Abstract
By alternative use of four RSL (reactive site loop) coding exon cassettes, the serpin (serine protease inhibitor) gene Spn4 from Drosophila melanogaster was proposed to enable the synthesis of multiple protease inhibitor isoforms, one of which has been shown to be a potent inhibitor of human furin. Here, we have investigated the inhibitory spectrum of all Spn4 RSL variants. The analyses indicate that the Spn4 gene encodes inhibitors that may inhibit serine proteases of the subtilase family (S8), the chymotrypsin family (S1), and the papain-like cysteine protease family (C1), most of them at high rates. Thus a cohort of different protease inhibitors is generated simply by grafting enzyme-adapted RSL sequences on to a single serpin scaffold, even though the target proteases contain different types and/or a varying order of catalytic residues and are descendents of different phylogenetic lineages. Since all of the Spn4 RSL isoforms are produced as intracellular residents and additionally as variants destined for export or associated with the secretory pathway, the Spn4 gene represents a versatile defence tool kit that may provide multiple antiproteolytic functions.
Keywords: cysteine protease inhibitor, endoplasmic reticulum (ER) retention signal, furin, serine protease inhibitor, serpin
Abbreviations: apoLp-II/I, apolipophorin-II/I; DMEM, Dulbecco's modified Eagle's medium; E-64, trans-epoxysuccinyl-L-leucylamido-(4-guanidino)-butane; ER, endoplasmic reticulum; GST, glutathione S-transferase; PC, proprotein convertase; RSL, reactive site loop; SI, stoichiometry of inhibition; TEV, tobacco etch virus; Z-FR-AMC, benzyloxycarbonyl-Phe-Arg-7-amido-4-methylcoumarin
INTRODUCTION
The serpins (serine protease inhibitors) constitute a superfamily of proteins with unusual conformational flexibility and functional diversity [1]. Most serpins inhibit members from one of several peptidase clans, including chymotrypsin-like enzymes, subtilase-like proteases, papain-related cysteine proteases and caspases [1,2]. These enzymes exert their function by using different orders and combinations of catalytic residues [3]. A few serpins function as cross-class inhibitors, as shown by their ability to block the activity of serine and papain-like cysteine proteases [4]. There are also non-inhibitory serpins that fulfil such diverse tasks as assisting in procollagen folding or chromatin organization, while still others serve as hormone carriers or play an as yet unknown physiological role. Serpins are involved in many important physiological processes, including blood coagulation, fibrinolysis, inflammation, tumour cell invasion and innate immunity [5].
Inhibitory serpins employ a suicide reaction mechanism by presenting an exposed sequence, the RSL (reactive site loop), to the proteolytic enzyme. The target specificity of serpins is co-determined by the sequence and structure of the RSL, although exosite contacts may play an important role in some cases [6]. After formation of a non-covalent encounter complex, a bait peptide bond in the RSL is attacked by the enzyme, followed by formation of an acyl-enzyme intermediate. In the inhibitory pathway, cleavage of the RSL results in a large-scale movement of the attached protease and its incorporation into a serpin–protease complex, in which both of the interaction partners is inactive [5,7,8].
All organisms have to cope with a plethora of extra- and intracellular proteolytic enzymes, either endogenous ones or ones associated with pathogens. Sometimes, a paradoxical situation seems to emerge, for instance, when intruders exploit endogenous enzymes with an important role in host physiology for their purposes. Mammalian furin, a subtilase-like enzyme, plays an eminent role as PC (proprotein convertase) that processes a large variety of intra- and extra-cellular proteins. However, many viruses and some bacteria use the endoproteolytic activity of this enzyme for their propagation [9]. Similarly, endosomal/lysosomal cysteine proteases can fulfil distinct host-specific functions [10]; however, their role as processing enzymes for proteins of a number of infectious viruses is just beginning to emerge [11]. Thus a finely tuned control of these Janus-faced enzymes is essential in order to maintain a balanced network of enzyme–inhibitor interactions that satisfies the complex regulatory requirements of the organism.
We and others have recently identified a serpin, Spn4A from Drosophila melanogaster, as a potent inhibitor of human furin [12–14] and Drosophila PC2 (amontillado) [14]. Spn4A is a product of the Spn4 gene [15] that has been inferred to enable the synthesis of numerous Spn4 variants with different cellular locations and target enzyme specificities, due to the presence (variants Spn4A–Spn4D) or absence of a signal peptide (variants Spn4E–Spn4H), and alternative use of four different RSL coding exon cassettes [16]. To gain more insight into the functions of the Spn4 gene, we have recombinantly expressed all RSL variants and investigated their biochemical properties. The results indicate that transcription of the Spn4 gene enables, in an economic manner, the synthesis of multiple protease inhibitors that may protect cells against the uncontrolled activities of a unique variety of extra- and intra-cellular proteases.
EXPERIMENTAL
Materials
Human neutrophil elastase [cat. no. 324681], cathepsin B (cat. no. 219364), cathepsin L (cat. no. 19402), recombinant human cathepsin S (cat. no. 219343) and MeOSuc-Ala-Ala-Pro-Val-AMC (cat. no. 324740) were obtained from Calbiochem. E-64 [trans-epoxysuccinyl-L-leucylamido-(4-guanidino)butane; proteinase inhibitor] (E 3132) was from Sigma. Z-FR-AMC (benzyloxycarbonyl-Phe-Arg-7-amido-4-methylcoumarin) and (Z-FR)2-R110 were from Alexis and Molecular Probes respectively. Bovine chymotrypsin [tosylphenylalanylchloromethane (‘TPCK’) treated, modified] was purchased from Princeton Separations.
Polyclonal rabbit IgGs directed against Drosophila Dfurin1 and Dfurin2 [17] respectively were obtained from Dr J. Creemers (Department for Human Genetics, Flanders Interuniversity Institute for Biotechnology, Leuven, Belgium). A rabbit antiserum directed against recombinant GST (glutathione S-transferase)–Spn4 that recognizes all four RSL variants was partially purified by affinity chromatography on GST covalently coupled with glutathione–Sepharose 4B (Amersham Biosciences) via dimethylpimelinedi-imidate-dihydrochloride [6]. The rabbit antiserum raised against apoLp-II (apolipophorin-II) has been described previously [18].
Production of Spn4 variants in Escherichia coli
From the genomic organization, eight Spn4 isoforms with four different RSL sequences (Figure 1) that may contain or lack an N-terminal signal sequence have been deduced [16]. For biochemical characterization, all four RSL variants (residue 1 to the individual isoform-specific C-terminal end, numbering refers to isoforms E–H) were expressed as GST fusion proteins in E. coli. After purification by glutathione affinity chromatography and cleavage with TEV (tobacco etch virus) protease, His6-tag-containing con-taminants (residual fusion protein, His-tagged GST and His6–TEV) were removed on Protino® Ni 2000 columns, as outlined previously [12]. The recombinant Spn4 variants contain a two-residue-extension (Gly-Ala) at their N-terminus, due to the presence of a TEV cleavage site. Considerably varying amounts of the different Spn4 fusion proteins that differ solely in the 40–58 residue-long C-terminal RSL domains were consistently obtained from the soluble fraction of the bacterial extracts. The purity of the preparations was assessed by staining with Coomassie Brilliant Blue. Despite various efforts, the Spn4F and Spn4G preparations always contained cleaved inhibitor molecules (∼20% as estimated from Coomassie Brilliant Blue staining of SDS gels). The concentration of the purified proteins was determined by a standard Bradford assay.
Figure 1. Alignment of the C-terminal sequences of human α1-antitrypsin (A1AT) and the four RSL variants of Spn4.
Isoforms Spn4A–Spn4D differ from variants Spn4E–Spn4H by the presence of an N-terminal signal peptide. Conserved positions are represented by white-on-black printing. An arrow marks the P1 position of A1AT.
Cultivation and transfection of insect cells and COS-7 cells
Drosophila S2 cells were maintained at 26 °C in Schneider's Drosophila medium. The construction of the pIZ/V5-His (Invitrogen)-based expression plasmid coding for the truncated apoLp-II/I-38 protein has been described [18]. Plasmid pIZ-Spn4A contains the cDNA coding for Spn4A, including the genuine signal peptide and the C-terminal ER (endoplasmic reticulum) retention signal [12]. Spodoptera frugiperda Sf9 cells, adapted to growth in serum-free Insect-Xpress medium (Cambrex), were transiently transfected with linear poly(ethyleneimine) (25 kDa; Polysciences Europe GmbH, cat. no. 23966) in 12-well Nunclon Surface Plates (3.5 cm2 per well) with a mixture consisting of 1 μg of plasmid DNA and 5 μg of poly(ethyleneimine) [19], using various molar ratios of the plasmids (4:1; 1:1 and 1:5) coding for Spn4A and apoLp-II/I-38. After 2 days, supernatants were precipitated by the addition of trichloroacetic acid [final concentration: 5% (v/v)]. Concentrated supernatants were analysed by Western blotting, using antibodies directed against Spn4 and apoLp-II at a dilution of 1:10000 and 1:4000 respectively.
For expression in COS-7 cells, the cDNAs coding for the eight individual Spn4 variants were subcloned into the expression vector pcDNA3.1(+) as outlined in [12]. COS-7 cells were cultivated in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) foetal calf serum. The cells were transfected in 25 cm2 T-flasks (Nunc) with Lipofectamine™ 2000 (Invitrogen) following the supplier's instructions. After 4 h, the transfection medium was replaced by DMEM supplemented with insulin and transferrin, and aspirated after 24 h of further cultivation.
Enzymatic assays and kinetic methods
All assays (100 μl) were conducted in 96-well microtitre plates at 30 °C using a FLUOstar/POLARstar Galaxi spectrometer (BMG LabTechnologies). To determine the rate constants kassoc for the inhibition of elastase, cathepsin L or cathepsin S by the Spn4 variants, progress curves were recorded under pseudo first-order conditions using the serpin concentrations [I] specified in Figure 4. The data were fitted to eqn (1) for slow tight binding inhibition [20,21]:
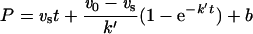 |
(1) |
where P is fluorescence, ν0 is initial velocity, νs is steady-state velocity, t is time, k′ is apparent first order rate constant and b is fluorescence at time t=0. Non-linear regression (EnzFitter) provided k′ for each serpin concentration [I].
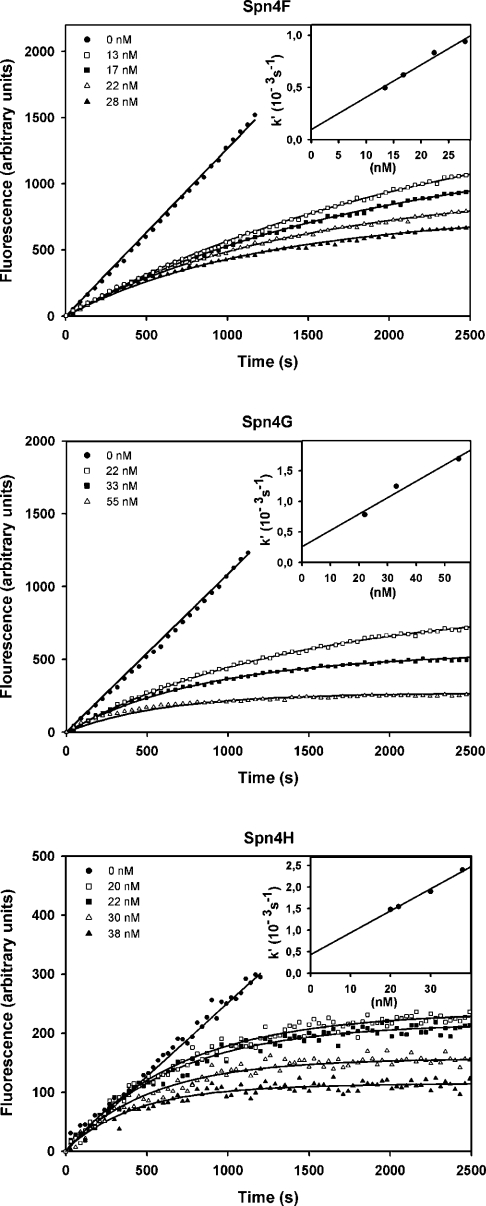
Figure 4. Inhibition of human neutrophil elastase by variants Spn4F–Spn4H.
Progress curves were recorded by measuring the cleavage of MeOSuc-AAPV-AMC (91 μM) by human neutrophil elastase in the presence of Spn4F, Spn4G or Spn4H. The apparent first order rate constant k′ for each curve was plotted against the inhibitor concentration (inset). From the slope of the linear regression line kassoc was calculated using eqn (2). The enzyme was used at concentrations of 1 (Spn4F and Spn4G) or 0.5 nM (Spn4H).
By plotting k′ versus [I], the rate constant kassoc was obtained from linear regression according to eqn (2):
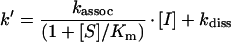 |
(2) |
To record progress curves, human neutrophil elastase (0.5 or 1 nM, determined by absorption at 280 nm) was assayed in 50 mM Tris/HCl, 150 mM NaCl, 0.05% Triton X-100 and 2% (v/v) DMSO (pH 7.5) using 91 μM MeOSuc-AAPV-AMC as a substrate. Cleavage of the fluorogenic substrate was monitored at 460 nm (λex=380 nm). Using substrate concentrations between 10 and 400 μM and fitting the usual Michaelis–Menten equation to the data, a Michaelis constant Km of 178 μM was obtained.
The concentration of catalytically active cathepsin L or cathepsin S was determined by titrating with E-64 [22]. After incubation with the inhibitor (0–500 nM) for 1 h at 30 °C in cathepsin buffer (50 mM sodium acetate, 1 mM EDTA, 4 mM dithiothreitol and 0.05% Brij35, pH 5.5), the residual enzyme activity was measured using 18 μM Z-FR-AMC (λex=380 nm and λem=460 nm) or 10 μM (Z-FR)2-R110 (λex=485 nm and λem=520 nm) respectively. To analyse the SI (stoichiometry of inhibition), the cathepsins were titrated likewise using Spn4F or Spn4G. Substrate concentrations between 0.5 and 50 μM were used for Km determination. The cleavage of Z-FR-AMC by cathepsin L followed usual Michaelis–Menten kinetics with Km=6.4 μM. For the cleavage of (Z-FR)2-R110 by cathepsin S, inhibition of hydrolysis was observed at higher substrate concentrations. To account for substrate inhibition the data were fitted to eqn (3) [23] and a Km of 2.9 μM was determined:
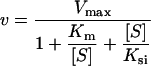 |
(3) |
The variables ν, Vmax and Ksi represent the rate of substrate hydrolysis, the maximal rate and the equilibrium constant for substrate inhibition respectively. To apply the progress curve method, the enzymatic activity of cathepsin L (0.45 nM) and cathepsin S (2.5 nM) was measured at the substrate concentrations indicated above.
Other methods
The generation of complexes between Spn4E and Dfurin1 or Dfurin2 was initiated by adding recombinant Spn4E to the culture medium of S2 cells as indicated in the legend to Figure 2. Elastase (25 nM) or chymotrypsin (50 nM) was incubated with Spn4 variants (molar ratio, 1:3 to 1:5) for 5 min at 30 °C in 50 mM Tris/HCl and 150 mM NaCl (pH 7.5 or 8.0 respectively), before samples were boiled (4 min) in Laemmli buffer and analysed by Western blotting. The formation of inhibitor–cysteine protease adducts was initiated by adding Spn4 variants to cathepsin L (50 nM) or cathepsin S (67 nM) in 50 mM sodium acetate buffer and 1 mM EDTA (pH 5.5). After 5 min at 30 °C, E-64 was added (final concentration: 3 μM). SDS sample buffer without reducing agent was added, and after boiling for 4 min, the samples were resolved on 10% NuPAGE® gels (Invitrogen). Western blotting and immunodetection were performed as described in [24].
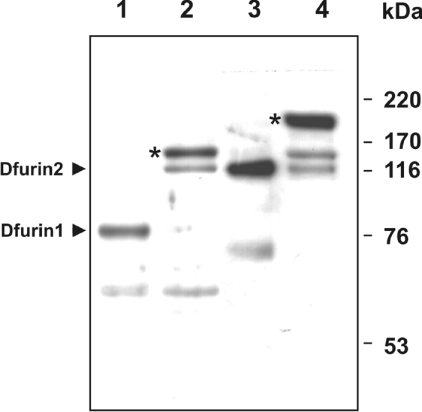
Figure 2. Formation of SDS-stable complexes between recombinant Spn4E and Dfurin1 or Dfurin2.
Supernatants (20 μl) from S2 cells were incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of 50 ng of recombinant Spn4E. After reducing SDS/PAGE (10% gels), the reaction products were analysed by Western blotting, using antibodies directed against Dfurin1 (lanes 1 and 2) or Dfurin2 (lanes 3 and 4) respectively. An asterisk marks Spn4E–Dfurin1 and Spn4E–Dfurin2 complexes depicting the expected sizes. The molecular masses of marker proteins are indicated on the right.
RESULTS
Spn4E forms SDS-stable complexes with Dfurin1 and Dfurin2 and the signal peptide containing isoform Spn4A inhibits the processing of apoLp-II/I in Sf9 cells
Previous investigations have demonstrated that Spn4A/E is a potent inhibitor of human furin [12–14]. In order to analyse the potential of the inhibitor in its natural context, the effects of Spn4E towards fruitfly PCs were examined. The D. melanogaster genome contains two genes, Dfur1 and Dfur2 respectively, that encode furin-like PCs [17,25]. In transfected mammalian cells, several isoforms of these enzymes are produced, some of which are released into the medium [26].
To study the PC–Spn4E interaction in the fruit fly system, we first explored the potential secretion of Dfurins into the medium of Drosophila S2 cells. The culture medium of these cells possessed a major band at approx. 80 kDa and a minor band at approx. 60 kDa, as indicated by Western-blot analysis with anti-Dfurin1 antibodies (Figure 2). The medium of S2 cells also reacted with an anti-Dfurin2 antiserum, revealing the presence of a major approx. 130 kDa Dfurin2 band, indicating that S2 cells express both types of furin genes and that the insect cells can release the enzymes into the medium.
Addition of purified recombinant Spn4E to the medium of the insect cells resulted in the appearance of a novel major immunoreactive band with a size of approx. 145 kDa after incubation with anti-Dfurin1 antibodies with concurrent loss of the Dfurin1 signal, demonstrating the formation of SDS-stable Dfurin1–Spn4E complexes (Figure 2, lane 2). Dfurin2–Spn4E complexes (molecular mass ∼180 kDa) were identified in a similar manner (Figure 2, lane 4). We suspect that two minor bands appearing after Spn4E addition represent partially degraded Dfurin1–Spn4E and Dfurin2–Spn4E complexes respectively (∼130 kDa band, lane 2; ∼145 kDa band, lane 4). The nature of the approx. 60 kDa band in lanes 1 and 2 respectively is not known. The generation of complexes between Spn4E and Dfurin1 or Dfurin2 was corroborated by using anti-Spn4 antibodies (results not shown). Due to high background levels we were not able to demonstrate the presence of Dfurin1 or Dfurin2 or its complexes with Spn4E in S2 cell lysates. We conclude that Spn4A/E could function as a regulator of both fruit fly Dfurins.
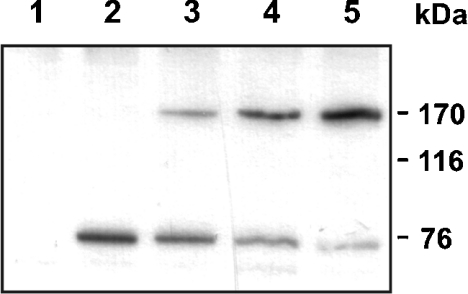
To characterize the role of the furin-inhibiting Spn4 variants further, the processing of apoLp-II/I, an insect homologue of mammalian apolipoprotein B, was investigated. ApoLp-II/I (3380 amino acids) is cleaved post-translationally after the sequence RQKR↓ [18,27,28], resulting in the formation of two apolipoproteins, apoLp-I and apoLp-II. To study the effect of variant Spn4A on apoLp-II/I processing, Sf9 cells from S. frugiperda which express a furin homologue [29] and which are able to process apoLp-II/I were used. The cells were transiently transfected with a vector coding for a truncated version of apoLp-II/I from the locust Locusta migratoria encompassing the N-terminal part of the protein, including the presumed furin recognition/cleavage site, and a plasmid expressing Spn4A. Cells transfected with the apoLp-II/I construct alone secreted correctly processed apoLp-II into the medium, as demonstrated by the appearance of an approx. 76 kDa immunoreactive apoLp-II band (Figure 3). Spn4A impaired cleavage in a dose-dependent manner as indicated by the secretion of the unprocessed truncated apoLp-II/I (molecular mass ∼170 kDa) in the medium of cells co-transfected with the plasmid encoding the PC inhibitor. These data show that Spn4A can regulate the PC-mediated processing of apoLp-II/I in insect cells.
Figure 3. Spn4A inhibits processing of apoLp-II/I.
Sf9 cells were transiently transfected with a plasmid coding for a truncated version of apoLp-II/I (∼170 kDa) either in the absence or presence of various amounts of an Spn4A expression plasmid. Two days post-transfection, supernatants from transfected cells were inspected for apoLp-II/I processing, using an antiserum raised against apoLp-II (∼76 kDa). Lane 1, cells transfected only with the Spn4A expression plasmid; lane 2, cells transfected with a plasmid coding for truncated apoLp-II/I; lanes 3–5, cells co-transfected with a mixture of plasmids coding for apoLp-II/I and Spn4A at various ratios (from left to right: 4:1, 1:1 and 1:5). The masses of marker proteins are indicated on the right.
Interaction of isoforms Spn4F–Spn4H with enzymes of the S1 peptidase family
The RSL regions of variants Spn4F–Spn4H depict sequences quite different from those of isoform Spn4E. To examine their inhibitory profiles, the interaction of these variants with enzymes of the S1 serine protease (chymotrypsin) family that constitute the classical targets for serpins was investigated. The region preceding the putative scissile bond of the RSLs from variants Spn4F–Spn4H consists primarily of hydrophobic and/or small aliphatic residues (Figure 1) that provide several metazoan serpins with activity towards elastase and/or chymotrypsin. Accordingly, the interaction with these enzymes was studied.
Incubation of human neutrophil elastase with each of the variants Spn4F–Spn4H resulted in formation of SDS-stable enzyme–inhibitor adducts that reacted with anti-Spn4 antibodies, as determined by immunoblotting (results not shown). However, most of the variant molecules served as substrate for the enzyme as shown by a significant increase in cleaved inhibitor molecules, indicated by their increased mobility. There were no complexes in the samples containing variant Spn4E and elastase. In analogous experiments with bovine chymotrypsin, a part of the enzyme molecules was found to be trapped in complexes when exposed to a 3-fold excess of Spn4F or Spn4G, but again most of the Spn4F and Spn4G served as substrate. The complexes had a size of approx. 55 kDa, as expected for binding of the serpin to the C-chain of chymotrypsin, which contains the catalytic serine residue. No complexes were observed in the Spn4H/chymotrypsin reaction.
Kinetic parameters for interaction of these Spn4 isoforms with furin and neutrophil elastase were recorded with the progress-curve method. Spn4F, Spn4G and Spn4H did not inhibit furin. The curves for elastase inhibition are shown in Figure 4. By non-linear regression, second order association rate constants (kassoc) between 3.9×104 and 7.6×104 M−1·s−1 were calculated. A synopsis of the kinetic data obtained is given in Table 1.
Table 1. Inhibitory profile of Spn4 isoforms.
The association rate constants kassoc and the SI values were determined as described in the Experimental section. –, no inhibition.
| Spn4E | Spn4F | Spn4G | Spn4H | |||||
|---|---|---|---|---|---|---|---|---|
| Enzyme | kassoc (M−1·s−1) | SI | kassoc (M−1·s−1) | SI | kassoc (M−1·s−1) | SI | kassoc (M−1·s−1) | SI |
| Furin (human) | (5.5±0.5)×106* | 3.5* | – | – | – | |||
| Neutrophil elastase | – | (4.6±0.4)×104 | >4‡ | (3.9±0.2)×104 | >4‡ | (7.6±0.9)×104 | >4‡ | |
| Cathepsin L | – | (1.0±0.1)×106 | 1.0 | (2.6±0.8)×105 | 4.3 | – | ||
| Cathepsin S | – | (1.8±0.4)×105 | 2.7 | (9.2±0.8)×104 | 3.6 | – | ||
*Data from [12].
‡Estimated from Western blotting experiments.
Spn4F and Spn4G are cross-class serpins and potent inhibitors of papain-like cysteine proteases
The presence of a bulky hydrophobic residue at the putative P2 position (Leu340) and the presence of a proline (position 344) at the P3′ position (for numbering, see [16]) suggested that Spn4G might inhibit papain-like cysteine proteases, since these features have been reported to favour interaction of serpins with such enzymes [30,31]. Consequently, the inhibitory potential of Spn4 variants against cysteine proteases was examined. In this initial enzyme screening, Spn4F and Spn4G appeared to inhibit cathepsins L and S respectively, but not cathepsin B. Spn4E and Spn4H were not active towards any of these enzymes. In order to investigate whether the decreased activities of the cysteine proteases in the presence of Spn4F or Spn4G were due to enzyme inhibition rather than to competitive substrate reactions, the ability of the proteases to form SDS-stable complexes with their counterparts was assessed. Each of the two Spn4 RSL isoforms initially appearing as cysteine protease inhibitors was able to form complexes with cathepsin S as detected by Western blotting after non-reducing SDS/PAGE (Figure 5). Under these conditions, some of the inhibitor molecules migrated in the 97 kDa region. We attribute these series of signals to (hetero)dimer formation of cleaved and uncleaved serpin molecules. We also consistently observed seemingly larger amounts of cleaved Spn4F and Spn4G respectively in comparison with Coomassie Brilliant Blue-stained SDS gels of the same preparations (results not shown), suggesting that the polyclonal antiserum used reacts more avidly with cleaved inhibitor molecules in Western blots.
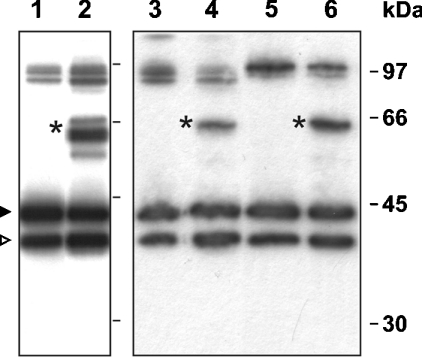
Figure 5. Spn4F and Spn4G may be captured in SDS-stable complexes following exposure to cysteine proteases.
Isoforms Spn4F and Spn4G were incubated at 30 °C for 5 min with cathepsin L or cathepsin S respectively. Complexes were detected after non-reducing SDS/PAGE (10% gels) using anti-Spn4 antibodies. Lane 1, Spn4F; lane 2, Spn4F exposed to cathepsin L; lane 3, Spn4F; lane 4, Spn4F exposed to cathepsin S; lane 5, Spn4G; lane 6, Spn4G exposed to cathepsin S. Inhibitor–enzyme complexes are marked by an asterisk. Uncleaved and cleaved forms of serpins are marked by black and open arrowheads respectively. The double band with a mobility corresponding to that of the 97 kDa size marker (right) is attributed to dimer formation of uncleaved and cleaved forms respectively of the serpin variants.
Spn4F also formed complexes with cathepsin L (Figure 5). In the case of the Spn4G–cathepsin L interaction, only trace amounts of complexes were detected (results not shown). No complexes between these serpins and the cysteine proteases were identified in the presence of reducing agents, suggesting that the thioester bond is not stable under the standard conditions commonly used for analysis of serpin–protease complexes.
To determine the SI, constant amounts of E-64 titrated cathepsins L and S respectively were incubated with various concentrations of Spn4F or Spn4G, and the residual enzyme activity was monitored by measuring the cleavage of the appropriate fluorogenic substrates. The SI values extrapolated for interaction of cathepsin L with Spn4F and Spn4G were 1.0 and 4.3 respectively. The corresponding values for the reactions between cathepsin S and these variants were 2.7 and 3.6 (Table 1) respectively. The rates of enzyme inhibition were determined under pseudo-first order conditions, applying the progress-curve method. The apparent association rate constants (kassoc) calculated for inhibition of cathepsin S by Spn4F and Spn4G were 1.8×105 and 9.2×104 M−1·s−1 respectively. The apparent association rate constants determined for the reaction between cathepsin L and Spn4F or Spn4G were 1.0×106 and 2.6×105 M−1·s−1 respectively. We conclude from these experiments that Spn4F and Spn4G are the first cross-class inhibitors identified in D. melanogaster that can inactivate papain-like cysteine proteases and members of the S1 family of serine proteases.
Localization of Spn4 variants in transfected COS-7 cells
The encounter with interaction partners at their homing compartment(s) is a prerequisite for a regulatory role of an inhibitor towards a target enzyme. Mammalian cysteine proteases of the papain family are usually transferred via their signal peptides into the lumen of the ER and then to lysosomes [32] and other organelles where they may exert various functions [33]. Cathepsin L, for instance, is involved in prohormone processing in secretory vesicles of neuroendocrine cells [34], and bovine serpin 2C, an inhibitor of cathepsin L, has been predicted to co-localize with its target in membrane-containing organelles [35]. We therefore investigated the cellular association of the cysteine protease inhibitors Spn4B and Spn4C and variants Spn4A and Spn4D (controls), all of which contain an N-terminal signal peptide [16]. Western blots of supernatants of the transfected COS-7 cells demonstrated that variants Spn4B–Spn4D are secreted into the medium (Figure 6). Variant Spn4A, however, was not exported to a major extent, in contrast with a deletion mutant (Spn4AΔHDEL) that lacks the C-terminal His-Asp-Glu-Leu sequence. Control experiments indicated that variants Spn4E–Spn4H that are devoid of the N-terminal 22 amino acids are not secreted (results not shown). Confocal immunofluorescence microscopy of transfected COS-7 cells individually expressing variants Spn4A–Spn4D provided no evidence for a significant signal overlap for the lysosomal marker Lysotracker Red DND99, and the antibody-mediated FITC fluorescence of the Spn4 isoforms (results not shown). Together, these data indicate (i) that the hydrophobic N-terminal sequence functions as a bona fide export signal and (ii) that the intracellular retention of variant Spn4A is mediated by the C-terminal ER retrieval signal. However, the data do not exclude that Spn4 variants may be associated with lysosomal structures in cells equipped with a regulated secretory pathway or under certain (patho)physiological conditions.
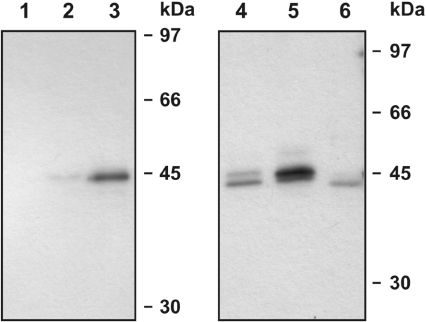
Figure 6. Variants Spn4B–Spn4D are exported, in contrast with variant Spn4A.
COS-7 cells were transfected individually with expression plasmids coding for isoforms Spn4A–Spn4D. After 24 h, the cellular supernatants were collected, resolved by SDS/PAGE (10% gels), and analysed by Western blotting for the presence of Spn4, using a polyclonal anti-Spn4 antiserum. Lane 1, control cells transfected with the empty vector; lanes 2 and 3, cells expressing Spn4A or Spn4AΔHDEL respectively; lanes 4–6, cells expressing variants Spn4B–Spn4D.
DISCUSSION
The exploration of the target specificities of the Spn4 RSL variants has revealed antiproteolytic activities directed towards subtilase-like and chymotrypsin-like serine proteases and towards papain-related cysteine proteases. The products of the Spn4 gene thus can exert protective effects against a broad spectrum of different types of proteolytic enzymes. These findings illuminate a remarkable general feature of serpins as protease inhibitors. Based on an economic splicing strategy, various inhibitors are generated from a single gene that targets proteases of different peptidase clans from different evolutionary lineages. Using a single scaffold, serpin variants are generated that belong to the most potent inhibitors of papain-like cysteine proteases and subtilase-like enzymes known, generated simply by exchange of the approx. 50 residues long RSL domain. There are some other genes from insects that use a multi-RSL strategy to generate serpin isoforms directed against different proteases, including serpin-1 from Manduca sexta [36] and SRPN10 from Anopheles gambiae [37]. The targets of these inhibitor variants include chymotrypsin-like enzymes and subtilisins of microbial origin; however, inhibition of cysteine proteases was not investigated. It remains to be determined whether some of the RSL variants from these or other serpin genes will inhibit enzymes from peptidase families other than those identified up to now.
The inhibitory activity of the RSL isoforms towards several classes of peptidases suggests that the Spn4 gene is designed as a broad-spectrum defence and control tool. Apart from the different target specificities of the individual RSL isoforms observed in vitro, several other features suggest such a role. First, each RSL isoform exists either as a variant that is exported or associated with the secretory pathway or as intracellular variant, potentially enabling the control of pathogens within and outside of cells. Secondly, previous investigations have shown that fruit flies challenged with microbes express transiently increased levels of Spn4 mRNAs [38,39]; however, the different Spn4 splice forms were not individually analysed. The presumed roles of Spn4 variants in defence might include inactivation of proteolytic enzymes from intruders or, alternatively, they could limit the spatial and/or temporal extent of the innate immune response. The precursors of several antimicrobial peptides, like diptericins or attacins that are induced after immune challenge, contain furin-like cleavage/consensus sites [40].
Little is known about the role of cysteine protease-inhibiting serpins in insects. However, cysteine proteases belong to the armory of many pathogens [41], and hosts use cysteine protease inhibitors to counteract the proteolytic activities of intruders [42,43], and vice versa. We note that isoforms Spn4B and Spn4C are equipped with an N-terminal signal peptide that mediates their export, in contrast with most other cysteine protease-inhibiting serpins known. Since the fruit fly has no adaptive immune system, the simultaneous expression of a set of effective protease inhibitors targeted against a wide spectrum of proteolytic enzymes might contribute to the survival strategy of insects. Further work is required to identify targets for Spn4 variants, both within the hitherto little characterized spectrum of natural Drosophila pathogens and the host's proteolytic complement [44,45]. A screening of endogenous cysteine proteases and similar enzymes of fruit fly pathogens is needed to fully elucidate the role of each Spn4 variant. In this context a comparison of the Spn4 isoforms with the products of the SRPN10 gene from A. gambiae [37] is interesting. The SRPN10 gene is the mosquito orthologue of Spn4; however, only the presumably furin-inhibiting RSL cassette depicts sequence similarity to variant A/E of Spn4, implying different functions for the other SRPN10 RSL variants. Moreover, SRPN10 transcripts coding for variants equipped with an obvious N-terminal signal peptide have not been detected. This is not surprising, since each of these insects has to cope with different environmental conditions. A. gambiae females are malaria vectors that can feed on blood; D. melanogaster lives on decaying fruit and is exposed to a different world of microbes and fungi. Recent findings indicate little overlap in the immune responses evoked by different pathogens [46].
Acknowledgments
We thank Dr J. Creemers for anti-Dfurin antibodies. The help of A. Strathmann and Y. Wang in some experiments is gratefully acknowledged.
References
- 1.Gettins P. G. W. Serpin structure, mechanism, and function. Chem. Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 2.Komiyama T., Gron H., Pemberton P. A., Salvesen G. S. Interaction of subtilisins with serpins. Protein Sci. 1996;5:874–882. doi: 10.1002/pro.5560050509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlings N. D., Barrett A. J. Evolutionary families of peptidases. Biochem. J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komiyama T., Ray C., Pickup D., Howard A., Thornberry N., Peterson E., Salvesen G. Inhibition of interleukin-1 beta converting enzyme by the cowpox virus serpin CrmA. An example of cross-class inhibition. J. Biol. Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 5.Silverman G. A., Bird P. I., Carrell R. W., Church F. C., Coughlin P. B., Gettins P. G., Irving J. A., Lomas D. A., Luke C. J., Moyer R. W., et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 6.Brinkmeyer S., Eckert R., Ragg H. Reformable intramolecular cross-linking of the N-terminal domain of heparin cofactor II: effects on enzyme inhibition. Eur. J. Biochem. 2004;271:4275–4283. doi: 10.1111/j.1432-1033.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 7.Huntington J. A., Read R. J., Carrell R. W. Structure of a serpin–proteinase complex shows inhibition by deformation. Nature. 2000;407:923–926. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 8.Dementiev A., Dobó J., Gettins P. G. W. Active site distortion is sufficient for proteinase inhibition by serpins. J. Biol. Chem. 2006;281:3452–3457. doi: 10.1074/jbc.M510564200. [DOI] [PubMed] [Google Scholar]
- 9.Garten W., Hallenberger S., Ortmann D., Schafer W., Vey M., Angliker H., Shaw E., Klenk H. D. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 10.Reinheckel T., Deussing J., Roth W., Peters C. Towards specific functions of lysosomal cysteine peptidases: phenotypes of mice deficient for cathepsin B or cathepsin L. Biol. Chem. 2001;382:735–741. doi: 10.1515/BC.2001.089. [DOI] [PubMed] [Google Scholar]
- 11.Chandran K., Sullivan N. J., Felbor U., Whelan S. P., Cunningham J. M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oley M., Letzel M. C., Ragg H. Inhibition of furin by serpin Spn4A from Drosophila melanogaster. FEBS Lett. 2004;577:165–169. doi: 10.1016/j.febslet.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 13.Osterwalder T., Kuhnen A., Leiserson W. M., Kim Y. S., Keshishian H. Drosophila serpin 4 functions as a neuroserpin-like inhibitor of subtilisin-like proprotein convertases. J. Neurosci. 2004;24:5482–5491. doi: 10.1523/JNEUROSCI.5577-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richer M. J., Keays C. A., Waterhouse J., Minhas J., Hashimoto C., Jean F. The Spn4 gene of Drosophila encodes a potent furin-directed secretory pathway serpin. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10560–10565. doi: 10.1073/pnas.0401406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han J., Zhang H., Min G., Kemler D., Hashimoto C. A novel Drosophila serpin that inhibits serine proteases. FEBS Lett. 2000;468:194–198. doi: 10.1016/s0014-5793(00)01224-2. [DOI] [PubMed] [Google Scholar]
- 16.Krüger O., Ladewig J., Köster K., Ragg H. Widespread occurrence of serpin genes with multiple reactive centre-containing exon cassettes in insects and nematodes. Gene. 2002;293:97–105. doi: 10.1016/s0378-1119(02)00697-2. [DOI] [PubMed] [Google Scholar]
- 17.Roebroek A. J., Creemers J. W., Pauli I. G., Kurzik-Dumke U., Rentrop M., Gateff E. A., Leunissen J. A., Van de Ven W. J. Cloning and functional expression of Dfurin2, a subtilisin-like proprotein processing enzyme of Drosophila melanogaster with multiple repeats of a cysteine motif. J. Biol. Chem. 1992;267:17208–17215. [PubMed] [Google Scholar]
- 18.Smolenaars M. M. W., Kasperaitis M. A. M., Richardson P. E., Rodenburg K. W., Van der Horst D. J. Biosynthesis and secretion of insect lipoprotein: involvement of furin in cleavage of the apoB homolog, apolipophorin-II/I. J. Lipid Res. 2005;46:412–421. doi: 10.1194/jlr.M400374-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Boussif O., Lezoualc'h F., Zanta M. A., Mergny M. D., Scherman D., Demeneix B., Behr J. P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morrison J. F., Walsh C. T. The behavior and significance of slow-binding enzyme inhibitors. Adv. Enzymol. Relat. Areas Mol. Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 21.Dufour E. K., Denault J. B., Bissonnette L., Hopkins P. C., Lavigne P., Leduc R. The contribution of arginine residues within the P6-P1 region of alpha 1-antitrypsin to its reaction with furin. J. Biol. Chem. 2001;276:38971–38979. doi: 10.1074/jbc.M102959200. [DOI] [PubMed] [Google Scholar]
- 22.Barrett A. J., Kembhavi A. A., Brown M. A., Kirschke H., Knight C. G., Tamai M., Hanada K. L-trans-epoxysuccinyl-leucylamido(4-guanidino)butane (E-64) and its analogues as inhibitors of cysteine proteinases including cathepsins B, H and L. Biochem. J. 1982;201:189–198. doi: 10.1042/bj2010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasiljeva O., Dolinar M., Turk V., Turk B. Recombinant human cathepsin H lacking the mini chain is an endopeptidase. Biochemistry. 2003;42:13522–13528. doi: 10.1021/bi035355k. [DOI] [PubMed] [Google Scholar]
- 24.Böhme C., Nimtz M., Grabenhorst E., Conradt H. S., Strathmann A., Ragg H. Tyrosine sulfation and N-glycosylation of human heparin cofactor II from plasma and recombinant Chinese hamster ovary cells and their effects on heparin binding. Eur. J. Biochem. 2002;269:977–988. doi: 10.1046/j.0014-2956.2001.02732.x. [DOI] [PubMed] [Google Scholar]
- 25.Hayflick J. S., Wolfgang W. J., Forte M. A., Thomas G. A unique Kex2-like endoprotease from Drosophila melanogaster is expressed in the central nervous system during early embryogenesis. J. Neurosci. 1992;12:705–717. doi: 10.1523/JNEUROSCI.12-03-00705.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Bie I., Savaria D., Roebroek A. J., Day R., Lazure C., Van de Ven W. J., Seidah N. G. Processing specificity and biosynthesis of the Drosophila melanogaster convertases dfurin1, dfurin1-CRR, dfurin1-X, and dfurin2. J. Biol. Chem. 1995;270:1020–1028. doi: 10.1074/jbc.270.3.1020. [DOI] [PubMed] [Google Scholar]
- 27.Kutty R. K., Kutty G., Kambadur R., Duncan T., Koonin E. V., Rodriguez I. R., Odenwald W. F., Wiggert B. Molecular characterization and developmental expression of a retinoid- and fatty acid-binding glycoprotein from Drosophila. A putative lipophorin. J. Biol. Chem. 1996;271:20641–20649. doi: 10.1074/jbc.271.34.20641. [DOI] [PubMed] [Google Scholar]
- 28.Bogerd J., Babin P. J., Kooiman F. P., André M., Ballagny C., van Marrewijk W. J. A., Van der Horst D. J. Molecular characterization and gene expression in the eye of the apolipophorin II/I precursor from Locusta migratoria. J. Comp. Neurol. 2000;437:546–558. doi: 10.1002/1096-9861(20001127)427:4<546::aid-cne4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 29.Cieplik M., Klenk H. D., Garten W. Identification and characterization of Spodoptera frugiperda furin: a thermostable subtilisin-like endoprotease. Biol. Chem. 1998;379:1433–1440. doi: 10.1515/bchm.1998.379.12.1433. [DOI] [PubMed] [Google Scholar]
- 30.Luke C., Schick C., Tsu C., Whisstock J. C., Irving J. A., Brömme D., Juliano L., Shi G. P., Chapman H. A., Silverman G. A. Simple modifications of the serpin reactive site loop convert SCCA2 into a cysteine proteinase inhibitor: a critical role for the P3′ proline in facilitating RSL cleavage. Biochemistry. 2000;39:7081–7091. doi: 10.1021/bi000050g. [DOI] [PubMed] [Google Scholar]
- 31.Al-Khunaizi M., Luke C. J., Askew Y. S., Pak S. C., Askew D. J., Cataltepe S., Miller D., Mills D. R., Tsu C., Brömme D., et al. The serpin SQN-5 is a dual mechanistic-class inhibitor of serine and cysteine proteinases. Biochemistry. 2002;41:3189–3199. doi: 10.1021/bi015999x. [DOI] [PubMed] [Google Scholar]
- 32.Pillay C. S., Elliot E., Dennison C. Endolysosomal proteolysis and its regulation. Biochem. J. 2002;363:417–429. doi: 10.1042/0264-6021:3630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chapman H. A., Riese R. J., Shi G.-P. Emerging roles for cysteine proteases in human biology. Annu. Rev. Physiol. 1997;59:63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- 34.Yasothornsrikul S., Greenbaum D., Medzihradszky K. F., Toneff T., Bundey R., Miller R., Schilling B., Petermann I., Dehnert J., Logvinova A., et al. Cathepsin L in secretory vesicles functions as a prohormone-processing enzyme for production of the enkephalin peptide neurotransmitter. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9590–9595. doi: 10.1073/pnas.1531542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hwang S. R., Stoka V., Turk V., Hook V. Y. The novel bovine serpin endopin 2C demonstrates selective inhibition of the cysteine protease cathepsin L compared to the serine protease elastase, in cross-class inhibition. Biochemistry. 2005;44:7757–7767. doi: 10.1021/bi050053z. [DOI] [PubMed] [Google Scholar]
- 36.Jiang H., Kanost M. R. Characterization and functional analysis of 12 naturally occurring reactive site variants of serpin-1 from Manduca sexta. J. Biol. Chem. 1997;272:1082–1087. doi: 10.1074/jbc.272.2.1082. [DOI] [PubMed] [Google Scholar]
- 37.Danielli A., Kafatos F. C., Loukeris T. G. Cloning and characterization of four Anopheles gambiae serpin isoforms, differentially induced in the midgut by Plasmodium berghei invasion. J. Biol. Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. [DOI] [PubMed] [Google Scholar]
- 38.Irving P., Troxler L., Heuer T. S., Belvin M., Kopczynski C., Reichhart J. M., Hoffmann J. A., Hetru C. A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2001;98:15119–15124. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vodovar N., Vinals M., Liehl P., Basset A., Derouard J., Spellman P., Boccard F., Lemaitre B. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11414–11419. doi: 10.1073/pnas.0502240102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedengren M., Borge K., Hultmark D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem. Biophys. Res. Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- 41.Turk B., Turk D., Salvesen G. S. Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr. Pharm. Design. 2002;8:1623–1637. doi: 10.2174/1381612023394124. [DOI] [PubMed] [Google Scholar]
- 42.Sakata Y., Arima K., Takai T., Sakurai W., Masumoto K., Yuyama N., Suminami Y., Kishi F., Yamashita T., Kato T., et al. The squamous cell carcinoma antigen 2 inhibits the cysteine proteinase activity of a major mite allergen, Der p 1. J. Biol. Chem. 2004;279:5081–5087. doi: 10.1074/jbc.M311585200. [DOI] [PubMed] [Google Scholar]
- 43.Dubin G. Proteinaceous cysteine protease inhibitors. Cell. Mol. Life Sci. 2005;62:653–669. doi: 10.1007/s00018-004-4445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubin G. M., Yandell M. D., Wortman J. R., Gabor Miklos G. L., Nelson C. R., Hariharan I. K., Fortini M. E., Li P. W., Apweiler R., Fleischmann W., et al. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross J., Jiang H., Kanost M. R., Wang Y. Serine proteases and their homologs in the Drosophila melanogaster genome: an initial analysis of sequence conservation and phylogenetic relationships. Gene. 2003;304:117–131. doi: 10.1016/s0378-1119(02)01187-3. [DOI] [PubMed] [Google Scholar]
- 46.Aguilar R., Jedlicka A. E., Mintz M., Mahairaki V., Scott A. L., Dimopoulos G. Global gene expression analysis of Anopheles gambiae responses to microbial challenge. Insect Biochem. Mol. Biol. 2005;35:709–719. doi: 10.1016/j.ibmb.2005.02.019. [DOI] [PubMed] [Google Scholar]