Abstract
Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a genetically heterogeneous heart-muscle disorder characterized by progressive fibrofatty replacement of right ventricular myocardium and an increased risk of sudden cardiac death. Mutations in desmosomal proteins that cause ARVC have been previously described; therefore, we investigated 88 unrelated patients with the disorder for mutations in human desmosomal cadherin desmocollin-2 (DSC2). We identified a heterozygous splice-acceptor–site mutation in intron 5 (c.631-2A→G) of the DSC2 gene, which led to the use of a cryptic splice-acceptor site and the creation of a downstream premature termination codon. Quantitative analysis of cardiac DSC2 expression in patient specimens revealed a marked reduction in the abundance of the mutant transcript. Morpholino knockdown in zebrafish embryos revealed a requirement for dsc2 in the establishment of the normal myocardial structure and function, with reduced desmosomal plaque area, loss of the desmosome extracellular electron-dense midlines, and associated myocardial contractility defects. These data identify DSC2 mutations as a cause of ARVC in humans and demonstrate that physiologic levels of DSC2 are crucial for normal cardiac desmosome formation, early cardiac morphogenesis, and cardiac function.
Arrhythmogenic right ventricular cardiomyopathy/dysplasia (ARVC/D [MIM 107970]) is a myocardial disorder characterized by progressive loss of cardiomyocytes and fibrofatty replacement.1,2 The disease commonly affects the right ventricle, but left ventricular involvement can also arise. ARVC often presents with ventricular arrhythmias originating in the right ventricle and is a major cause of sudden death in the young.1–3
Familial forms of ARVC exist, although their prevalence varies widely.4,5 Several genomic loci and six disease genes have been identified in autosomal dominant and recessive forms of ARVC. These genes primarily encode desmosomal proteins, but mutations in the cardiac ryanodine receptor (RYR2) have been described in some families with distinctive clinical phenotypes, and there are limited data that perturbed transforming growth-factor β3 (TGFβ3) signaling may result in ARVC.6–9 Mutations in plakoglobin (JUP) and desmoplakin (DSP) genes cause autosomal recessive ARVC in syndromes involving skin and hair, as well as heart.10,11 More recently, dominant mutations in DSP have been described, while mutations in plakophilin-2 (PKP2) have been reported to be responsible for a major proportion of ARVC cases.12–16 JUP, DSP, and PKP2 are all components of the desmosomal intercellular junction complex known to be essential for maintaining tissue integrity and, increasingly, implicated in cell signaling. Other major constituents of the desmosomal plaque are the desmosomal cadherins—desmocollin (DSC) and desmoglein (DSG). These are type I integral membrane glycoproteins that contain four highly conserved extracellular subdomains (ECs), a more variable extracellular anchor domain, a single transmembrane domain, an intracellular anchor domain, and additional cytoplasmatic subdomains. Within the first extracellular domain is a highly conserved cell adhesion–recognition (CAR) sequence thought important for heterophilic interaction in desmosomal cadherin-based adhesion.17–19 In addition to calcium-dependent cell adhesion, these proteins are regulators of tissue morphogenesis and also may participate in intracellular signaling processes.17–21 Mutations in DSG2 have recently been identified in ARVC,14,22 so we hypothesized that mutations in DSC2 may account for some cases of the disease. Using a zebrafish model system, we also have determined a requirement for DSC2 in the emergence of normal myocardial structure and function.
After giving their written informed consent under an institutional review board–approved protocol, a total of 88 unrelated white patients who were given diagnoses of ARVC, according to European Society of Cardiology and International Society and Federation of Cardiology Task Force criteria,23 and who were without mutations in the PKP2 gene were screened for mutations in all exons and flanking intronic sequences of DSC2 and DSG2, by direct bidirectional sequencing of PCR-amplified genomic fragments. In one 58-year-old male patient, we identified an A→G transition in DSC2 that affects the highly conserved 3′ splice-acceptor site of intron 5 (c.631-2A→G) (fig. 1A). This DSC2 variant (GenBank accession number NM_024422.2) was not present in 500 white control chromosomes. No other mutations were found in DSC2 or DSG2 in the 88 probands.
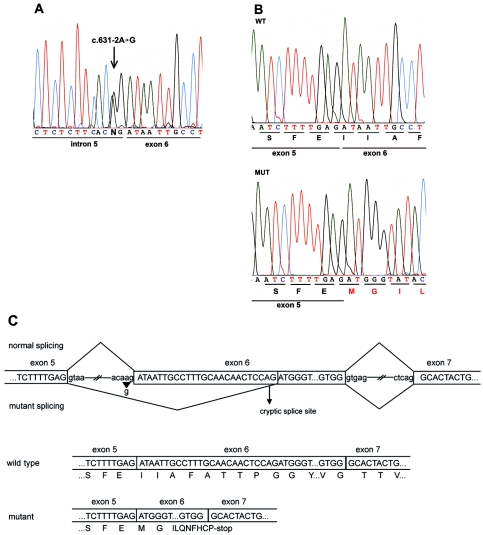
Figure 1. .
DSC2 mutation c.631-2A→G causes ARVC. A, Partial nucleotide sequence of DSC2 intron 5–exon 6 junction. The genomic sequence shows a heterozygous A→G transition in the splice-acceptor site position of intron 5. B, Partial nucleotide sequence of cloned wild-type (WT) and mutant (MUT) transcripts derived from DSC2 cDNA of the mutation carrier (pCR2.1 plasmid) (TOPO-TA cloning [Invitrogen]). The mutant transcript shows a deletion of the first 25 bp of exon 6, which causes a frameshift and a premature stop codon after 10 novel aa residues (MGILQNFHCP*). C, Schematic view of the splicing mechanism in normal and mutant DSC2. The mutation c.631-2A→G (arrowhead) affects the splice-acceptor site of intron 5 and activates an alternative cryptic splice site (arrow) in exon 6. The consequence is a 25-bp deletion that shifts the reading frame and generates a premature stop codon.
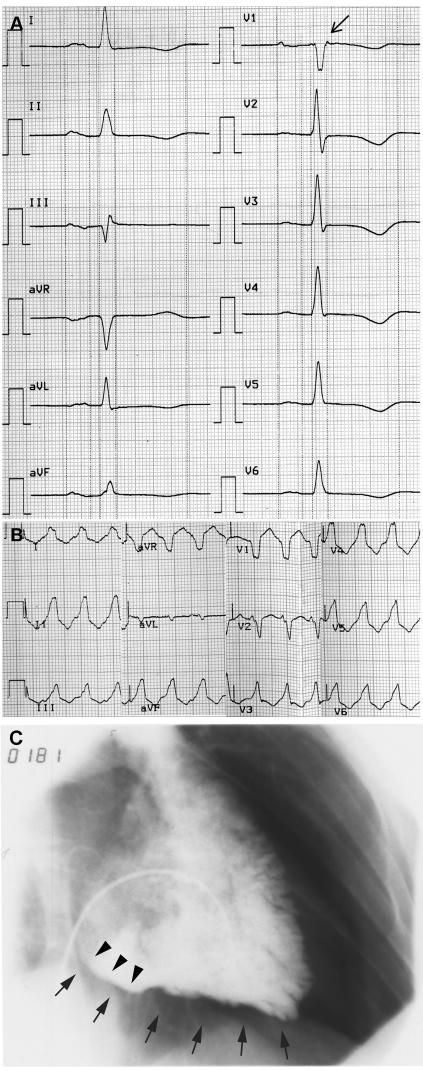
Clinical evaluation of the patient with the c.631-2A→G mutation (table 1) revealed an electrocardiogram (ECG) with inverted T waves across the precordial leads (V1–V6), an epsilon wave in lead V1 (fig. 2A), and echocardiographic as well as angiographic evidence of a severely dilated, hypokinetic right ventricle with diastolic bulging and trabecular disarrangement (fig. 2C). Left ventricular size and function were normal. Ventricular arrhythmias with left bundle branch block morphology (fig. 2B) were apparent by age 43 years and led to the implantation of a cardiac defibrillator and the initiation of antiarrhythmic medication. The family history was negative for other potentially clinically affected individuals. The father of the patient died of pneumonia at age 64 years, and the patient's mother died of cancer at age 71 years. The clinically unaffected 61-year-old sister was not available for genetic analyses.
Table 1. .
Clinical Findings of the Patient with the DSC2 Mutation
| Metric and Clinical Findings | Task Force Criteriaa (Major/Minor) |
| Twelve-lead ECG: | |
| Epsilon wave in V1 | 1/0 |
| Inverted T waves in precordial leads (V1–V6) | 0/1 |
| Holter monitoring: | |
| Nonsustained left bundle branch block–type tachycardia | 0/1 |
| 2D echocardiogram/right ventricular angiogram: | |
| Severe dilatation and hypokinesia of the right ventricle, trabecular disarrangement, diastolic bulging, no left ventricular involvement | 1/0 |
| Tissue characteristics of a right ventricular biopsy: | |
| Mild interstitial fibrosis and fatty infiltration of myocardium | 0/0 |
| Family history: | |
| No additional family members available | 0/0 |
| Diagnostic score | 2/2 |
For the diagnosis of ARVC to be established, the subject must meet two major criteria, one major and two minor criteria, or four minor criteria in different categories.23
Figure 2. .
Clinical features of the mutation carrier. A, Twelve-lead resting ECG (50 mm/s). Note the T-wave inversions in the leads V1–V6 and the epsilon wave (arrow) in V1. B, ECG (25 mm/s) of a ventricular tachycardia with left bundle branch block morphology. C, Image of an angiogram showing an enlarged right ventricle in 30° right-anterior oblique, with evidence of diastolic bulging (arrowheads) at the right ventricle inflow tract and diffuse inferior hypokinesia (arrows).
To further investigate the c.631-2A→G mutation, we performed RT-PCR with exon-linking oligonucleotides (c607F/c1073R) (table 2) of DSC2 mRNA from both lymphocytes and with a cardiac biopsy sample obtained from the proband. In lymphocytes, in addition to the transcript of normal size, there was evidence of a shorter DSC2 transcript. RT-PCR products were cloned, and sequencing analysis revealed aberrant splicing of the mutant transcript, with activation of a cryptic splice-acceptor site in exon 6 and deletion of 25 nt. The mutant transcript encoded a frameshift resulting in 10 novel aa residues and a premature termination codon (fig. 1B and 1C). The predicted truncated DSC2 molecule lacks the highly conserved CAR sequence of DSC2 thought to be required for heterophilic adhesive interactions. RT-PCR from cardiac mRNA of the patient did not reveal evidence of the truncated transcript. In addition, we performed allele-specific real-time PCR to quantify the amounts of mRNA of wild-type and mutant transcripts in cardiac tissue. We designed allele-specific forward primers (c631F/c800R and cMutF/c800R) (table 2) of DSC2 and performed real-time PCR experiments on individual RNA samples from the cardiac biopsy of the patient and control samples, by monitoring the increase of fluorescence through the binding of SYBR green to double-stranded DNA (iCycler [Biorad]). mRNA expression data were normalized to β2-microglobulin and 28S-RNA content. We found a marked reduction of the mutant DSC2 transcript (3% mRNA abundance), and the specimen of the patient contained less wild-type DSC2 mRNA (60%) compared with control myocardium (fig. 3A). These mRNA expression data were further supported by western-blot analysis of the cardiac biopsy sample performed with DSC2 (Progen and gift from W. W. Franke [Heidelberg], 1:10,000) antibodies. The specimen of the patient contained less wild-type DSC2 protein than did control myocardium (fig. 3B).
Table 2. .
Oligonucleotide Primer Sequences
| Primer | Primer Sequence (5′→3′) |
| c607F | CGTGAGCAGTATGAATCTTTTGAG |
| c800R | GCACACACTTGTCCCACAGTAGTG |
| c1073R | GTACGAGTAAATGTTGGCAAGTG |
| c631F | ATAATTGCCTTTGCAACAACTCCAG |
| cMutF | CAGTATGAATCTTTTGAGATGGG |
Figure 3. .

Expression analysis of the mutant DSC2. A, Allele-specific quantitative real-time PCR of DSC2 wild-type (WT) and mutant (MUT) mRNA of cardiac tissue derived from patient (P) and control (C) samples. Note the reduced expression (3%) of the mutant allele in cardiac tissue in the patient. B, Western-blot analysis of a right ventricular biopsy sample obtained from the patient with the DSC2 mutation c.631-2A→G. Note that the amount of DSC2 in the patient is reduced when compared with control myocardium. Blots were probed with antibodies for DSC2 (Progen, 1:10,000) and α-tubulin (Sigma-Aldrich, 1:5,000) as loading control.
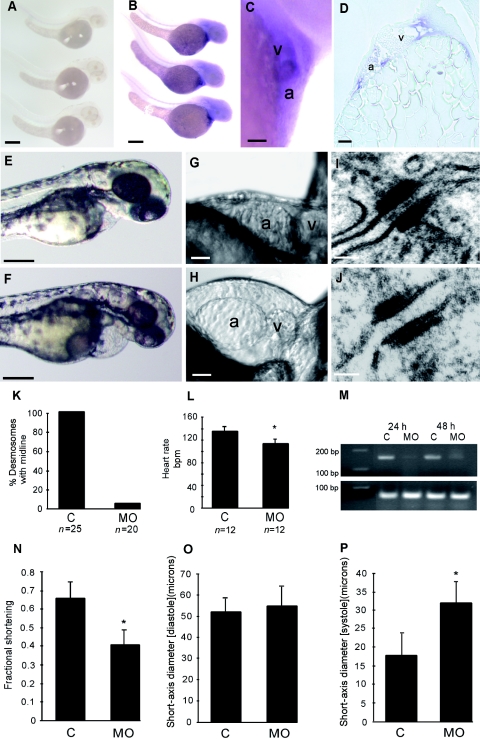
To explore the in vivo effects of the mutated DSC2 on cardiac structure and function, we recapitulated the effects of this splice-site mutation in zebrafish. We first cloned the zebrafish ortholog of human DSC2 by PCR by homology with public genomic data. The full-length ORF encodes an 892-aa protein sharing 34% identity with the human protein, and it contains all the major DSC domains. Whole-mount in situ hybridization throughout development, with use of sense or antisense riboprobe generated from the full-length clone (GenBank accession number BX649302) and standard protocols, revealed that this zebrafish dsc2 was expressed at relatively low levels in the brain and heart. The most obvious expression in the heart was seen at 48 h post fertilization (hpf) (fig. 4A–4C). Cross-sections of the heart confirmed staining in the atrium, ventricle, and both the inflow and outflow tracts (fig. 4D).
Figure 4. .
Zebrafish dsc2 expression and knockdown. A–D, Zebrafish dsc2 expression. Whole-mount in situ hybridizations with use of full-length sense (A) and antisense (B) zebrafish dsc2 riboprobe show weak staining in the heart and brain at 48 hpf with the antisense probe, whereas the sense control riboprobe does not reveal any staining. Scale bars indicate 100 μM. Higher magnification image of the heart (C) and longitudinal section through the ventricle (D) of whole-mount in situ stained embryos confirm cardiac expression. Scale bars indicate 25 μM. a = Atrium; v = ventricle. E–P, Knockdown of dsc2 in zebrafish. E, Phenotype at 48 hpf of an embryo injected with mismatch control oligo compared with a morphant (injected with 25 ng of morpholino dsc2-1) embryo at the same stage (F). Scale bars indicate 100 μM. High-resolution pictures of the heart of morphant embryo (H) compared with control-injected embryo (G). Scale bars indicate 25 μM. Transmission EM of cardiac desmosomes from a control embryo (I) and a morphant embryo (J) at 48 hpf. Scale bars indicate 50 nm. K, Percentage of desmosomes with midline in control (C) and morphant (MO) embryos. L, Heart rate measured at room temperature in morphant embryos compared with control injected embryos (P<.05). M, RT-PCR of zebrafish dsc2, to determine morpholino efficiency. N, Fractional shortening of the ventricular chamber of control and morphant fish at 48 hpf. Diastolic (O) and systolic (P) short-axis diameter measured in controls and morphant embryos (P<.05).
Using antisense morpholino oligos, we were able to target not only the translation start site but also the splice-acceptor sites for exon 6 and exon 11 of zebrafish dsc2. Each of the three morpholinos resulted in marked reduction in the levels of dsc2 mRNA in the morphant embryos (fig. 4M). The morpholino phenotypes revealed significant dose-dependent bradycardia, chamber dilatation, and abnormal cardiac contractility with progressive pericardial edema developing by 48 hpf in morphant fish (fig. 4E–4H). No abnormalities were evident in embryos injected with mismatch control morpholino over a broad range of doses. The mean ± SD heart rate in control fish was 136.7 ± 8.3 beats per min (bpm) (n=12) at 22°C versus 114.4 ± 9 bpm (n=12) in morphant embryos (P<.05) (fig. 4L). Cardiac-chamber size and contractility were measured using high-speed video microscopy (fig. 4N). Fractional shortening was reduced from 0.66 ± 0.09 to 0.41 ± 0.07 in control versus morphant embryos, respectively, at 48 hpf (P<.05). The ventricular-end systolic short-axis diameter was increased from 18.1 ± 5.7 μm in control fish to 32.2 ± 5.5 μm in morphant fish (fig. 4O and 4P). In contrast, no significant difference in the ventricular-end diastolic diameter was observed. These findings were associated with minor focal CNS necrosis (correlating with the brain expression domain of zebrafish dsc2). Electron microscopy (EM) revealed that the normal desmosomal extracellular electron-dense midline failed to form in the myocardial tissue of the ventricle in morphant embryos at 48 hpf but was present in control embryos (fig. 4I–4K).
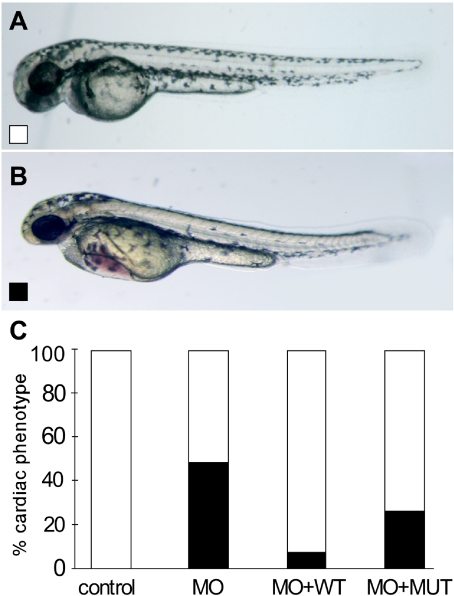
Rescue of the dsc2 morphant cardiac phenotypes was possible with coinjection of wild-type human DSC2 mRNA but not with mRNA encoding the mutant human DSC2 (fig. 5A–5C). Using a dose of dsc2 morpholino resulting in a cardiac phenotype (bradycardia and edema) in 48.4% of morphant embryos, we observed a substantial reduction in the proportion (7.8%) with cardiac dysfunction in embryos injected with morpholino and wild-type DSC2 mRNA but no significant reduction in the proportion (26.4%) of embryos injected with morpholino and mutant DSC2 mRNA (fig. 5C). Taken together, these findings suggest that the dose of DSC2 is critical for normal cardiac function and that the c.631-2A→G mutation encodes an mRNA incapable of rescuing the in vivo effects of gene-dose reduction.
Figure 5. .
Rescue of dsc2 morphants (MO). A, Wild-type phenotype. B, Morphant phenotype. C, Rescue of dsc2 morphant phenotype (blackened bars) is seen with wild-type (WT) (unblackened bars) but not with mutant (MUT) DSC2 mRNA. Results are based on data from three different experiments with 25 ng of morpholino and 1 pg of the respective capped mRNAs.
DSC2—together with other desmosomal cadherins, armadillo proteins, and plakins—form cell-adhesion complexes that provide mechanical strength in tissues by anchoring intermediate filaments. It has been suggested that DSCs and DSGs, in addition to their cell-adhesion function, act as molecular sensors regulating cellular behavior and transducing cellular signals.17–19 The dominant DSC2 mutation (c.631-2A→G) described here predicts a truncated DSC2 protein lacking the highly conserved CAR sequence located in the first EC1 domain, which is present in all classic and desmosomal cadherins. Studies of classic cadherins suggest that the CAR sequence forms part of an adhesive interface between cadherin molecules on opposite cell surfaces and led to the postulation of an “adhesion zipper” model.24 Dominant negative mutants (deletion of EC1–EC4) of different desmosomal cadherins transfected into cultured keratinocytes with adenoviral vector disrupt cell-cell adhesion.25,26 The truncated protein encoded by the mutant allele in this case may have a dominant negative effect disrupting cell-cell adhesion, but the markedly decreased levels of mRNA observed with the mutant DSC2 allele suggest that nonsense-mediated mRNA decay and consequent haploinsufficiency likely contribute to the disease mechanism.
Knockdown of dsc2 in zebrafish results in dose-dependent bradycardia, impaired contractility, and cardiac edema, associated with consistent loss of the desmosomal extracellular electron-dense midlines. This structure containing the five extracellular subdomains of DSC2 is important for homophilic dimerization in cis and trans orientation as well as for stable cell-cell contacts. Importantly, we were able to rescue the dsc2 knockdown cardiac phenotype with wild-type human DSC2 mRNA but not with mutant (c.631-2A→G) mRNA. These studies indicate that physiologic levels of DSC2 are crucial for early cardiac morphogenesis, desmosome formation, and normal cardiac function, as well as independently confirming the in vivo loss of function of the truncated mutant protein.
Mutations in other desmosomal cadherins have been described in different human skin diseases. An N-terminal deletion in DSG1 causes autosomal dominant striate palmoplantar keratoderma (PPKS1 [MIM 148700]),26 and DSG4 mutations result in defective hair follicle differentiation (LAH [MIM 607903]).27 DSC2 and DSG2 are expressed ubiquitously in desmosomal-containing tissues,21 but DSG1 and -3 and DSC1 and -3 are restricted to stratified epithelia.18 Although DSC2 is expressed in skin epithelium as well as in many other tissues, no additional phenotypic features could be detected in the affected patient. It is possible that absence of cutaneous abnormalities in our patient with a DSC2 mutation could be due to compensation by other desmosomal cadherins in skin but not in cardiac tissue. DSC1 and DSC3, which are also mainly expressed in the basal layer of the epidermis, are excellent candidates to compensate for DSC2 function. This hypothesis is supported by in vitro studies, which have demonstrated adhesive interactions between different isoforms of cadherins that occur in the same desmosome.28–30
Recently, it was reported that mutations in DSG2 account for a significant proportion of ARVC in Italian and U.S. populations,14,22 but we were unable to confirm this result in our cohort of 88 patients with ARVC. Possible reasons for such a variable prevalence of DSG2 mutations in cohorts with ARVC include the geographic or ethnic origins of the populations, selection biases, and the relatively small sizes of the original cohorts studied.
Whereas the involvement of multiple desmosomal protein genes has led to speculation regarding the sensitivity of myocardium to mechanical disruption, the pathogenic mechanisms leading to ARVC in humans are not clear. Mice null for various junctional protein genes (Dsp, Jup, Pkp2, and Cdh2) expressed in cardiac tissue exhibit embryonic lethality, with multiple developmental abnormalities, including severe cardiovascular defects.31–34 Recently, Garcia-Gras et al.35 reported that heterozygous cardiac-restricted deletion of Dsp in mice partially recapitulates the phenotype of human ARVC. These researchers also suggested a novel molecular mechanism for adipocytic and fibrous replacement of myocytes in the pathogenesis of ARVC, controlled by the canonical Wnt/β-catenin signaling pathway.35 Further in vivo and in vitro studies of DSC2 and other desmosomal molecules involved in the disease process will provide new insights into the molecular mechanisms of ARVC.
In conclusion, we describe the first evidence, to our knowledge, of disease-causing mutation in the human DSC2 gene: a heterozygous mutation (c.631-2A→G) resulting in ARVC. Expression analyses in the affected patient suggest that DSC2 gene dose may contribute to the disease pathogenesis. This is further supported by evidence that graded knockdown of dsc2 in zebrafish embryos profoundly perturbs myocardial desmosome structure and contractile function.
Acknowledgments
The authors are grateful to the patients, family members, and their referring physicians. We thank S. Milan and C. Nandy for technical assistance. This project was supported by German Research Council research grant Ge 1222/1–2 (to B.G.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for DSC2 transcript variant [accession number NM_024422.2] and zebrafish DNA from clone [accession number BX649302])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for ARVC/D 1–10, DSC2, DSG2, PPKS1, and LAH)
References
- 1.Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C, Grosgogeat Y (1982) Right ventricular dysplasia: a report of 24 adult cases. Circulation 65:384–398 [DOI] [PubMed] [Google Scholar]
- 2.Fontaine G, Fontaliran F, Hebert JL, Chemla D, Zenati O, Lecarpentier Y, Frank R (1999) Arrhythmogenic right ventricular dysplasia. Annu Rev Med 50:17–35 10.1146/annurev.med.50.1.17 [DOI] [PubMed] [Google Scholar]
- 3.Thiene G, Nava A, Corrado D, Rossi L, Pennelli N (1988) Right ventricular cardiomyopathy and sudden death in young people. N Engl J Med 318:129–133 [DOI] [PubMed] [Google Scholar]
- 4.Nava A, Thiene G, Canciani B, Scognamiglio R, Daliento L, Buja G, Martini B, Stritoni P, Fasoli G (1988) Familial occurrence of right ventricular dysplasia: a study involving nine families. J Am Coll Cardiol 12:1222–1228 [DOI] [PubMed] [Google Scholar]
- 5.Wlodarska EK, Konka M, Kepski R, Zaleska T, Ploski R, Ruzyllo W, Janion M, Jaworska K, Rydlewska-Sadowska W, Hoffman P (2004) Familial form of arrhythmogenic right ventricular cardiomyopathy. Kardiol Pol 60:1–14 [PubMed] [Google Scholar]
- 6.Paul M, Schulze-Bahr E, Breithardt G, Wichter T (2003) Genetics of arrhythmogenic right ventricular cardiomyopathy—status quo and future perspectives. Z Kardiol 92:128–136 10.1007/s00392-003-0892-9 [DOI] [PubMed] [Google Scholar]
- 7.Sen-Chowdhry S, Syrris P, McKenna WJ (2005) Genetics of right ventricular cardiomyopathy. J Cardiovasc Electrophysiol 16:927–935 10.1111/j.1540-8167.2005.40842.x [DOI] [PubMed] [Google Scholar]
- 8.Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, Danieli GA, Rampazzo A (2005) Regulatory mutations in transforming growth factor-β3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res 65:366–373 10.1016/j.cardiores.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 9.Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, Muriago M, Basso C, Thiene G, Danieli GA, Rampazzo A (2001) Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2). Hum Mol Genet 10:189–194 10.1093/hmg/10.3.189 [DOI] [PubMed] [Google Scholar]
- 10.Norgett EE, Hatsell SJ, Carvajal-Huerta L, Cabezas JC, Common J, Purkis PE, Whittock N, Leigh IM, Stevens HP, Kelsell DP (2000) Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum Mol Genet 9:2761–2766 10.1093/hmg/9.18.2761 [DOI] [PubMed] [Google Scholar]
- 11.McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ (2000) Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease). Lancet 355:2119–2124 10.1016/S0140-6736(00)02379-5 [DOI] [PubMed] [Google Scholar]
- 12.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, Towbin JA, Danieli GA (2002) Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet 71:1200–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermott DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, Peters S, Grossmann KS, Drenckhahn J, Michely B, Sasse-Klaassen S, Birchmeier W, Dietz R, Breithardt G, Schulze-Bahr E, Thierfelder L (2004) Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet 36:1162–1164 10.1038/ng1461 [DOI] [PubMed] [Google Scholar]
- 14.Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A, Frigo G, Vettori A, Valente M, Towbin J, Thiene G, Danieli GA, Rampazzo A (2006) Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation 113:1171–1179 10.1161/CIRCULATIONAHA.105.583674 [DOI] [PubMed] [Google Scholar]
- 15.van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld AC, Wilde AA, van der Smagt J, Boven LG, Mannens MM, van Langen IM, Hofstra RM, Otterspoor LC, Doevendans PA, Rodriguez LM, van Gelder IC, Hauer RN (2006) Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation 113:1650–1658 10.1161/CIRCULATIONAHA.105.609719 [DOI] [PubMed] [Google Scholar]
- 16.Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL, Elliott PM, McKenna WJ (2006) Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation 113:356–364 10.1161/CIRCULATIONAHA.105.561654 [DOI] [PubMed] [Google Scholar]
- 17.Green KJ, Gaudry CA (2000) Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol 1:208–216 10.1038/35043032 [DOI] [PubMed] [Google Scholar]
- 18.Garrod DR, Merritt AJ, Nie Z (2002) Desmosomal cadherins. Curr Opin Cell Biol 14:537–455 10.1016/S0955-0674(02)00366-6 [DOI] [PubMed] [Google Scholar]
- 19.Jamora C, Fuchs E (2002) Intercellular adhesion, signalling and the cytoskeleton. Nat Cell Biol 4:E101–E108 10.1038/ncb0402-e101 [DOI] [PubMed] [Google Scholar]
- 20.Ko KS, Arora PD, McCulloch CA (2001) Cadherins mediate intercellular mechanical signaling in fibroblasts by activation of stretch-sensitive calcium-permeable channels. J Biol Chem 276:35967–35977 10.1074/jbc.M104106200 [DOI] [PubMed] [Google Scholar]
- 21.Nuber UA, Schafer S, Schmidt A, Koch PJ, Franke WW (1995) The widespread human desmocollin Dsc2 and tissue-specific patterns of synthesis of various desmocollin subtypes. Eur J Cell Biol 66:69–74 [PubMed] [Google Scholar]
- 22.Awad MM, Dalal D, Cho E, Amat-Alarcon N, James C, Tichnell C, Tucker A, Russell SD, Bluemke DA, Dietz HC, Calkins H, Judge DP (2006) DSG2 mutations contribute to arrhythmogenic right ventricular dysplasia/cardiomyopathy. Am J Hum Genet 79:136–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKenna WJ, Thiene G, Nava A, Fontaliran F, Blomstrom-Lundqvist C, Fontaine G, Camerini F, on behalf of the Task Force of the Working Group Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology (1994) Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Br Heart J 71:215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Overduin M, Harvey TS, Bagby S, Tong KI, Yau P, Takeichi M, Ikura M (1995) Solution structure of the epithelial cadherin domain responsible for selective cell adhesion. Science 267:386–389 10.1126/science.7824937 [DOI] [PubMed] [Google Scholar]
- 25.Hanakawa Y, Amagai M, Shirakata Y, Sayama K, Hashimoto K (2000) Different effects of dominant negative mutants of desmocollin and desmoglein on the cell-cell adhesion of keratinocytes. J Cell Sci 113:1803–1811 [DOI] [PubMed] [Google Scholar]
- 26.Rickman L, Simrak D, Stevens HP, Hunt DM, King IA, Bryant SP, Eady RA, Leigh IM, Arnemann J, Magee AI, Kelsell DP, Buxton RS (1999) N-terminal deletion in a desmosomal cadherin causes the autosomal dominant skin disease striate palmoplantar keratoderma. Hum Mol Genet 8:971–976 10.1093/hmg/8.6.971 [DOI] [PubMed] [Google Scholar]
- 27.Kljuic A, Bazzi H, Sundberg JP, Martinez-Mir A, O’Shaughnessy R, Mahoney MG, Levy M, Montagutelli X, Ahmad W, Aita VM, Gordon D, Uitto J, Whiting D, Ott J, Fischer S, Gilliam TC, Jahoda CA, Morris RJ, Panteleyev AA, Nguyen VT, Christiano AM (2003) Desmoglein 4 in hair follicle differentiation and epidermal adhesion: evidence from inherited hypotrichosis and acquired pemphigus vulgaris. Cell 113:249–260 10.1016/S0092-8674(03)00273-3 [DOI] [PubMed] [Google Scholar]
- 28.North AJ, Chidgey MA, Clarke JP, Bardsley WG, Garrod DR (1996) Distinct desmocollin isoforms occur in the same desmosomes and show reciprocally graded distributions in bovine nasal epidermis. Proc Natl Acad Sci USA 93:7701–7705 10.1073/pnas.93.15.7701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chitaev NA, Troyanovsky SM (1997) Direct Ca2+-dependent heterophilic interaction between desmosomal cadherins, desmoglein and desmocollin, contributes to cell-cell adhesion. J Cell Biol 138:193–201 10.1083/jcb.138.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii K, Norvell SM, Bannon LJ, Amargo EV, Pascoe LT, Green KJ (2001) Assembly of desmosomal cadherins into desmosomes is isoform dependent. J Invest Dermatol 117:26–35 10.1046/j.0022-202x.2001.01400.x [DOI] [PubMed] [Google Scholar]
- 31.Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W (2004) Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J Cell Biol 167:149–160 10.1083/jcb.200402096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallicano GI, Kouklis P, Bauer C, Yin M, Vasioukhin V, Degenstein L, Fuchs E (1998) Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J Cell Biol 143:2009–2022 10.1083/jcb.143.7.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruiz P, Brinkmann V, Ledermann B, Behrend M, Grund C, Thalhammer C, Vogel F, Birchmeier C, Gunthert U, Franke WW, Birchmeier W (1996) Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J Cell Biol 135:215–225 10.1083/jcb.135.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kostetskii I, Li J, Xiong Y, Zhou R, Ferrari VA, Patel VV, Molkentin JD, Radice GL (2005) Induced deletion of the N-cadherin gene in the heart leads to dissolution of the intercalated disc structure. Circ Res 96:346–354 10.1161/01.RES.0000156274.72390.2c [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Gras E, Lombardi R, Giocondo MJ, Willerson JT, Schneider MD, Khoury DS, Marian AJ (2006) Suppression of canonical Wnt/β-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest 116:2012–2021 10.1172/JCI27751 [DOI] [PMC free article] [PubMed] [Google Scholar]