Abstract
A central means by which mammalian cells respond to low oxygen tension is through the activation of the transcription factor HIF-1 (hypoxia-inducible factor-1). Under normoxic conditions, HIF-1α (the α subunit of HIF-1) is targeted for rapid degradation by the ubiquitin–proteasome pathway. Under hypoxic conditions, this degradation is inhibited, thereby leading to the stabilization and activation of HIF-1α. Here, we report the identification of IOP1 (iron-only hydrogenase-like protein 1), a protein homologous with enzymes present in anaerobic organisms that contain a distinctive iron–sulfur cluster. IOP1 is present in a broad range of cell types. Knockdown of IOP1 using siRNA (small interfering RNA) in mammalian cells increases protein levels of HIF-1α under both normoxic and hypoxic conditions, and augments hypoxia-induced HRE (hypoxia response element) reporter gene and endogenous HIF-1α target gene expressions. We find that IOP1 knockdown up-regulates HIF-1α mRNA levels, thereby providing a mechanism by which knockdown induces the observed effects. The results collectively provide evidence that IOP1 is a component of the protein network that regulates HIF-1α in mammalian cells.
Keywords: hypoxia-inducible factor (HIF), iron-only hydrogenase-like protein 1 (IOP1), nuclear prelamin A recognition factor (Narf), proline hydroxylase domain-containing protein (PHD), yeast
Abbreviations: GST, glutathione S-transferase; HA, haemagglutinin; HEK-293 cell, human embryonic kidney cell; HIF, hypoxia-inducible factor; FIH, factor inhibiting HIF; HRE, hypoxia response element; IκBα, inhibitory κBα; IOP, iron-only hydrogenase-like protein; JNK1, c-Jun N-terminal kinase 1; MBP, maltose-binding protein; Narf, nuclear prelamin A recognition factor; PHD, proline hydroxylase domain-containing protein; poly(A)+, polyadenylated; siRNA, small interfering RNA; VHL, von Hippel–Lindau protein
INTRODUCTION
The transcription factor HIF (hypoxia inducible factor) is a global mediator of the mammalian transcriptional response to hypoxia [1,2]. HIF is a heterodimeric complex consisting of α and β subunits, both of which are members of the PAS (Per/Arnt/Sim) family of proteins. The α subunits are the primary oxygen-responsive component of the HIF complex. Three α subunits of HIF have been identified, HIF-1α, -2α and -3α [3–6]. These α subunits contain an oxygen-dependent degradation domain that confers a striking oxygen-dependent lability to these proteins [7,8]. Specifically, two proline residues (Pro402 and Pro564 in the case of human HIF-1α) are hydroxylated in the presence of molecular oxygen by a family of PHDs [proline hydroxylase domain-containing proteins; also known as HIF proline hydroxylases or EGLNs (egg-laying-defective nine homologues)] [9–15]. These hydroxylated proline residues provide a recognition motif for VHL (von Hippel–Lindau protein), a component of an E3 ubiquitin ligase complex that targets hydroxylated HIF-1α for degradation by the ubiquitin–proteasome pathway [16,17]. Thus, under normoxic conditions, HIF-1α is constitutively degraded and maintained at low steady-state levels. Under hypoxic conditions, the degree of proline hydroxylation is diminished, and HIF-1α protein levels rise [18–21]. HIF-1α then heterodimerizes with the β subunit, which is the aryl hydrocarbon nuclear translocator, whose stability, unlike that of HIF-1α, is not particularly regulated by oxygen tension [1].
The HIF complex then binds to the promoters and enhancers of a multitude of genes involved in cellular, local and systemic responses to hypoxia. These include genes encoding proteins involved in glucose uptake (such as GLUT1), glycolysis, angiogenesis (vascular endothelial growth factor) and erythropoiesis (erythropoietin) [2]. Numerous studies have implicated HIF-1α as the α subunit regulating most of the HIF-inducible genes, including most of those involved in glucose uptake, glycolysis and pH regulation, among others. HIF-2α-specific genes appear to be far more restricted, and even less is known about HIF-3α targets [22–24]. Interestingly, a splice variant of HIF-3α is an inhibitor of the HIF pathway in the cornea [25,26].
While protein turnover provides the main mechanism by which HIF-1α is controlled by oxygen tension, it is clear that it can be regulated at multiple levels. Thus FIH (factor inhibiting HIF) hydroxylates Asn803 in the transactivation domain of HIF-1α in an oxygen-dependent manner, thereby blocking its interaction with the transcriptional co-activator CBP [CREB (cAMP-response-element-binding protein)-binding protein]/p300 [27–30]. The transcriptional activity of HIF-1α is also subject to regulation by phosphorylation, one example being phosphorylation of Thr796 potentiating HIF-1α transcriptional activity [31,32]. Growth factors such as Her/Neu2 induce HIF-1α by increasing the rate of HIF-1α translation [33]. Yet other proteins, such as OS-9, directly facilitate PHD-induced hydroxylation of HIF [34]. Hence, HIF is positioned at the centre of an enlarging network of proteins that govern its activity, either directly or indirectly.
In the present paper, we describe a novel protein, which we name IOP1 (iron-only hydrogenase-like protein 1). This protein is homologous with an enzyme originally identified in anaerobic bacteria that plays a central role in redox homoeostasis under anaerobic conditions. In mammalian cells, we find that knockdown of this protein induces HIF-1α at both the mRNA and protein level, potentiating hypoxia-induced activation of both HIF reporter genes and endogenous HIF target genes. We therefore propose that this protein is a new component of the HIF-1α regulatory network.
MATERIALS AND METHODS
Yeast two-hybrid screen
The yeast two-hybrid screen [35] was performed with a Matchmaker GAL4 Two-Hybrid System 3 kit (BD Biosciences). The bait vector, pGBKT7-PHD2 R383A, which encodes a catalytically inactive PHD2, was constructed from pGBKT7 and pcDNA3-FlagPHD2 R383A [36]. Saccharomyces cerevisiae strain AH109 transformed with pGBKT7-PHD2 R383A was mated with S. cerevisiae strain Y187 pretransformed with a human adult kidney Matchmaker Library (BD Biosciences), and positives were selected on −Ade/−His/−Leu/−Trp/+X-α-Gal (5-bromo-4-chloroindol-3-yl α-D-galactopyranoside) medium.
Plasmids
pGEX-IOP1 was constructed by subcloning the 1.8 kb EcoRI/SmaI fragment of IMAGE clone 4660895 (A.T.C.C.) into the EcoRI/SmaI site of pGEX-5X-1. Results from 5′-RACE (5′-rapid amplification of cDNA ends) performed on HeLa cell total RNA are consistent with the first ATG within this clone being the physiological initiator codon. pcDNA3-IOP1 and pcDNA3-HA-IOP1 were constructed by subcloning the 1.8 kb BamHI/XhoI fragment of pGEX-IOP1 into the BamHI/XhoI sites of pcDNA3 and pcDNA3-HA respectively. pcDNA3-HA-IOP1R, which codes for an IOP1 cDNA resistant to IOP1-A siRNA (small interfering RNA) (see below), was constructed by QuikChange® mutagenesis (Stratagene) using the following two oligonucleotides (silent nucleotide changes are underlined): 5′-CGCATTGAAGATGACGGGTCGTACTTCCAAATTAACCAAG-3′ and 5′-CTTGGTTAATTTGGAAGTACGACCCGTCATCTTCAATGCG-3′.
pENTR-IOP2 was constructed by first subcloning the 1.1 kb EcoRI/XhoI fragment of IMAGE clone 3447710 (A.T.C.C.) into the EcoRI/XhoI site of pENTR3C. Then, into the BamHI (blunt)/XmaI site of the product was subcloned the 0.6 kb SfoI/XmaI fragment of IMAGE clone 2820621, yielding pENTR-IOP2. pDEST-HA-IOP2 was prepared by GATEWAY LR Clonase reactions (Invitrogen) using pENTR-IOP2 as the entry vector and pDEST-HA as the destination vector.
pGL3-HIF-1 promoter was constructed by first amplifying by PCR a 1.4 kb DNA sequence from BAC (bacterial artificial chromosome) clone RPCI-11 618G20 (Invitrogen) using the following primers: 5′-GTTGTAGATCTGAAAAAACAAAAGTAGCG-3′ and 5′-ACTAAAGCTTCTGTGCACTGAGGAGCTGAG-3′. This DNA sequence encodes nucleotides −1395 to −1 relative to the human HIF-1α gene transcription start site and contains the HIF-1α gene promoter [37]. The PCR product was digested with BglII and HindIII, subcloned into the BglII/HindIII site of pGL3-Basic (Promega), and verified by sequencing. The sources of all other plasmids have been described [11,36,38,39].
Antibodies
The IOP1-(417–476) coding sequence was subcloned into pMAL-c2X (New England Biolabs) or pGEX-5X-1 (Amersham Biosciences), and subsequently employed for purification of the corresponding MBP (maltose-binding protein) or GST (glutathione S-transferase) fusion proteins respectively from Escherichia coli using affinity chromatography. Polyclonal antibodies to MBP–IOP1-(417–476) were raised in rabbits and then affinity purified on GST–IOP1-(417–476) coupled with agarose (Alpha Diagnostic International). Anti-FLAG (M2), anti-HIF-1α (Clone 54) and anti-PHD2 (NB-137) antibodies were from Sigma, BD Biosciences and Novus Biologicals respectively. Anti-HA (haemagglutinin; F-7), anti-HIF-2α (H-310), anti-GST (Z-5), anti-(cytochrome c) (7HB), anti-α-tubulin (B-7), anti-β-tubulin (D-10, H-235), anti-IκBα (inhibitory κBα; C-21) and anti-JNK1 (c-Jun N-terminal kinase 1) (FL) antibodies were from Santa Cruz Biotechnology.
Northern blotting
pBS-IOP1 (359–714) was constructed by subcloning the 0.4 kb XbaI/BglII fragment of pcDNA3-HA-IOP1 containing the indicated nucleotides of the IOP1 cDNA into the XbaI/BamHI site of pBS-SK. A radiolabelled antisense RNA probe for IOP1 was prepared from linearized pBS-IOP1 (359–714) with T7 RNA polymerase and 50 μCi of [α-32P]ATP using a Strip-EZ RNA probe synthesis kit (Ambion). The probe was then hybridized to a Multiple Tissue Northern Blot membrane (BD Biosciences) for 18 h at 65 °C, washed with low- and high-stringency wash solutions (Ambion Northern Max-Gly kit), exposed for autoradiography, stripped, and then reprobed with a radiolabelled antisense RNA probe for γ-actin [40]. A probe for PHD2 was prepared by subcloning the 0.7 kb XhoI/XbaI fragment of pcDNA3-FlagPHD2 [36] into the XhoI/XbaI site of pBS-KS. A probe for HIF-1α was prepared by subcloning the 0.7 kb BglII/SalI fragment of pcDNA3-HA-HIF-1α [38] into the BamHI/SalI site of pBS-SK. A probe for GLUT1 (a gift from Dr Celeste Simon and Dr Cheng-Jun Hu, University of Pennsylvania School of Medicine) was subcloned into pBS-KS. For these probes, radiolabelled antisense probes were prepared using a Maxiscript T7 kit (Ambion). Poly(A)+ (polyadenylated) RNA was isolated from cells using an Oligotex Direct kit (Qiagen).
siRNA
Short hairpin loop vectors were prepared in pShag1 [41] (a gift from Dr Gregory Hannon, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, U.S.A.). Shag-IOP1-7, which targets bp 376–404 of the IOP1 coding sequence, was constructed by subcloning into the BseRI/BamHI site of pShag1 a duplex consisting of the following two oligonucleotides: 5′-TATCTGTAGGATTCAGCTGAAACCGTGCGAAGCTTGGCACGGTTTCAGCTGAATCCTACAGATACTGTTTTTT-3′ and 5′-GATCAAAAAACAGTATCTGTAGGATTCAGCTGAAACCGTGCCAAGCTTCGCACGGTTTCAGCTGAATCCTACAGATACG-3′. pShag-mIOP1-7 was constructed using the following two oligonucleotides: 5′-TGTCTGTGGGGTCCAGCCGAAACCTGGCGAAGCTTGGCCAGGTTTCGGCTGGACCCCACAGACACTGTTTTTT-3′ (relevant mismatches to IOP1-7 sequence are high-lighted) and 5′-GATCAAAAAACAGTGTCTGTGGGGTCCAGCCGAAACCTGGCCAAGCTTCGCCAGGTTTCGGCTGGACCCCACAGACACG-3′.
pShag-IOP2-4, which targets bp 751–779 of the IOP2 coding sequence, was constructed in an analogous manner with the following two oligonucleotides: 5′-CAATTTCACCTGATGTTAACACGCAGTCGAAGCTTGGACTGCGTGTTAACATCAGGTGAAATTCCTCTTTTTT-3′ and 5′-GATCAAAAAAGAGGAATTTCACCTGATGTTAACACGCAGTCCAAGCTTCGACTGCGTGTTAACATCAGGTGAAATTGCG-3′. The IOP1 nucleotide sequences targeted by the other short hairpin loop vectors were as follows: 1, 1175–1203; 2, 557–585; 3, 710–738; 4, 793–821; 5, 995–1023; 6, 1088–1115; 8, 1032–1060; 9, 65–87; 10, 281–303. The IOP2 nucleotide sequences targeted by the other short hairpin loop vectors were as follows: 1, 1115–1143; 2, 501–529; 3, 662–690; 5, 905–933; 6, 1041–1069.
pShag-PHD2 was constructed based on published PHD2 siRNA sequence information [42] using the following two oligonucleotides: 5′-ATAACAAGCAACCATGGCTTTCGTCCGGGAAGCTTGCCGGACGAAAGCCATGGTTGCTTGTTATCCGTTTTTT-3′ and 5′-GATCAAAAAACGGATAACAAGCAACCATGGCTTTCGTCCGGCAAGCTTCCCGGACGAAAGCCATGGTTGCTTGTTATCG-3′.
The sequences for synthetic siRNA duplexes (Dharmacon) were as follows. IOP1-A: GACGGGAGCUACUUCCAAAUU and UUUGGAAGUAGCUCCCGUCUU. IOP1-B: GCAUCAAGCCUGUCAAAGUUU and ACUUUGACAGGCUUGAUGCUU. PHD2-A: CAAGGUAAGUGGAGGUAUAdTdT and UAUACCUCCACUUACCUUGdTdT. PHD2-B: UGUACGUCAUGUUGAUAAUdTdT and AUUAUCAACAUGACGUACAdTdT. Control-A: Non-specific Control Duplex IX (Dharmacon catalogue no. D001206-09-05). Control-B: siCONTROL Non-targeting siRNA no. 1 (Dharmacon catalogue no. D-001210-01-05).
Cell culture and transfection
HeLa, COS, 786-O and Hep3B cells were obtained from A.T.C.C. The source of N2a cells and HEK-293 cells (human embryonic kidney cells) has been described in [43]. Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 100 units/ml penicillin and 100 μg/ml streptomycin. A Hep3B cell line stably transfected with pcDNA3-HA-IOP1R was obtained by selection with 1000 μg/ml G418. Hep3B cells stably transfected with pcDNA3 were selected under the same conditions and pooled. For normoxic conditions, cells were maintained in a humidified 5%-CO2 tissue-culture incubator. For experiments involving 1% O2, cells were placed in either a Billups-Rothenberg modular incubator or an In Vivo 200 hypoxia workstation (Ruskinn Technology) perfused with 1% O2/5% CO2/balance N2. For other experiments, cells were placed in an InVivo 200 hypoxia workstation perfused with 10% H2/5% CO2/balance N2 in the presence of a palladium catalyst. In this case, measurements using GC reveal an oxygen concentration of 0.2%.
Cells were typically seeded in 6-well or 12-well plates for synthetic siRNA studies, or in 24-well plates for luciferase assays, and transfected using Lipofectamine® 2000 (synthetic siRNA) or FuGENE™ 6 (luciferase assays). For synthetic siRNA transfections, the final siRNA concentration was 20 nM. For experiments involving Western blotting, medium was replaced 24 h post-transfection and cells were stimulated 72 h post-transfection. For experiments involving real-time PCR analysis, cells were transfected a second time 24 h post-transfection, re-fed 48 h after the initial transfection, and stimulated 72 h after the initial transfection.
Luciferase assays, Western blotting and proline hydroxylase assays
For luciferase assays, cells were stimulated 48 h post-transfection and then harvested 18 h later. Typically, 100 ng of pShag vector, 50 ng of pRL-TK (internal transfection control), and 25–100 ng of (eHRE)3-Luc or pGL3-HIF-1 promoter were transfected. Luciferase activities were measured using a Dual-luciferase Reporter Assay System (Promega) and a Wallac LB9507 luminometer. For Western blotting, cellular lysates were prepared as described in [38], except that dithiothreitol was omitted. Protein concentrations of extracts were determined using a Bio-Rad DC protein assay kit. For a given experiment, equal quantities of cellular lysates (typically 10 μg) were subjected to Western blotting. Assays for endogenous PHD activity were performed using GST–HIF-1α-(531–575) as a substrate and 35S-labelled VHL capture as described in [36], except that exogenous iron was omitted and the ascorbate and 2-oxoglutarate concentrations employed were 1 and 0.5 mM respectively.
Real-time PCR
Total RNA was harvested from cells using TRIzol® reagent (Invitrogen). Reverse-transcription reactions were performed using TaqMan reverse-transcription reagents (ABI). Real-time PCR reactions were performed on 20 ng equivalents of cDNA using an ABI 7300 Real-time PCR machine and TaqMan probes (ABI). Relative quantification was performed employing the ΔΔCt method and 18 S RNA as the endogenous control. For HIF-1α mRNA stability experiments, SYBR Green Master mix (ABI) was employed in conjunction with the following oligonucleotides. hHIF1 5′: 5′-TTTTACCATGCCCCAGATTCA-3′; hHIF1 3′: 5′-AGTGCTTCCATCGGAAGGACT-3′; hIOP1 5′: 5′-GAAAAAAGGGCGGGAAGTG-3′; and hIOP1 3′: 5′-GCTCCCGTCATCTTCAATGC-3′. The endogenous control was β-actin using the following oligonucleotides. hβ-actin 5′: 5′-GCCCTGAGGCACTCTTCCA-3′; hβ-actin 3′: 5′-ATGCCACAGGACTCCATGC-3′. Dissociation curve analysis revealed single peaks for each set of probes. Actinomycin D was obtained from Calbiochem. HIF-1α mRNA stability data were analysed by exponential regression using Excel software.
Gas analysis
Hydrogen and certain oxygen measurements were performed on an Agilent 2890A micro gas chromatograph equipped with a MolSieve 5A column and a thermal conductivity detector, and argon as the carrier gas. Gases were analysed directly from the headspace of flasks containing transiently transfected mammalian cells, or headspace vials containing extracts prepared under hypoxic conditions and supplemented with 20 mM sodium dithionate and 5 mM Methyl Viologen as the electron donor. To assay for hydrogen-dependent reducing activity, cell extracts were measured for their capacity to reduce 5 mM Methyl Viologen in the presence of 10% hydrogen (90% nitrogen).
RESULTS
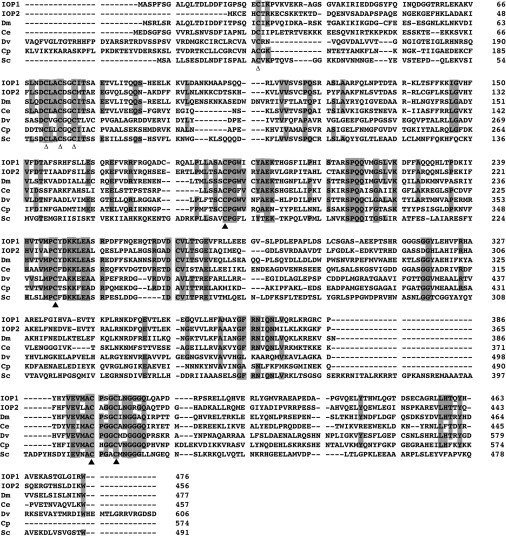
We performed a yeast two-hybrid screen of a human adult kidney library using PHD2 as a bait to screen a human adult kidney cDNA library, and we isolated two independent partial clones (residues 171–476 and 198–476) of a novel protein corresponding to Unigene Hs.513247 (Figure 1). This protein is homologous with the iron-only hydrogenases, a family of oxygen-labile proteins found in anaerobic organisms that catalyses the reversible reduction of protons to molecular hydrogen gas, a reaction critical for maintaining redox balance under anaerobic conditions [44–46]. Within the hydrogenase domain, it is 36 and 30% identical with iron-only hydrogenases from Desulfovibrio vulgaris and Clostridium pasteurianum respectively (P<10−18 and P<10−14 respectively). In addition, this protein is 47% identical in this domain with Narf (nuclear prelamin A recognition factor), a human protein previously found to bind prelamin A [47].
Figure 1. IOP1 is homologous with bacterial iron-only hydrogenases.
Comparison of amino acid sequences of human IOP1, human IOP2; homologues from D. melanogaster (Dm; GenBank® accession no. NP_652122), C. elegans (Ce; GenBank® accession no. AAF60782), S. cerevisiae (Sc; GenBank® accession no. NP_014159); and iron-only hydrogenases from Desulfovibrio vulgaris (Dv; GenBank® accession no. CAA40970) and Clostridium pasteurianum (Cp; GenBank® accession no. AAA23248). Numbers indicate amino acid residues. Shaded residues are identical in at least five of the seven sequences. Closed triangles denote cysteine residues that are ligands of the H-cluster in the bacterial iron-only hydrogenases. Open triangles denote cysteine residues that are ligands of the [4Fe-4S] cluster in the ferredoxin-like domain of bacterial iron-only hydrogenases.
Because of this homology to the iron-only hydrogenases, we name the novel factor iron-only hydrogenase-like protein 1 (IOP1), and propose renaming Narf as IOP2. Bacterial iron-only hydrogenases contain a distinctive active-site iron–sulfur cluster, termed the H-cluster [48–50], and an additional iron–sulfur cluster present in a ferredoxin-like domain. The H-cluster contains six iron atoms, four of which are present as a [4Fe-4S] cubane subcluster that is bridged to a [2Fe] subcluster. The eight cysteine residues involved in chelating the H-cluster and the ferredoxin Fe cluster in bacterial hydrogenases are conserved in IOP1 and IOP2, as well as in homologues from other species that include Drosophila melanogaster, Caenorhabditis elegans and S. cerevisiae (Figure 1). Homologues are also present in other multicellular organisms that include Gallus gallus (GenBank® accession no. XP_414836.1), Xenopus laevis (GenBank® accession no. AAH73323.1) and Danio rerio (GenBank® accession no. XP_683711.1).
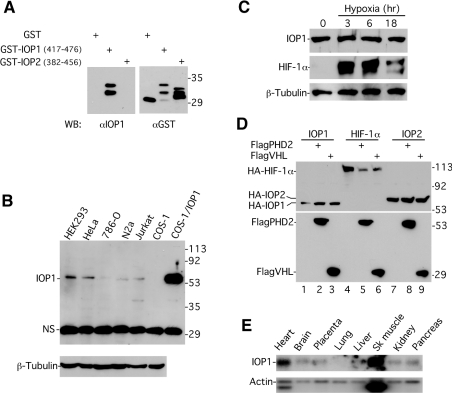
We have not been able to co-immunoprecipitate full-length IOP1 with full-length PHD2, despite extensive efforts under a number of conditions, and in spite of the fact that fragments of IOP1 can readily co-immunoprecipitate with full-length PHD2 (results not shown). Nonetheless, the homology of IOP1 to a protein involved in anaerobic bacterial metabolism compelled us to study this protein in more detail. We prepared antibodies that react with IOP1 but not IOP2 (Figure 2A). Western blotting using these antibodies reveals the presence of a 55 kDa protein in a variety of cell lines that co-migrates with that obtained from COS-1 cells overexpressing IOP1 (Figure 2B). Hypoxic exposure of HEK-293 cells, which leads to elevated HIF-1α protein levels, failed to reveal any appreciable change in that of IOP1 (Figure 2C). Consistent with this, co-expression in COS-1 cells of either PHD2 or VHL with IOP1, or for that matter IOP2, did not diminish the expression level of either IOP (Figure 2D, top panel, lanes 1–3 and 7–9). This is in contrast with the level of co-expressed HIF-1α, which is markedly decreased by both (top panel, lanes 4–6). Northern blotting reveals an IOP1 mRNA of 2.4 kb with a wide tissue distribution, particularly high in heart and skeletal muscles (Figure 2E). The distribution is similar to that seen for PHD2 on Northern-blot analysis of human tissues [51], as well as that of IOP2/Narf [47].
Figure 2. IOP1 protein levels are not regulated by the PHD2–VHL pathway.
(A) A 20 ng sample of GST, GST–IOP1-(417–476) or GST–IOP2-(382–456) was subjected to Western blotting using either anti-IOP1 or anti-GST antibodies. The positions of molecular-mass markers (in kDa) are shown to the right. The two bands observed in the anti-IOP1 Western blot of GST–IOP1-(417–476) correspond to bands observed in the anti-GST Western blot of the same protein preparation, and probably represent the full-length protein and a proteolytic product of it. (B) Extracts (10 μg) prepared from various human (HEK-293, HeLa, 786-0 and Jurkat), mouse (N2a) and monkey (COS-1) cell lines were examined by Western blotting using anti-IOP1 (top) or anti-β-tubulin (bottom) antibodies. Also shown is an extract prepared from COS-1 cells transiently transfected with a full-length IOP1 expression vector (pcDNA3-IOP1). The positions of molecular-mass markers (in kDa) are shown to the right. NS denotes a non-specific band. (C) HEK-293 cells were subjected to hypoxia (0.2% O2) for the indicated periods of time, extracts were prepared, and then aliquots (10 μg) were examined by Western blotting using antibodies against IOP1, HIF-1α or β-tubulin. (D) COS-1 cells were co-transfected with expression vectors for HA-tagged IOP1, IOP2 or HIF-1α, and ones for either FLAG-tagged PHD2 or VHL. The cells were lysed, and 10 μg of extracts was then examined for expression levels of the tagged proteins using either anti-HA (top) or anti-FLAG (bottom) antibodies. The positions of molecular-mass markers (in kDa) are shown to the right. (E) A multiple tissue Northern blot was hybridized with a 32P-labelled antisense probe to IOP1, stripped, and then reprobed with one for γ-actin. The sizes of the mRNAs for IOP1 and γ-actin are 2.4 and 1.9 kb respectively.
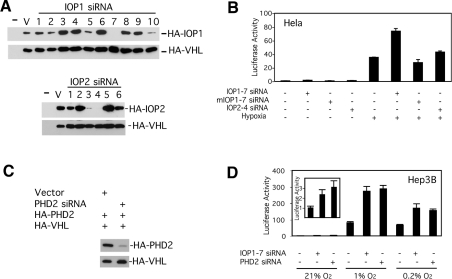
We examined potential functions of IOP1. We first considered the possibility that IOP1 might be a regulated protein that, for example, might be catalytically active as a hydrogenase. However, we have so far been unable to demonstrate hydrogenase activity using recombinant IOP1 or IOP2 under either normoxic or hypoxic conditions (results not shown). We next considered the possibility that instead of being a regulated protein, IOP1 might in fact regulate the HIF pathway itself. We employed an siRNA approach, and first screened a series of short hairpin loop-derived siRNAs for their capacity to reduce co-expressed IOP1 protein levels, identifying one that was particularly potent (designated IOP1-7, Figure 3A). A comparably effective one was found for IOP2 (IOP2-4, Figure 3A). The specificity of these siRNA vectors is supported by the fact that they do not affect the levels of co-expressed VHL (Figure 3A), nor that of the other IOP isoform (results not shown). We next co-transfected HeLa cells with these constructs and an HRE (hypoxia response element) reporter gene, and as shown in Figure 3(B), found that the IOP1-7 siRNA significantly enhanced hypoxia-induced activation of an HRE reporter gene (sixth column). IOP2-4 siRNA or a mutant version of IOP1-7 siRNA (mIOP1-7), which differs at six nucleotides compared with wild-type, failed to produce this effect (Figure 3B, eighth and seventh columns respectively). The mutant siRNA is, as expected, defective in silencing human IOP1 (results not shown). The effect of the IOP1 siRNA was also seen with HEK-293 cells subjected to hypoxia (results not shown).
Figure 3. Regulation of HRE activity by IOP1.
(A, C) COS cells were co-transfected with an expression vector for HA-tagged proteins, and ones for short hairpin loops or the pShag1 vector (V). At 2 days post-transfection, cellular extracts were examined by Western blotting using anti-HA antibodies. (B, D) The indicated cells were co-transfected with an (eHRE)3-Luc reporter gene, pRL-TK, and short hairpin loop expression vectors encoding the indicated siRNAs. At 2 days later, cells were maintained under 21% O2 (B, D), or subjected to 0.2% O2 (B, D) or 1% O2 (D) for 18 h, and harvested. Firefly luciferase activities were normalized to that of Renilla luciferase. Inset of (D) shows results for 21% O2, with the columns corresponding to the first three columns of (D), and with a different scale for the y-axis. (B) and (D) show triplicate measurements with means±S.D., and are representative of two to four independent experiments.
Knockdown of PHD2 increases HIF1-α protein levels in many mammalian cell lines [42,52], thereby implicating PHD2 as the isoform that maintains HIF-1α at low levels under normoxia. We prepared an siRNA vector directed against PHD2 that was effective in decreasing co-expressed PHD2 protein levels (Figure 3C). In Hep3B cells, this siRNA augmented HRE reporter gene activity under normoxia (Figure 3D, inset), consistent with a previous report [42]. Importantly, we find the effects of IOP1 siRNA to be comparable with that of PHD2 siRNA under both moderate (1% O2) and severe (0.2% O2) hypoxia (Figure 3D), as well as under normoxia (Figure 3D, inset).
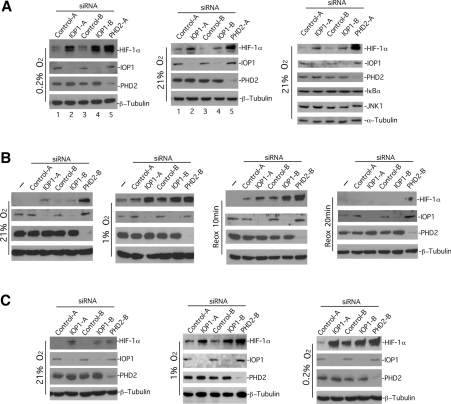
We examined the possibility that IOP1 siRNA-induced potentiation of HRE reporter gene activity might be due to increased HIF-1α protein levels. We employed a different siRNA delivery method, synthetic siRNA, and as a further demonstration of specificity, we examined two IOP1 siRNAs that targeted sequences distinct from that targeted by the IOP1-7 short hairpin loop just described. As negative controls, we used two non-specific siRNAs, and as a positive control, PHD2 siRNA. As shown in Figure 4(A), the two different IOP1 siRNAs were both effective in decreasing IOP1 protein levels in Hep3B cells (left, second panel from top, lanes 2 and 4), just as a PHD2 siRNA was in decreasing PHD2 protein levels (left, third panel from top, lane 5). These results confirm the authenticity of the 55 kDa band recognized by the anti-IOP1 antibodies. Importantly, under conditions in which PHD2 knockdown increases hypoxia-induced HIF-1α levels (lane 5), we find that IOP1 knockdown augments it as well (lanes 2 and 4). These effects are also seen under normoxia (Figure 4A, middle), conditions under which siRNA to PHD2 augments HIF-1α levels (lane 5), as seen previously [42,52]. As additional controls, IOP1 knockdown does not affect the protein levels of either IκBα or JNK1 (Figure 4A, right). We also note modest increases in PHD2 protein level upon IOP1 knockdown in these cells (for example, compare lanes 1 and 2, third panel from top, Figure 4A, left), consistent with previously published data indicating that PHD2 is a HIF gene target [23,53–55].
Figure 4. Regulation of HIF-1α protein level by IOP1.
Hep3B (A, B) or HEK-293 (C) cells were transfected with the indicated synthetic siRNAs. Some cells were exposed, 72 h post-transfection, to 0.2 or 1% O2 for 3 h. In (B), some cells were exposed to 1% O2 for 3 h and then reoxygenated (Reox) in 21% O2 for the indicated times. For a given experiment, equal quantities of cellular lysates were subjected to Western blotting using the indicated antibodies.
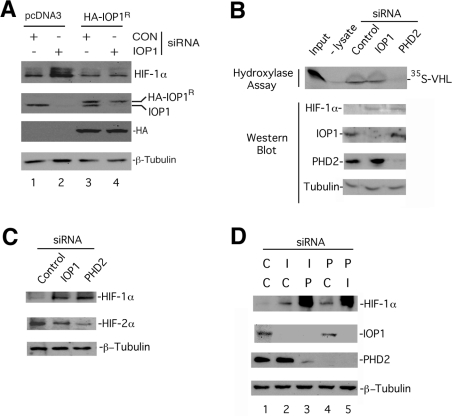
The augmentation of HIF-1α levels by IOP1 knockdown is seen at 3, 6 and 24 h of 0.2% O2 (results not shown). This effect is also seen under milder hypoxic conditions, 1% O2 (Figure 4B, second set of panels from the left), conditions under which comparable effects are seen with PHD2 knockdown. We also treated cells with siRNA, subjected them to 1% O2, and then reoxygenated them, which resulted in the rapid degradation of HIF-1α (Figure 4B, two sets of panels on the right), thus excluding the possibility that IOP1 knockdown simply abolishes the HIF-1α degradation pathway. We find that the effects of both IOP1 and PHD2 siRNAs are similar in HEK-293 cells, under normoxia as well as under both modest (1% O2) and severe (0.2% O2) hypoxia (Figure 4C). To confirm the specificity of the siRNA effect, we generated a Hep3B cell line stably transfected with a vector expressing an HA-tagged siRNA-resistant version of IOP1 (HA–IOP1R). This HA-tagged IOP1 migrates with a slower mobility than endogenous IOP1 on SDS/PAGE, and Western blotting confirms siRNA resistance (Figure 5A, second panel from top, compare lanes 3 and 4). Importantly, expression of this siRNA-resistant IOP1 reverses the IOP1 knockdown-induced augmentation of HIF-1α levels (top panel, compare lanes 2 and 4).
Figure 5. Evidence for PHD2-independent effects of IOP1 knockdown on HIF-1α.
(A) Hep3B cells stably transfected with either pcDNA3 or pcDNA3-HA-IOP1R and maintained under normoxia were treated with either Control-B (CON) or IOP1-A (IOP1) siRNA. Cells were lysed, 72 h post-transfection, and subjected to Western blotting. Positions of HA-IOP1R and endogenous IOP1 are as indicated. (B) Hep3B cells maintained under normoxia were treated with the indicated siRNAs. Cells were lysed, 72 h post-transfection, and equal quantities of cellular lysates were subjected to HIF proline hydroxylase assay using 35S-labelled VHL capture, or to Western blotting using the indicated antibodies. Input represents 5% of the total. (C) Hep3B cells maintained under normoxia were treated with the indicated siRNAs. Cells were lysed, 72 h post-transfection, and subjected to Western blotting. (D) Hep3B cells maintained under normoxia were transfected with one siRNA and then 24 h later with a second siRNA, as indicated (C, control siRNA; I, IOP1 siRNA; P, PHD2 siRNA). Cells were lysed 72 h post-transfection. In (A–D), equal quantities of cellular lysates were subjected to Western blotting using the indicated antibodies.
The identification of IOP1 from a two-hybrid screen using PHD2 as bait would seemingly make a compelling case for IOP1 acting on HIF-1α through PHD2. In this regard, the yeast homologue of iron-only hydrogenase (Nar1p) plays an essential role in the assembly of cytoplasmic iron–sulfur clusters [56], raising the possibility that IOP1 might facilitate the insertion of the active-site ferrous iron into the active site of PHD2. To test this, we treated Hep3B cells with siRNA to either IOP1 or PHD2, and assayed for endogenous HIF proline hydroxylase activity against HIF-1α-(531–575). If IOP1 was essential for iron incorporation into PHD2, one might expect that IOP1 knockdown would diminish HIF proline hydroxylase activity in this assay, which is performed in the absence of exogenous iron. In Hep3B cells, as with most cell lines, PHD2 is the predominant PHD isoform [52], and consistent with this, PHD2 knockdown substantially diminishes HIF proline hydroxylase activity (Figure 5B, top panel). Under these same conditions, we find that IOP1 knockdown does not appreciably change HIF proline hydroxylase activity (Figure 5B, top panel). These results therefore do not support the hypothesis that IOP1 is involved in iron assembly into PHD2. In additional experiments, we find the IOP1 does not affect total non-haem iron levels in these cells (see Supplemental Figure 1 at http://www.BiochemJ.org/bj/401/bj4010341add.htm). We also note that under conditions where IOP1 knockdown increases HIF-1α protein levels, those of HIF-2α are not (Figure 5C).
We next examined whether PHD2 knockdown attenuates the effect of IOP1 knockdown on HIF-1α levels, which would be the expectation if IOP1 affected HIF-1α in a PHD2-dependent manner. Towards this end, we treated cells with an initial round of transfection of siRNA to either IOP1 or PHD2 followed by a second round of transfection of siRNA to either PHD2 or IOP1. As shown in Figure 5(D), we find that knockdown of PHD2 does not attenuate IOP1 knockdown-induced HIF-1α protein levels. In fact, it augments it (for example, compare lanes 2 with 3), thereby further supporting the possibility that IOP1 knockdown acts to increase HIF-1α levels in a PHD2-independent manner. Consistent with this, we have so far been unable to potentiate PHD2 hydroxylase activity in vitro using recombinant IOP1, or affect either HIF-1α protein levels or HIF-dependent reporter gene activity by overexpressing IOP1 (see Supplemental Figure 2 at http://www.BiochemJ.org/bj/401/bj4010341add.htm, and results not shown). In addition, we have not observed changes in HIF-1α protein half-life upon IOP1 knockdown, nor does IOP1 knockdown alter a PHD2 knockdown-induced increase in HIF-1α protein half-life (see Supplemental Figure 3 at http://www.BiochemJ.org/bj/401/bj4010341add.htm).
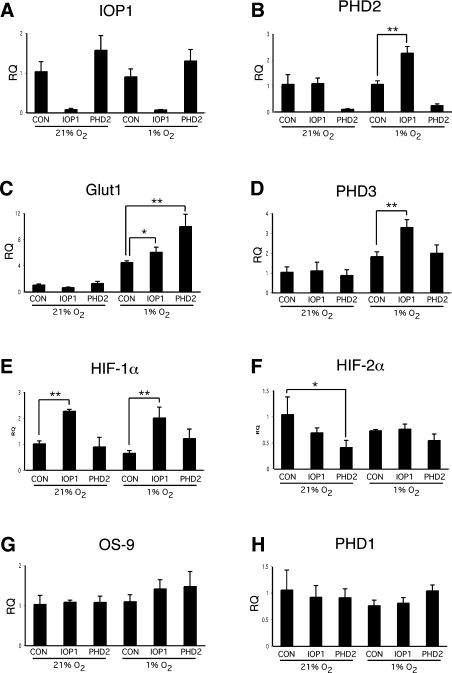
While it remains possible that IOP1 acts directly on PHD2, the data prompted us to consider the possibility that IOP1 might regulate other aspects of HIF-1α, such as mRNA levels. To test this, we treated Hep3B cells with IOP1, PHD2 or control siRNA, subjected some cells to hypoxia, isolated total RNA from these cells and then performed real-time PCR. Examination of the results for IOP1 and PHD2 establishes that mRNAs for both are substantially diminished by their respective siRNA treatments (to levels of 7 and 9% respectively under normoxia, and 7 and 23% under hypoxia respectively; Figures 6A and 6B). Moreover, examination of the results for GLUT1 and PHD3 confirms the known hypoxia-inducibility of these genes (Figures 6C and 6D respectively, compare first and fourth columns).
Figure 6. Regulation of HIF-1α mRNA and HIF target genes by IOP1.
Hep3B cells were transfected with Control-B (CON), IOP1-A (IOP1) or PHD2-A (PHD2) siRNA. At 72 h post-transfection, some cells were exposed to 1% O2 for 8 h. RNA was extracted from the cells and reverse transcribed. Real-time PCR was then performed using TaqMan probes for (A) IOP1, (B) PHD2, (C) GLUT1, (D) PHD3, (E) HIF-1α, (F) HIF-2α, (G) OS-9 or (H) PHD1. RQ indicates relative quantification. For each treatment, three samples were analysed, and means+S.D. are shown. *P<0.05, **P<0.01, using Student's t test.
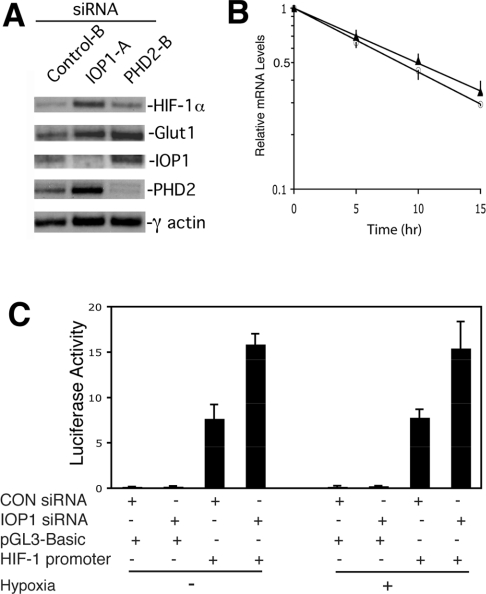
Importantly, we find that IOP1 knockdown significantly up-regulates HIF-1α mRNA under both normoxia and hypoxia (2.3- and 3.1-fold over the control siRNA respectively; Figure 6E). This effect is selective because it is not seen with HIF-2α, which is not significantly affected or even diminished by either IOP1 or PHD2 knockdown (Figure 6F). Consistent with this, IOP1 knockdown does not increase HIF-2α protein levels (Figure 5C). OS-9 mRNA is not affected either, suggesting that the effects of IOP1 knockdown on HIF-1α protein levels are not mediated through changes in OS-9 (Figure 6G). PHD1 mRNA provides an additional negative control (Figure 6H). Conversely, IOP1 knockdown augments hypoxic induction of PHD2 and PHD3 mRNA (Figures 6B and 6D respectively), both of which are known HIF target genes [23,53–55,57–59]. In addition, both IOP1 and PHD2 knockdowns both augment hypoxia-induced induction of the known HIF target gene GLUT1 (Figure 6C). Northern blotting supports the notion that IOP1 knockdown increases HIF-1α mRNA levels, as well as that of the HIF target genes GLUT1 and PHD2 under hypoxic conditions (Figure 7A).
Figure 7. Effects of IOP1 knockdown on HIF-1α mRNA stability and HIF-1α gene transcription.
(A) Northern-blot analyses. Hep3B cells were transfected with the indicated siRNAs. At 3 days post-transfection, cells were exposed to 0.2% O2 for 8 h and harvested. Poly(A)+ RNA (0.2 μg) was subjected to agarose-gel electrophoresis and Northern blotting was then performed with the indicated probes. (B) Analysis of HIF-1α mRNA stability. Hep3B cells were transfected with the indicated siRNAs. At 2 days post-transfection, cells were treated with actinomycin D (5 μg/ml), and at various time points total RNA was harvested, reverse transcribed and subjected to real-time PCR analysis for HIF-1α mRNA using SYBR Green. Each point is the mean±S.D. for three independent samples. The measured HIF-1α mRNA half-life in the presence of control siRNA (○) is 8.6 h, whereas that in the presence of IOP1 siRNA (▲) is 9.8 h. (C) HIF-1α promoter gene analysis. Hep3B cells were transfected with either Control-B (CON) or IOP1-A (IOP1) siRNA, and then 24 h later with the indicated reporter gene and pRL-TK. At 48 h after the initial transfection, cells were maintained under 21% O2 or subjected to 0.2% O2 for 16 h, and harvested. Firefly luciferase activities were normalized to that of Renilla luciferase. Experiments were performed in triplicate, and means+S.D. are shown.
Two possible mechanisms by which IOP1 knockdown might increase HIF-1α mRNA are through (i) changes in HIF-1α mRNA stability or (ii) changes in HIF-1α gene transcription. We examined the former by treating Hep3B cells with control or IOP1 siRNA and then measuring the HIF-1α mRNA half-life following actinomycin D treatment. While we find that IOP1 knockdown (to 9%) increases HIF-1α levels (2.0-fold), it does not significantly affect HIF-1α mRNA half-life (Figure 7B). We examined the possibility that the effects might be mediated through changes in gene transcription by first subcloning a 1.4 kb fragment of the HIF-1α gene containing the promoter into a luciferase reporter construct. We then knocked down IOP1 in Hep3B cells and examined the activity of this reporter gene under either normoxic or hypoxic conditions. The HIF-1α promoter reporter gene displays activity (Figure 7C, compare first and third columns). Importantly, IOP1 knockdown augments HIF-1α gene promoter activity under normoxic as well as hypoxic conditions (for example, compare third and fourth columns). This therefore provides a mechanism by which IOP1 regulates HIF-1α mRNA levels.
DISCUSSION
Following the discovery of proline hydroxylation as the key event in oxygen-dependent regulation of HIF-1α activity, much recent research on HIF has focused on the enzymes that catalyse this distinctive modification [18,19,21,60,61]. At the same time, it has become increasingly clear that HIF sits at the centre of a network of proteins that govern its activity. Aside from the PHDs, this expanding network includes VHL, FIH, Siah2, OS-9, CSN5/Jab1, ING4 (inhibitor of growth 4) and Hsp90 (heat-shock protein 90), as well as TBP-1 (TATA-box-binding protein 1), ARD1 (arrest at start of cell cycle defective 1), VDU2 (VHL-interacting deubiquitinating enzyme 2) and p53 among others [16,27,34,62–71]. The results presented here provide evidence that IOP1 is a novel member of this growing network of proteins. We find that knockdown of IOP1 elevates HIF-1α mRNA levels and HIF-1α protein levels, and augments hypoxia-induced HIF target gene expression. We therefore conclude that IOP1 normally serves to maintain HIF-1α mRNA levels at low steady-state levels. The knockdown studies furthermore imply that the function of IOP1 cannot be substituted for by IOP2/Narf, which, like IOP1, is homologous with the iron-only hydrogenases. The only known function of IOP2/Narf is binding to prelamin A, a component of the nuclear envelope [47].
Since their discovery in anaerobic bacteria, iron-only hydrogenases have been the subject of intensive investigations from biological, mechanistic and bioengineering perspectives. These enzymes play a central role in maintaining redox balance under anaerobic conditions. In contrast with mammalian cells, in which the electron acceptor for pyruvate oxidation is NAD+, in many anaerobic bacteria such as Clostridium pasteurianum it is ferredoxin [72]. Reduced ferredoxin, in turn, is then oxidized by iron-only hydrogenase to yield hydrogen gas and oxidized ferredoxin. While iron-only hydrogenases catalyse this seemingly simple chemical reaction, the reduction of protons to produce hydrogen gas, the underlying chemical mechanism continues to remain a matter of debate [73]. At their active site, these enzymes contain six iron atoms with a distinctive arrangement, the H-cluster, and X-ray crystallographic studies have revealed that in addition to sulfide, both cyanide and carbon monoxide constitute iron ligands [74,75]. They characteristically are oxygen-labile, and this has been attributed to redox changes at a distal Fe atom within the H-cluster [73].
Intriguingly, homologues of iron-only hydrogenases have been identified not only in anaerobic bacteria, but also in eukaryotes. In certain amitochondriate anaerobes such as Trichomonas vaginalis, they are present in a distinctive hydrogen-producing organelle, the hydrogenosome [76]. In green algae such as Chlamydomonas reinhardtii, they are induced under anaerobic conditions, so as to preserve the integrity of the oxygen-labile H-cluster [77]. In both situations, the iron-only hydrogenase is catalytically active and serves, as in anaerobic bacteria, to remove excess reducing equivalents. Additional insights into their function have recently emerged. For example, the iron-only hydrogenase of T. vaginalis is complexed to components of NADH dehydrogenase, thereby coupling malate dehydrogenation with hydrogen evolution [78,79]. As previously mentioned, in the baker's yeast S. cerevisiae, the homologue of iron-only hydrogenase, Nar1p, is a cytoplasmic iron–sulfur cluster protein that serves a distinct function, playing an essential role in the maturation of cytoplasmic iron–sulfur clusters [56]. Nar1p contains iron–sulfur clusters [56], but it is not yet known whether it is active as a hydrogenase.
In contrast with green algal iron-only hydrogenases, IOP1 is not induced at either the mRNA or protein level by hypoxia (Figures 2C and 6A). Indeed, we have so far not been able to detect hydrogenase activity with IOP1 (results not shown). In addition, the knockdown studies provide strong evidence that IOP1 is functional under both hypoxic and normoxic conditions, in contrast with bacterial iron-only hydrogenases, which are typically inactivated by exposure to oxygen. While this might be interpreted as indicating that IOP1 is not an oxygen-labile protein, it remains possible that IOP1 function may display a more subtle oxygen dependence. For example, by Western blotting (Figure 4), reporter gene assays (Figure 3), and real-time PCR analyses (Figure 6), the functional effects of IOP1 knockdown consistently appear to be more significant under hypoxic than normoxic conditions. Further investigation is required to find out whether this relates to an inherent oxygen sensitivity of IOP1 or to the experimental methodologies employed.
Mammalian cells possess two iron-only hydrogenase homologues, IOP1 and IOP2, whereas yeast possesses one, Nar1p. We did not find evidence that IOP1 is necessary for iron incorporation into the active site of PHD2 (Figure 5B). It should be noted, however, that the active-site iron of PHD2 is not in an iron–sulfur cluster, therefore leaving open the possibility that IOP1, and conceivably IOP2, may constitute functional homologues of Nar1p in the assembly of cytoplasmic iron–sulfur clusters. Should IOP1 fulfil this function, it will be of further interest to determine whether the effects of IOP1 on HIF-1α gene transcription are mediated in a manner dependent or independent of the cytoplasmic iron–sulfur cluster assembly pathway, which, like the hypoxia response pathway, fulfils an ancient cellular function.
Online data
Acknowledgments
We are grateful to Dr Gregory Hannon for the gift of pShag1, and Dr Celeste Simon and Dr Cheng-Jun Hu for the gift of the GLUT1 Northern-blot probe. We thank Dr Celeste Simon, Dr Zissimos Mourelatos and Dr Zheng Tu for critical readings of this paper, and Dr Brad Johnson for advice on the two-hybrid screening. This work was supported by American Heart Association Grant-in-Aid 0465447U, a University of Pennsylvania Cancer Center Pilot Project grant and NIH (National Institutes of Health) grants R01-CA090261 and R01-GM071459 to F.S.L.
References
- 1.Semenza G. L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- 2.Semenza G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 3.Wang G. L., Jiang B. H., Rue E. A., Semenza G. L. Hypoxia-inducible factor 1 is a basic-helix–loop–helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ema M., Taya S., Yokotani N., Sogawa K., Matsuda Y., Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hogenesch J. B., Chan W. K., Jackiw V. H., Brown R. C., Gu Y. Z., Pray-Grant M., Perdew G. H., Bradfield C. A. Characterization of a subset of the basic-helix–loop–helix-PAS superfamily that interacts with components of the dioxin signaling pathway. J. Biol. Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 6.Gu Y. Z., Moran S. M., Hogenesch J. B., Wartman L., Bradfield C. A. Molecular characterization and chromosomal localization of a third α-class hypoxia inducible factor subunit, HIF3α. Gene Expr. 1998;7:205–213. [PMC free article] [PubMed] [Google Scholar]
- 7.Huang L. E., Gu J., Schau M., Bunn H. F. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin–proteasome pathway. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard M. A., Qi H., Chung J., Lee E. H., Kondo Y., Hara S., Conaway R. C., Conaway J. W., Ohh M. Multiple splice variants of the human HIF-3 α locus are targets of the von Hippel–Lindau E3 ubiquitin ligase complex. J. Biol. Chem. 2003;278:11032–11040. doi: 10.1074/jbc.M208681200. [DOI] [PubMed] [Google Scholar]
- 9.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. G., Jr HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 10.Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., von Kriegsheim A., Hebestreit H. F., Mukherji M., Schofield C. J., et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 11.Yu F., White S. B., Zhao Q., Lee F. S. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc. Natl. Acad. Sci. U.S.A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O'Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., et al. C elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 14.Bruick R. K., McKnight S. L. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 15.Ivan M., Haberberger T., Gervasi D. C., Michelson K. S., Gunzler V., Kondo K., Yang H., Sorokina I., Conaway R. C., Conaway J. W., et al. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13459–13464. doi: 10.1073/pnas.192342099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell P. H., Wiesener M. S., Chang G. W., Clifford S. C., Vaux E. C., Cockman M. E., Wykoff C. C., Pugh C. W., Maher E. R., Ratcliffe P. J. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 17.Salceda S., Caro J. Hypoxia-inducible factor 1alpha (HIF-1α) protein is rapidly degraded by the ubiquitin-proteasome system under normoxic conditions: its stabilization by hypoxia depends on redox-induced changes. J. Biol. Chem. 1997;272:22642–22647. doi: 10.1074/jbc.272.36.22642. [DOI] [PubMed] [Google Scholar]
- 18.Schofield C. J., Ratcliffe P. J. Signalling hypoxia by HIF hydroxylases. Biochem. Biophys. Res. Commun. 2005;338:617–626. doi: 10.1016/j.bbrc.2005.08.111. [DOI] [PubMed] [Google Scholar]
- 19.Kaelin W. G. Proline hydroxylation and gene expression. Annu. Rev. Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 20.Hirota K., Semenza G. L. Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem. Biophys. Res. Commun. 2005;338:610–616. doi: 10.1016/j.bbrc.2005.08.193. [DOI] [PubMed] [Google Scholar]
- 21.Dann C. E., III, Bruick R. K. Dioxygenases as O2-dependent regulators of the hypoxic response pathway. Biochem. Biophys. Res. Commun. 2005;338:639–647. doi: 10.1016/j.bbrc.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 22.Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. Differential roles of hypoxia-inducible factor 1α (HIF-1α) and HIF-2α in hypoxic gene regulation. Mol. Cell. Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vengellur A., Woods B. G., Ryan H. E., Johnson R. S., LaPres J. J. Gene expression profiling of the hypoxia signaling pathway in hypoxia-inducible factor 1α null mouse embryonic fibroblasts. Gene Expr. 2003;11:181–197. doi: 10.3727/000000003108749062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warnecke C., Zaborowska Z., Kurreck J., Erdmann V. A., Frei U., Wiesener M., Eckardt K. U. Differentiating the functional role of hypoxia-inducible factor (HIF)-1α and HIF-2α (EPAS-1) by the use of RNA interference: erythropoietin is a HIF-2α target gene in Hep3B and Kelly cells. FASEB J. 2004;18:1462–1464. doi: 10.1096/fj.04-1640fje. [DOI] [PubMed] [Google Scholar]
- 25.Makino Y., Cao R., Svensson K., Bertilsson G., Asman M., Tanaka H., Cao Y., Berkenstam A., Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 26.Makino Y., Kanopka A., Wilson W. J., Tanaka H., Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia- inducible factor-3α locus. J. Biol. Chem. 2002;277:32405–32408. doi: 10.1074/jbc.C200328200. [DOI] [PubMed] [Google Scholar]
- 27.Mahon P. C., Hirota K., Semenza G. L. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lando D., Peet D. J., Whelan D. A., Gorman J. J., Whitelaw M. L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 29.Lando D., Peet D. J., Gorman J. J., Whelan D. A., Whitelaw M. L., Bruick R. K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewitson K. S., McNeill L. A., Riordan M. V., Tian Y. M., Bullock A. N., Welford R. W., Elkins J. M., Oldham N. J., Bhattacharya S., Gleadle J. M., et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 31.Lancaster D. E., McNeill L. A., McDonough M. A., Aplin R. T., Hewitson K. S., Pugh C. W., Ratcliffe P. J., Schofield C. J. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem. J. 2004;383:429–437. doi: 10.1042/BJ20040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gradin K., Takasaki C., Fujii-Kuriyama Y., Sogawa K. The transcriptional activation function of the HIF-like factor requires phosphorylation at a conserved threonine. J. Biol. Chem. 2002;277:23508–23514. doi: 10.1074/jbc.M201307200. [DOI] [PubMed] [Google Scholar]
- 33.Laughner E., Taghavi P., Chiles K., Mahon P. C., Semenza G. L. HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell. Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek J. H., Mahon P. C., Oh J., Kelly B., Krishnamachary B., Pearson M., Chan D. A., Giaccia A. J., Semenza G. L. OS-9 interacts with hypoxia-inducible factor 1α and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1α. Mol. Cell. 2005;17:503–512. doi: 10.1016/j.molcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 35.Fields S., Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 36.Huang J., Zhao Q., Mooney S. M., Lee F. S. Sequence determinants in hypoxia-inducible factor-1α for hydroxylation by the prolyl hydroxylases PHD1, PHD2, and PHD3. J. Biol. Chem. 2002;277:39792–39800. doi: 10.1074/jbc.M206955200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iyer N. V., Leung S. W., Semenza G. L. The human hypoxia-inducible factor 1α gene: HIF1A structure and evolutionary conservation. Genomics. 1998;52:159–165. doi: 10.1006/geno.1998.5416. [DOI] [PubMed] [Google Scholar]
- 38.Yu F., White S. B., Zhao Q., Lee F. S. Dynamic, site-specific interaction of hypoxia-inducible factor-1α with the von Hippel–Lindau tumor suppressor protein. Cancer Res. 2001;61:4136–4142. [PubMed] [Google Scholar]
- 39.Lee F. S., Peters R. T., Dang L. C., Maniatis T. MEKK1 activates both IκB kinase α and IκB kinase β. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9319–9324. doi: 10.1073/pnas.95.16.9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu Z., Kelley V. R., Collins T., Lee F. S. IκB kinase is critical for TNF-α-induced VCAM1 gene expression in renal tubular epithelial cells. J. Immunol. 2001;166:6839–6846. doi: 10.4049/jimmunol.166.11.6839. [DOI] [PubMed] [Google Scholar]
- 41.Paddison P. J., Caudy A. A., Bernstein E., Hannon G. J., Conklin D. S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berra E., Benizri E., Ginouves A., Volmat V., Roux D., Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Q., Lee F. S. The transcriptional activity of the APP intracellular domain–Fe65 complex is inhibited by activation of the NF-κB pathway. Biochemistry. 2003;42:3627–3634. doi: 10.1021/bi027117f. [DOI] [PubMed] [Google Scholar]
- 44.Horner D. S., Heil B., Happe T., Embley T. M. Iron hydrogenases: ancient enzymes in modern eukaryotes. Trends Biochem. Sci. 2002;27:148–153. doi: 10.1016/s0968-0004(01)02053-9. [DOI] [PubMed] [Google Scholar]
- 45.Frey M. Hydrogenases: hydrogen-activating enzymes. ChemBioChem. 2002;3:153–160. doi: 10.1002/1439-7633(20020301)3:2/3<153::AID-CBIC153>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 46.Vignais P. M., Billoud B., Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- 47.Barton R. M., Worman H. J. Prenylated prelamin A interacts with Narf, a novel nuclear protein. J. Biol. Chem. 1999;274:30008–30018. doi: 10.1074/jbc.274.42.30008. [DOI] [PubMed] [Google Scholar]
- 48.Peters J. W. Structure and mechanism of iron-only hydrogenases. Curr. Opin. Struct. Biol. 1999;9:670–676. doi: 10.1016/s0959-440x(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 49.Nicolet Y., Lemon B. J., Fontecilla-Camps J. C., Peters J. W. A novel FeS cluster in Fe-only hydrogenases. Trends Biochem. Sci. 2000;25:138–143. doi: 10.1016/s0968-0004(99)01536-4. [DOI] [PubMed] [Google Scholar]
- 50.Rees D. C., Howard J. B. The interface between the biological and inorganic worlds: iron–sulfur metalloclusters. Science. 2003;300:929–931. doi: 10.1126/science.1083075. [DOI] [PubMed] [Google Scholar]
- 51.Freeman R. S., Hasbani D. M., Lipscomb E. A., Straub J. A., Xie L. SM-20, EGL-9, and the EGLN family of hypoxia-inducible factor prolyl hydroxylases. Mol. Cell. 2003;16:1–12. [PubMed] [Google Scholar]
- 52.Appelhoff R. J., Tian Y. M., Raval R. R., Turley H., Harris A. L., Pugh C. W., Ratcliffe P. J., Gleadle J. M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 53.Marxsen J. H., Stengel P., Doege K., Heikkinen P., Jokilehto T., Wagner T., Jelkmann W., Jaakkola P., Metzen E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-α-prolyl-4-hydroxylases. Biochem. J. 2004;381:761–767. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aprelikova O., Chandramouli G. V., Wood M., Vasselli J. R., Riss J., Maranchie J. K., Linehan W. M., Barrett J. C. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J. Cell. Biochem. 2004;92:491–501. doi: 10.1002/jcb.20067. [DOI] [PubMed] [Google Scholar]
- 55.Greijer A., van der Groep P., Kemming D., Shvarts A., Semenza G., Meijer G., van de Wiel M., Belien J., van Diest P., van der Wall E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1) J. Pathol. 2005;206:291–304. doi: 10.1002/path.1778. [DOI] [PubMed] [Google Scholar]
- 56.Balk J., Pierik A. J., Netz D. J., Muhlenhoff U., Lill R. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron–sulphur proteins. EMBO J. 2004;23:2105–2115. doi: 10.1038/sj.emboj.7600216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metzen E., Stiehl D. P., Doege K., Marxsen J. H., Hellwig-Burgel T., Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem. J. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.del Peso L., Castellanos M. C., Temes E., Martin-Puig S., Cuevas Y., Olmos G., Landazuri M. O. The von Hippel Lindau/hypoxia-inducible factor (HIF) pathway regulates the transcription of the HIF-proline hydroxylase genes in response to low oxygen. J. Biol. Chem. 2003;278:48690–48695. doi: 10.1074/jbc.M308862200. [DOI] [PubMed] [Google Scholar]
- 59.Pescador N., Cuevas Y., Naranjo S., Alcaide M., Villar D., Landazuri M. O., Del Peso L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005;390:189–197. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaelin W. G., Jr The von Hippel–Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem. Biophys. Res. Commun. 2005;338:627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 61.Schofield C. J., Ratcliffe P. J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 62.Nakayama K., Frew I. J., Hagensen M., Skals M., Habelhah H., Bhoumik A., Kadoya T., Erdjument-Bromage H., Tempst P., Frappell P. B., et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1α abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 63.Bae M. K., Ahn M. Y., Jeong J. W., Bae M. H., Lee Y. M., Bae S. K., Park J. W., Kim K. R., Kim K. W. Jab1 interacts directly with HIF-1α and regulates its stability. J. Biol. Chem. 2002;277:9–12. doi: 10.1074/jbc.C100442200. [DOI] [PubMed] [Google Scholar]
- 64.Bemis L., Chan D. A., Finkielstein C. V., Qi L., Sutphin P. D., Chen X., Stenmark K., Giaccia A. J., Zundel W. Distinct aerobic and hypoxic mechanisms of HIF-α regulation by CSN5. Genes Dev. 2004;18:739–744. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ozer A., Wu L. C., Bruick R. K. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc. Natl. Acad. Sci. U.S.A. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isaacs J. S., Jung Y. J., Mimnaugh E. G., Martinez A., Cuttitta F., Neckers L. M. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 α-degradative pathway. J. Biol. Chem. 2002;277:29936–29944. doi: 10.1074/jbc.M204733200. [DOI] [PubMed] [Google Scholar]
- 67.Corn P. G., McDonald E. R., III, Herman J. G., El-Deiry W. S. Tat-binding protein-1, a component of the 26S proteasome, contributes to the E3 ubiquitin ligase function of the von Hippel–Lindau protein. Nat. Genet. 2003;35:229–237. doi: 10.1038/ng1254. [DOI] [PubMed] [Google Scholar]
- 68.Jeong J. W., Bae M. K., Ahn M. Y., Kim S. H., Sohn T. K., Bae M. H., Yoo M. A., Song E. J., Lee K. J., Kim K. W. Regulation and destabilization of HIF-1α by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 69.Li Z., Wang D., Messing E. M., Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.An W. G., Kanekal M., Simon M. C., Maltepe E., Blagosklonny M. V., Neckers L. M. Stabilization of wild-type p53 by hypoxia-inducible factor 1α. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 71.Bardos J. I., Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta. 2005;1755:107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 72.Meyer J. Clostridial iron–sulphur proteins. J. Mol. Microbiol. Biotechnol. 2000;2:9–14. [PubMed] [Google Scholar]
- 73.Armstrong F. A. Hydrogenases: active site puzzles and progress. Curr. Opin. Chem. Biol. 2004;8:133–140. doi: 10.1016/j.cbpa.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 74.Peters J. W., Lanzilotta W. N., Lemon B. J., Seefeldt L. C. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- 75.Nicolet Y., Piras C., Legrand P., Hatchikian C. E., Fontecilla-Camps J. C. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- 76.Hackstein J. H., Akhmanova A., Boxma B., Harhangi H. R., Voncken F. G. Hydrogenosomes: eukaryotic adaptations to anaerobic environments. Trends Microbiol. 1999;7:441–447. doi: 10.1016/s0966-842x(99)01613-3. [DOI] [PubMed] [Google Scholar]
- 77.Happe T., Hemschemeier A., Winkler M., Kaminski A. Hydrogenases in green algae: do they save the algae's life and solve our energy problems? Trends Plant Sci. 2002;7:246–250. doi: 10.1016/s1360-1385(02)02274-4. [DOI] [PubMed] [Google Scholar]
- 78.Dyall S. D., Yan W., Delgadillo-Correa M. G., Lunceford A., Loo J. A., Clarke C. F., Johnson P. J. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004;431:1103–1107. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- 79.Hrdy I., Hirt R. P., Dolezal P., Bardonova L., Foster P. G., Tachezy J., Embley T. M. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.