Abstract
The IGF-1R [type 1 IGF (insulin-like growth factor) receptor] is activated upon binding to IGF-I and IGF-II leading to cell growth, survival and migration of both normal and cancerous cells. We have characterized the binding interaction between the IGF-1R and its ligands using two high-affinity mouse anti-IGF-1R mAbs (monoclonal antibodies), 7C2 and 9E11. These mAbs both block IGF-I binding to the IGF-1R but have no effect on IGF-II binding. Epitope mapping using chimaeras of the IGF-1R and insulin receptor revealed that the mAbs bind to the CR (cysteine-rich) domain of IGF-1R. The epitope was finely mapped using single point mutations in the IGF-1R. Mutation of Phe241, Phe251 or Phe266 completely abolished 7C2 and 9E11 binding. The three-dimensional structure showed that these residues cluster on the surface of the CR-domain. BIAcore analyses revealed that IGF-I and a chimaeric IGF-II with the IGF-I C-domain competed for the binding of both mAbs with the IGF-1R, whereas neither IGF-II nor a chimaeric IGF-I with the IGF-II C-domain affected antibody binding. We therefore conclude the IGF-I C-domain interacts with the CR (cysteine-rich) domain of the receptor at the cluster of residues Phe241, Phe251 and Phe266. These results allow precise orientation of IGF-I within the IGF-I–IGF-1R complex involving the IGF-I C-domain binding to the IGF-1R CR domain. In addition, mAbs 7C2 and 9E11 inhibited both IGF-I- and IGF-II-induced cancer cell proliferation, migration and IGF-1R down-regulation, demonstrating that targeting the IGF-1R is an effective strategy for inhibition of cancer cell growth.
Keywords: binding epitope, cancer, insulin-like growth factor (IGF), type 1 insulin-like growth factor receptor (IGF-1R), mono-clonal antibody
Abbreviations: BHK21, baby-hamster kidney 21; CR, cysteine-rich; Eu–IGF-I, europium-labelled receptor grade human insulin-like growth factor-I; HBS, Hepes-buffered saline; HEK-293-EBNA cells, human embryonic kidney 293 cells expressing EBNA-1; HRP, horseradish peroxidase; IGF, insulin-like growth factor; IGF-1R, type 1 IGF receptor; IGF-ICII, IGF-I with the IGF-II C-domain; IGF-IICI, IGF-II with the IGF-I C-domain; IR, insulin receptor; ka, association rate; kd, dissociation rate; mAb, monoclonal antibody; P6 cells, Balb/c/3T3 cells overexpressing human IGF-1R; R− cells, mouse 3T3-like cells with a targeted ablation of the IGF-1R gene; RU, response units; s-IGF-1R, the soluble extracellular part of the human IGF-1R1–906
INTRODUCTION
The IGF-1R [type 1 IGF (insulin-like growth factor) receptor] is a transmembrane protein tyrosine kinase that plays important roles in both normal and malignant growth [1]. The IGF-1R and IR (insulin receptor) share 70% sequence identity [2,3]. Both receptors are glycoproteins and consist of two α- and two β-subunits. The α-subunits are entirely extracellular and are involved in ligand binding, whereas the β-subunits contain transmembrane and intracellular domains [2]. Binding of the ligands IGF-I or IGF-II to the α-subunits of the IGF-1R induces conformational changes in the receptor leading to autophosphorylation of three tyrosine residues in the kinase catalytic C-domains of the β-subunits [4] and activation of downstream signalling pathways [5,6]. Activation of the IGF-1R results in cell proliferation, survival and migration.
IGF-1R overexpression or increased IGF-1R kinase activity is associated with a broad range of human cancers and therefore the IGF-1R is widely considered as a very promising target for cancer treatment [1,7,8]. Generation of IGF-1R inhibitors would be greatly assisted by a good understanding of how the IGF-I–IGF-1R complex is formed, but so far a structure of the IGF-I–IGF-1R complex has not been solved. The only IGF-1R ectodomain structure available is of the first three domains [L1, CR (cysteine-rich), L2] and this receptor fragment is unable to bind ligand [9]. Alanine scanning mutagenesis of the IGF-1R [10] has located IGF-I-binding sites to the L1 domain and to the region between residues 692 and 702 of the insert domain. In addition, residues Arg240, Phe241, Glu242 and Phe251 of the CR domain form a small patch together with Trp79 in the L1 domain for binding to IGF-I [10]. In contrast, none of these residues is involved in binding to IGF-II [11]. Studies using IR/IGF-1R chimaeras also demon-strated the importance of the IGF-1R CR domain for IGF-I binding [12,13].
Mutagenesis has also been used to map the IGF-I-binding sites for the IGF-1R (as reviewed in [14]). IGF-I residues important for IGF-1R binding include Phe23, Tyr24, Tyr31, Arg36, Arg37, Val44 and Tyr60. Recently we demonstrated that the C- and D-domains of IGF-I and IGF-II contribute to their differences in IGF-1R binding specificity and play a role in its activation [15]. IGF-I with the IGF-II C- and D-domains (IGF-I CIIDI) bound to IGF-1R with an affinity equal to IGF-II, whereas IGF-II with the IGF-I C and D domains (IGF-II CIDI) bound with an affinity equal to IGF-I.
We still have relatively little information regarding the orient-ation of the ligand in the IGF-I–IGF-1R complex. While it is tempting to use the information revealed from recent cross-linking studies between insulin and the IR [16–19] as a model for the IGF-I–IGF-1R interaction, there are differences in the binding curves from the two interactions suggesting different modes of interaction. Insulin exhibits a bell-shaped dose–response curve for accelerated dissociation from the IR, whereas IGF-I has a sigmoid curve of dissociation from the IGF-1R [3,20]. Therefore in order to generate specific inhibitors we need to understand how the IGF-I–IGF-1R complex was formed. Using two high affinity IGF-1R mAbs (monoclonal antibodies), 7C2 and 9E11 (described in [21]), we have characterized the binding epitope at the individual amino acid level and demonstrated the interaction of the IGF-I C-domain with the IGF-1R CR-domain thereby allowing the orientation of IGF-I in the IGF-1R-binding pocket. These mAbs also inhibit IGF-I binding, cancer survival and migration and lead to IGF-1R down-regulation, demonstrating that targeting the IGF-1R is an effective strategy for inhibition of cancer cell growth.
EXPERIMENTAL
Materials
All chemicals were from Sigma, except Lipofectamine™ 2000 (Life Technologies) and BSA (BovoStar grade; Bovogen). The αIR-3 mAb and control IgG1 were purchased from Calbiochem and Chemicon respectively, and mAb 24-60 was a gift from Professor K. Siddle (Department of Clinical Biochem-istry, University of Cambridge, Cambridge, U.K.). Anti-IGF-1R antibody C-20 was from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.). mAbs 24-55, 16-13 and receptor grade IGF-I and IGF-II were from GroPep Ltd (Adelaide, SA, Australia). The chimaeric ligands IGF-I CII (IGF-I with the IGF-II C-domain) and IGF-II CI (IGF-II with the IGF-I C-domain) were made by Dr A. Denley (School of Molecular and Biomedical Science, The University of Adelaide, Adelaide, SA, Australia) [15]. Biotinyl-ated mAb 16−13 was provided by Dr F. Occhiodoro [CSIRO (Commonwealth Scientific and Industrial Research Organisation), Clayton South, VIC, Australia]. MagicMark™ XP molecular-mass markers were from Invitrogen. Expression vectors encoding secreted alanine mutants of the IGF-1R or the chimaeric IGF-1R/IR256–266 were generated as described previously [10].
Cell lines and cell culture conditions
The MCF-7 breast cancer cell line and the HT-29 colon cancer cell line were obtained from the American Type Culture Collection (Manassas, VA, U.S.A.). HEK-293-EBNA cells (human embryo-nic kidney 293 cells expressing EBNA-1) were from Invitrogen. BHK21 (baby-hamster kidney 21) cells producing recombinant s-IGF-1R (the soluble extracellular part of the human IGF-1R1–906) were provided by Dr K. Surinya (School of Mole-cular and Biomedical Science, The University of Adelaide, SA, Australia). BHK21 cells producing s-IGF-1R (BHK21+IGF-1R) were grown in GMEM-S (Glasgow minimal essential medium for methionine sulphoximine selection) containing 10% (v/v) dialysed fetal bovine serum, 2% (v/v) glutamine synthetase (50×) and 25 μM methionine sulfoximine. P6 cells (Balb/c/3T3 cells overexpressing human IGF-1R) and R− cells (mouse 3T3-like cells with a targeted ablation of the IGF-1R gene) were provided kindly by Professor R. Baserga (Kimmel Cancer Center, Thomas Jefferson University, Philadelphia, PA, U.S.A.) [22]. NIH 3T3 cells stably expressing chimaeric IGF-1R/IR receptors were gifts from Professor A. Ullrich (Max Planck Institute of Biochemistry, Martinsried, Germany) [23,24].
BIAcore determination of mAb affinities for the IGF-1R
The affinities of anti-IGF-1R mAbs for the ectodomain of IGF-IR were determined using a BIACORE 2000 (BIAcore, Uppsala, Sweden) as basically described previously [25]. Anti-mouse IgG1 polyclonal antibody (BIAcore) was immobilized on to a CM5 chip via amine group linkage using standard coupling procedures as described in [26]. Purified mAbs (7C2, 9E11 or αIR-3; 2 μg/ml; 5 μl) in HBS (Hepes-buffered saline) running buffer (10 mM Hepes, pH 7.4, 150 mM NaCl, 3.4 mM EDTA and 0.005% Tween 20) were captured on the anti-mouse IgG1 surface at a flow rate 3 μl/min to give a response of ∼50 RU (response units). s-IGF-1R (75 μl) was immediately injected at various concen-trations (12.5, 6.25, 3.12, 1.56 or 0.76 nM) at 3 μl/min for 25 min and was then allowed to dissociate for 50 min. All flow cells were regenerated for 4 mins with 10 mM glycine/HCl (pH 1.7). Reference flow cell data were subtracted from all runs to account for bulk refractive index due to the buffer. The response to HBS injection (without s-IGF-1R) over the mAb was subtracted from all runs to account for bulk refractive index due to the buffer in the second association phase. Rate constants were derived using BIA Evaluation 3.2 software. Interactions were fitted globally across all concentrations to a 1:1 Langmuir binding model, which describes 1:1 binding between analyte (A) and ligand (B) (A+B↔A−B). The affinity constant for each mAb, KD, was calculated from the ratio of the rate constants kd (dissociation rate)/ka (association rate). Mass transfer control experiments were conducted according to the BIAcore 2000 instrument handbook.
BIAcore analysis of mAb competition with ligand for IGF-1R binding
Solutions containing a fixed concentration of s-IGF-1R (12.5 nM) and a range of concentrations of IGF-I or IGF-II (0.01, 0.1, 1, 10, 100 and 250 nM) in HBS running buffer were equilibrated for 3 h at room temperature. They were passed over the captured anti-IGF-1R mAbs as described above. The same experiment was carried out for the chimaeric ligands, IGF-I CII and IGF-II CI, except the fixed concentration of the s-IGF-1R was 6.25 nM.
Europium competition assays
The abilities of the mAbs 9E11 and 7C2 to inhibit the interaction between europium-labelled IGFs and the IGF-1R were measured in competition-binding assays conducted as described generally in [15]. Competition between other anti-IGF-1R mAbs (24-60 and αIR-3) and europium-labelled Fab domains of mAbs 9E11 or 7C2 for binding to the IGF-1R was measured using the same assay. IGF-1R was captured on white Greiner Lumitrac 600 plates coated with the mAb 24-31, an anti-IGF-1R mAb that did not interfere with ligand binding, or mAb 24-55, which does not bind to the same epitope as mAbs 24-60 and αIR-3 (see Table 2) [13,27]. IGF-1R was either isolated from P6 cell lysates [15] or from culture supernatants from mammalian cells expressing s-IGF-1R (BHK21+sIGF-1R) or HEK-293-EBNA cells transiently expressing IGF-1R alanine mutants (see below). Eu–IGF-I (europium-labelled receptor grade human IGF-I) and Eu–IGF-II were prepared by Mr P. A. Hoyne (CSIRO Health Sciences and Nutrition, Parkville, Australia) as outlined by the manufacturer and described previously [15]. The final concentration of the Eu–IGF-I or -II solutions was ∼10 nM in each well of the plate (300000 counts). Fab domains of 9E11 and 7C2 mAbs were labelled using the same method and were used at a final concentration of 3.3 nM (300000 counts). Fab domains were prepared for labelling by first digesting the mAbs with papain in the presence of the reducing agent cysteine, as described previously [28] and then purifying them using a Protein A column as described previously [29]. In all assays, the background fluorescence detected in wells containing no IGF-1R was subtracted from all other fluorescence values. Prism 3.03 software was used to calculate IC50 values employing curve-fitting with a one-site competition model [15]. Paired Student's t tests were used for all statistical analyses. Significance was accepted at P<0.05.
Table 2. Summary of epitopes of murine anti-IGF-1R mAbs and their effect on IGF-I binding to the IGF-1R.
Epitope mapping with chimaeric receptors
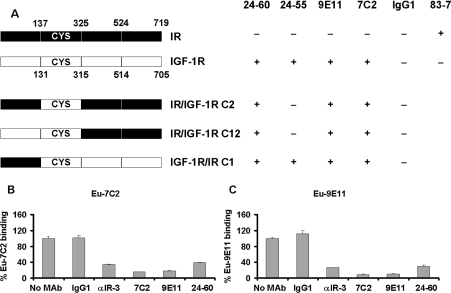
The three chimaeric receptors, IR/IGF-1R C2, IR/IGF-1R C12 and IGF-1R/IR C1 [23,24], were used to broadly define the 9E11 and 7C2 epitopes (Figure 2). Binding of both antibodies (10 μg/ml) to the NIH 3T3 stable cell lines expressing the chimaeric receptors was assessed by flow cytometry analysis. The anti-IgG1 antibody was used as a negative isotype control and the mAbs 24-60 and 24-55 were applied as positive controls. mAbs bound to the chimaeric receptors were detected by FITC-conjugated sheep anti-mouse IgG (Chemicon). Flow cytometry acquisition was carried out with a FACScan Flow Cytometer using CellQuest Pro software (Becton Dickinson).
Figure 2. Epitope mapping using IGF-1R/IR chimaeric receptors and competition with other mAbs.
(A) Schematic summary of epitope mapping by FACS analysis using chimaeric IGF-1R/IR receptors. Antibodies binding to IGF-1R, IR and chimaeric receptors are shown as +, whereas lack of binding is denoted by −. The ability of europium labelled mAbs 7C2 (B) and 9E11 (C) to compete with 25 nM unlabelled 7C2, 9E11, 24-60, αIR-3 and control IgG1 for binding to the sIGF-1R was measured as described in Materials and methods section. Results are expressed as a percentage of europium label bound in the absence of competing mAb, and are means±S.D. of three independent experiments performed in triplicate.
Epitope mapping using alanine mutants of the IGF-1R
Alanine mutants of the IGF-1R or chimaeric IGF-1R/IR256–266 (residues 256–266 of the IGF-1R replaced with amino acids 262–277 of IR-A) [30] were expressed transiently in HEK-293-EBNA cells following transfection of the recombinant cDNAs using Lipofectamine™ 2000 reagent according to the manufac-turer's instructions. Culture supernatants were harvested after 72 h and an ELISA was used to measure expression of the chimaeras. Biotinylated mAb 16-13 detected receptor captured on a 96-well plate coated with mAb 24-55 (0.25 μg/well). The epitope for the mAb 16-13 is near the N-terminus of the IGF-1R (between residues 62 and 184) [27], which is intact in all of the recombi-nant constructs. The plate was then washed and the binding was detected with streptavidin–HRP (horseradish peroxidase; diluted 1:200; Chemicon) and ABTS [2,2′-azinobis-(3-ethylbenzo-thiazoline-6-sulfonic acid)] reagent (Roche) following the manufacturer's instructions.
The supernatant for each mutant of IGF-1R was diluted to give the same absorbance as s-IGF-1R (0.28 mg/ml) as detected in the ELISA and 100 μl of each diluted supernatant was added to the europium binding assay as described above. This allowed a direct comparison of mAb binding between wild-type and mutant s-IGF-1R.
Cell viability assay
A total of 12000 HT-29 cells were seeded per well into 96-well flat-bottom plates and cultured for 48 h. Prior to treatment the cells were washed and serum-starved for 5 h. Different treatment solutions in serum-free growth medium containing 0.5% (w/v) BSA were added to wells for a further 48 h. Cell proliferation was measured using the CellTiter-Glo luminescent cell viability assay (Promega) following the manufacturer's instructions [31]. Luminescence was recorded on a POLARstar Galaxy microplate reader (BMG Lab Technologies) and FLUOstar Galaxy PC software. The background luminescence for the wells containing no cells was subtracted from all other luminescence counts.
Migration assay
Migration assays were conducted as described previously [32]. Briefly, a 96-well modified Boyden chamber (Neuro Probe) and a 12 μm polycarbonate filter coated with type 1 collagen were used. Cells (60000/well) were prelabelled with 1 μg/ml calcein (Molecular Probes) and then incubated with 25 nM mAbs 7C2, 9E11 or αIR-3, or IgG1 (as a negative control) for 1 h at 37 °C in 5% CO2/95% air atmosphere. Cells migrated toward IGFs or IGF chimaeras for 5 h.
Receptor down-regulation analysis
Down-regulation of IGF-1R by mAbs 9E11 and 7C2 was demonstrated in MCF-7 cells using the method essentially as described previously [33]. Cells (7×105) seeded into each well of six-well plates were incubated in serum-free growth medium for 20 h at 37 °C in a 5% CO2/95% air atmosphere. Treatment solutions (50 nM IGF-I or 25 nM mAbs 9E11, 7C2 or αIR-3) were added and the cells were incubated for 24 h at 37 °C in a 5% CO2/95% air atmosphere. Total protein (15 μg) for each treated MCF-7 cell lysate and R− cell lysate (negative control) were separated on a SDS/10% (w/v) PAGE gel under reducing conditions and transferred on to a nitrocellulose membrane (Hybond™ P, Amersham Pharmacia Biotech). Total protein was determined using the BCA (bicinchoninic acid) assay (Pierce Biotechnology). Membranes were blocked with 5% (w/v) skimmed milk in PBS for 2 h at room temperature, and the IGF-1R was detected using an anti-IGF-1R antibody (C-20; diluted 1:1000) followed by an donkey anti-rabbit HRP-conjugated antibody (diluted 1:10000; Rockland) and enhanced chemiluminescence detection.
RESULTS
mAbs 7C2 and 9E11 bind IGF-1R with high affinity
Two high-affinity mAbs (7C2 and 9E11) were selected from a hybridoma screen and were shown to be specific for the IGF-1R [21]. Both mAbs 7C2 and 9E11 bind the s-IGF-1R with high affinity (0.5–2.1 nM) resulting from fast association and slow dissociation rates as measured by BIAcore analysis (Table 1). Kinetic analyses suggest that 7C2 has a slightly higher affinity than αIR-3 for binding to the receptor, although the KD values for the three mAbs were not significantly different (P>0.05).
Table 1. BIAcore kinetic analysis of binding of mabs to s-IGF-1R.
The results generated are from three separate runs and the average of ka, kd and KD for each mAb is shown. Results are means (±S.D.) from three independent experiments. The dissociation constant KD=kd/ka.
| mAb | ka (1/Ms) (×105) | kd (1/s) (×10−4) | KD (M) (×10−9) |
|---|---|---|---|
| 7C2 | 0.66 (±1.8) | 0.31 (±0.2) | 0.5 (±1.6) |
| 9E11 | 0.8 (±0.2) | 1.7 (±0.1) | 2.1 (±0.4) |
| αIR-3 | 1.0 (±0.3) | 1.2 (±0.5) | 1.3 (±0.5) |
mAbs 7C2 and 9E11 compete with IGF-I but not IGF-II for binding to the IGF-1R
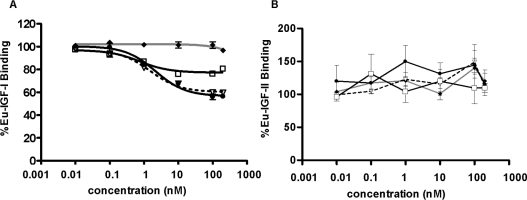
mAbs 9E11 and 7C2 competed with Eu–IGF-I for binding to cap-tured solubilized IGF-1R in competition-binding assays (Fig-ure 1A). Similar concentrations of mAbs 7C2 and 9E11 were able to inhibit s-IGF-1R binding, with 7C2 being slightly more potent than 9E11 as was seen in the BIAcore experiments. While mAb 24-60 has been reported to inhibit IGF-I binding with an IC50 of 0.5 nM [27] it does not compete as effectively as mAbs 7C2 and 9E11 in our assay, suggesting that the epitope for these antibodies differ slightly (Figure 1A). All mAbs failed to inhibit Eu–IGF-II binding (Figure 1B).
Figure 1. Inhibition of IGF-I binding to the IGF-1R by anti-IGF-1R mAbs.
Increasing concentrations of mAbs 7C2 (●), 9E11 (▽) and 24-60 (□) and a control IgG1 (◆) were tested for their ability to inhibit Eu–IGF-I (A) and Eu–IGF-II (B) binding to immunocaptured IGF-1R from solubilized P6 cell extracts. IGF-I and IGF-II (10 nM) were also used in the competition assay. Results are expressed as a percentage of europium label bound in the absence of competing mAb or ligand. The P values were calculated by comparing the data for no treatment (buffer) with other data.
mAbs 7C2 and 9E11 bind to the CR domain of the IGF-1R
Cells expressing IGF-1R/IR chimaeras were used to define the epitopes of mAbs 7C2 and 9E11. mAbs 7C2, 9E11 and mAb 24-60 bound to all three chimaeras of IGF-1R/IR, whereas mAb 24-55 only bound to the IGF-1R/IR C1 expressing cells (Fig-ure 2A). It can therefore be concluded that the epitope for the mAbs 7C2, 9E11 and 24-60 are in the CR domain of the IGF-1R between amino acids 131 and 315. The reported epitopes for mAbs 24-60 and 24-55 are between amino acids 184 and 283 and 440 and 586 of the IGF-1R respectively [27].
Competition of europium-labelled 7C2 and 9E11 with other mAbs
Further confirmation of the epitope mapping was obtained by testing the ability of mAbs 9E11 and 7C2 to compete with previously characterized mAbs 24-60 and αIR-3 in a europium competition-binding assay. As the flow cytometry analysis of chimaeric receptors showed that the epitope for mAbs 7C2 and 9E11 was between residues 131 and 315 of IGF-1R, the mAb 24-55, which binds to IGF-1R residues 440–586, was used to capture s-IGF-1R. Europium-labelled mAbs 7C2 and 9E11 both significantly competed with 24-60, αIR-3 and each other for binding to the IGF-1R (Figures 2B and 2C). This indicates that all four antibodies share an overlapping epitope on the IGF-1R.
Identification of IGF-1R residues involved in binding
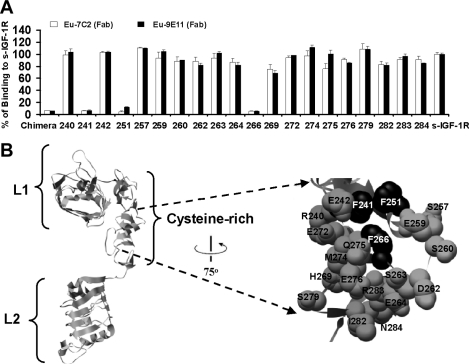
To define the epitope of mAbs 7C2 and 9E11 further, the binding of europium-labelled mAb 9E11 and 24-60 Fab domains to alanine mutants of the IGF-1R CR domain was investigated. Also, the role of the IGF-1R mobile loop between residues 255–265 [34] on mAb binding was tested using the chimaeric IGF-1R/IR256–266. ELISA detection in culture supernatants was shown for transient expression of all recombinant soluble IGF-1R alanine mutant receptors, except IGF-1RI255A. Supernatants were diluted to give the same s-IGF-1R concentration as the wild-types-IGF-1R (0.28 mg/ml). Binding assays using europium-labelled 9E11 and 7C2 (Fab fragments) revealed that the mAbs could bind all but three of the s-IGF-1R alanine mutants (IGF-1RF241A, IGF-1RF251A and IGF-1RF266A; Figure 3A). It is unlikely that the alanine mutagenesis is causing gross structural perturbation leading to the lack of mAb binding, as the F266A mutation does not affect IGF-I or IGF-II binding, and the mutation of F241A and F251A do not affect IGF-II binding [10,11]. Also, the chimaeric secreted receptor IGF-1R/IR256–266 bound poorly to both europium-labelled mAbs (Figure 3A). This is consistent with the fact that IGF-1RF266A also binds poorly. The defined epitope therefore consists of a hydrophobic patch including residues Phe241, Phe251 and Phe266, and is shown mapped on to the IGF-1R structure in Figure 3(B). This represents the first example of an IGF-1R monoclonal being mapped at this level of resolution.
Figure 3. Epitope mapping using IGF-1R alanine mutants and the chimaeric IGF-1R/IR256–266 receptor.
(A) Alanine mutants, the chimaeric IGF-1R/IR256–266 receptor and wild-type IGF-1R (s-IGF-1R) were expressed as soluble receptors in culture medium. Levels of receptor in all supernatants were measured by ELISA and adjusted to 0.28 mg/ml prior to performing binding assays using europium-labelled mAbs 7C2 and 9E11 (Fab domains). Binding to culture supernatants is expressed as a percentage of binding to s-IGF-1R. Residue number is indicated below for alanine mutants. Chimaeric refers to the chimaeric IGF-1R/IR256–266. The graph shown is representative of three experiments and bars are means±S.D. of triplicates. (B) Ribbon diagram of the IGF-1R L1, CR and L2 domains based on the structure reported by Garrett et al [9] highlighted space-filled using black alanine mutants, which disrupt binding of europium-labelled 7C2 and 9E11 binding (residues Phe241, Phe251 and Phe266). The Figure was created using the UCSF Chimera molecular graphics program [45].
IGF-I and the IGF-I C-domain compete with the mAbs 9E11 and 7C2
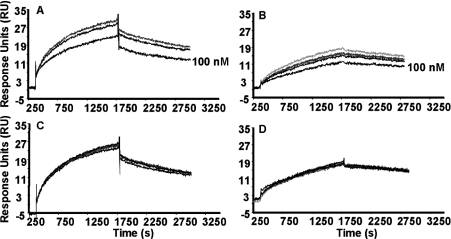
BIAcore binding studies revealed that pre-incubation of the s-IGF-1R with increasing concentrations of IGF-I and IGF-II CI caused a reduction in the response when binding to mAb 9E11 (Figures 4A and 4B). At the highest concentration (100 nM) there was a 74±7% reduction in the amount of s-IGF-1R binding to the 9E11 sensor surface compared with the response to s-IGF-1R alone. Interestingly, IGF-II and IGF-I CII did not cause any reduc-tion in response compared with s-IGF-1R alone, with a 107±6% response to the same concentration (Figures 4C and 4D). A similar effect was seen with mAb 7C2 (results not shown). Hence it can be concluded that the IGF-I C-domain is responsible for compet-ition of IGF-I with mAbs 7C2 and 9E11 for binding to the s-IGF-1R.
Figure 4. Effect of IGF-I, IGF-II and chimaeras on binding of s-IGF-1R to mAb 9E11.
BIAcore analysis was used to detect the effect of (A) IGF-I, (B) IGF-II CI, (C) IGF-II or (D) IGF-I CII on the association and dissociation of s-IGF-1R with captured mAb 9E11. Increasing concentrations of ligands (from top to bottom, 0, 0.1, 1, 10 and 100 nM) were pre-incubated with s-IGF-1R prior to injection over a mAb 9E11 sensor surface. IGFs and chimaeric ligands were incubated with 12.5 or 6.25 nM of s-IGF-1R respectively. Each panel is representative of two or three experiments.
Inhibition of cell viability and migration
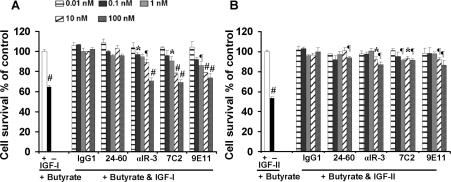
Proliferation assays revealed that mAbs αIR-3, 7C2 and 9E11 caused a small but statistically significant inhibition of survival of cultured HT-29 cells induced by 10 nM IGF-I in the presence of 5 mM butyrate, a potent apoptosis-inducing agent (Figure 5A). In addition, mAbs αIR-3, 7C2 and 9E11 inhibited the IGF-II-induced cell proliferation significantly (Figure 5B). At a concen-tration of 100 nM, mAbs 7C2, 9E11 and αIR-3 blocked IGF-in-duced cell proliferation to the same extent. However, when HT-29 cells were rescued to a similar extent by IGF-I or IGF-II (using 10 or 50 nM concentrations respectively), none of the mAbs inhibited the effect of IGF-II as potently as they did for IGF-I (Figure 5B). Interestingly, despite previous reports of mAb 24-60 being able to inhibit IGF-I binding [27], in this assay mAb 24-60 had little or no effect on IGF-I- or IGF-II-mediated proliferation.
Figure 5. Inhibition of IGF-I action through the IGF-1R by anti-IGF-1R mAbs.
Proliferation of HT-29 colon cancer cells was measured in the presence of 5mM butyrate and 10 nM IGF-I (A) or 50 nM IGF-II (B) with increasing amounts of mAbs 7C2, 9E11, 24-60 and αIR-3 or a control IgG1. Results are expressed as a percentage of proliferation in the presence of ligand alone. Results are means±S.D. of triplicate samples and are representative of three separate experiments. The P values were calculated by comparing the results for the treatment of cells with IGF-I or IGF-II (but not mAb) with other treatments. *0.01<P<0.05; ¶0.001<P<0.01; #P<0.001.
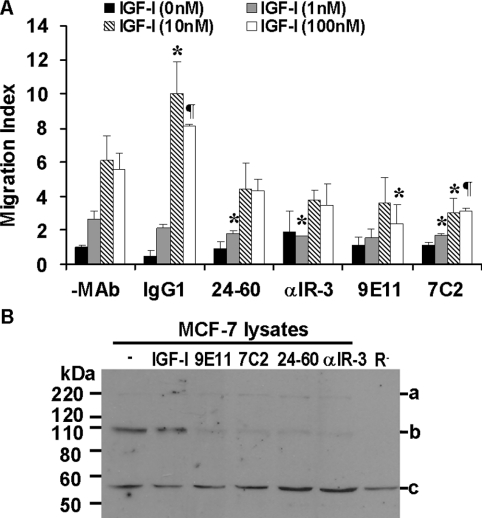
mAbs 9E11 and 7C2 (25 nM) inhibited migration of MCF-7 breast cancer cells towards IGF-I (Figure 6A) and IGF-II (results not shown). Interestingly, mAbs αIR-3 (25 nM) and 24-60 (25 nM) appear only slightly less potent in this assay, with significant inhibition of migration seen only at the lowest IGF-I concentration used. This observation would suggest that inhibition of migration is dependent predominantly on the ability of the antibodies to promote receptor down-regulation (see below) rather than a direct inhibitory effect via ligand binding.
Figure 6. Inhibition of MCF-7 breast cancer cell migration and stimulation of IGF-1R down-regulation by anti-IGF-1R mAbs.
(A) MCF-7 cell migration towards increasing concentrations of IGF-I was measured in the presence of 25 nM mAbs 7C2, 9E11, 24-60 and αIR-3 or a control IgG1. P values represent significance compared with no mAb at the matched concentration (*0.01<P<0.05; ¶ 0.001<P<0.01). Results are means±S.D. of triplicates and a representative of three separate experiments is shown. (B) Immunoblot of MCF-7 cell lysates from untreated cells or MCF-7 cells treated with IGF-I, 9E11, 7C2, 24-60 or αIR-3 and R− cell lysates probed with anti-IGF-1R C-20. Bands represent (a) pro-IGF-1R, (b) IGF-1R β subunit and (c) a non-specific band (used here as loading control).
mAbs 7C2 and 9E11 down-regulate IGF-1R
Treatment of the MCF-7 breast cancer cell line with mAbs 7C2 and 9E11 resulted in down-regulation of the IGF-1R. Following 24 h of treatment of MCF-7 cells with 25 nM 7C2, 9E11, 24-60 or αIR-3, the level of IGF-1R in cell lysates was dramatically reduced compared with untreated cells, and all antibodies at this concentration promoted down-regulation to a similar extent (Figure 6B). In contrast, the treatment with IGF-I (50 nM) did not down-regulate the IGF-1R (Figure 6B). A non-specific band at 55 kDa was detected in lysates of cells not expressing the IGF-1R (R− cells) using the anti-IGF-1R antibody (C-20).
DISCUSSION
In the present study, two IGF-1R mAbs directed against the human IGF-1R (mAbs 7C2 and 9E11) were used to characterize the interaction of IGF-I with its receptor and to orientate the IGF-I molecule in the IGF-1R-binding pocket. Both antibodies interact with the IGF-1R with high affinity (0.5–2.1 nM) and are able to inhibit IGF-I but not IGF-II binding (Figure 1A). Epitope mapping using IGF-1R/IR chimaeras located the mAb 7C2- and 9E11-binding site in the CR domain (Figure 2). Interestingly this corresponds to the known epitopes of mAbs 24-60 and αIR-3 (Table 2). Fine epitope mapping with alanine mutants of the IGF-1R CR domain showed that mAbs 7C2 and 9E11 do not bind to IGF-1RF241A, IGF-1RF251A and IGF-1RF266A. Therefore we can conclude that residues Phe241, Phe251 and Phe266 form the binding epitope for both mAbs 7C2 and 9E11. When mapped on to the IGF-1R structure [9] (Figure 3B) it can be seen that these residues form a continuous hydrophobic patch. Previously, an IGF-I-binding site was identified by alanine scanning mutagenesis of the CR domain, which involves residues Arg240, Phe241, Glu242 and Phe251 [10]. These residues form a small patch on the CR domain, which together with Trp79 in the L1 domain bind IGF-I but are not involved in IGF-II binding [10]. Interestingly, the binding epitope of mAbs 7C2 and 9E11 overlap the IGF-I-binding site involving residues Phe241 and Phe251 of IGF-1R. Hence it can be concluded that binding of mAbs 7C2 and 9E11, in particular to amino acids Phe241 and Phe251, is responsible for their specific inhibition of IGF-I binding to the receptor. The difference in binding epitopes for IGF-I and the antibodies most probably reflects a different mechanism of interaction with antibody–epitope interactions, mediated generally through small thermodynamic hot-spots [35].
Furthermore, using IGF-I/IGF-II chimaeras we showed that the mAbs inhibit IGF-I binding by inhibiting the interaction of the IGF-I C-domain with the IGF-1R CR domain. Both IGF-I and IGF-II CI inhibited s-IGF-1R binding to mAbs 7C2 and 9E11, whereas IGF-II and IGF-I CII were unable to compete for the mAb-binding site. As we have precisely defined the mAb epitopes, we can conclude that the IGF-I C-domain interacts with the IGF-1R CR domain.
Therefore the present study has precisely located the site of interaction between the IGF-1R CR domain and the IGF-I C-domain. This has greatly improved our understanding of the interaction through the CR region, which was implicated previously using chimaeric receptors [36]. Although we have shown previ-ously that the C-domains of IGF-I and IGF-II confer their binding specificity for the IGF-1R [15], the present study shows that it is the C-domain interaction with the residues Phe241 and Phe251 of the CR region, which provides additional binding energy resulting in a higher affinity of IGF-I than IGF-II for the IGF-1R. Previous studies using site-directed mutagenesis have identified residues Arg36, Arg37 and Tyr31 in the IGF-I C-domain as being important for IGF-1R binding [36–38]. A recently published model [39] con-structed using the constraint of Arg36 of IGF-I-contacting residue Glu242 of the CR of IGF-1R provides a framework for discussion and future experimentation, although it is unable to represent the entire complex as it is based on only the first three N-terminal domains, which, by themselves, are unable to bind ligand. The model does encompass some experimentally derived observ-ations. For example, residues Asp8, Tyr28, His30, Leu33, Phe58, Trp79, Phe90 and Glu242 of the IGF-1R are in contact with IGF-I as is suggested by alanine scanning mutagenesis [10]. We believe in keeping with this model, and the data in the present study, it is most likely that Tyr31 is involved in the interaction with the hy-drophobic residues Phe241 and Phe251, which form part of the mAb epitope.
In addition, we have shown that while mAbs 7C2 and 9E11 only inhibit binding of IGF-I (and not IGF-II) to the IGF-1R, the mAbs are able to inhibit proliferation and migration stimulated by both ligands. This result can be explained by our observation that both mAbs induce down-regulation of the IGF-1R, thus indirectly inhibiting the action of IGF-II by lowering the receptor concentration at the cell surface. IGF-1R down-regulation has been observed with several described mAbs, which are currently being developed as therapies for the treatment of IGF-I-responsive cancers [40–44]. Receptor down-regulation is believed to provide an additional benefit to such mAb therapies allowing not only the inhibition of ligand binding, but also a reduction in receptor levels resulting in inhibition of downstream signalling events.
In summary, we have generated two high-affinity mAbs directed against the human IGF-1R. These antibodies are able to block IGF-I but not IGF-II binding and by IGF-1R down-regulation can inhibit biological responses stimulated by both ligands. Signi-ficantly, we have mapped precisely the mAb epitopes to the CR domain of the IGF-1R and can now orientate the IGF-I C-domain in the IGF-1R-binding pocket to make contact with the IGF-1R CR domain through residues Phe241 and Phe251. This information provides the first step in the understanding of how the IGF-I–IGF-1R complex is formed. It highlights the fact that the interaction of insulin with the IR is different from the IGF-I–IGF-1R interaction, and that there is still a great deal to be understood that about the IGF-I–IGF-1R interaction.
Acknowledgments
We thank Ms Carlie Delaine and Ms Kerrie McNeil for their technical assistance, and Dr Leah Cosgrove for her useful discussions throughout. The research in J.W. laboratory was supported by an NIH (National Institutes of Health) grant (RO1 DK065890). M.K. received an Iranian postgraduate scholarship from the Ministry of Science, Research and Technology. The research was supported by funding from the University of Adelaide RIBG (Research Infrastructure Block Grants).
References
- 1.Pollak M. N., Schernhammer E. S., Hankinson S. E. Insulin-like growth factors and neoplasia. Nat. Rev. Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 2.Adams T. E., Epa V. C., Garrett T. P., Ward C. W. Structure and function of the type 1 insulin-like growth factor receptor. Cell. Mol. Life Sci. 2000;57:1050–1093. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Meyts P., Whittaker J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 2002;1:769–783. doi: 10.1038/nrd917. [DOI] [PubMed] [Google Scholar]
- 4.Kato H., Faria T. N., Stannard B., Roberts C. T., Jr, Le Roith D. Essential role of tyrosine residues 1131, 1135, and 1136 of the insulin-like growth factor-I (IGF-I) receptor in IGF-I action. Mol. Endocrinol. 1994;8:40–50. doi: 10.1210/mend.8.1.7512194. [DOI] [PubMed] [Google Scholar]
- 5.O'Connor R. Regulation of IGF-I receptor signaling in tumor cells. Horm. Metab. Res. 2003;35:771–777. doi: 10.1055/s-2004-814166. [DOI] [PubMed] [Google Scholar]
- 6.Adams T. E., McKern N. M., Ward C. W. Signalling by the type 1 insulin-like growth factor receptor: interplay with the epidermal growth factor receptor. Growth Factors. 2004;22:89–95. doi: 10.1080/08977190410001700998. [DOI] [PubMed] [Google Scholar]
- 7.Yee D. Targeting insulin-like growth factor pathways. Br. J. Cancer. 2006;94:465–468. doi: 10.1038/sj.bjc.6602963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller B. S., Yee D. Type I insulin-like growth factor receptor as a therapeutic target in cancer. Cancer Res. 2005;65:10123–10127. doi: 10.1158/0008-5472.CAN-05-2752. [DOI] [PubMed] [Google Scholar]
- 9.Garrett T. P., McKern N. M., Lou M., Frenkel M. J., Bentley J. D., Lovrecz G. O., Elleman T. C., Cosgrove L. J., Ward C. W. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature. 1998;394:395–399. doi: 10.1038/28668. [DOI] [PubMed] [Google Scholar]
- 10.Whittaker J., Groth A. V., Mynarcik D. C., Pluzek L., Gadsboll V. L., Whittaker L. J. Alanine scanning mutagenesis of a type 1 insulin-like growth factor receptor ligand binding site. J. Biol. Chem. 2001;276:43980–43986. doi: 10.1074/jbc.M102863200. [DOI] [PubMed] [Google Scholar]
- 11.Sorensen H., Whittaker L., Hinrichsen J., Groth A., Whittaker J. Mapping of the insulin-like growth factor II binding site of the type I insulin-like growth factor receptor by alanine scanning mutagenesis. FEBS Lett. 2004;565:19–22. doi: 10.1016/j.febslet.2004.03.077. [DOI] [PubMed] [Google Scholar]
- 12.Hoyne P. A., Elleman T. C., Adams T. E., Richards K. M., Ward C. W. Properties of an insulin receptor with an IGF-1 receptor loop exchange in the cysteine-rich region. FEBS Lett. 2000;469:57–60. doi: 10.1016/s0014-5793(00)01237-0. [DOI] [PubMed] [Google Scholar]
- 13.Gustafson T. A., Rutter W. J. The cysteine-rich domains of the insulin and insulin-like growth factor I receptors are primary determinants of hormone binding specificity: evidence from receptor chimeras. J. Biol. Chem. 1990;265:18663–18667. [PubMed] [Google Scholar]
- 14.Denley A., Cosgrove L. J., Booker G. W., Wallace J. C., Forbes B. E. Molecular interactions of the IGF system. Cytokine Growth Factor Res. 2005;16:421–439. doi: 10.1016/j.cytogfr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Denley A., Bonython E. R., Booker G. W., Cosgrove L. J., Forbes B. E., Ward C. W., Wallace J. C. Structural determinants for high-affinity binding of insulin-like growth factor II to insulin receptor (IR)-A, the exon 11 minus isoform of the IR. Mol. Endocrinol. 2004;18:2502–2512. doi: 10.1210/me.2004-0183. [DOI] [PubMed] [Google Scholar]
- 16.Kurose T., Pashmforoush M., Yoshimasa Y., Carroll R., Schwartz G. P., Burke G. T., Katsoyannis P. G., Steiner D. F. Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the α-subunit of the insulin receptor: identification of a new insulin-binding domain in the insulin receptor. J. Biol. Chem. 1994;269:29190–29197. [PubMed] [Google Scholar]
- 17.Wan Z., Xu B., Huang K., Chu Y. C., Li B., Nakagawa S. H., Qu Y., Hu S. Q., Katsoyannis P. G., Weiss M. A. Enhancing the activity of insulin at the receptor interface: crystal structure and photo-cross-linking of A8 analogues. Biochemistry. 2004;43:16119–16133. doi: 10.1021/bi048223f. [DOI] [PubMed] [Google Scholar]
- 18.Xu B., Hu S. Q., Chu Y. C., Huang K., Nakagawa S. H., Whittaker J., Katsoyannis P. G., Weiss M. A. Diabetes-associated mutations in insulin: consecutive residues in the B chain contact distinct domains of the insulin receptor. Biochemistry. 2004;43:8356–8372. doi: 10.1021/bi0497796. [DOI] [PubMed] [Google Scholar]
- 19.Huang K., Xu B., Hu S. Q., Chu Y. C., Hua Q. X., Qu Y., Li B., Wang S., Wang R. Y., Nakagawa S. H., et al. How insulin binds: the B-chain α-helix contacts the L1 β-helix of the insulin receptor. J. Mol. Biol. 2004;341:529–550. doi: 10.1016/j.jmb.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 20.Christoffersen C. T., Bornfeldt K. E., Rotella C. M., Gonzales N., Vissing H., Shymko R. M., ten Hoeve J., Groffen J., Heisterkamp N., De Meyts P. Negative cooperativity in the insulin-like growth factor-I receptor and a chimaeric IGF-I/insulin receptor. Endocrinology. 1994;135:472–475. doi: 10.1210/endo.135.1.8013387. [DOI] [PubMed] [Google Scholar]
- 21.Keyhanfar M., Forbes B. E., Cosgrove L. J., Wallace J. C., Booker G. W. Production and characterisation of monoclonal antibodies against insulin-like growth factor type 1 receptor (IGF-1R) Hybridoma. 2006;25:230–237. doi: 10.1089/hyb.2006.25.230. [DOI] [PubMed] [Google Scholar]
- 22.Sell C., Rubini M., Rubin R., Liu J. P., Efstratiadis A., Baserga R. Simian virus 40 large tumor antigen is unable to transform mouse embryonic fibroblasts lacking type 1 insulin-like growth factor receptor. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11217–11221. doi: 10.1073/pnas.90.23.11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher R., Mosthaf L., Schlessinger J., Brandenburg D., Ullrich A. Insulin and insulin-like growth factor-1 binding specificity is determined by distinct regions of their cognate receptors. J. Biol. Chem. 1991;266:19288–19295. [PubMed] [Google Scholar]
- 24.Schumacher R., Soos M. A., Schlessinger J., Brandenburg D., Siddle K., Ullrich A. Signaling-competent receptor chimaeras allow mapping of major insulin receptor binding domain determinants. J. Biol. Chem. 1993;268:1087–1094. [PubMed] [Google Scholar]
- 25.Fagerstam L. G., Frostell A., Karlsson R., Kullman M., Larsson A., Malmqvist M., Butt H. Detection of antigen-antibody interactions by surface plasmon resonance: application to epitope mapping. J. Mol. Recognit. 1990;3:208–214. doi: 10.1002/jmr.300030507. [DOI] [PubMed] [Google Scholar]
- 26.Forbes B. E., Hartfield P. J., McNeil K. A., Surinya K. H., Milner S. J., Cosgrove L. J., Wallace J. C. Characteristics of binding of insulin-like growth factor (IGF)-I and IGF-II analogues to the type 1 IGF receptor determined by BIAcore analysis. Eur. J. Biochem. 2002;269:961–968. doi: 10.1046/j.0014-2956.2001.02735.x. [DOI] [PubMed] [Google Scholar]
- 27.Soos M. A., Field C. E., Lammers R., Ullrich A., Zhang B., Roth R. A., Andersen A. S., Kjeldsen T., Siddle K. A panel of monoclonal antibodies for the type I insulin-like growth factor receptor. Epitope mapping, effects on ligand binding, and biological activity. J. Biol. Chem. 1992;267:12955–12963. [PubMed] [Google Scholar]
- 28.Raychaudhuri G., McCool D., Painter R. H. Human IgG1 and its Fc fragment bind with different affinities to the Fc receptors on the human U937, HL-60 and ML-1 cell lines. Mol. Immunol. 1985;22:1009–1019. doi: 10.1016/0161-5890(85)90104-x. [DOI] [PubMed] [Google Scholar]
- 29.Harlow E., Lane D. Using Antibodies: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 30.Mynarcik D. C., Williams P. F., Schaffer L., Yu G. Q., Whittaker J. Identification of common ligand binding determinants of the insulin and insulin-like growth factor 1 receptors: insights into mechanisms of ligand binding. J. Biol. Chem. 1997;272:18650–18655. doi: 10.1074/jbc.272.30.18650. [DOI] [PubMed] [Google Scholar]
- 31.Crouch S. P., Kozlowski R., Slater K. J., Fletcher J. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J. Immunol. Methods. 1993;160:81–88. doi: 10.1016/0022-1759(93)90011-u. [DOI] [PubMed] [Google Scholar]
- 32.Denley A., Brierley G. V., Carroll J. M., Lindenberg A., Booker G. W., Cosgrove L. J., Wallace J. C., Forbes B. E., Roberts C. T., Jr Differential activation of insulin receptor isoforms by insulin-like growth factors is determined by the C-domain. Endocrinology. 2006;147:1029–1036. doi: 10.1210/en.2005-0736. [DOI] [PubMed] [Google Scholar]
- 33.Hailey J., Maxwell E., Koukouras K., Bishop W. R., Pachter J. A., Wang Y. Neutralizing anti-insulin-like growth factor receptor 1 antibodies inhibit receptor function and induce receptor degradation in tumor cells. Mol. Cancer Ther. 2002;1:1349–1353. [PubMed] [Google Scholar]
- 34.Ward C. W., Hoyne P. A., Flegg R. H. Insulin and epidermal growth factor receptors contain the cysteine repeat motif found in the tumor necrosis factor receptor. Proteins. 1995;22:141–153. doi: 10.1002/prot.340220207. [DOI] [PubMed] [Google Scholar]
- 35.Sundberg E. J., Mariuzza R. A. Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 2002;61:119–160. doi: 10.1016/s0065-3233(02)61004-6. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W., Gustafson T. A., Rutter W. J., Johnson J. D. Positively charged side chains in the insulin-like growth factor-1 C- and D-regions determine receptor binding specificity. J. Biol. Chem. 1994;269:10609–10613. [PubMed] [Google Scholar]
- 37.Jansson M., Andersson G., Uhlen M., Nilsson B., Kordel J. The insulin-like growth factor (IGF) binding protein 1 binding epitope on IGF-I probed by heteronuclear NMR spectroscopy and mutational analysis. J. Biol. Chem. 1998;273:24701–24707. doi: 10.1074/jbc.273.38.24701. [DOI] [PubMed] [Google Scholar]
- 38.Bayne M. L., Applebaum J., Chicchi G. G., Miller R. E., Cascieri M. A. The roles of tyrosines 24, 31, and 60 in the high affinity binding of insulin-like growth factor-I to the type 1 insulin-like growth factor receptor. J. Biol. Chem. 1990;265:15648–15652. [PubMed] [Google Scholar]
- 39.Epa V. C., Ward C. W. Model for the complex between the insulin-like growth factor I and its receptor: towards designing antagonists for the IGF-1 receptor. Protein Eng. Des. Sel. 2006;19:377–384. doi: 10.1093/protein/gzl022. [DOI] [PubMed] [Google Scholar]
- 40.Lu D., Zhang H., Ludwig D., Persaud A., Jimenez X., Burtrum D., Balderes P., Liu M., Bohlen P., Witte L., Zhu Z. Simultaneous blockade of both the epidermal growth factor receptor and the insulin-like growth factor receptor signaling pathways in cancer cells with a fully human recombinant bispecific antibody. J. Biol. Chem. 2004;279:2856–2865. doi: 10.1074/jbc.M310132200. [DOI] [PubMed] [Google Scholar]
- 41.Lu D., Zhang H., Koo H., Tonra J., Balderes P., Prewett M., Corcoran E., Mangalampalli V., Bassi R., Anselma D., et al. A fully human recombinant IgG-like bispecific antibody to both the epidermal growth factor receptor and the insulin-like growth factor receptor for enhanced antitumor activity. J. Biol. Chem. 2005;280:19665–19672. doi: 10.1074/jbc.M500815200. [DOI] [PubMed] [Google Scholar]
- 42.Maloney E. K., McLaughlin J. L., Dagdigian N. E., Garrett L. M., Connors K. M., Zhou X. M., Blattler W. A., Chittenden T., Singh R. An anti-insulin-like growth factor I receptor antibody that is a potent inhibitor of cancer cell proliferation. Cancer Res. 2003;63:5073–5083. [PubMed] [Google Scholar]
- 43.Sachdev D., Li S. L., Hartell J. S., Fujita-Yamaguchi Y., Miller J. S., Yee D. A chimaeric humanized single-chain antibody against the type I insulin-like growth factor (IGF) receptor renders breast cancer cells refractory to the mitogenic effects of IGF-I. Cancer Res. 2003;63:627–635. [PubMed] [Google Scholar]
- 44.Goetsch L., Gonzalez A., Leger O., Beck A., Pauwels P. J., Haeuw J. F., Corvaia N. A recombinant humanized anti-insulin-like growth factor receptor type I antibody (h7C10) enhances the antitumor activity of vinorelbine and anti-epidermal growth factor receptor therapy against human cancer xenografts. Int. J. Cancer. 2005;113:316–328. doi: 10.1002/ijc.20543. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]