Abstract
Trypanosoma cruzi, the human parasite that causes Chagas disease, contains a functional pentose phosphate pathway, probably essential for protection against oxidative stress and also for R5P (ribose 5-phosphate) production for nucleotide synthesis. The haploid genome of the CL Brener clone of the parasite contains one gene coding for a Type B Rpi (ribose 5-phosphate isomerase), but genes encoding Type A Rpis, most frequent in eukaryotes, seem to be absent. The RpiB enzyme was expressed in Escherichia coli as a poly-His tagged active dimeric protein, which catalyses the reversible isomerization of R5P to Ru5P (ribulose 5-phos-phate) with Km values of 4 mM (R5P) and 1.4 mM (Ru5P).
4-Phospho-D-erythronohydroxamic acid, an analogue to the reaction intermediate when the Rpi acts via a mechanism involving the formation of a 1,2-cis-enediol, inhibited the enzyme competi-tively, with an IC50 value of 0.7 mM and a Ki of 1.2 mM. Site-directed mutagenesis allowed the demonstration of a role for His102, but not for His138, in the opening of the ribose furanosic ring. A major role in catalysis was confirmed for Cys69, since the C69A mutant was inactive in both forward and reverse directions of the reaction. The present paper contributes to the know-ledge of the mechanism of the Rpi reaction; in addition, the absence of RpiBs in the genomes of higher animals makes this enzyme a possible target for chemotherapy of Chagas disease.
Keywords: Chagas disease, pentose phosphate pathway, reaction mechanism, ribose 5-phosphate isomerase (Rpi), Trypanosoma cruzi, Type B ribose 5-phosphate isomerase (RpiB)
Abbreviations: EcRpiB, Escherichia coli ribose 5-phosphate isomerase B; MESNA, sodium 2-mercapto-ethanesulfonate; MtRpiB, Mycobacterium tuberculosis ribose 5-phosphate isomerase B; 4PEA, 4-phospho-D-erythronate; 4PEAm, 4-phospho-D-erythronamide; 4PEH, 4-phospho-D-erythronohydroxamic acid; 4PEHz, 4-phospho-D-erythronhydrazide; 5PRH, 5-phospho-D-ribonohydroxamic acid; PPP, pentose phosphate pathway; R5P, ribose 5-phosphate; Rpi, ribose 5-phosphate isomerase; Ru5P, ribulose 5-phosphate; TBS, tris-buffered saline; TcRpiB, Trypanosoma cruzi ribose 5-phosphate isomerase B
INTRODUCTION
Trypanosoma cruzi, the parasitic protozoan which causes American trypanosomiasis (also known as Chagas disease) has a functional PPP (pentose phosphate pathway). A crucial role for the PPP in the protection of trypanosomatids against oxidative stress and in the provision of nucleotide precursors has been suggested [1,2]. All seven enzymes of the PPP are present in the four major stages in the biological cycle of the parasite, namely the epimastigote and the metacyclic trypomastigote present in the insect vector, and the intracellular amastigote and the bloodstream trypomastigote present in the infected mammal [1].
The PPP consists of two branches, the oxidative branch leading from glucose 6-phosphate to Ru5P (ribulose 5-phosphate), with the reduction of two molecules of NADP, and the non-oxidative or sugar interconversion branch, which ultimately leads back to glycolytic intermediates. Rpi (ribose 5-phosphate isomerase; EC 5.3.1.6) is a key enzyme of the PPP which catalyses the reversible aldose-ketose isomerization between R5P (ribose 5-phosphate) and Ru5P. Two completely unrelated enzyme proteins are able to catalyse this reaction [3,4]. In Escherichia coli, one of them, RpiA, is a constitutively expressed 23 kDa protein [5,6], whereas the other, RpiB, is a 16 kDa protein, whose expression is regulated by a repressor [7]. Expression of either enzyme allows normal growth of E. coli, but the double mutant rpiA−/rpiB− is severely impaired for growth under all experimental conditions tested, showing that the reaction itself is very important for the bacterium [7].
RpiAs are broadly distributed in the three kingdoms of life, including most eukaryotic organisms, fungi and some bacteria. On the other hand, inspection of the protein family data base, Pfam (accession number PF02502) showed that most RpiBs are found in prokaryotic organisms; there are few exceptions in lower eukaryotes, such as Giardia lamblia, Entamoeba histolytica and some fungi, and in the insect Anopheles gambiae.
Looking for Rpi homologues in the T. cruzi genome, we found that while the RpiB enzymes seemed to be present in the parasite, RpiA homologues were completely absent. Thus TcRpiB (T. cruzi RpiB) turned out to be the only enzyme of the T. cruzi PPP which does not have its own counterpart in higher eukaryotes.
As with most sugar phosphate isomerases, both RpiAs and RpiBs seem to act via a cis-enediol(ate) intermediary mechanism instead of a hydride transfer mechanism which has been reported to occur for some sugar isomerases such as D-xylose isomerase [8] and L-rhamnose isomerase [9]. Prior to isomerization, it is believed that RpiAs and RpiBs take from solution furanosic R5P which represents the majority in solution; however the true substrate for the isomerization step is the far less abundant R5P in the free aldehyde form. Furthermore, R5P isomerization gives the acyclic product Ru5P, which is also the substrate of the enzyme. For these reasons it has been proposed that the enzyme first catalyses the opening of the furanose ring of R5P. Structural data on RpiBs from Mycobacterium tuberculosis and E. coli gave strong insights about the RpiB mechanism and the roles of some amino acid residues were proposed [10–12], but no definitive proof was given.
In the present paper, we describe the overexpression of the TcRpiB and some general properties of the purified recombinant enzyme. Two key amino acid residues, one of them involved in the isomerization step and the other involved in the ring opening step, have been identified by site-directed mutagenesis. The demon-stration of the fact that the RpiB is the only Rpi present in the parasite, and has no homologue in higher animals, opens up the possibility of considering it as a new potential target for the chemotherapy of Chagas disease.
MATERIALS AND METHODS
Chemicals and reagents
R5P was purchased from Sigma. MESNA (sodium 2-mercapto-ethanesulfonate) and Ru5P were purchased from Fluka Chemie GmbH. Oligonucleotide primers were from Gibco, Life Technologies. dNTPs (deoxynucleoside-5′triphosphates) were from Promega. Restriction endonucleases were from New England Biolabs. E. coli strain BL21 codon Plus (DE3) and the QuickChange™ site-directed mutagenesis kit were purchased from Stratagene. Polyclonal antibodies against TcRpiB were obtained in rabbits inoculated with purified recombinant His- tagged TcRpiB.
Parasites and culture
Epimastigotes of the CL Brener strain of T. cruzi were grown in axenic medium, harvested and washed as previously described [13]. Metacyclic trypomastigotes were obtained by spontaneous differentiation of epimastigotes of the CL Brener clone at 28 °C, followed by purification using DEAE-cellulose chromatography [14]. Amastigotes and trypomastigotes were obtained by infection of Vero cell monolayers with trypomastigotes of the CL Brener strain [15]. Trypomastigotes were obtained free of cellular debris by leaving them to swim off the centrifuged pellet by incubating for 1 h at 37 °C [15]. Cell-free extracts were obtained in 50 mM Tris/HCl buffer (pH 7.8) containing 0.5 mM TLCK (tosyl-lysyl-chloromethyl ketone) and 4 mM dithiothreitol, by three cycles of freezing at −20 °C and thawing, followed by sonication in a Branson 450 Sonifier, by three continuous pulses (20 s each) at 60% of maximal power. The suspensions were centrifuged at 20000 g for 10 min.
Cloning of the Rpi gene
TcRpiB was obtained by performing PCR on genomic DNA from T. cruzi epimastigotes. Primers were designed according to the sequence data obtained from the T. cruzi Genome Project database search. The sequences of the primers were as follows: sense primer 5′-GCTAGCATGACGCGCCGAGTCG-3′ and antisense primer 5′-TGCATTCCTGTACATCATTTCTCG-3′. PCR conditions were as follows: initial denaturation (5 min at 94 °C), 25 cycles of denaturation (30 s at 94 °C), annealing (30 s at 55 °C), elongation (60 s at 72 °C) and a final extension step (10 min at 72 °C). The PCR products were isolated from a 1% agarose gel, purified by the Qiaex II protocol (Qiagen), and cloned into a pGEM-T Easy vector (Promega). Sequencing of the products was performed using an ABI Prism® 377 DNA se-quencer (PerkinElmer).
Expression and purification of poly-His tagged recombinant TcRpiB
The TcRpiB gene was excized as a NheI/SalI fragment from the pGEM-T Easy vector, gel purified and subcloned into the NheI and XhoI sites of the pET28a(+) expression vector (Novagen). The resulting construct presented a poly-His tag at the N-terminal and was transformed into E. coli BL21 codon Plus (DE3) cells. The recombinant protein was expressed by induction of log-phase cultures (200 ml; D600=0.6) with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) for 3 h at 37 °C with shaking at 250 rev./min. Bacteria were harvested by centrifugation (5000 g for 30 min at 4 °C), resuspended in 20 ml of 50 mM Tris/HCl (pH 7.6) containing 150 mM NaCl, 0.1% Triton X-100, 1 mM PMSF and 0.2 mg/ml lysozyme, and centrifuged (12000 g for 30 min at 4 °C) to obtain the bacterial crude extract. The recombinant enzyme was purified in one step using Ni2+ resin (ProBond) pre-equilibrated in 50 mM Tris/HCl (pH 7.6) containing 500 mM NaCl. The column was washed sequentially with 50 ml of the equilibration buffer, 50 ml of the equilibration buffer plus 50 mM imidazole and 2 ml of the equilibration buffer plus 100 mM imidazole. TcRpiB was eluted with the equilibration buffer plus 300 mM imidazole. Depending on the assay method to be used with the sample, dialysis was performed against 50 mM Tris/HCl (pH 7.6), 150 mM NaCl containing either 1 mM EDTA and 0.5 mM 2-mercaptoethanol or 5 mM MESNA.
Molecular mass determination
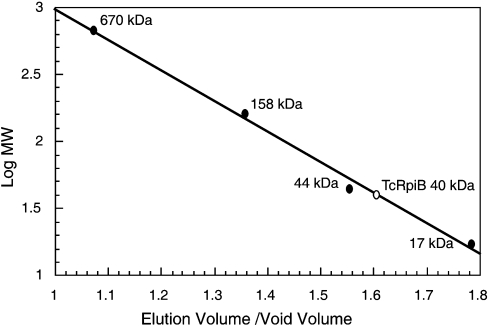
The TcRpiB apparent subunit molecular mass was estimated by SDS/PAGE [16]. The active TcRpiB molecular mass was determined by gel-filtration in a Bio-Sil SEC 250 HPLC column (Bio-rad) eluted with 50 mM Tris/HCl buffer (pH 7.6) containing 150 mM NaCl. Column calibration was performed using bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa) and horse myoglobin (17 kDa) as molecular mass standards.
Western blot analysis
For Western blots, soluble T. cruzi proteins (30 μg) were resolved on SDS/PAGE (15% gels), transferred on to a nitrocellulose Hybond ECL® membrane (Amersham Biosciences), blocked in 3% (w/v) non-fat dried skimmed milk in TBS [Tris-buffered saline; 2% glycine and 150 mM NaCl in 50 mM Tris/HCl (pH 7.6)] (blocking buffer) followed by incubation with an anti-TcRpiB antibody (1:500 for 1.5 h). Blots were washed with TBS (2×10 min), 0.05% Nonidet P-40 in TBS (1×10 min) and TBS (2×10 min). Peroxidase-conjugated goat anti-rabbit IgG (Sigma) (1:10000 for 1 h) in blocking buffer was used as the secondary antibody. The membranes were washed again and developed using SuperSignal WestPico Chemiluminescent Substrate (Pierce).
Enzyme assay
Rpi activity was assayed by three different methods. First, to determine the Km for R5P, to test inhibitors and to evaluate the pH effect over the enzymatic activity a direct spectro-photometric method at 290 nm was used [17] in 100 mM Tris/HCl (pH 7.6). Inhibitors were tested in the presence of 3.12 mM R5P. Km determination was performed at R5P concentrations in a range between 3.1 and 50 mM. An absorbance of 0.072 at 290 nm was considered for 1 mM Ru5P. Secondly, to determine the Km for Ru5P, and to test metals and metal chelators and the effect of iodoacetamide, a modification of Dische's cysteine-carbazole method was used [18]. When using Ru5P as a substrate the incubation mixture contained 5 μl of the diluted enzyme in buffer A [100 mM Tris/HCl (pH 8.4), 1 mM EDTA and 0.5 mM 2-mer- captoethanol] plus 5 μl of Ru5P giving final concentrations between 1 and 10 mM. The final enzyme concentrations in the reaction mixtures typically ranged between 2.5 and 8 ng/μl for wild-type and mutant H102A, and up to 200 ng/μl for the mutant C69A. Incubation was for 15 min at room temperature (25 °C). Following incubation, 15 μl of 0.5% cysteinium chloride, 125 μl of 75% (v/v) sulfuric acid and 5 μl of a 0.1% solution of carbazole in ethanol were added. After 30 min standing at room temperature (25 °C), the A546 was determined. A blank without enzyme was run for each substrate concentration. Reaction linearity was checked in every experiment varying enzyme concentration and time. To estimate the remaining Ru5P a calibration curve was generated; in the assay conditions 0.1 μmol of Ru5P gave an A546 of 2.9 in a final reaction volume of 155 μl.
Assays with R5P were performed in an incubation mixture containing 17 μl of R5P at 10 mM concentration plus 50 μl of the enzyme diluted in buffer A; the next steps were as for Ru5P, except for the volumes of cysteinium chloride, sulfuric acid and carbazole, which were increased by 6.7-fold. Thirdly, to determine the enzyme activity on cell-free extracts of the parasite, a coupled enzyme assay following consumption of NADH at 340 nm was used in a mixture containing 50 mM triethanolamine (pH 7.5), 5 mM MgCl2, 0.2 mM NADH, 0.025 mM thiamine pyrophosphate, 1 unit of transketolase (EC 2.2.1.1), 1 unit of ribulose 5-phosphate epimerase (EC 5.1.3.1), 20 units of triose-phosphate isomerase (EC 5.3.1.1), 2 units of α-glycerophosphate dehydrogenase (EC 1.1.1.8) and 1 mM R5P.
pH profile
The TcRpiB pH profile was determined by incubating the enzyme [5 min at room temperature (25 °C)] with 5 mM MESNA in the following buffers at 100 mM: sodium acetate/acetic acid (pH 4.5–5.5), Mes/NaOH (pH 5.5–6.5), Hepes/HCl (pH 6.3–8.5) and glycine/NaOH (pH 8.6–11.3). Reactions were started by the addition of R5P at 12.5 mM. The A290 of the commercially available Ru5P was checked between 0.12 and 1.3 mM over the whole pH range tested and did not change significantly.
Site-directed mutagenesis
Site-directed mutagenesis of the TcRpiB gene was performed using the QuickChange™ site-directed mutagenesis kit and pET28a(+)-TcRpiB as template, according to the instructions provided by the manufacturer. Following mutagenesis, the plas-mids were digested with DpnI to destroy the template DNA, E. coli BL21 codon Plus (DE3) cells were transformed, and the recombinant proteins produced and purified as described above.
RESULTS
Identification of the TcRpiB gene
In order to identify Rpi homologues in T. cruzi, a BLAST search over the parasite Genome Project database was performed. For this purpose, sequences of RpiAs and RpiBs from diverse sources were used as the query. Although no match against RpiA was found, two open reading frames encoding putative proteins with sequence similarity to RpiB were identified (last searched in February 2006). Both RpiB open reading frames were 98% identical with each other and the eleven base replacements found were either silent or conservative except for a histidine to tyrosine residue replacement. Based on this search, specific primers were designed as described in the Materials and methods section. After cloning and sequencing the PCR products, we were able to confirm the existence of one of these open reading frames, which was depo-sited in GenBank® under the accession number DQ782334 and which we have named TcRpiB. The coding sequence predicts a 17.4 kDa protein consisting of 159 amino acid residues. Figure 1 shows the alignment of TcRpiB with the sequences deduced from homologous genes obtained from the T. brucei and Leishmania major Genome Projects, as well as those from G. lamblia, E. histolytica, E. coli and M. tuberculosis. The Figure also depicts the amino acid residues known to be involved in the active-site formation. RpiBs are homodimeric proteins, the active-site pocket being formed by amino acid residues from both subunits (Figure 1) [6,10,12,19]. Active-site amino acid residues are highly conserved among the family although there is a remarkable change inside the active site of the M. tuberculosis enzyme, where the highly conserved Cys66 (using E. coli numbering) is replaced by a glycine residue. Interestingly, structural studies showed that although farther away in the sequence, the side chain of Glu75 assumes the same position as Cys66 in the active site of the enzyme [10].
Figure 1. Comparison of the sequence of TcRpiB with those of other Protozoa and prokaryotes.
Sequences are shown for the three related trypanosomatids T. cruzi, T. brucei (gi: 70834348) and Leishmania major (gi: 68127548), and two other parasites Giardia lamblia (gi: 29248748) and Entamoeba histolítica (gi: 56468369) together with the sequence of the crystalized RpiBs from M. tuberculosis H37Rv (gi: 57116993) and E. coli (gi: 85676843 W3110). Similar residues according to the BLOSUM62 matrix are indicated by different tones of grey, according to the degree of similarity (darker grey indicates more similar residues). Mutated amino acid residues are underlined. M. tuberculosis Glu75 is indicated with an asterisk. Residues that lie within the Rpi active-site pocket are annotated with 1 for the first subunit and with 2 for the second.
Expression, purification and properties of recombinant TcRpiB
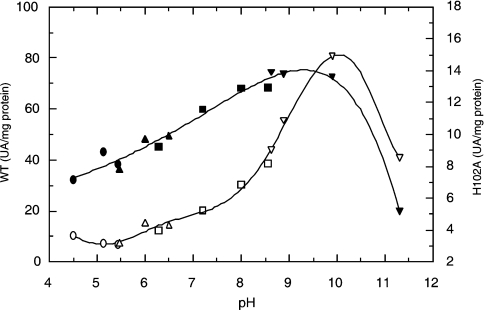
Recombinant TcRpiB was expressed and purified as described in the Materials and methods section. Gel filtration of TcRpiB showed an active protein peak with an apparent molecular mass of 39.6 kDa, which indicated that the enzyme was a homodimer (Figure 2). In a similar way to other RpiB enzymes, TcRpiB was completely inhibited by 5 mM iodoacetamide, probably due to the reactivity of the highly conserved active site Cys69 (results not shown). The optimal pH with R5P as the substrate was broad, from 8 to 10 (Figure 3). The Km and turnover values for R5P were 4 mM and 12 s−1 respectively. When using Ru5P as the substrate, the Km was 1.4 mM and turnover was 4.7 s−1.
Figure 2. Molecular mass of recombinant TcRpiB as determined by gel-filtration chromatography.
Molecular mass markers: bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa) and horse myoglobin (17 kDa).
Figure 3. Specific activity versus pH curves for the wild-type (WT) TcRpiB and H102A mutant.
●, WT acetate/acetic; ○, H102A acetate/acetic; ▲, WT Mes/NaOH; △, H102A Mes/NaOH; ■, WT Hepes/HCl; (□) H102A Hepes/HCl; ▼, WT glycine/NaOH; ▽, H102A glycine/NaOH.
Rpi activity in T. cruzi
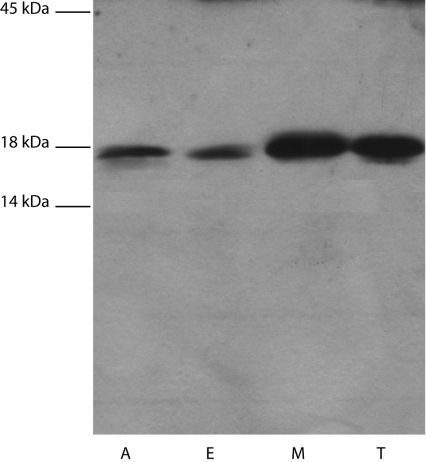
Rpi activity was detected in cell-free extracts of amastigotes, epimastigotes, metacyclic trypomastigotes and cell-culture trypo-mastigotes, being 13±4, 45±5, 43±6 and 40±12 nmol/min per mg of protein respectively. The expression of TcRpiB in the different forms of the parasite was confirmed by Western blotting with an anti-recombinant TcRpiB antibody. All the cell-free extracts showed a single band with an apparent molecular mass of approx. 18 kDa (Figure 4). Although the specific activity was similar in the latter three parasite stages, it did not correlate in the Western blot, where less protein was observed for the epimastigote stage than for the other stages. An explanation for this could be that, since the enzyme assays were performed in cell-free extracts, we cannot rule out the possibility of the presence of phosphatases or other enzymes which might interfere with the assay and which might have different levels in the parasite stages tested (for further analysis of interferences in the Rpi assay see reference [20]).
Figure 4. Differential expression of TcRpiB in different life cycle stages of T. cruzi.
Western blot of soluble extracts (30 μg) of amastigotes (A), epimastigotes (E), metacyclic trypo-mastigotes (M) and cell-derived trypomastigotes (T). Molecular mass markers: egg ovalbumin (45 kDa), bovine milk β-lactoglobulin (18 kDa) and bovine milk α-lactalbumin (14 kDa).
TcRpiB mechanism
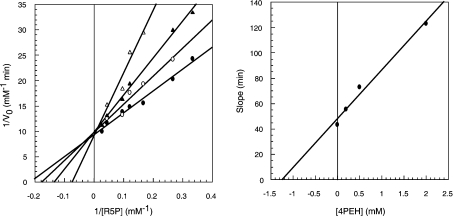
In an aldose-ketose isomerization reaction, the hydrogen atom which is transferred between C1 and C2 can move by one of two possible mechanisms: a hydride shift or a proton transfer via a 1,2-cis-enediol intermediate [21,22]. Enzymes reported to act by the former mechanism require a metal acting as an electrophilic centre so as to promote hydride transfer [23,24]. We might expect that if TcRpiB acts this way, then activity would be affected by metals and metal chelators. Both divalent (Cd2+, Mg2+ and Zn2+) and monovalent (Na+ and K+) cations, at ranges from 10 nM to 100 μM and 50 to 250 mM respectively, were assayed. Further-more, enzyme samples were treated with EDTA and o-phenan-throline at final concentrations of 0.1, 1 and 10 mM. TcRpiB activity was not affected in any case, which indicated that TcRpiB was not a metal-dependent enzyme. Because no non-metal dependent aldose-ketose isomerases have been reported to act through a hydride shift mechanism, our results strongly disfavour the possibility of such a mechanism for TcRpiB, giving the way to the cis-enediol intermediate mechanism. If TcRpiB acts by an acid-base mechanism involving a cis-enediol intermediate, then we would expect that compounds that mimic this intermediate should inhibit the reaction. The possible inhibitory effects of five compounds homologous to the cis-enediol intermediary [5PRH (5-phospho-D-ribonohydroxamic acid), 4PEAm (4-phospho-D-erythronamide), 4PEA (4-phospho-D-erythronate), 4PEH (4-phospho-D-erythronohydroxamic acid) and 4PEHz 4-phospho-D-erythronhydrazide)] were tested. The best inhibitor was 4PEH which behaved as a competitive inhibitor, with an IC50 value of 0.7 mM and a Ki of 1.2 mM (Figure 5). 4PEA, 5PRH and 4PEHz showed a weaker inhibition, and so for these compounds only IC50 values were obtained (5 mM for 4PEA and higher than 10 mM for 5PRH and 4PEHz). The inhibitory effect of most of these compounds contrary to the null effect of metals and metal chelators, strongly suggests a cis-enediol intermediary mechanism instead of a hydride transfer.
Figure 5. Inhibition of TcRpiB.
Left-hand panel, double reciprocal plot of initial reaction velocity versus R5P concentration obtained at various concentrations of the inhibitor 4PEH. ●, no inhibitor; ○, 0.2mM 4PEH; ▲, 0.5 mM 4PEH; and △, 2.0 mM 4PEH. Right-hand panel, secondary plot of slopes against inhibitor concentrations.
Site-directed mutagenesis
Isomerization of R5P to Ru5P requires the open chain form of the sugar instead of the furanose form of R5P, which in fact represents the majority in solution (99%) [25]. Thus it has been proposed that prior to isomerization the enzyme should bind the β-furanose form of R5P and catalyse the opening of the ring. From the analysis of X-ray structures of MtRpiB (M. tuberculosis RpiB) in complex with the inhibitors 4PEH and 4PEA, and docking of the substrates, a main role for His102 and His138 in the furanose ring opening was suggested. Glu75 in the M. tuberculosis enzyme and Cys66 from E. coli (Cys69 in T. cruzi numbering) have been proposed as the catalytic base for the isomerization [10,11]. Another highly conserved amino acid residue proposed as the catalytic base was His10 (His11 in T. cruzi numbering) [12].
Based on these considerations, we tested the potential function-ality of His11, His102, His138 and Cys69 by site-directed mutagenesis. Kinetic parameters are summarized in Table 1.
Table 1. Kinetic parameters of the R5P to Ru5P and Ru5P to R5P isomerizations catalysed by wild-type (WT) and mutant TcRpiBs.
ND, not detectable; −, not determined.
| R5P → Ru5P | Ru5P → R5P | |||
|---|---|---|---|---|
| Km (mM) | kcat (s−1) | Km (mM) | kcat (s−1) | |
| WT | 4±1 | 12±5 | 1.4±1.3 | 4.7±1.5 |
| C69A | ND | ND | ND | ND |
| H102A | 2±1 | 1 | 2.5±0.7 | 10.7±3.4 |
| H138A | 10±3 | 10±3 | − | − |
| H11A | 23±1 | 97±16 | − | − |
The H11A mutant was a protein with a Km for R5P 6-fold higher than that for the wild-type enzyme, whereas kcat was improved by 8-fold. As the stability of the H11A mutant to freezing and thawing in solution was considerably lower than that of the wild-type enzyme (results not shown) it is possible to suggest that His11 may play a role in the stabilization of the protein. A role of His11 in catalysis might be excluded, but the effect on Km for R5P may be due either to the protein destabilization, or to a direct effect of the mutation itself on binding. In contrast with the H11A mutant, we were not able to detect activity in the C69A mutant when using either R5P or Ru5P as substrates, which confirms its major role in catalysis. Mutant H138A did not show a difference in kcat while the Km for R5P doubled. As activity was substantially retained in the forward reaction, a main role for His138 in the ring opening step seems rather unlikely in the case of the TcRpiB enzyme. In contrast with the H138A mutant, the H102A mutant protein showed a 10-fold decrease in kcat and the Km for R5P was little affected, indicating a role of the targeted amino acid residue in catalysis. In order to clarify its responsibility in the ring opening step, kinetic parameters were measured in the reverse reaction, i.e. using Ru5P as the substrate. In this reverse reaction, the Km was little affected and kcat was increased approx. 2-fold over that of the wild-type enzyme. As the His102 mutant showed an activity severely impaired in the forward reaction but was not affected when using Ru5P as the substrate, a main role in the ring opening step can be assigned to this amino acid residue. When determined with R5P as the substrate, the optimal pH was also changed for the mutant, becoming 10, with a sharp decrease on the acidic side of the curve, in contrast with the curve for the wild-type enzyme (Figure 3). This decrease in the acidic side might be due to the loss of a catalytically important residue in this sense of the reaction, but a conformational change induced by the mutation cannot be excluded, although the Km values for both R5P (at pH 7.6) and Ru5P (at pH 8.4) did not change significantly.
DISCUSSION
Expression of TcRpiB in the main cellular stages of T. cruzi was followed by activity and Western blot assays showing that TcRpiB is constitutively expressed. As in T. cruzi, Rpi activity was reported in the different forms of the trypanosomatids T. brucei [26], Leishmania tropica and L. major [27]. Therefore Rpi is not only expressed in the extracellular organisms but also in the intracellular stages of T. cruzi and Leishmania spp. These trypanosomatids do not have an RpiA homologue in their genome whereas they have an RpiB homologue. In consequence, unless there is another type of Rpi, as yet unknown, the Rpi activity in these organisms corresponds exclusively to RpiB.
TcRpiB is the first protein from the sugar isomerases family LacA/B_RpiB, which in addition comprises galactose isomerase subunits A and B (Pfam accession number PF02502), charac-terized from an eukaryotic organism. Solving its reaction mechan-ism may give insights to be extended to other members of the LacA/B family.
The inhibitory effect of the enediol intermediate homologues, and the lack of effect of metal ions and metal chelators, in addition to previous structural data of the similar MtRpiB complexed with the enediol homologues 4PEH and 4PEA [11] enabled us to assume an isomerization reaction which develops through an enediol intermediary. The inhibition pattern shown by 4PEH, 4PEA, 4PEAm and 4PEHz is identical with that produced by the same compounds on MtRpiB.
To clarify the role of key amino acid residues in the reaction mechanism of TcRpiB, we took advantage from the data available for the more studied EcRpiB (E. coli RpiB) and MtRpiB. It has been previously proposed that MtRpiB binds the β-furanose form of R5P. After the sugar is bound, the enzyme catalyses the ring opening step in which His102 and His138 would be involved. His102 has been proposed to donate a proton to ring oxygen atom O4, and the second one would act as a base that accepts a proton from O1H [11]. We think that the present study validates the main role of His102 in the ring opening step, since the H102A mutant was severely affected in its catalytic properties when using R5P as substrate, but not when using Ru5P.
C69A is a protein devoid of activity in both the forward and reverse reaction proving that Cys69 is strictly necessary for cata-lysis. This result is in good agreement with the structural data for MtRpiB and EcRpiB that show Glu75 in MtRpiB or the corresponding Cys66 in EcRpiB is well positioned to transfer a proton between C1 and C2 [10–12]. Based on the distance between Asp9 and His10 from EcRpiB (Asp10 and His11 in TcRpiB numbering) it has been proposed that His10 could act in concert with Asp9 as the catalytic base of isomerization [12]. However, subsequent studies with the MtRpiB crystallized with the inhibitors 4PEA and 4PEH show the corresponding histidine residue at a hydrogen bond distance of the phosphate moiety of the inhibitors. Our results clearly ruled out a catalytic role for TcRpiB His11, but we cannot discard its participation in substrate binding.
The results in the present paper add to our knowledge of the reaction mechanism of RpiBs. In addition, as there are no homo-logues of this type of enzyme reported in upper eukaryotic organ-isms, TcRpiB may be considered a potentially interesting target for the development of new chemotherapeutic drugs.
Acknowledgments
A.L.S. is a Research Fellow of ANPCyT, SECYT (Agencia Nacional de Promoción Científica y Tecnológica), and Fundacion RepsolYPF Argentina, and J.J.C. is a member of CONICET (Consejo Nacional de Investigaciones Científicas y Técnicas). This work was financed by grant PICT2000 08149, from ANPCyT, SECYT. Financial support from the Ministère de l'Education Nationale et de la Recherche (E.B., 2001–2004) is gratefully acknowledged.
References
- 1.Maugeri D. A., Cazzulo J. J. The pentose phosphate pathway in Trypanosoma cruzi. FEMS Microbiol. Lett. 2004;234:117–123. doi: 10.1016/j.femsle.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Maugeri D. A., Cazzulo J. J., Burchmore R. J., Barrett M. P., Ogbunude P. O. Pentose phosphate metabolism in Leishmania mexicana. Mol. Biochem. Parasitol. 2003;130:117–125. doi: 10.1016/s0166-6851(03)00173-7. [DOI] [PubMed] [Google Scholar]
- 3.Poulsen T. S., Chang Y. Y., Hove-Jensen B. D-Allose catabolism of Escherichia coli: involvement of alsI and regulation of als regulon expression by allose and ribose. J. Bacteriol. 1999;181:7126–7130. doi: 10.1128/jb.181.22.7126-7130.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim C., Song S., Park C. The D-allose operon of Escherichia coli K-12. J. Bacteriol. 1997;179:7631–7637. doi: 10.1128/jb.179.24.7631-7637.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner A. J., Cooper R. A. The regulation of ribose 5-phosphate isomerisation in Escherichia coli K12. FEBS Lett. 1971;12:293–296. doi: 10.1016/0014-5793(71)80202-8. [DOI] [PubMed] [Google Scholar]
- 6.Essenberg M. K., Cooper R. A. Two ribose 5-phosphate isomerases from Escherichia coli K12: partial characterisation of the enzymes and consideration of their possible physiological roles. Eur. J. Biochem. 1975;55:323–332. doi: 10.1111/j.1432-1033.1975.tb02166.x. [DOI] [PubMed] [Google Scholar]
- 7.Sorensen K. I., Hove-Jensen B. Ribose catabolism of Escherichia coli: characterization of the rpiB gene encoding ribose phosphate isomerase B and of the rpiR gene, which is involved in regulation of rpiB expression. J. Bacteriol. 1996;178:1003–1011. doi: 10.1128/jb.178.4.1003-1011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitlow M., Howard A. J., Finzel B. C., Poulos T. L., Winborne E., Gilliland G. L. A metal-mediated hydride shift mechanism for xylose isomerase based on the 1.6 Å Streptomyces rubiginosus structures with xylitol and D-xylose. Proteins. 1991;9:153–173. doi: 10.1002/prot.340090302. [DOI] [PubMed] [Google Scholar]
- 9.Korndorfer I. P., Fessner W. D., Matthews B. W. The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution. J. Mol. Biol. 2000;300:917–933. doi: 10.1006/jmbi.2000.3896. [DOI] [PubMed] [Google Scholar]
- 10.Roos A. K., Andersson C. E., Bergfors T., Jacobsson M., Karlen A., Unge T., Jones T. A., Mowbray S. L. Mycobacterium tuberculosis ribose-5-phosphate isomerase has a known fold, but a novel active site. J. Mol. Biol. 2004;335:799–809. doi: 10.1016/j.jmb.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 11.Roos A. K., Burgos E., Ericsson D. J., Salmon L., Mowbray S. L. Competitive inhibitors of Mycobacterium tuberculosis ribose-5-phosphate isomerase B reveal new information about the reaction mechanism. J. Biol. Chem. 2005;280:6416–6422. doi: 10.1074/jbc.M412018200. [DOI] [PubMed] [Google Scholar]
- 12.Zhang R. G., Andersson C. E., Skarina T., Evdokimova E., Edwards A. M., Joachimiak A., Savchenko A., Mowbray S. L. The 2.2 Å resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction. J. Mol. Biol. 2003;332:1083–1094. doi: 10.1016/j.jmb.2003.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cazzulo J. J., Franke de Cazzulo B. M., Engel J. C., Cannata J. J. End products and enzyme levels of aerobic glucose fermentation in trypanosomatids. Mol. Biochem. Parasitol. 1985;16:329–343. doi: 10.1016/0166-6851(85)90074-x. [DOI] [PubMed] [Google Scholar]
- 14.de Sousa M. A. A simple method to purify biologically and antigenically preserved bloodstream trypomastigotes of Trypanosoma cruzi using DEAE-cellulose columns. Mem. Inst. Oswaldo Cruz. 1983;78:317–333. doi: 10.1590/s0074-02761983000300009. [DOI] [PubMed] [Google Scholar]
- 15.Andrews N. W., Colli W. Adhesion and interiorization of Trypanosoma cruzi in mammalian cells. J. Protozool. 1982;29:264–269. doi: 10.1111/j.1550-7408.1982.tb04024.x. [DOI] [PubMed] [Google Scholar]
- 16.Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J. Biol. Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 17.Wood T. Spectrophotometric assay for D-ribose-5-phosphateketol-isomerase and for D-ribulose-5-phosphate 3-epimerase. Anal. Biochem. 1970;33:297–306. doi: 10.1016/0003-2697(70)90300-3. [DOI] [PubMed] [Google Scholar]
- 18.Domagk G. F., Alexander W. R., Doering K. M. Protein structure and enzymatic activity, XIV: purification and properties of ribosephosphate isomerase from skeletal muscle. Hoppe-Seyler's Z. Physiol. Chem. 1974;355:781–786. doi: 10.1515/bchm2.1974.355.2.781. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q., Schwarzenbacher R., McMullan D., von Delft F., Brinen L. S., Canaves J. M., Dai X., Deacon A. M., Elsliger M. A., Eshagi S., et al. Crystal structure of a ribose-5-phosphate isomerase RpiB (TM1080) from Thermotoga maritima at 1.90 Å resolution. Proteins. 2004;56:171–175. doi: 10.1002/prot.20129. [DOI] [PubMed] [Google Scholar]
- 20.Bublitz C., Steavenson S. The pentose phosphate pathway in the endoplasmic reticulum. J. Biol. Chem. 1988;263:12849–12853. [PubMed] [Google Scholar]
- 21.Nagorski R. W., Richard J. P. Mechanistic imperatives for aldose-ketose isomerization in water: specific, general base- and metal ion-catalyzed isomerization of glyceraldehyde with proton and hydride transfer. J. Am. Chem. Soc. 2001;123:794–802. doi: 10.1021/ja003433a. [DOI] [PubMed] [Google Scholar]
- 22.Rose I. A. Mechanism of the aldose-ketose isomerase reactions. Adv. Enzymol. Relat. Areas Mol. Biol. 1975;43:491–517. doi: 10.1002/9780470122884.ch6. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Y. J., Merz K. M., Jr, Farber G. K. Theoretical examination of the mechanism of aldose-ketose isomerization. Protein Eng. 1993;6:479–484. doi: 10.1093/protein/6.5.479. [DOI] [PubMed] [Google Scholar]
- 24.Fenn T. D., Ringe D., Petsko G. A. Xylose isomerase in substrate and inhibitor michaelis states: atomic resolution studies of a metal-mediated hydride shift. Biochemistry. 2004;43:6464–6474. doi: 10.1021/bi049812o. [DOI] [PubMed] [Google Scholar]
- 25.Jung C. H., Hartman F. C., Lu T. Y., Larimer F. W. D-ribose-5-phosphate isomerase from spinach: heterologous overexpression, purification, characterization, and site-directed mutagenesis of the recombinant enzyme. Arch. Biochem. Biophys. 2000;373:409–417. doi: 10.1006/abbi.1999.1554. [DOI] [PubMed] [Google Scholar]
- 26.Cronin C. N., Nolan D. P., Voorheis H. P. The enzymes of the classical pentose phosphate pathway display differential activities in procyclic and bloodstream forms of Trypanosoma brucei. FEBS Lett. 1989;244:26–30. doi: 10.1016/0014-5793(89)81154-8. [DOI] [PubMed] [Google Scholar]
- 27.Al-Mulla Hummadi Y. M., Al-Bashir N. M., Najim R. A. Leishmania major and Leishmania tropica: II. Effect of an immunomodulator, S(2) complex on the enzymes of the parasites. Exp. Parasitol. 2006;112:85–91. doi: 10.1016/j.exppara.2005.09.006. [DOI] [PubMed] [Google Scholar]