonB1 mutants at 42°. Interestingly, sonB1 mutants displayed a higher level of tyrosine-phosphorylated cdc2 compared to wild-type strains (Figure 3, A and B). As this was observed at 32° as well as at 42°, this effect is likely not related to the DNA damage sensitivity of sonB1 mutants at 42° but may reflect the demonstrated defect of sonB1 mutants in G2/M regulation (De Souza et al. 2003).
Figure 3.—
sonB1 mutants undergo tyrosine phosphorylation of Cdc2 and arrest in G2 in response to DNA damage. (A and B) Log-phase wild-type (GR5), sonB1 (CDS40), nimA5 (SO6), and nimA5 sonB1 (CDS119) cultures were shifted to 42° for 3 hr. Cultures were then divided into two and grown for another 2 hr in the presence or absence of 0.04% MMS. The relative levels of tyrosine 15-phosphorylated Cdc2 were determined by immunoblotting with an antibody specific for this epitope at the indicated time points. Levels of tubulin are shown as a loading control. (C) Chromosome mitotic index (CMI) of nimA5 (SO54), nimA5 cdc2AF (AT27), and nimA5 sonB1 (CDS119) germlings treated or not with 0.025% MMS during a nimA5 G2 arrest (42°) prior to release to nimA5 permissive temperature (32°) in the absence of MMS. Note that nimA5 and nimA5 + sonB1 cells did not display an increase in CMI percentage even 30 min following release from nimA5 arrest in the presence of MMS. (D) Wild-type (GR5) and sonB1 (CDS40) conidiospores were germinated in the presence or absence of 0.01% MMS and the CMI was determined by DAPI staining. Nocodazole (5 μg/ml) was included to prevent mitotic exit once cells entered mitosis.
While the above data demonstrate that sonB1 mutants are able to tyrosine phosphorylate Cdc2 in response to DNA damage, it is still possible that just enough Cdc2/cyclinB is able to enter the nucleus, be activated, and allow inappropriate mitotic entry. To determine if this is the case, we next examined if sonB1 mutants enter mitosis prematurely if DNA is damaged. To do this, we first synchronized cells at the nimA5 G2 arrest point (Ye et al. 1997). These G2-arrested cells were either treated or not treated with MMS to elicit DNA damage. Cells were then released to the nimA5 permissive temperature of 32° by media exchange and mitotic entry followed by examining cells for condensed DNA at time points after release. As expected, the nimA5 control delayed mitotic entry in the presence of DNA damage while cells also containing the cdc2AF mutation entered mitosis similarly in the presence or absence of DNA damage (Figure 3C). Cells containing the sonB1 mutation delayed mitotic entry in the presence of DNA damage (Figure 3C), consistent with these cells having an intact G2 DNA damage checkpoint. However, as sonB1 mutants are not DNA damage sensitive at 32° (Figure 1), it is possible that the delay in mitotic entry of the nimA5 sonB1 mutant in these experiments was due to a rapid reactivation of sonB1 upon shifting cultures to 32°. To determine if sonB1 mutants delay mitotic entry if cells are maintained at 42°, we germinated wild-type or sonB1 conidiospores at 42° in the presence or absence of MMS and followed entry into the first mitosis. Under these conditions, sonB1 mutants delayed entry into mitosis in the presence of DNA damage similarly to a wild-type strain (Figure 3D). Together, these results indicate that sonB1 mutants have a functional G2 DNA damage checkpoint, even though sonB1 germlings are more sensitive to UV irradiation than are sonB1 quiescent conidiospores.
sonB1 mutants display synthetic lethality with scaANBS1 mutants at 42° without DNA damage:
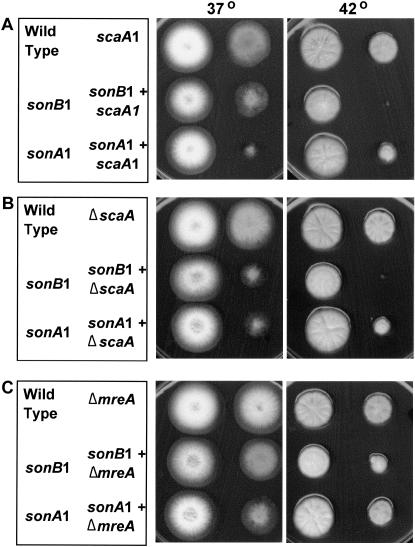
The above indicates that the sonB1 mutant is defective in some part of the DNA damage response other than the G2 DNA damage checkpoint. To further investigate this, we determined if sonB1 mutants genetically interacted with mutants defective in different aspects of the DNA damage response in A. nidulans (Kafer and Mayor 1986; Yoon et al. 1995; Kafer and May 1997; van Heemst et al. 1997; Ye et al. 1997; De Souza et al. 1999; Hofmann and Harris 2000; Bruschi et al. 2001; Semighini et al. 2003; Malavazi et al. 2006; Nayak et al. 2006). Most striking were the genetic interactions between sonB1 and mutants of the A. nidulans NBS1 ortholog scaANBS1 (Bruschi et al. 2001). Notably, sonB1 mutants were not viable at 42° when combined with the scaA1NBS1 mutation, even without addition of any genotoxic agent (Figure 4A). Although sonB1 scaA1NBS1 double mutants formed smaller colonies at 32° and 37° than either single mutant, the synthetic lethality was restricted to 42°, the same temperature at which sonB1 mutants display DNA damage sensitivity (Figure 4A). The scaA1 mutation is predicted to result in a truncated SCAANBS1 protein due to a single base transversion introducing a stop codon in the scaA1 reading frame (Bruschi et al. 2001). To determine if the temperature-dependent synthetic lethality between sonB1 and scaA1NBS1 was due to complete loss of SCAANBS1 function, we constructed a double mutant between sonB1 and a null allele of scaA (Semighini et al. 2003). The sonB1 ΔscaANBS1 and sonB1 scaA1NBS1 double mutants behaved identically, confirming that SCAANBS1 function is required for sonB1 survival at 42°. SCAANbs1 is a component of the MRN complex, which consists of MRE11, RAD50, and NBS1 (Semighini et al. 2003; for review see D'Amours and Jackson 2002; d'adda di Fagagna et al. 2004; Stavridi and Halazonetis 2005; Zhang et al. 2006). We next determined if sonB1 displayed a similar synthetic lethal interaction with a disrupted allele of mreAMRE11, the A. nidulans ortholog of MRE11 (Semighini et al. 2003). In contrast to the synthetic lethality observed with scaANBS1 mutants, sonB1 mreAMRE11 double mutants were able to form a colony at 42° (Figure 4C). The fact that MRE11 encodes the DNA nuclease, strand annealing, and strand dissociation activity of the MRN complex (Semighini et al. 2003; for review see D'Amours and Jackson 2002) suggests that the synthetic lethality between sonB1 and the scaANBS1 mutants at 42° is independent of these MRN complex activities.
Figure 4.—
The sonB1 mutant is synthetically lethal with scaANBS1 mutants at 42°. Wild type and the indicated single or double mutants were inoculated on plates and grown at either 37° (2 days) or 42° (3 days). Note that sonB1 displays synthetic lethality with either (A) scaA1NBS1 or (B) the scaANBS1 null at 42° but not with the (C) mreAMRE11 mutant. Strains used were CDS323, CDS324, CDS326, CDS350, CDS351, CDS364, CDS365, CDS373, CDS374, CDS375, MKF11, and R153 and are listed in supplemental Table S1 at http://www.genetics.org/supplemental/.
Both the sonA1 and sonB1 NPC mutants suppress the nimA1 G2 arrest at 42°. SONAGle2 and SONBnNup98 directly bind each other and the mutations in these genes that suppress nimA1 likely do so by similar mechanisms (De Souza et al. 2003). However, these NPC mutants differ in that only sonB1, and not sonA1, is sensitive to DNA-damaging agents at 42° (Figure 1). We therefore determined whether the sonA1 mutation genetically interacted with MRN complex mutants in a similar manner to sonB1 at 42°. As with sonB1 mutants, sonA1 mutants did not display synthetic lethality with the mreAMRE11 disruption at 42° (Figure 4C). Additionally, and in contrast to the synthetic lethality between sonB1 and scaANBS1 mutants at 42°, both the sonA1 scaANBS1 null and sonA1 scaA1NBS1 double mutants were viable at 42° (Figure 4, A and B). Together, these data suggest that sonB1 synthetic lethality with the scaANbs1 mutants at 42° is likely independent of the sonB1 mutant defect that suppresses a nimA1 G2 arrest. Rather, sonB1 scaANBS1 mutant synthetic lethality at 42° may result from combining the DNA damage response defect of sonB1 with lack of SCAANBS1 function. However, this interpretation should be viewed with caution as the sonA1 mutant displayed synthetic sickness with scaANBS1 and mreAMRE11 mutants at 37° (Figure 4), perhaps suggesting a more general genetic interaction between the MRN complex and the SONAGle2/SONBnNup98 NPC subcomplex.
Synthetic lethality between sonB1 and scaANBS1 mutants at 42° is independent of the scaANBS1 checkpoint functions:
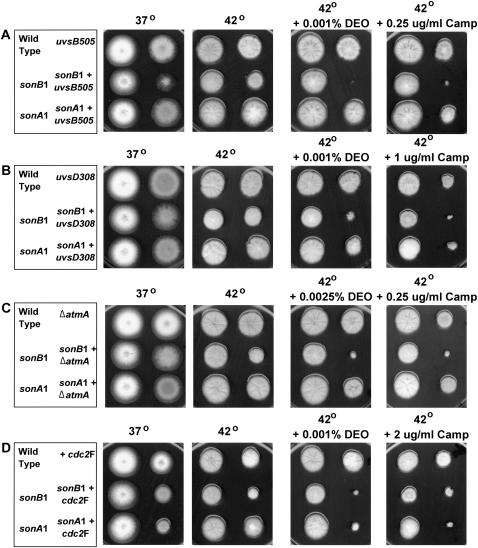
One of the functions of NBS1 is to regulate checkpoint pathways in response to DNA damage and this function is conserved for A. nidulans SCAANBS1 (Semighini et al. 2003). The role of NBS1 in checkpoint regulation involves the ATR/ATRIP complex and/or ATM and ultimately prevents mitotic entry by tyrosine 15 phosphorylation of Cdc2 (for review see Zhou and Elledge 2000). We rationalized that if loss of checkpoint function in scaANBS1 mutants was causing synthetic lethality with sonB1 mutants, sonB1 mutants should also show synthetic lethality with other checkpoint-deficient mutants. However, in contrast to sonB1 synthetic lethality with scaANBS1 mutants at 42°, sonB1 was viable at 42° when combined with the uvsB505ATR, uvsD308ATRIP, cdc2F, or ΔatmAATM checkpoint-deficient mutants (Figure 5). Therefore, it is unlikely that loss of checkpoint function in scaANBS1 mutants causes synthetic lethality with sonB1 at 42°.
Figure 5.—
The sonB1 mutant is not synthetically lethal with checkpoint-deficient mutants at 42°. (A–D) Wild type and the indicated single or double mutants were inoculated on plates and grown at either 37° (2 days) or 42° (3 days). Colony formation of strains in the presence of the DNA-damaging agents DEO or camptothecin was also evaluated as indicated. Strains used were A574, CDS204, CDS207, CDS293, CDS314, CDS319, CDS320, CDS353, CDS364, CDS365, CDS366, CDS367, CDS369, FRY24, and R153 and are listed in supplemental Table S1 at http://www.genetics.org/supplemental/.
Although the sonB1 mutant was viable in combination with the above checkpoint-deficient mutants, sonB1 did significantly increase the DNA damage sensitivity of uvsB505ATR, uvsD308ATRIP, cdc2F, and ΔatmAATM at 42° (Figure 5). This is consistent with the sonB1 mutation affecting a different part of the DNA damage response from the G2 DNA damage checkpoint, supporting our earlier conclusion (Figure 2).
The sonA1 nucleoporin mutant was viable at 42° in combination with all checkpoint mutants tested (Figure 5). However, interestingly, the sonA1 mutation increased the DNA damage sensitivity of the cdc2F mutant (Figure 5D). Given that the sonA1 nucleoporin mutation allows Cdc2/cyclinB into the nucleus at 42° during a nimA1 arrest (Wu et al. 1998), it is likely that increased nuclear access of active Cdc2F/cyclinB in the sonA1 cdc2F double mutant leads to increased DNA damage sensitivity. Somewhat surprisingly, the sonA1 mutation did not increase the DNA damage sensitivity of uvsB505ATR, uvsD308ATRIP, or ΔatmAATM mutants, which are defective in the pathway leading to tyrosine phosphorylation of Cdc2. This may reflect the relative levels of nontyrosine-phosphorylated Cdc2 in uvsB505ATR, uvsD308ATRIP, or ΔatmAATM mutants compared with that of the cdc2F mutant under these conditions.
The sonB1 mutation does not cause DNA damage:
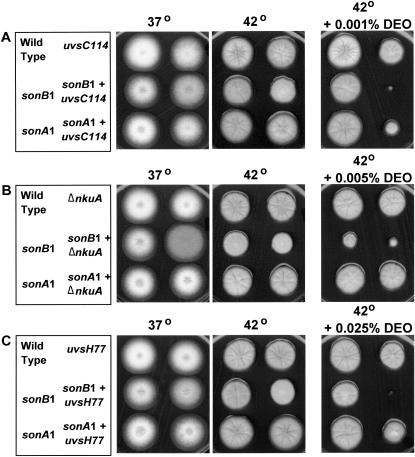
Another possibility to explain the synthetic lethal interaction between sonB1 and scaANBS1 mutants is that the sonB1 mutation itself leads to DNA double-strand breaks, which are not repaired in the absence of SCAANBS1 function. If this were the case, sonB1 mutants should also display synthetic lethality at 42° with mutants deficient in the repair of DNA double-strand breaks. DNA double-strand breaks are repaired by either homologous recombination or nonhomologous end joining (NHEJ). In A. nidulans, the uvsCRad51 gene encodes a Rad51 ortholog and uvsCRad51 mutants are sensitive to DNA double-strand breaks and display defects in homologous recombination (Chae and Kafer 1997; Seong et al. 1997; van Heemst et al. 1997; Ichioka et al. 2001). We generated the sonB1 uvsC114Rad51 double mutant, which was viable at 42° (Figure 6A), arguing that the sonB1 mutation does not lead to DNA double-strand breaks. Similarly, deletion of nkuAKu70, the A. nidulans ortholog of the Ku70 gene that functions in NHEJ in other systems (for review see Hopfner et al. 2002), had no effect on sonB1 viability at 42° (Figure 6B). However, surprisingly, the nkuAKu70 null displays no sensitivities to DNA-damaging agents, indicating that Ku70-mediated NHEJ may play only a minor role in the DNA damage response in A. nidulans or that there is a second NHEJ pathway functioning without nkuAKu70 (Nayak et al. 2006). We next tested the viability of the sonB1 mutation when combined with the A. nidulans uvsH77Rad18 postreplication repair-deficient mutant (Yoon et al. 1995). The sonB1 uvsH77Rad18 double mutant was viable at 42°, providing further evidence that the sonB1 mutation does not cause DNA damage (Figure 6C).
Figure 6.—
The sonB1 mutant is not synthetically lethal with DNA-repair-deficient mutants at 42°. (A–C) Wild type and the indicated single or double mutants were inoculated on plates and grown at either 37° (2 days) or 42° (3 days). Colony formation of strains in the presence of the DNA-damaging agent DEO was also evaluated as indicated. Strains used were CDS260, CDS261, CDS311, CDS315, CDS330, CDS352, CDS368, CDS370, CDS364, CDS365, R153, and TN02 are listed in supplemental Table S1 at http://www.genetics.org/supplemental/.
Interestingly, sonB1 uvsC114Rad51 and sonB1 uvsH77Rad18 double mutants were not viable at 42° in the presence of concentrations of DEO at which the respective single mutants formed viable colonies (Figure 6, A and C). Moreover, this effect was specific to the sonB1 mutant and was not observed with the sonA1 mutant (Figure 6). The sonB1 nkuA null double mutant did not display any significant increase in sensitivity to DEO or camptothecin (Figure 6; data not shown) in comparison to the single mutants. These data provide further evidence that SONBnNup98 has a role in the DNA damage response and indicate that this function is likely on a different pathway than either UVSCRad51 or UVSHRad18.
γ-H2AX phosphorylation has a role in the DNA damage response independent of SONBnNup98:
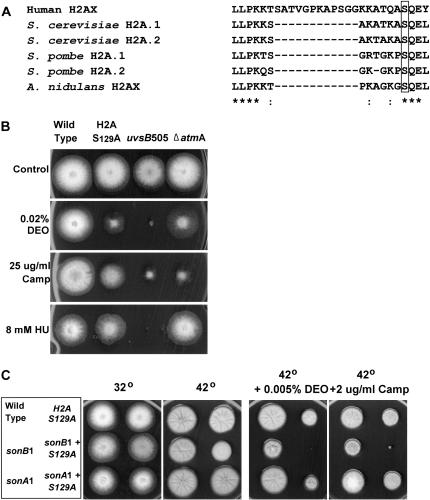
We have previously shown that the histone H2A/H2B gene pair acts as a copy-number suppressor of sonB1 cold sensitivity and sensitivity to hydroxyurea at 32°, but has no effect on the sonA1 mutant (De Souza et al. 2003). Phosphorylation of a conserved serine in the histone H2AX variant (to generate γ-H2AX) in nucleosomes located near sites of DNA double-strand breaks has important roles in the DNA damage response, including regulating MRN complex localization to sites of DNA damage (Kobayashi et al. 2002; Celeste et al. 2003; for review see Vidanes et al. 2005). This conserved serine of histone H2AX isoforms is present near the C terminus of the single A. nidulans histone H2A gene (Figure 7A) (May and Morris 1987), which we term H2AX. Given the genetic interaction between sonB1 and histone H2AX/H2B, and the role of γ-H2AX phosphorylation for MRN complex function, we determined the effect of preventing γ-H2AX phosphorylation on sonB1 mutants at 42°. We endogenously replaced the conserved serine in the C-terminal of H2AX with alanine to generate a H2AX-S129A mutant that was viable. The DNA damage sensitivity of the H2AX-S129A mutant was compared to mutants in the uvsBATR and atmAATM PIKK kinases, orthologs of which phosphorylate H2AX in other systems (Downs et al. 2000; Celeste et al. 2003; Nakamura et al. 2004). The H2AX-S129A mutant was sensitive to DEO and camptothecin (Figure 7B), demonstrating for the first time in A. nidulans that serine 129 is required for the DNA damage response and is likely phosphorylated in response to DNA damage. The H2AX-S129A mutant displayed no sensitivity to hydroxyurea (Figure 7B), indicating that serine 129 is not essential for the response to a slow S-phase in A. nidulans. The H2AX-S129A mutant was less sensitive than the uvsB505ATR mutant to DNA-damaging agents, consistent with UVSBATR having roles in the DNA damage response in addition to γ-H2AX phosphorylation. In contrast to uvsB505ATR, the atmAATM null was more sensitive than the H2AX-S129A mutant only in the presence of camptothecin, which leads to the formation of DNA double-strand breaks (Figure 7B). This is consistent with ATM function being more specific in response to DNA double-strand breaks and with ATM being required for events in addition to H2AX-S129 phosphorylation during double-strand break repair (Malavazi et al. 2006).
Figure 7.—
γ-H2AX phosphorylation is required for the DNA damage response in A. nidulans but not for viability of the sonB1 mutant at 42°. (A) Alignment (ClustalW, Biology Workbench at http://workbench.sdcs.edu/) of the C-terminal tail of H2AX from the indicated organisms. The serine in the conserved SQE motif is boxed. Identical (*) and conserved groups (:) are indicated. (B) DNA damage sensitivities of wild-type (R153), H2AX S129A (CDS198), uvsB505ATR (CDS314), and atmAATM null (CDS320) strains at 32°. (C) Colony formation of wild type and the indicated single or double mutants at 32° and 42° in the presence or absence of 0.005% DEO or 2 μg/ml camptothecin. Strains used were CDS198, CDS200, CDS201, CDS364, CDS365, and R153 and are listed in supplemental Table S1 at http://www.genetics.org/supplemental/.
We next assessed whether preventing H2AX-S129A phosphorylation had any effect on the sonB1 mutant. The sonB1 H2AX-S129A double mutant was not synthetically lethal at 42° and therefore H2AX phosphorylation is not essential for viability of sonB1 mutants at 42°. Notably, sonB1 H2AX-S129A double mutants displayed a much greater sensitivity to both DEO and camptothecin at 42° compared to either single mutant (Figure 7C). This effect was specific for sonB1 as sonA1 H2AX-S129A double mutants did not display any such additive effects (Figure 7C). These data are consistent with sonB1 functioning on a different pathway of the DNA damage response from that leading to H2AX-S129 phosphorylation.
DISCUSSION
Here we report that the sonB1 nucleoporin mutant is highly sensitive to DNA-damaging agents specifically at 42°, indicating that this mutation causes a defect in the DNA damage response at 42°. As the sonB1 mutation also suppresses a nimA1 G2 arrest at 42° (De Souza et al. 2003), we hypothesized that sonB1 mutants may have a defective G2 DNA damage checkpoint resulting in cells entering mitosis without repair of DNA damage. Supporting this, we found that sonB1 germlings that had entered the cell cycle were sensitive to UV irradiation but sonB1 quiescent conidiospores were not. This differential in the UV sensitivity of germlings and conidiospores is similar to that seen for A. nidulans mutants, which are unable to tyrosine phosphorylate Cdc2, causing a defective G2 DNA damage checkpoint (Ye et al. 1997; De Souza et al. 1999). However, we surprisingly found that in sonB1 mutants Cdc2 is tyrosine phosphorylated and cells arrest in G2 normally in response to DNA damage, indicating that the G2 DNA damage checkpoint is functional. Further, sonB1 cdc2F double mutants were more sensitive than the respective single mutants to DNA-damaging agents, providing genetic evidence that SONBnNup98 functions on a different pathway in the DNA damage response from that leading to Cdc2 tyrosine phosphorylation. Therefore, the DNA damage sensitivity of the sonB1 mutant at 42° is independent of the G2 DNA damage checkpoint.
It has become clear that the MRN complex is a key player in the DNA damage response (for review see D'Amours and Jackson 2002; Stavridi and Halazonetis 2005; Zhang et al. 2006). The MRN complex initially functions early in the DNA damage response, together with the ATM and ATR kinases, to regulate signaling and checkpoint pathways and then later in the DNA damage response to help facilitate homologous recombination and NHEJ (Uziel et al. 2003; Difilippantonio et al. 2005; Lee and Paull 2005; Stiff et al. 2005; You et al. 2005; Jazayeri et al. 2006; for review see Abraham and Tibbetts 2005; Stavridi and Halazonetis 2005; Zhang et al. 2006). The most striking finding of this study is the temperature-dependent, synthetic lethal interaction between sonB1 and either the scaA1NBS1 mutant or the scaANBS1 null. This synthetic lethality was restricted to 42°, the same temperature at which sonB1 displays high DNA damage sensitivity. Although sonB1 mutants are defective in some aspect of G2 regulation at 42°, we do not believe that this is the defect causing synthetic lethality with scaANBS1 mutants. This is because, while both the sonA1 and sonB1 nucleoporin mutants suppress a nimA1 G2 arrest at 42° (Wu et al. 1998; De Souza et al. 2003), only sonB1, and not sonA1, displays DNA damage sensitivity and is completely dead in combination with scaANBS1 mutants at 42°. This argues that sonB1 synthetic lethality with scaANBS1 mutants is likely not due to sonB1 G2 regulation defects at 42°, but rather due to the defect of the sonB1 mutant in the DNA damage response. Our data therefore suggest that it is a combination of the sonB1 DNA damage response defect and the lack of SCAANBS1 function that causes synthetic lethality.
One potential link between sonB1 and the MRN complex is the histone H2AX/H2B gene pair. We previously isolated histone H2AX/H2B as a copy-number suppressor of sonB1 cold sensitivity and hydroxyurea sensitivity at 32° (De Souza et al. 2003). However, the high level of DNA damage sensitivity of sonB1 mutants is not suppressed by extra-copy histone H2AX/H2B (data not shown). Phosphorylation of H2AX by the ATM/ATR kinases to generate γ-H2AX is important for the localization of the NBS1 to sites of DNA damage (Kobayashi et al. 2002; Celeste et al. 2003). This function is likely conserved in A. nidulans as we found that the H2AX-S129A mutant was sensitive to DNA-damaging agents and it has been shown that SCAANBS1 accumulates in the nucleus in an UVSBATR-dependent manner in response to DNA damage (Fagundes et al. 2005). However, we found that sonB1 H2AX-S129A double mutants were viable at 42°, suggesting that it is not a defect in NBS1 localization to γ-H2AX that causes synthetic lethality between sonB1 and the scaANBS1 mutants. Further, the marked increase in DNA damage sensitivity of sonB1 H2AX-S129A double mutants relative to the respective single mutants suggests that SONBnNup98 and γ-H2AX phosphorylation are on different pathways of the DNA damage response.
Intriguingly, the sonB1 mutant was viable at 42° when combined with a disrupted allele of mreAMRE11, the A. nidulans ortholog of mre11 that encodes the DNA nuclease, strand annealing, and strand dissociation activities of the MRN complex (Semighini et al. 2003; for review see D'Amours and Jackson 2002; Zhang et al. 2006). These MRE11 DNA-modifying activities are likely important for the DNA repair functions of the MRN complex. Therefore, sonB1 synthetic lethality is likely independent of the DNA repair functions of the MRN complex, which is further supported by our finding that the sonB1 mutant was not synthetically lethal with other DNA repair mutants at 42°. One NBS1 function that is independent of MRE11 is NBS1 binding to the γ-H2AX phosphoserine epitope via the FHA/BRCT domain located in the N-terminal of NBS1 (Kobayashi et al. 2002; for review see Zhang et al. 2006). While we have shown that γ-H2AX phosphorylation is not required for survival of sonB1 mutants at 42°, it is likely that the NBS1 FHA/BRCT domain also binds other as yet unidentified phosphoserine epitopes. Given our data, it is tempting to speculate that SCAANBS1 binding to phosphoserine epitopes may be required for survival of sonB1 mutants at 42°. Although the FHA/BRCT domain of NBS1 orthologs is not well conserved, a recent bioinformatics study suggests that SCAANBS1 contains the conserved residues required for binding to phosphoserine epitopes (Becker et al. 2006).
Our data suggest that the loss of cell cycle checkpoint functions that occurs in scaANBS1 mutants (Semighini et al. 2003) is not the defect in these mutants causing synthetic lethality in combination with sonB1 mutants at 42°. This is because the sonB1 mutation is not synthetically lethal in combination with mutations in other cell cycle checkpoint regulators, including the A. nidulans orthologs of ATR, ATRIP, ATM, or the cdc2F mutant (Ye et al. 1997; De Souza et al. 1999; Hofmann and Harris 2000; Malavazi et al. 2006). In addition, given that the sonB1 mutant was viable when combined with these cell cycle checkpoint mutants or the uvsCRad51 or uvsHRad18 DNA repair mutants, it is unlikely that sonB1 in itself causes DNA damage at 42°. Notably, however, as double mutants between sonB1 and uvsCRad51 or uvsHRad18 mutants were more sensitive to DEO than were the respective single mutants, it is likely that SONBnNup98 functions on a different pathway in the DNA damage response from that with UVSCRad51 or UVSHRad18.
Interestingly, the NPC has been demonstrated to have roles in tethering telomeres to the nuclear periphery in budding yeast (Galy et al. 2000; Feuerbach et al. 2002; Therizols et al. 2006). Further, a recent study has shown that anchoring of telomeres to the nuclear periphery is required for efficient DNA double-strand break repair (Therizols et al. 2006). These authors found that nucleoporin mutants that failed to properly tether telomeres to the nuclear periphery display a decreased efficiency in the repair of DNA double-strand breaks induced proximal to telomeres. It will therefore be interesting to determine if telomeric localization and/or function is disrupted in sonB1 mutants and whether this contributes to the DNA damage sensitivity of sonB1 mutants. Further, given that the MRN complex functions in telomere regulation (Verdun et al. 2005; for review see d'Adda di Fagagna et al. 2004), it is possible that the synthetic lethality between sonB1 and scaANBS1 mutants may be due to combining different defects in telomere biology caused by these mutations. Another explanation for the DNA-damage-sensitive phenotype of sonB1 is that some aspect of nucleocytoplasmic transport required for the damage response does not function in this mutant at 42°. We consider this unlikely, given that nuclear transport of a nuclear localization sequence reporter construct is normal in sonB1 mutants at 42° (C. De Souza and S. A. Osmani, unpublished observations) and that the sonA1 nucleoporin mutant does not display similar DNA damage sensitivities. However, given that the recruitment of the MRN complex to sites of DNA damage has been reported to require relocation of MRE11 and RAD50 from the cytoplasm to the nucleus (Tauchi et al. 2001; Kobayashi et al. 2002), we cannot rule out the involvement of SONBnNup98 in regulating specific nuclear transport pathways during the DNA damage response.
Null alleles of certain budding yeast nucleoporins display sensitivity to DNA-damaging agents (Galy et al. 2000; Bennett et al. 2001; Chang et al. 2002; Loeillet et al. 2005; Therizols et al. 2006); however, we know of no single amino acid substitutions in yeast nucleoporins that cause sensitivity to DNA-damaging agents. Similar to the case for sonB1, the DNA damage sensitivity of budding yeast nucleoporin nulls does not appear to be caused by general defects in DNA repair (Loeillet et al. 2005; Therizols et al. 2006). Interestingly, genomewide screens have revealed that null alleles of the budding yeast nup120 and nup133 nucleoporins, which are sensitive to DNA-damaging agents, display synthetic lethality with null alleles of MRN (MRX in budding yeast) coding genes (Loeillet et al. 2005). While the mechanism of this interaction has not been established, it suggests that the genetic interaction between the NPC and the MRN complex is likely conserved.
Notably, of the 13 nucleoporins that are nonessential in A. nidulans, none display obvious sensitivities to DNA-damaging agents (Osmani et al. 2006). This makes it even more intriguing that a single amino acid substitution in the essential sonBNup98 nucleoporin gene causes conditional DNA damage sensitivity at 42°. Most DNA-damage-sensitive mutations identified to date in simple organisms occur in nonessential genes. This is because the genetic screens used to identify them require mutations to be viable but display sensitivity to DNA-damaging agents. This fact has selected for DNA-damage-sensitive mutations in nonessential genes. Screens for conditional DNA-damage-sensitive mutations, similar to the heat-dependent DNA damage sensitivity of the sonB1 mutation, may therefore identify essential genes that function in novel aspects of the DNA damage response.
Acknowledgments
We thank all members of the Osmani Laboratory for their helpful discussions and input into this work. We also thank Gustavo Goldman and Iran Malavazi (Universidade de São Paulo, São Paulo, Brazil) for strains and especially for sending the atmA null prior to publication. We thank Etta Kafer for helpful discussions and her tireless contribution to the field. This work was supported by National Institutes of Health grant GM 042564.
References
- Abraham, R. T., and R. S. Tibbetts, 2005. Cell biology: guiding ATM to broken DNA. Science 308: 510–511. [DOI] [PubMed] [Google Scholar]
- Becker, E., V. Meyer, H. Madaoui and R. Guerois, 2006. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics 22: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin et al., 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29: 426–434. [DOI] [PubMed] [Google Scholar]
- Bruschi, G. C., C. C. de Souza, M. R. Fagundes, M. A. Dani, M. H. Goldman et al., 2001. Sensitivity to camptothecin in Aspergillus nidulans identifies a novel gene, scaA+, related to the cellular DNA damage response. Mol. Genet. Genomics 265: 264–275. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus et al., 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Casolari, J. M., C. R. Brown, D. A. Drubin, O. J. Rando and P. A. Silver, 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 19: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt et al., 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5: 675–679. [DOI] [PubMed] [Google Scholar]
- Chae, S. K., and E. Kafer, 1997. Two uvs genes of Aspergillus nidulans with different functions in error-prone repair: uvsI, active in mutation-specific reversion, and uvsC, a recA homolog, required for all UV mutagenesis. Mol. Gen. Genet. 254: 643–653. [DOI] [PubMed] [Google Scholar]
- Chang, M., M. Bellaoui, C. Boone and G. W. Brown, 2002. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc. Natl. Acad. Sci. USA 99: 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Adda di Fagagna. F., S. H. Teo, and S. P. Jackson, 2004. Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18: 1781–1799. [DOI] [PubMed] [Google Scholar]
- D'Amours, D., and S. P. Jackson, 2002. The Mre11 complex: at the crossroads of DNA repair and checkpoint signaling. Nat. Rev. Mol. Cell Biol. 3: 317–327. [DOI] [PubMed] [Google Scholar]
- De Souza, C. P. C., X. Ye and S. A. Osmani, 1999. Checkpoint defects leading to premature mitosis also cause endoreplication of DNA in Aspergillus nidulans. Mol. Biol. Cell 10: 3661–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, C. P., A. H. Osmani, L. P. Wu, J. L. Spotts and S. A. Osmani, 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102: 293–302. [DOI] [PubMed] [Google Scholar]
- De Souza, C. P., K. P. Horn, K. Masker and S. A. Osmani, 2003. The SONBNUP98 nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza, C. P., A. H. Osmani, S. B. Hashmi and S. A. Osmani, 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14: 1973–1984. [DOI] [PubMed] [Google Scholar]
- Difilippantonio, S., A. Celeste, O. Fernandez-Capetillo, H. T. Chen, S. M. Reina et al., 2005. Role of Nbs1 in the activation of the Atm kinase revealed in humanized mouse models. Nat. Cell Biol. 7: 675–685. [DOI] [PubMed] [Google Scholar]
- Dilworth, D. J., A. J. Tackett, R. S. Rogers, E. C. Yi, R. H. Christmas et al., 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs, J. A., N. F. Lowndes and S. P. Jackson, 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Fagundes, M. R. C. K., C. P. Semighini, I. Malavazi, M. Savoldi, J. F. de Lima et al., 2005. Aspergillus nidulans uvsBATR and scaANBS1 genes show genetic interactions during recovery from replication stress and DNA damage. Eukaryot. Cell 4: 1239–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerbach, F., V. Galy, E. Trelles-Sticken, M. Fromont-Racine, A. Jacquier et al., 2002. Nuclear architecture and spatial positioning help establish transcriptional states of telomeres in yeast. Nat. Cell Biol. 4: 214–221. [DOI] [PubMed] [Google Scholar]
- Galy, V., J. C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou et al., 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 403: 108–112. [DOI] [PubMed] [Google Scholar]
- Goldman, G. H., and E. Kafer, 2004. Aspergillus nidulans as a model system to characterize the DNA damage response in eukaryotes. Fungal Genet. Biol. 41: 428–442. [DOI] [PubMed] [Google Scholar]
- Goldman, G. H., S. L. McGuire and S. D. Harris, 2002. The DNA damage response in filamentous fungi. Fungal Genet. Biol. 35: 183–195. [DOI] [PubMed] [Google Scholar]
- Hetzer, M. W., T. C. Walther and I. W. Mattaj, 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21: 347–380. [DOI] [PubMed] [Google Scholar]
- Hofmann, A. F., and S. D. Harris, 2000. The Aspergillus nidulans uvsB gene encodes an ATM-related kinase required for multiple facets of the DNA damage response. Genetics 154: 1577–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner, K. P., C. D. Putnam and J. A. Tainer, 2002. DNA double-strand break repair from head to tail. Curr. Opin. Struct. Biol. 12: 115–122. [DOI] [PubMed] [Google Scholar]
- Ichioka, D., T. Itoh and Y. Itoh, 2001. An Aspergillus nidulans uvsC null mutant is deficient in homologous DNA integration. Mol. Gen. Genet. 264: 709–715. [DOI] [PubMed] [Google Scholar]
- Ishii, K., G. Arib, C. Lin, G. Van Houwe and U. K. Laemmli, 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell 109: 551–562. [DOI] [PubMed] [Google Scholar]
- Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith et al., 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8: 37–45. [DOI] [PubMed] [Google Scholar]
- Kafer, E., and G. May, 1997. The uvsF gene region in Aspergillus nidulans codes for a protein with homology to DNA replication factor C. Gene 191: 155–159. [DOI] [PubMed] [Google Scholar]
- Kafer, E., and O. Mayor, 1986. Genetic analysis of DNA repair in Aspergillus: evidence for different types of MMS-sensitive hyperrec mutants. Mutat. Res. 161: 119–134. [DOI] [PubMed] [Google Scholar]
- Kobayashi, J., H. Tauchi, S. Sakamoto, A. Nakamura, K. Morishima et al., 2002. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 12: 1846–1851. [DOI] [PubMed] [Google Scholar]
- Lee, J. H., and T. T. Paull, 2005. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science 308: 551–554. [DOI] [PubMed] [Google Scholar]
- Loeillet, S., B. Palancade, M. Cartron, A. Thierry, G. F. Richard et al., 2005. Genetic network interactions among replication, repair and nuclear pore deficiencies in yeast. DNA Rep. 4: 459–468. [DOI] [PubMed] [Google Scholar]
- Malavazi, I., C. P. Semighini, M. R. Z. Kress, S. D. Harris and G. H. Goldman, 2006. Regulation of hyphal morphogenesis and the DNA damage response by the Aspergillus nidulans ATM homolog AtmA. Genetics 173: 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May, G. S., and N. R. Morris, 1987. The unique histone H2A gene of Aspergillus nidulans contains three introns. Gene 58: 59–66. [DOI] [PubMed] [Google Scholar]
- McGowan, C. H., and P. Russell, 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16: 629–633. [DOI] [PubMed] [Google Scholar]
- Menon, B. B., N. J. Sarma, S. Pasula, S. J. Deminoff, K. A. Willis et al., 2005. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 102: 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T. M., L. L. Du, C. Redon and P. Russell, 2004. Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol. Cell. Biol. 24: 6215–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak, T., E. Szewczyk, C. E. Oakley, A. H. Osmani, L. Ukil et al., 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. R., and S. A. Osmani, 1993. Cell-cycle analysis using the filamentous fungus Aspergillus nidulans, pp. 127–142 in The Cell Cycle: A Practical Approach, edited by P. Fantes and R. Brooks. IRL Press/Oxford University Press, Oxford/New York.
- Osmani, A. H., S. L. McGuire and S. A. Osmani, 1991. Parallel activation of the NIMA and p34cdc2 cell cycle-regulated protein kinases is required to initiate mitosis in A. nidulans. Cell 67: 283–291. [DOI] [PubMed] [Google Scholar]
- Osmani, A. H., N. van Peij, M. Mischke, M. J. O'Connell and S. A. Osmani, 1994. A single p34cdc2 protein kinase (nimXcdc2) is required at G1 and G2 in Aspergillus nidulans. J. Cell Sci. 107: 1519–1528. [DOI] [PubMed] [Google Scholar]
- Osmani, A. H., J. Davies, H. L. Liu, A. Nile and S. A. Osmani, 2006. Systemic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Osmani, S. A., and P. M. Mirabito, 2004. The early impact of genetics on our understanding of cell cycle regulation in Aspergillus nidulans. Fungal Genet. Biol. 41: 401–410. [DOI] [PubMed] [Google Scholar]
- Osmani, S. A., and X. S. Ye, 1996. Cell cycle regulation in Aspergillus by two protein kinases. Biochem. J. 317: 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, S. A., G. S. May and N. R. Morris, 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104: 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M., G. Arib, C. Laemmli, J. Nishikawa, T. Durussel et al., 2006. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell 21: 379–391. [DOI] [PubMed] [Google Scholar]
- Semighini, C. P., M. R. Z. K. Fagundes, J. C. Ferreira, R. C. Pascon, M. H. De Souza Goldman et al., 2003. Different roles of the Mre11 complex in the DNA damage response in Aspergillus nidulans. Mol. Microbiol. 48: 1693–1709. [DOI] [PubMed] [Google Scholar]
- Seong, K. Y., S. K. Chae and H. S. Kang, 1997. Cloning of an E. coli RecA and yeast RAD51 homolog, radA, an allele of the uvsC in Aspergillus nidulans and its mutator effects. Mol. Cell 7: 284–289. [PubMed] [Google Scholar]
- Stavridi, E. S., and T. D. Halazonetis, 2005. Nbs1 moving up in the world. Nat. Cell Biol. 7: 648–650. [DOI] [PubMed] [Google Scholar]
- Stiff, T., C. Reis, G. K. Alderton, L. Woodbine, M. O'Driscoll et al., 2005. Nbs1 is required for ATR-dependent phosphorylation events. EMBO J. 24: 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi, H., J. Kobayashi, K. Morishima, S. Matsuura, A. Nakamura et al., 2001. The forkhead-associated domain of NBS1 is essential for nuclear foci formation after irradiation but not essential for hRAD50·hMRE11·NBS1 complex DNA repair activity. J. Biol. Chem. 276: 12–15. [DOI] [PubMed] [Google Scholar]
- Therizols, P., C. Fairhead, G. G. Cabal, A. Genovesio, J. C. Olivo-Marin et al., 2006. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, E. J., and S. R. Wente, 2006. Dynamic nuclear pore complexes: life on the edge. Cell 125: 1041–1053. [DOI] [PubMed] [Google Scholar]
- Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten et al., 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16: 991–1002. [DOI] [PubMed] [Google Scholar]
- Uziel, T., Y. Lerenthal, L. Moyal, Y. Andegeko, L. Mittelman et al., 2003. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 22: 5612–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heemst, D., K. Swart, E. F. Holub, R. van Dijk, H. H. Offenberg et al., 1997. Cloning, sequencing, disruption and phenotypic analysis of uvsC, an Aspergillus nidulans homologue of yeast Rad51. Mol. Gen. Genet. 254: 654–664. [DOI] [PubMed] [Google Scholar]
- Verdun, R. E., L. Crabbe, C. Haggblom and J. Karlseder, 2005. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell 23: 551–561. [DOI] [PubMed] [Google Scholar]
- Vidanes, G. M., C. Y. Bonilla and D. P. Toczyski, 2005. Complicated tails: histone modifications and the DNA damage response. Cell 121: 973–976. [DOI] [PubMed] [Google Scholar]
- Wu, L., S. A. Osmani and P. M. Mirabito, 1998. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 141: 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L, L. Ukil, A. Osmani, F. Nahm, J. Davies et al., 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3: 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X. S., R. R. Fincher, A. Tang, K. O'Donnell and S. A. Osmani, 1996. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 15: 3599–3610. [PMC free article] [PubMed] [Google Scholar]
- Ye, X. S., R. R. Fincher, A. Tang and S. A. Osmani, 1997. The G2/M DNA damage checkpoint inhibits mitosis through Tyr15 phosphorylation of p34cdc2 in Aspergillus nidulans. EMBO J. 15: 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, X. S., G. Xu, P. T. Pu, R. R. Fincher, S. L. McGuire et al., 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, J. H., B. J. Lee and H. S. Kang, 1995. The Aspergillus uvsH gene encodes a product homologous to yeast Rad18 and Neurospora UVS-2. Mol. Gen. Genet. 248: 174–181. [DOI] [PubMed] [Google Scholar]
- You, Z., C. Chahwan, J. Bailis, T. Hunter and P. Russell, 2005. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 25: 5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., J. Zhou and C. U. Lim, 2006. The role of NBS1 in DNA double strand break repair, telomere stability, and cell cycle checkpoint control. Cell Res. 16: 45–54. [DOI] [PubMed] [Google Scholar]
- Zhou, B. B., and S. J. Elledge, 2000. The DNA damage response: putting checkpoints in perspective. Nature 408: 433–439. [DOI] [PubMed] [Google Scholar]