Abstract
The HBx (X protein of hepatitis B virus) is a promiscuous transactivator implicated to play a key role in hepatocellular carcinoma. However, HBx-regulated molecular events leading to deregulation of cell cycle or establishment of a permissive environment for hepatocarcinogenesis are not fully understood. Our cell culture-based studies suggested that HBx had a profound effect on cell cycle progression even in the absence of serum. HBx presence led to an early and sustained level of cyclin–cdk2 complex during the cell cycle combined with increased protein kinase activity of cdk2 heralding an early proliferative signal. The increased cdk2 activity also led to an early proteasomal degradation of p27Kip1 that could be reversed by HBx-specific RNA interference and blocked by a chemical inhibitor of cdk2 or the T187A mutant of p27. Further, our co-immunoprecipitation and in vitro binding studies with recombinant proteins suggested a direct interaction between HBx and the cyclin E/A–cdk2 complex. Interference with different signalling cascades known to be activated by HBx suggested a constitutive requirement of Src kinases for the association of HBx with these complexes. Notably, the HBx mutant that did not interact with cyclin E/A failed to destabilize p27Kip1 or deregulate the cell cycle. Thus HBx appears to deregulate the cell cycle by interacting with the key cell cycle regulators independent of its well-established role in transactivation.
Keywords: cell cycle, cyclin-dependent kinase 2 (cdk2), cyclin E, p27Kip1, Src kinase, X protein of hepatitis B virus (HBx)
Abbreviations: cdk2, cyclin-dependent kinase 2; CSK, C-terminal Src kinase; DN, dominant negative; EGFP, enhanced green fluorescent protein; ERK, extracellular-signal-regulated kinase; GST, glutathione S-transferase; HA, haemagglutinin; HBV, hepatitis B virus; HBx, X protein of HBV; HCC, hepatocellular carcinoma; HEK-293 cell, human embryonic kidney cell; IFN, interferon; IRES, internal ribosome entry site; mAb, monoclonal antibody; MAPK, mitogen-activated protein kinase; PI3K, phosphoinositide 3-kinase; Rb, retinoblastoma protein; rHBx, recombinant HBx; RT, reverse transcriptase; SCF, Skp1/cullin/F-box; shRNA, small-hairpin RNA
INTRODUCTION
Cell cycle control by oncogenic viruses usually involves disruption of the normal restraints on cellular proliferation and transformation of cells towards malignancy [1,2]. The virus-encoded regulatory proteins usually play an important role in cell proliferation via the modulation of signal transduction pathways or interference with apoptotic cell death [3,4]. Increased cell proliferation along with enhanced viral gene expression may promote carcinogenesis [5].
HBV (hepatitis B virus) is an etiological factor for HCC (hepatocellular carcinoma). Overwhelming epidemiological evidence has shown a strong correlation between chronic HBV infection and development of HCC [6,7]. The pleiotropic HBx (X protein of HBV) has gained attention due to its presumptive role in oncogenesis. It is known to activate a wide range of viral and cellular promoters through interaction with a number of transcription factors [8,9]. It can also activate different cell signalling cascades including Ras–Raf–MAPK (mitogen-activated protein kinase), p38 MAPK, JAK (Janus kinase)/STAT (signal transducer and activator of transcription) pathway, c-Jun N-terminal kinase, PI3K (phosphoinositide 3-kinase), Src tyrosine kinases and calcium-dependent proline-rich tyrosine kinase-2 (reviewed in [8,10]) either to favour cell proliferation or to induce apoptotic cell death depending on the prevailing physiological conditions [11–14]. HBx can stimulate the growth-arrested (G0) cells to transit through G1-phase by activating Src kinases and inducing the cyclin A promoter and cyclin A–cdk2 (cyclin-dependent kinase 2) complexes [15,16]. Further, the HBx-transformed AML12 cells or Chang liver cells expressing HBx also show G1-to-S and G2-to-M progression, which has been interpreted to be due to increased mitogenic signalling coupled with activation of cyclins and CDC2 (cell division cycle 2) kinases [14,15]. However, the molecular mechanisms of increased rate of entry to S-phase and breakdown of the G1/S-phase checkpoint by HBx have not been addressed in these studies. Since the rate of entry to S-phase of the cell cycle is not dependent on the mitogenic signalling itself and rather dependent on the activity of cyclin E, cdk2 and its inhibitors p21Cip1 and p27Kip1 [17,18], we studied the mechanism of checkpoint deregulation by HBx in a human hepatoma cell line. Here, we provide first evidence for a novel nuclear function of HBx where it directly binds to the cyclin E/A–cdk2 complex and results in increased cdk2 activity, early proteasomal degradation of p27Kip1 and ultimately shortening of the cell cycle.
EXPERIMENTAL
DNA recombinants
Construction of the eukaryotic expression vectors for wild-type HBx (X0) and its deletion mutant (X9) was as described previously [19]. The prokaryotic expression vector (pET-X) for HBx and production of recombinant protein in Escherichia coli has also been reported previously [20]. The X0-IRES-EGFP (where IRES is internal ribosome entry site and EGFP is enhanced green fluorescent protein) expression vector was constructed by cloning the 475 bp EcoRI fragment of X0 in pIRES2-EGFP plasmid (Clontech Laboratories). Details of other expression vectors can be found elsewhere: shRNA (small-hairpin RNA) against HBx (X-D) [21]; DN (dominant negative) construct for Ras (N17Ras) [22]; negative regulator of CSK (C-terminal Src kinase) and DN-PI3K [23]; HA (haemagglutinin)-tagged cdk2 (cdk2–HA) and DN-cdk2–HA [24]; p27–HA and p27T187A–HA [25]; and HA-tagged ubiquitin (pUb, MT123) [26]. All the expression vectors were validated prior to use.
Chemical inhibitors and antibodies
The chemical inhibitors used and their working concentrations were: MEK1 inhibitor PD98059 (10 and 40 μM; Cell Signaling Technology); PI3K inhibitor wortmannin (1, 10 and 25 nM); protein tyrosine kinase inhibitor genistein (3 μM); proteasome inhibitors lactacystin (10 μM) and MG-132 (carbobenzoxy-L-leucycl-L-leucyl-leucinal; 20 μM); cdk2 inhibitor olomoucine (7 μM) (all from Calbiochem). Unless stated, cells were pretreated for 90 min with the respective inhibitors prior to preparation of lysates.
Specificity of the mAb (monoclonal antibody) (B-8/2/8) against HBx has been reported previously [20]. The polyclonal antiserum was raised in rabbits using rHBx (recombinant HBx) produced in E. coli [20]. All other antibodies were purchased from Santa Cruz Biotechnology.
Cell transfection and flow cytometry
The human hepatoma Huh7 cells and HEK-293 cells (human embryonic kidney cells) (CRL-2784) were grown in DMEM (Dulbecco's modified Eagle's medium) containing 10% (v/v) FBS (fetal bovine serum). Cells (5×105 in a 60 mm dish) were transfected with different plasmids (2.5 μg of DNA) using Lipofectin® (Invitrogen) as per the manufacturer's protocol. For expression/interaction studies, cells were harvested 48 h post-transfection. For pulse–chase experiments, cells were harvested at the indicated time intervals 30 h post-transfection. Cells were synchronized using serum-free medium for 24 h and stimulated by replacing complete medium. The cell-cycle kinetics were performed on cells grown in the absence or presence of serum using a FACS Calibur flow cytometer (Becton Dickinson) after staining with propidium iodide [27]. In addition to FL2 height, FL2 area (A) and width (W) were also measured to permit doublet discrimination [28]. Analysis of the multivariate data was performed with CELLQuest software (Becton Dickinson) and cell-cycle analysis was performed using FlowJo software (Tree Star). Results were compiled with Microsoft Excel and plotted with Sigma Plot software (SPSS).
RNA isolation and RT (reverse transcriptase)–PCR
Total RNA was isolated from transfected Huh7 cells using TRIzol® reagent as per supplier's instructions (Gibco BRL) and reverse-transcribed (5 μg of RNA) using oligo-dT primer and MuLV RT as per the supplier's instructions (Promega). Aliquots of RT samples were used for semi-quantitative PCR for mRNAs of cyclin E, cyclin A, HBx and β-actin. The following sets of primers were used: human β-actin, forward (actin-F) 5′-GCTATCCAGGCTGTGCTAT-3′ and reverse (actin-R) 5′-GATGGAGTTGAAGGTAGTTT-3′; cyclin A, (A-RT-F) 5′-AGCCAGTGAGTGTTAATGAAGTACC-3′ and (A-RT-R) 5′-GATGCAGAAAGTATTGGGTAAGAAA-3′; cyclin E, (E-RT-F) 5′-CAGCACTTTCTTGAGCAACACCCTC-3′ and (E-RT-R) 5′-TCTCTATGTCGCACCACTGATACCC-3′; HBx, HBx-17 (F) 5′-GCTGCTAGGCTGTACTGC-3′; and HBx-18, (R) 5′-TTAGGCAGAGGTGAAAAAG-3′. The PCR products of 446, 586 and 462 bp were obtained for β-actin, cyclin A and cyclin E respectively. X0 and X9 resulted in products of 465 bp and 360 bp respectively.
In vitro expression of proteins and in vitro binding studies
The X0 and X9 recombinant proteins (0.5 μg) were also produced in vitro using TNT T7 coupled reticulocyte lysate system, as per the supplier's instructions (Promega), and were used in binding studies with cell extracts or recombinant cyclin A–cdk2 complex, as per supplier's instructions (Cell Signaling Technology).
Immunoprecipitation and Western blotting
For immunoprecipitation, the transfected Huh7 cells were lysed in buffer A [50 mM Tris/HCl (pH 7.6), 120 mM NaCl and 0.5% (v/v) Nonidet P40] and the extracts were incubated with different primary antibodies (1 μg) for 2 h at 4 °C. Protein A–Sepharose beads (Amersham Biosciences) were added to each sample (10%, v/v) and incubated further for 1 h at 4 °C. After four washes with buffer A, samples were recovered in 25 μl of SDS gel-loading buffer by heat treatment [19], resolved by SDS/PAGE (12% gel) and electrotransferred on to a nitrocellulose membrane. Protein bands were visualized by ECL (enhanced chemiluminescence) detection system (Cell Signaling Technology). For Western blotting, equal amounts of protein samples were directly processed and visualized as above.
Pulse–chase analysis
To find the turnover of cyclin E and p27Kip1, the X0- or X9-transfected Huh7 cells were metabolically labelled in the presence of 110 μCi of [35S]cysteine/methionine mix (NEN Life Science Products) for 2 h, followed by incubation with an excess of unlabelled methionine and cysteine for different time periods. After washing with ice-cold PBS, cell lysates were prepared in buffer A, immunoprecipitated using anti-cyclin E or anti-p27 antibodies, resolved by SDS/PAGE (12% gel) and autoradiographed.
In vitro phosphorylation
Affinity-purified recombinant HBx, GST (glutathione S-transferase)–Rb (retinoblastoma protein) (Cell Signaling Technology), γ-IFN (γ-interferon) and histone H1 (Roche) were incubated in buffer C [50 mM Tris/HCl (pH 7.5), 10 mM MgCl2 and 1 mM dithiothreitol] in the presence of 10 μCi of [γ-32P]ATP and immunoprecipitated cdk2 for 30 min to 1 h at 30 °C. Samples were resolved by SDS/PAGE (15% gel) and autoradiographed. In vitro phosphorylation studies of p27Kip1 were carried out as described by Hengst and Reed [29].
RESULTS
Regulation of the cell cycle by HBx
Since HBx is known to deregulate the cell cycle through mitogenic signalling, we first studied the distribution of cells in different phases of the cell cycle in a population of cells co-expressing EGFP and HBx. Both non-hepatoma and hepatoma cells were transiently transfected with X0-IRES-EGFP and the HBx-positive cells (gated for EGFP expression) were analysed by FACS. HBx expression itself did not result in blockade of the cell cycle at any restriction point thus ruling out the possibility of spontaneous checkpoint activation. The overall cell cycle had the typical pattern of G1→S→G2/M phase progression (see Supplementary Figure 1 at http://www.BiochemJ.org/bj/401/bj4010247add.htm). Nevertheless, HBx had a profound alteration of the cell cycle transitions even in the absence of serum (Figure 1). The ‘slope’ of sinusoidal curves indicated the ‘rate of cycling’ events between individual stages in the presence of HBx. The control cells maintained a uniform slope, while the HBx-positive cells cultured for 18 h or longer showed rapid cycling with progressive deregulation (Figure 1A). Although the number of control as well as of the HBx-expressing cells gradually increased at G0/G1 block in the absence of mitogens (serum), the S- and G2/M-phases still progressed to the next phase, albeit slowly. Further, in the presence of serum more HBx-expressing cells appeared to undergo cycling as compared with control cells (Figure 1B). Interestingly, the HBx deletion mutant X9 (Δ85–119), which lacks the ability to induce mitogenic pathways [30], also failed to induce cell cycle progression (Table 1), suggesting the involvement of multiple signalling cascades or breakdown of anti-proliferative signals in the deregulation of the cell cycle by HBx [31].
Figure 1. Effect of HBx on the kinetics of cell cycle.
HEK-293 cells were transfected with X0-IRES-EGFP vector and grown for indicated time periods in the absence (A) or presence (B) of 10% serum. The EGFP-positive cells were gated to ensure the selection of HBx-positive cells and analysed by flow cytometry. The multivariate data were analysed as described in the Experimental section. Cells (%) in different phases of the cell cycle from the EGFP-gated population (+HBx) were plotted against indicated time intervals. At each time point, the EGFP-negative cells (–HBx) from the same sample were used as control.
Table 1. Flow cytometric analysis of HBx-expressing Huh7 cells.
Huh7 cells were transfected with X0 or X9 (2.5 μg) and, 4 h post-transfection, changed to serum-free medium for 24 h. After scraping, cells were stained with propidium iodide and analysed by flow cytometry (n=3). asyn, asynchronous; strv, serum starvation. Analysis was performed using Watson pragmatic algorithm.
| Distribution of cells during cell cycle (%) | |||
|---|---|---|---|
| Condition | G0/G1 | S | G2/M |
| Control (asyn) | 25.0 | 36.9 | 35.0 |
| Control (24 h strv) | 65.6 | 5.05 | 16.4 |
| Control (24 h strv, 3 h release) | 28.5 | 37.3 | 11.3 |
| X0 (24 h strv) | 15.5 | 46.0 | 36.3 |
| X9 (24 h strv) | 53.1 | 6.3 | 33.7 |
| X9 (24 h strv, 3 h release) | 30.9 | 23.4 | 4.6 |
HBx destabilizes p27Kip1
As our FACS data suggested increased rate of entry into S-phase in the presence of HBx, we next focused on the molecular constituents of the cyclin E–cdk2 complex that drives the G1/S-phase transition of the cell cycle [32]. Semi-quantitative RT–PCR was carried out to find out whether the proliferative advantage of HBx-expressing cells was related to high levels of cyclins. As shown in Figure 2(A), there was no major difference in the levels of cyclin E and cyclin A transcripts in the presence of X0 or X9 despite their differential regulation of the cell cycle. However, a marginal accumulation of cyclin E and A transcripts was observed in these as compared with controls. Next we analysed the composition of the cyclin–cdk2 complexes in the X0- and X9-transfected cells at protein level. The HBx- and mock-transfected cells were immunoprecipitated with cyclin E antibody and the immune complexes were sequentially probed for the presence of cyclin E, cdk2, p27 and HBx. Figure 2(B) shows that while the cyclin E and cdk2 levels remained unchanged in all cells, p27 was conspicuously absent from the X0-expressing cells (lane 2). The level of cyclin E-associated p27 was, however, maintained in the X9-transfected and control cells (lanes 3 and 4). Here, the expression of X0 and X9 was confirmed using the polyclonal antiserum because mutant X9 lacks the epitope for the mAb B8/2/8 [20].
Figure 2. Destabilization of p27Kip1 in the presence of HBx.
Huh7 cells were transfected either with the wild-type HBx expression vector (X0), mutant X9 or X0 along with the small hairpin construct X-D, and the RNA or protein levels of cyclins and/or cdk inhibitors were measured. (A) RT–PCR analysis of cyclin E, cyclin A, HBx and actin. (B, C) Immunoblot assay of the cyclin E-bound p27Kip1 and HBx in the absence or presence of X-D. (D) Western-blot analysis of the total pool of p27Kip1, p21Waf1, cyclin E and c-Jun in synchronized X0- and mock-transfected cells following serum stimulation for different time periods. IP, immunoprecipitation; PI, pre-immune control; WB, Western blotting.
The specificity of p27 regulation by X0 was further confirmed using an shRNA against HBx (X-D) [21]. Expression of X-D along with X0 stabilized the levels of p27 and restored its interaction with cyclin E (Figure 2C, upper panel, lane 2). The stability of p27 was further investigated in the synchronized X0-transfected cells. There was a marked reduction in the cellular pool of p27 as early as 6 h after serum stimulation as compared with control (12 h). The levels of p21Cip1 remained unchanged under these conditions (Figure 2D). Interestingly, an early appearance coupled with increased stability of cyclin E was also observed in the presence of HBx (Figure 2D).
Destabilization of p27Kip1 is signalled by a post-translational modification
Recent studies suggest an extensive role of translational and post-translational processes in the regulation of p27 [33]. Our studies along these lines suggested no influence of HBx on the translational stability of p27 and there was only a marginal increase in its overall ubiquitination status (see Supplementary Figure 2 at http://www.BiochemJ.org/bj/401/bj4010247add.htm). However, pulse–chase analysis demonstrated a rapid destabilization of p27 in the presence of HBx, reducing its half-life to nearly 3 h as against approx. 9 h in control cells (Figure 3A). Under similar conditions, the stability of p27 was not affected in the X9-transfected cells. The HBx-related destabilization of p27 could be blocked in the presence of olomoucine and proteasome inhibitors, suggesting the role for cdk2 and proteasome in the regulation of p27 [34,35]. Interestingly, there was a marked increase in the stability of cyclin E in the presence of X0 and X9 as well as lactacystin (Figure 3B).
Figure 3. Post-translational stability of p27Kip1 and cyclin E in the presence of HBx.
Huh7 cells were transfected with X0 or X9 and pulse–chased for indicated periods and immunoprecipitated (IP) for endogenous p27 (A) or cyclin E (B). Stability of the two regulators was also monitored in the presence of lactacystin (LC) or olomoucine (Olo).
Only cdk2-bound p27Kip1 is destabilized by HBx
During G1/S transition, p27 is known to associate with both cdk2–cyclin and cdk4–cyclin complexes. Therefore the levels of both cdks were monitored in the p27 immunoprecipitates of X0-transfected cells. As shown in Figure 4, even low levels of HBx could destabilize the cdk2-bound p27, while the cdk4-bound p27 remained unaffected. Since Skp2 (p45) of the SCF (Skp1/cullin/F-box) ubiquitin–protein ligase complex regulates the entry of cell cycle into S-phase by inducing the degradation of phosphorylated p27Kip1 [36], the dynamics of p27 interaction with Skp2 and cdk2 was studied. In the presence of HBx, this interaction appeared to initiate at 4 h but destabilized within a short span as compared with control (results not shown). The initial decline in the p27–Skp2 interaction could mark the end of G1-phase [37].
Figure 4. Abrogation of cdk2–p27Kip1 interaction in the presence of HBx.
Huh7 cells were transfected with p27–HA (2 μg) and increasing amounts (1–3 μg) of X0. The cell extracts were immunoprecipitated with anti-HA antibody followed by Western blotting for cdk2 and cdk4.
Phosphorylation of p27Kip1 at Thr187 is critical for destabilization
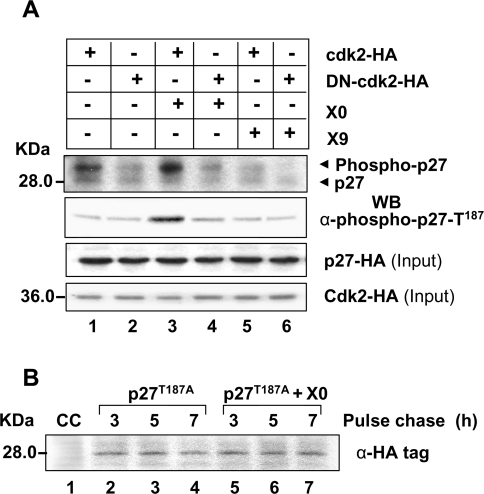
Since X0 and X9 differentially regulated p27, and the X0-related destabilization of p27 could be blocked by cdk2 inhibitor olomoucine, in vitro phosphorylation of p27 was studied using cdk2 immunoprecipitates from the X0- and X9-transfected cells. There was a marked increase in the phosphorylation of recombinant p27 in the presence of X0 (Figure 5A, lane 3) which was blocked in the presence of a DN mutant of cdk2 (DN-cdk2–HA) (lane 4). No change in the phosphorylation status of p27 was observed in the presence of X9 (lanes 5 and 6). Phosphorylation of p27 occurred exclusively at Thr187 which is a signature for its destabilization [34]. As expected, p27T187A mutant was not destabilized in the presence of X0 (Figure 5B).
Figure 5. Phosphorylation of p27Kip1 at Thr187 in the presence of HBx.
(A) In vitro phosphorylation of immunopurified p27–HA by cdk2 immunoprecipitates from X0- and X9-transfected cells. After autoradiography (top panel), the above blot was Western blotted using anti-phospho-p27-Thr187 antibody. One-tenth of the cell extract was also probed for total p27 and cdk2. (B) Pulse–chase analysis of p27T187A mutant in the absence or presence of X0. CC, control cells.
HBx potentiates cdk2 activity
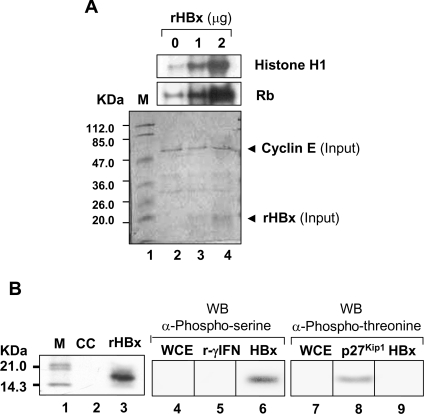
Increased p27 phosphorylation by the cdk2 immunoprecipitate from the X0-transfected cells prompted us to investigate possible change in phosphorylation potential of cdk2. In vitro phosphorylation of GST–Rb and histone H1 was carried out using the cdk2 or HBx immunoprecipitates from the X0- and X9-transfected cells. There was a marked increase in the phosphorylation of Rb and histone H1 in the presence of X0 that was inhibited by olomoucine (see Supplementary Figure 3 at http://www.BiochemJ.org/bj/401/bj4010247add.htm). However, like control cells, X9 did not potentiate the activity of cdk2. To establish the induction of cdk2 activity in the presence of HBx, histone H1 and GST–Rb were incubated with cdk2 immunoprecipitates and increasing amounts of rHBx. As shown in Figure 6(A), there was a dramatic increase in the cdk2 activity with increasing amounts of rHBx. Interestingly, rHBx was also observed to be a substrate of cdk2 with phosphorylation at its serine residue(s) (Figure 6B, lanes 3 and 6), suggesting a possible functional relationship between HBx and the active cyclin–cdk2 complex in a cellular environment.
Figure 6. In vitro activation of cdk2 activity and phosphorylation of HBx.
Extracts of untransfected Huh7 cells were immunoprecipitated with anti-cdk2 antibody and used for in vitro phosphorylation of different substrates. (A) Phosphorylation of histone H1 and GST–Rb in the presence of increasing amounts of purified rHBx. Coomassie-stained gel is shown below as loading control. (B) Phosphorylation of rHBx by cdk2. Phosphorylated HBx was further probed by anti-phosphoserine and anti-phosphothreonine antibodies. γ-IFN (r-γIFN) and p27Kip1 were used as substrate controls. CC, cell control; M, molecular mass markers; WB, Western blotting; WCE, whole cell extract.
HBx binds to the cyclin E/A–cdk2 complex
The interaction between HBx and cyclin–cdk2 complex was studied using the pull-down assay. Cell extracts were first immunoprecipitated with anti-HBx mAb followed by Western blotting for the key constituents of the cyclin–cdk2 complex. Our results suggested that HBx interacted with cyclin A, cyclin E as well as cdk2 (Figure 7A). Furthermore, antisera for cyclin A and cyclin E also co-precipitated HBx (Figure 7B, lane 6). Sequential re-probing of individual blots confirmed the presence of both HBx and cdk2, which suggested an association between HBx and cyclin E/A–cdk2 complex under in vivo conditions.
Figure 7. Interaction of HBx with cyclin A/E–cdk2 complex.
Huh7 cells were transfected with X0 or X9 and synchronized by serum starvation. Cell extracts were prepared for 6 h (for cyclin E), 8 h (for cdk2) or 15 h (for cyclin A) following serum stimulation. Cyclin–cdk complexes were immunoprecipitated (IP) either by anti-HBx antibody (A) or antisera against cyclin A and cyclin E (B), and analysed by Western blotting (WB) with indicated antibodies. To establish region D as site of interaction on HBx, cell extracts were immunoprecipitated with HBx polyclonal antisera followed by Western blotting for cyclin A, cyclin E and cdk2 (C). Domain structure of wild-type HBx (X0) and its deletion mutant X9 is shown schematically on top. (D) In vitro translated X0 and X9 proteins were incubated with control cell extracts, immunoprecipitated with anti-HBx antisera and Western blotted for cyclin E and cdk2. X0*, X0-transfected cell extract. (E) In vitro translated HBx (1 μg) was either directly incubated with recombinant cyclin A–cdk2 complex or after pre-incubation with anti-HBx mAb as described in the Experimental section. The samples were immunoprecipitated with anti-HBx mAb and Western blotted for cyclin A. PI, preimmune control; V, vector control; WCE, whole cell extract.
Sequence analysis of HBx predicts a conserved ‘R96XL98’ motif in its D region (amino acids 85–119) which is considered essential for interaction with G1 cyclins [38]. Immunoprecipitation of the X0- and X9-transfected cell extracts with polyclonal HBx antiserum followed by Western blotting for the constituents of cyclin E/A–cdk2 complex suggested that the D region of HBx was indeed involved in binding to both cyclin A and cyclin E and was not required for interaction with cdk2 (Figure 7C). Further, expression of X0 and X9 was confirmed in these extracts by Western blotting. To establish the specificity of HBx–cyclin interaction, X0 and X9 proteins produced in vitro were mixed with cell extracts and analysed for binding to cyclin E and cdk2 by sequential probing for cyclin E followed by cdk2. Only the X0 protein showed interaction with both cyclin E and cdk2, while X9 bound only to cdk2 (Figure 7D). Furthermore, X0 protein produced in vitro specifically bound to the recombinant cyclin A–cdk2 complex as this could be abolished after pre-incubation with the anti-HBx mAb and not observed with pre-immune sera (Figure 7E).
HBx interacts with cyclins in a cell cycle-dependent manner
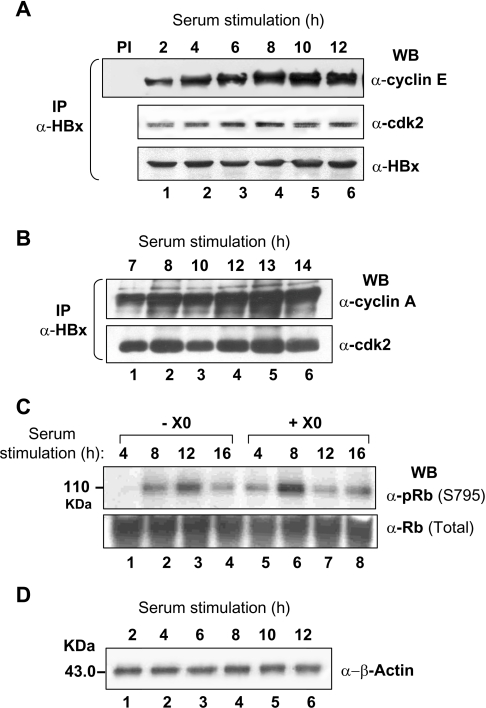
Since cyclin accumulation is a periodic event reflecting cell cycle progression [17], the interaction between HBx and the cyclin–cdk2 complex was further investigated in synchronized hepatoma cells. The levels of HBx-associated cyclin E, cyclin A and cdk2 were investigated by pull-down assay after releasing the G0/G1 block. Formation of an early ternary complex between HBx and cyclin E/A–cdk2 was apparent (Figure 8). While the HBx–cyclin E interaction could be observed as early as 2 h post-release (Figure 8A), that with cyclin A could be seen at 7 h (Figure 8B). These interactions appeared much earlier as compared with mock transfected cells (results not shown), increased over time and were sustained throughout the cell cycle. Here, the early appearance of active cyclin complexes was translated into early phosphorylation of endogenous retinoblastoma (Rb) at Ser795 (Figure 8C) which is an important measure of cell cycle progression [39].
Figure 8. Interaction between HBx and cyclin E/A–cdk2 complex during the cell cycle.
Extracts of the X0-transfected and serum synchronized Huh7 cells were immunoprecipitated (IP) with anti-HBx mAb at indicated time points after serum stimulation and the immune complexes were Western blotted (WB) for cyclin E (A), cyclin A (B) and cdk2 (A, B). The levels of total and phospho-Ser795-Rb were also measured in these extracts (C). One-tenth of extracts were used for measuring the levels of β-actin (D). PI, preimmune control.
Src kinases modulate the interaction of HBx with the cyclin E–cdk2 complex
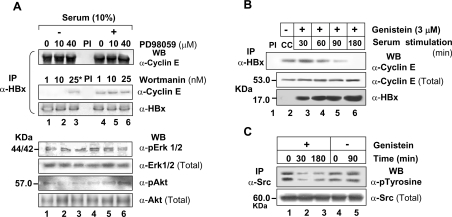
HBx is well known to modulate many signalling cascades [8,9]. To find the influence of these cascades on the association of HBx with active cyclin complex, co-immunoprecipitation studies were performed using pharmacological and physiological inhibitors of selected pathways. As shown in Figure 9, both MAPK inhibitor PD98059 and PI3K inhibitor wortmannin had no effect on the interaction between HBx and cyclin–cdk2 complex irrespective of the presence of serum (Figure 9A). However, Src kinase inhibitor genistein blocked the HBx–cyclin E interaction within 3 h (Figure 9B) by down-regulating tyrosine phosphorylation and suggested a specific requirement of these kinases in the formation of ternary complex (Figure 9C). Further, CSK, the physiological inhibitor of Src kinases, also abolished this interaction without affecting the cellular pool of active cyclin E–cdk2 complex. The DN constructs N17Ras and DN-PI3K showed no role for Ras–Raf–MAPK or PI3K pathways in this interaction (see Supplementary Figure 4 at http://www.BiochemJ.org/bj/401/bj4010247add.htm). The activation profiles of ERK1/2 (extracellular-signal-regulated kinase1/2) or Akt in these extracts confirmed the expression of these physiological inhibitors and pointed to an active maintenance of these pathways in the presence of HBx.
Figure 9. Regulation of HBx interaction with the cyclin–cdk complex by different signalling pathways.
X0-transfected Huh7 cells were grown in serum-free or serum-plus (10%) medium for 48 h and treated either with PD98059 (A), wortmannin (A) or genistein (B, C) for indicated time periods to inhibit endogenous MAPK, PI3K–Akt and Src kinases respectively. Cell extracts were immunoprecipitated with anti-HBx mAb followed by Western blotting for cyclin E (A, B). Extracts from the genistein-treated cells were also immunoprecipitated with anti-Src kinase antibody followed by Western blotting for phosphotyrosine (C). One-tenth of the cell extracts were also probed for the levels of active and total ERK1/2 and PI3K–Akt. The asterisk indicates treatment with wortmannin for 5 min. PI, preimmune control.
Thus HBx appears to use a constitutive Src kinase pathway to associate with the cyclin E–cdk2 complex leading to potentiation of cdk2 activity, early destabilization of p27Kip1 and deregulation of the cell cycle at G1/S-phase.
DISCUSSION
Oncogenic viruses are endowed with the ability to stimulate the cell cycle progression in order to facilitate their own replication. In doing so, viruses generally override the normal cell cycle restrictions or ‘checkpoints’ that are known to maintain the integrity of cell cycle transitions and in turn extend proliferative signals to host cells to establish a carcinogenic environment [2,7,40,41]. Driving the HBV-infected cells to grow continuously may be essential for active viral replication, persistent viral infection and full manifestation of the oncogenic potential of HBx leading to the development of HCC [6,7]. However, it is not clear how HBx signals the abrogation of the G1/S-phase checkpoint and arrival of an early S-phase. Although there are many reports on the shortening of the cell cycle in the presence of HBx, the molecular mechanism governing this intricate process has not been detailed so far. These reports put emphasis on mitogenic signalling by HBx as the main reason for the deregulation of the cell cycle [14,15] although the G1/S-phase transition is modulated exclusively by the ‘activity’ of cyclin E/cdk2, phosphorylation status of Rb and the stability of cdk inhibitors such as p21Cip1 and p27Kip1 [17,18]. Therefore the present study was focused at understanding the mechanism of the G1/S checkpoint deregulation by HBx. Our cell culture-based studies suggested that HBx can overcome the G0 and G1/S checkpoints even in the absence of serum and accelerated the overall pattern of G1→S→G2/M phase progression by increasing the rate of entry into S-phase. Note that mutant X9 that does not activate mitogenic signalling pathways [30] failed to bypass the G0 checkpoint, which is in accordance with absolute requirement of mitogenic stimulation for overcoming G0/quiescence state [17].
Since the early enhanced G1/S transition usually involves a combination of high levels of cyclins and low levels of cdk inhibitors as seen in many human malignancies [42,43] including HCC [44], the levels of cyclins and cdk inhibitors were monitored in the HBx-expressing cells. We observed an early specific degradation of p27Kip1 and stabilization of cyclin E in the presence of HBx. The reversal of p27 levels in the presence of shRNA against HBx suggested that p27 in the HBx-expressing cells were functional and not impaired for binding to the cyclin–cdk2 complex. The destabilization of p27 was initiated by cdk2-mediated phosphorylation of p27 at Thr187 which is known to signal its proteolytic degradation [34,45]. Interestingly, only cdk2-bound (and not cdk4-bound) p27 was targeted for degradation, suggesting extreme specificity of the regulatory mechanisms. The ubiquitination status of p27 was, however, not affected and HBx appeared to facilitate the distribution of phosphorylated p27 from cdk2-bound state to SCF-bound form probably via the PI3K pathway [37]. We were also intrigued by the stabilization of cyclin E in the presence of both X0 and X9. Since cyclin E regulation involves multi-site phosphorylation by cdk2 and GSK3-β (glycogen synthase kinase 3-β) [46], it is quite possible that the two kinases are differentially modulated by X0 and X9 leading to overall stabilization of cyclin E.
We also observed a dramatic increase in the cdk2 activity in the presence of HBx both in vitro and in vivo for its cognate substrates like histone H1 and Rb which is known to signal an early G1/S transition. This mechanism is reportedly shared by other viral proteins like Tax protein of human T-cell leukaemia type I [47] and frequently seen in HCC lesions [48]. Potentiation of cdk2 activity by HBx and presence of cdk2 activity in the HBx immunoprecipitates prompted us to explore whether HBx interacted with the cyclin–cdk complex and/or acted as a substrate of cdk2. Our in vitro phosphorylation studies with cdk2 revealed specific phosphorylation of HBx at serine residues. Although HBx lacks the consensus cdk2 recognition motifs (S/T)PX(K/R), its serine-rich N-terminal region (residues 21–57) could be a possible site for phosphorylation. Note that cdk2 does not have a strict requirement for this motif as cyclin E and the hOrc1p (human origin recognition complex protein) with mutation in the core motif could be phosphorylated [46]. Our in vitro and in vivo co-immunoprecipitation studies provide confirmatory evidence for HBx being an interacting partner of the cyclin E/A–cdk2 complex. While HBx appears to use its ‘RXL’ motif for its interaction with cyclins [38], the interaction with cdk2 was either indirect or through multiple sites.
The interaction between HBx and the cyclin–cdk2 complex was cell cycle dependent, where interaction with cyclin E appeared earlier than with cyclin A and persisted longer in the presence of HBx. An early accumulation of cyclin E could allow an early passage through G1/S restriction point and aid cdk2-independent functioning of cyclin E [49]. Besides, the sustained presence of cyclin–cdk2 complexes is likely to provide a proliferative advantage to cells through early dephosphorylation of Rb and other Rb family proteins [15]. Accordingly, we observed an early and increased phosphorylation of Rb at Ser795 in the presence of HBx which is well correlated with cdk2 activity and the hallmark of cells being either in G1/S-phase transition or in S-phase. Further, our findings on the constitutive requirement for Src kinases on the interaction between HBx and the cyclin–cdk complex itself are in tune with earlier observations on their involvement in cell cycle deregulation by HBx [16].
Thus HBx appears to deregulate the cell cycle by altering the rate of cycling events between individual stages. At molecular level, it involves destabilization of p27Kip1 and stabilization of cyclin E. The process is mediated by a direct interaction between HBx and cyclin E/A–cdk2 complexes and potentiation of cdk2 activity since mutant X9 that did not interact with cyclin E/A–cdk2 also failed to increase cdk2 activity, destabilize p27 or induce an early cell cycle progression. The enhanced cdk2 activity eventually regulates the activity of key cell cycle proteins such as Rb, p27Kip1 and cyclin E and affects its status. Our study also explains the reasons for shortening of the cell cycle by HBx. Since, viral HBx can transform cells in culture or induce HCC in transgenic mouse models [50,51], it will be interesting to know how low levels of HBx expressed from the HBV genome could induce cellular transformation. Note that HBx-expressing cells can transit through S- and G2/M-phases of the cell cycle even in complete absence of serum, suggesting not only a reduced requirement for exogenous factors but their insensitivity to anti-proliferative signals. It will be equally interesting to investigate the molecular mechanism of HBx-mediated breakdown of anti-proliferative signalling cascades in vivo.
Online data
Acknowledgments
The expression vectors for DN mutant N17Ras were from Dr S. Yonehara (Graduate School of Biostudies, Kyoto University, Kyoto, Japan), CSK and DN-PI3K were from Dr A. S. Baur (Department of Dermatology, University of Erlangen, Erlangen, Germany), cdk2 and DN-cdk2 were from Dr R. Schlegel (Department of Molecular and Cellular Toxicology, Harvard School of Public Health, Boston, MA, U.S.A.), p27Kip1 with HA tag and p27T187A were from Dr S. Meloche (Institut de Recherche en Immunovirologie et Cancérologie, Université de Montréal, Montreal, Quebec, Canada), and pUb was from Dr D. Bohmann (University of Rochester Medical Centre, New York, NY, U.S.A.). Help from R. Kumar in cell culture is acknowledged. This work was supported by a core grant from the International Centre for Genetic Engineering and Biotechnology, New Delhi, India. A.M. and V.C.J. are recipients of the senior research fellowship of the Council of Scientific and Industrial Research, New Delhi, India.
References
- 1.Swanton C., Jones N. Strategies in subversion: de-regulation of the mammalian cell cycle by viral gene products. Int. J. Exp. Pathol. 2001;82:3–13. doi: 10.1046/j.1365-2613.2001.00165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schang L. M. The cell cycle, cyclin-dependent kinases and viral infections: new horizons and unexpected connections. Progr. Cell Cycle Res. 2003;5:103–124. [PubMed] [Google Scholar]
- 3.O'Brien V. Viruses and apoptosis. J. Gen. Virol. 1998;79:1833–1845. doi: 10.1099/0022-1317-79-8-1833. [DOI] [PubMed] [Google Scholar]
- 4.Everett E., McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7:160–165. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 5.Butel J. S. Viral carcinogenesis: revelation of molecular mechanisms and etiology of human disease. Carcinogenesis. 2000;21:405–426. doi: 10.1093/carcin/21.3.405. [DOI] [PubMed] [Google Scholar]
- 6.Feitelson M. A., Pan J., Lian Z. Early molecular and genetic determinants of primary liver malignancy. Surg. Clin. North Am. 2004;84:339–354. doi: 10.1016/S0039-6109(03)00226-3. [DOI] [PubMed] [Google Scholar]
- 7.Ganem D., Prince A. M. Hepatitis B virus infection – natural history and clinical consequences. N. Engl. J. Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 8.Kumar V., Sarkar D. P. Hepatitis B virus X protein: structure-function relationships and role in viral pathogenesis. In: Triezenberg S. J., Kaufman J., Gossen M., editors. Handbook of Experimental Pharmacology. Berlin: Springer-Verlag; 2004. pp. 377–407. [Google Scholar]
- 9.Zhang X., Zhang H., Ye L. Effects of hepatitis B virus X protein on the development of liver cancer. J. Lab. Clin. Med. 2006;147:58–66. doi: 10.1016/j.lab.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard M. J., Schneider R. J. The enigmatic X gene of hepatitis B virus. J. Virol. 2004;78:12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim H., Lee H., Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J. Biol. Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 12.Arbuthnot P., Capovilla A., Kew M. Putative role of hepatitis B virus X protein in hepatocarcinogenesis: effects on apoptosis, DNA repair, mitogen-activated protein kinase and JAK/STAT pathways. J. Gastroenterol. Hepatol. 2000;15:357–368. doi: 10.1046/j.1440-1746.2000.02069.x. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y. I., Kang-Park S., Do S. I., Lee Y. I. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 2001;276:16969–16977. doi: 10.1074/jbc.M011263200. [DOI] [PubMed] [Google Scholar]
- 14.Lee S., Tarn C., Wang W. H., Chen S., Hullinder R. L., Andrisani O. M. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J. Biol. Chem. 2002;277:8730–8740. doi: 10.1074/jbc.M108025200. [DOI] [PubMed] [Google Scholar]
- 15.Benn J., Schneider R. J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. U.S.A. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouchard M., Giannakopoulosm S., Wang E. H., Tanese N., Schneider R. J. Hepatitis B virus HBx protein activation of cyclin A–cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J. Virol. 2001;75:4247–4257. doi: 10.1128/JVI.75.9.4247-4257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherr C. J., Roberts J. M. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 18.Murray A. W. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- 19.Kumar V., Jayasuryan N., Kumar R. A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc. Natl. Acad. Sci. U.S.A. 1996;93:5647–5652. doi: 10.1073/pnas.93.11.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar V., Jayasuryan N., Reddi H., Sahal D., Panda S. K. A monoclonal antibody against the X protein of hepatitis B virus: fine mapping of its epitope and application in a quantitative ELISA of the X protein in sera of hepatitis B patients. Hybridoma. 1998;17:157–164. doi: 10.1089/hyb.1998.17.157. [DOI] [PubMed] [Google Scholar]
- 21.Hung L., Kumar V. Specific inhibition of gene expression and transactivation functions of hepatitis B virus X protein and c-myc by small interfering RNAs. FEBS Lett. 2004;560:210–214. doi: 10.1016/S0014-5793(04)00113-9. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe S., Itoh T., Arai K. JAK2 is essential for activation of c-fos and c-myc promoters and cell proliferation through the human granulocyte-macrophage colony-stimulating factor receptor in BA/F3 cells J. Biol. Chem. 1996;271:12681–12686. doi: 10.1074/jbc.271.21.12681. [DOI] [PubMed] [Google Scholar]
- 23.Wolf D., Witte V., Laffert B., Blume K., Stromer E., Trapp S., D'Aloja P., Schurmann A., Baur A. S. HIV-1 Nef associated PAK and PI3-kinases stimulate Akt-independent Bad-phosphorylation to induce anti-apoptotic signals. Nat. Med. 2001;7:1217–1224. doi: 10.1038/nm1101-1217. [DOI] [PubMed] [Google Scholar]
- 24.van den Heuvel S., Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 25.Rodier G., Montagnoli A., Di Marcotullio L., Coulombe P., Draetta G. F., Pagano M., Meloche S. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 2001;20:6672–6682. doi: 10.1093/emboj/20.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 27.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu Y. W., Wang R., Schmid I., Sakamoto K. M. Analysis with flow cytometry of green fluorescent protein expression in leukemic cells. Cytometry. 1999;36:333–339. doi: 10.1002/(sici)1097-0320(19990801)36:4<333::aid-cyto8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 29.Hengst L., Reed S. I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 30.Nijhara R., Jana S. S., Goswami S. K., Kumar V., Sarkar D. P. An internal segment (residues 58–119) of the hepatitis B virus X protein is sufficient to activate MAP kinase pathways in mouse liver. FEBS Lett. 2001;504:59–64. doi: 10.1016/s0014-5793(01)02773-9. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 32.Harper J. W. Cyclin dependent kinase inhibitors. Cancer Surv. 1997;29:91–107. [PubMed] [Google Scholar]
- 33.Besson A., Gurian-West M., Chen X., Kelly-Spratt K. S., Kemp C. J., Roberts J. M. A pathway in quiescent cells that controls p27Kip1 stability, subcellular localization, and tumor suppression. Genes Dev. 2006;20:47–64. doi: 10.1101/gad.1384406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheaff R. J., Groudine M., Gordon M., Roberts J. M., Clurman B. E. Cyclin E–CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- 35.Vlach J., Hennecke S., Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sutterluty H., Chatelain E., Marti A., Wirbelauer C., Senften M., Muller U., Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- 37.Bashir T., Dorrello N. V., Amador V., Guardavaccaro D., Pagano M. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 2004;428:190–193. doi: 10.1038/nature02330. [DOI] [PubMed] [Google Scholar]
- 38.Brown N. R., Noble M. E., Endicott J. A., Johnson L. N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- 39.Connell-Crowley L., Harper J. W., Goodrich D. W. Cyclin D1/Cdk4 regulates retinoblastoma protein-mediated cell cycle arrest by site-specific phosphorylation. Mol. Biol. Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teodoro J. G., Branton P. E. Regulation of apoptosis by viral gene products. J. Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banks L., Pim D., Thomas M. Viruses and the 26S proteasome: hacking into destruction. Trends Biochem. Sci. 2003;28:452–459. doi: 10.1016/S0968-0004(03)00141-5. [DOI] [PubMed] [Google Scholar]
- 42.Bloom J., Pagano M. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 2003;13:41–47. doi: 10.1016/s1044-579x(02)00098-6. [DOI] [PubMed] [Google Scholar]
- 43.Hwang H. C., Clurman B. E. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 44.Hui A. M., Makuuchi M., Li X. Cell cycle regulators and human hepatocarcinogenesis. Hepatogastroenterology. 1998;45:1635–1642. [PubMed] [Google Scholar]
- 45.Tsvetkov L. M., Yeh K. H., Lee S. J., Sun H., Zhang H. p27Kip1 ubiquitination and degradation is regulated by the SCFSkp2 complex through phosphorylated Thr187 in p27. Curr. Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- 46.Welcker M., Singer J., Loeb K. R., Grim J., Bloecher A., Gurien-West M., Clurman B. E., Roberts J. M. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 47.Iwanaga R., Ohtani K., Hayashi T., Nakamura M. Molecular mechanism of cell cycle progression induced by the oncogene product Tax of human T-cell leukemia virus type I. Oncogene. 2001;20:2055–2067. doi: 10.1038/sj.onc.1204304. [DOI] [PubMed] [Google Scholar]
- 48.Ito Y., Matsuura N., Sakon M., Miyoshi E., Noda K., Takeda T., Umeshita K., Nagano H., Nakamori S., Dono K., et al. 1-S modulators in hepatocellular carcinoma: p27 independently predicts the recurrence. Hepatology. 1999;30:90–99. doi: 10.1002/hep.510300114. [DOI] [PubMed] [Google Scholar]
- 49.Ekholm S. V., Zickert P., Reed S. I., Zetterberg A. Accumulation of cyclin E is not a prerequisite for passage through the restriction point. Mol. Cell. Biol. 2001;21:3256–3265. doi: 10.1128/MCB.21.9.3256-3265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaefer S., Gerlich W. H. In vitro transformation by hepatitis B virus DNA. Intervirology. 1995;38:143–154. doi: 10.1159/000150425. [DOI] [PubMed] [Google Scholar]
- 51.Singh M., Kumar V. Transgenic mouse models of hepatitis B virus-associated hepatocellular carcinoma. Rev. Med. Virol. 2003;13:243–253. doi: 10.1002/rmv.392. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.