Abstract
Double-stranded RNA (dsRNA) can specifically inhibit gene expression in a variety of organisms by invoking post-transcriptional degradation of homologous mRNA. Here we show that dsRNA-mediated gene regulation also occurs in the fission yeast Schizosaccharomyces pombe. We present evidence that: (i) reporter gene silencing is significantly enhanced when additional non-coding sense RNA is co-expressed with antisense RNA; (ii) expression of a panhandle RNA also silences target gene expression; (iii) expression of dsRNA is associated with siRNAs; (iv) a novel host-encoded factor which enhances antisense RNA gene silencing also enhances panhandle RNA-mediated gene inhibition. Both the exogenously introduced lacZ and c-myc genes are shown to be susceptible to dsRNA- mediated gene silencing in this model. Taken together, these data indicate that RNA-mediated gene silencing can occur through a RNAi-like mechanism in fission yeast.
INTRODUCTION
The utility of double-stranded RNA (dsRNA)-mediated gene interference (known as RNAi in animals) (1) has been demonstrated in a variety of organisms, including Caenorhabditis elegans (2), plants (3), Drosophila (4), planaria (5), trypanosomes (6), Hydra (7), zebrafish (8,9) and, most recently, in mammalian tissue (10–12). Addition ally, another form of post-transcriptional gene silencing (PTGS) in which extra copies of a target gene suppress both the endogenous and introduced transgene (co-suppression or quelling) has been observed in Paramecium (13), Neurospora crassa (14), plants (15), Dictyostelium (16) and mammalian cells (17). It has been suggested that in the different categories of RNA-mediated gene silencing, the culprit dsRNA is formed either by (i) the cryptic transcription of antisense RNA in cells where extra copies of the sense gene have been introduced, (ii) the simultaneous expression of antisense and sense sequences, (iii) the formation of RNA hairpins from inverted repeats or (iv) the direct introduction of dsRNA (18). The dsRNA is thought to act as a substrate for dsRNA-dependent RNA polymerase that generates a complementary RNA (cRNA) (19,20). This cRNA may target mRNA for degradation or hybridise to antisense RNA to generate additional dsRNA. It has been shown that the long dsRNA is fragmented into small 21–25 nt dsRNA species (21) and that this is the primary mediator of gene silencing (22). This fragmentation process is mediated by the RNase III-like nuclease DICER (23), however, it has recently been shown that its role may not be essential in all circumstances (24).
In the case of plants and worms it has been shown that RNAi acts in a sub-stoichiometric fashion and has the ability to migrate between cells (2,25). However, the potency of PTGS seems to vary between organisms. For example, dsRNA appears to have an amplification or catalytic component in most of the organisms investigated, but was shown to be less robust in the vertebrate zebrafish where gene suppression was dependent on the concentration of introduced dsRNA (8). This suggests that either the level of dsRNA has not reached the threshold which may be required for activation of a catalytic event or that some of the factors involved in robust forms of RNA-mediated gene silencing are absent and/or inhibitors of the gene silencing phenomenon are present in that organism.
The proteins that are essential for the RNAi pathway are present in Schizosaccharomyces pombe, suggesting that this mechanism is also required in this yeast. Indeed, it has recently been shown that this RNAi machinery is required for heterochromatic gene silencing in fission yeast (26) and that this is associated with small interfering RNAs (siRNA) homologous to these centromeric regions (27). In addition, it has been shown that the formation of heterochromatin at the silent mating type region (28) and accurate chromosomal segregation during mitosis requires the RNAi machinery. However, it has not yet been directly demonstrated that dsRNAs are involved in gene regulation in this model yeast system.
The lacZ fission yeast model has previously been employed to investigate features of antisense RNA technology in vivo (29). It has been shown that gene inhibition is dependent on the dose of antisense RNA while co-localisation of antisense and target genes does not affect the level of target gene suppression in this system (30). Additionally, the size of the antisense transcript (31,32) and the region to which it is targeted (33) can affect the efficacy of target gene inhibition. We further demonstrated that overexpression of host-encoded factors can enhance antisense RNA-mediated gene silencing in this system (34). Here we demonstrate that antisense RNA-mediated gene regulation is enhanced by expression of additional sense RNA. Furthermore, it is shown that expression of panhandle RNA also inhibits target gene suppression, suggesting that the generation of dsRNA through either intermolecular or intramolecular hybridisation is central to antisense RNA-mediated gene silencing in S.pombe. The usefulness of this model is extended by co-expressing the antisense enhancing sequence aes2 to (i) further show that antisense RNA-mediated gene regulation is linked to dsRNA-mediated gene silencing and (ii) illustrate that concomitant expression of host-encoded factors can enhance different forms of RNA-mediated gene silencing. In addition, the presence of siRNAs is demonstrated in both antisense RNA- and panhandle RNA-expressing strains, indicating that both function through a common RNAi-like mechanism.
MATERIALS AND METHODS
Schizosaccharomyces pombe strain and plasmid construction
All yeast strains were maintained on standard YES or EMM medium (35). Yeast cells were transformed with plasmid DNA by electroporation (36). Repression of nmt1 transcription was achieved by the addition of thiamine to EMM medium at a final concentration of 4 µM. The ura4-antisense plasmid was generated by subcloning the full-length lacZ fragment contained in pGT2 (31) into the plasmid pREP4 (37) in the antisense orientation. The construction of the long lacZ antisense and the frameshifted sense lacZ containing episomal plasmids have been described (31). This non-coding lacZ fragment was generated by introducing a frameshift mutation at the ClaI restriction site (31). The functional lacZ sequence was subcloned into the BamHI site of pREP4 to generate pM85-1. The 792 bp BglII c-myc antisense fragment (CM-17) (33) was subcloned into pREP4 to generate the sense c-myc vector, pN12-1. Construction of the aes2-encoding plasmid has been described elsewhere (34). Construction of the antisense lacZ integrating vectors and stable antisense lacZ strains has also been described (30). A sense lacZ version of the integrating vector was made by subcloning the full-length non-coding BamHI lacZ fragment (31) into the BamHI site of pRIP2/s in the sense orientation (37). This was transformed into the strain SP41 (h+, ade6-704, ura4-D18), and a single-copy integrant was isolated (M60-3). M60-3 was then mated with RB3-2 to introduce the target lacZ gene.
The lacZ panhandle integration vector, pM30-8, was generated by first introducing a NotI site into the XmaI site of pRIP1/s (37) using the self-complementary linker 5′-CCG GGC GGC CGC-3′ to generate pL121-14. The 2.5 kb sequence of the 5′ end of the non-coding lacZ gene (31) was then PCR amplified using the forward primer 5′-ATG CGG CCG CAA TTC CCG GGG ATC GAA AGA-3′ and the reverse primer 5′-ATG CGG CCG CAA TGC GGG TCG CTT CAC TTA-3′ to produce NotI ends. This product was then subcloned into the NotI site of pL121-14 in the antisense orientation. The full-length frameshifted lacZ fragment was then introduced into the BamHI site of this vector in the sense orientation upstream of the 2.5 kb antisense fragment to generate pM30-8. The episomal lacZ inverted repeat vector, pM53-1, was generated by subcloning 2.5 kb of the 5′ lacZ frameshifted gene in the antisense orientation downstream of the full-length lacZ frameshifted sequence in pREP1 (37). The frameshifted sequence was replaced with functional lacZ sequence to generate a panhandle capable of expressing β-galactosidase (pM81-2). The control vector pM91-1 was generated by removing the 2.5 kb NotI lacZ fragment from pM81-2 and reintroducing it in the sense orientation.
β-Galactosidase assays
The expression of the lacZ gene-encoded product, β-galactosidase, was quantitated using a cell permeabilisation protocol as previously described (30). A semi-quantitative overlay assay was also employed for the in vivo lacZ panhandle assay (33).
Plasmid segregation
Raw data was normalised to account for plasmid segregation as previously described (34). Briefly, by plating strains on both selective and non-selective media we have found that ∼73% of the cell population contained LEU2-based plasmids while ∼69% of the cell population contained both ura4- and LEU2-based plasmids. These values were used to normalise the raw data obtained from the quantitative β-galactosidase assays.
Detection of small lacZ-specific RNAs
Yeast total RNA was isolated using the glass bead method as previously described (31). This RNA was then separated on a 15% 8 M urea TBE gel and RNAs spanning 20–30 bases in size were excised. The recovered RNA was ethanol precipitated, loaded onto a 15% 8 M urea TBE gel and transferred to nylon membrane. In addition to the RNA samples, lacZ-specific DNA oligonucleotides spanning 22–33 bases were included to confirm size and sequence specificity of detected RNAs.
To prepare the lacZ-specific RNA probe for detecting small RNAs, a pGem3Zf-based plasmid containing the 3.5 kb lacZ gene was linearised with EcoRI and used as a template for in vitro transcription. The radiolabelled lacZ sense RNA was generated using SP6 RNA polymerase and the MAXIScript kit according to the manufacturer’s instructions. The labelled transcripts were sheared to an average size of ∼50 nt by adding 300 µl of an alkaline buffer (80 mM sodium bicarbonate and 120 mM sodium carbonate) to the 20 µl reaction and incubating at 60°C for 3 h (38). Pre-hybridisation and hybridisation were performed at 42°C using ExpressHyb solution according to the manufacturer’s instructions. The membrane was then washed twice with 2× SSC, 0.2% SDS at 42°C and twice with 20 mM Tris–HCl (pH 7.5), 5 mM EDTA, 60 mM sodium chloride and 10 µg/ml RNase A at 37°C for 1 h to remove non-specific background.
RESULTS
Expression of additional sense RNA enhances antisense RNA-mediated target gene suppression
It has been demonstrated that co-expression of antisense RNA with additional sense RNA can enhance gene silencing (3). This is thought to be due to the potential formation of additional dsRNA that may induce an RNAi-like mechanism. To determine whether gene suppression in fission yeast is due to formation of an antisense RNA:target mRNA hybrid or an antisense RNA:sense RNA hybrid, a version of lacZ which is unable to be translated into functional β-galactosidase was co-expressed in strains expressing the target and antisense lacZ genes. If antisense RNA is required to hybridise to target mRNA for inhibition of the gene expression pathway, then overexpression of the sense RNA should compete with the target mRNA for the available antisense molecules, thereby producing a decrease in lacZ gene suppression.
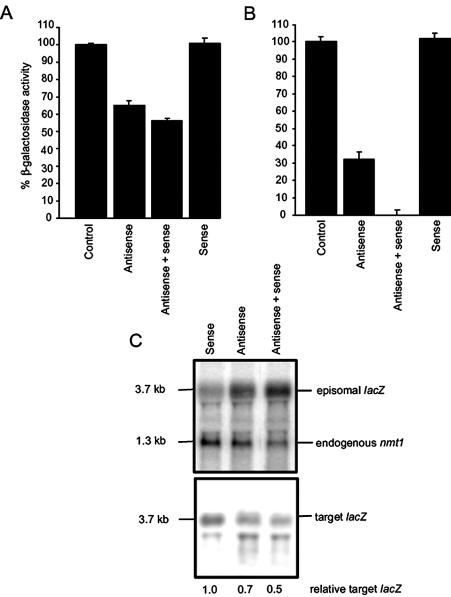
Initially, the nmt1-driven antisense lacZ gene was integrated in single copy into a strain (RB3-2) containing the chromosomally expressed adh1-driven target lacZ gene. The adh1 promoter is constitutively expressed while the nmt1 promoter is conditional and only active in the absence of thiamine. A crippled version of the sense lacZ gene (driven by nmt1) was also integrated in single copy into a separate target strain and then crossed with the antisense and target-containing strain. Strains were then isolated containing (i) the antisense and target lacZ genes, (ii) the sense and target lacZ genes or (iii) the antisense, sense and target lacZ genes. In the strain containing the antisense gene alone, β-galactosidase activity was ∼65% of the control strain, while there was no change in the strain expressing sense lacZ alone (Fig. 1A). Surprisingly, in the strain expressing both complementary transcripts, the level of gene silencing was not only maintained but moderately enhanced (Fig. 1A). To further increase the potential formation of intracellular dsRNA, an episomal sense lacZ plasmid was co-transformed with the episomal antisense lacZ plasmid into the target strain RB3-2. Significantly more stimulation of lacZ inhibition was observed in the presence of the episomally expressed complementary RNAs (Fig. 1B). When β-galactosidase activity was normalised for plasmid segregation (39) the strain expressing both complementary RNAs exhibited ∼100% lacZ inhibition, compared with 68% silencing by antisense alone. Expression of the sense lacZ RNA had no effect on β-galactosidase activity. Northern analysis demonstrated that the total relative level of episomally expressed nmt1-driven lacZ RNA in the strain transformed with both sense and antisense genes was approximately equal to the sum of that seen when either plasmid was expressed alone (Fig. 1C). Additionally, there was a concomitant reduction in the steady-state level of target mRNA with a lower signal present in the strain containing both sense and antisense genes compared with the strain containing antisense alone (Fig. 1C). However, since dividing cells unequally segregate episomal plasmids, the total sample population will contain cells that contain lacZ mRNA but no antisense RNA or dsRNA specific for this target. As a result, a complete reduction in target mRNA was not seen. These results demonstrate that the presence of additional sense RNA does not titrate the antisense RNA from the target mRNA and therefore indicates that increasing the potential formation of dsRNA, but not necessarily an antisense RNA:target mRNA hybrid, is required for efficient interference of target gene expression in S.pombe. Further, this effect seems to be dose-dependent.
Figure 1.
Effect of increasing target sense RNA in antisense RNA-expressing strains. (A) Strains containing single integrated copies of the target lacZ gene alone (control), the target and the antisense lacZ genes (antisense), the target and the sense lacZ genes (sense) and the target and both the antisense and sense lacZ genes (antisense + sense) were assayed for β-galactosidase activity in the absence of thiamine. (B) β-Galactosidase activity was determined in the strain RB3-2 transformed with the episomally expressed antisense lacZ plasmid (antisense), the episomally expressed sense lacZ plasmid (sense) or both antisense and sense lacZ plasmids (antisense + sense). For each strain three independent colonies were assayed in triplicate. (C) The 5′ and 3′ UTRs of the antisense and sense lacZ transcripts contain nmt1 sequences. RNA from episomally transformed RB3-2 strains was therefore probed with the nmt1 fragment to identify episomally expressed transgenes and the endogenous nmt1 gene (top). Membranes were then stripped and re-probed with the ura4 3′ fragment to identify target mRNA (bottom). The relative levels of target and episomally expressed lacZ RNA were normalised to endogenous nmt1 transcript and quantitated by phosphorimager analysis.
A lacZ panhandle RNA inhibits lacZ gene expression
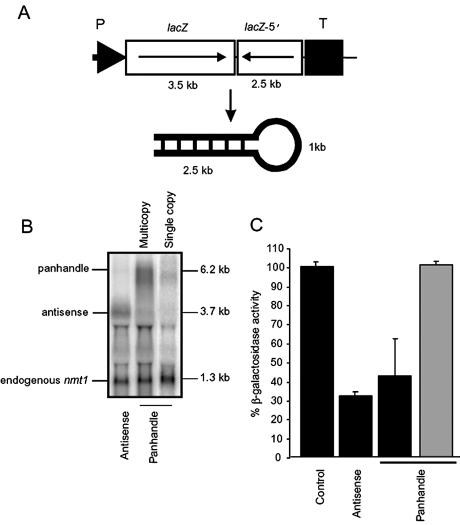
To confirm that dsRNA is central to efficient antisense RNA-mediated gene silencing in fission yeast a vector was engineered containing the full-length 3.5 kb frameshifted lacZ sequence with a 2.5 kb inverted repeat (Fig. 2A). This construct generates a panhandle transcript of ∼6.2 kb in length, with a 1 kb loop and 2.5 kb of self-complementarity which, predictably, will form a strong intramolecular RNA duplex. The panhandle construct was driven by the conditional nmt1 promoter. This gene was initially integrated into a fission yeast strain in single copy and the target lacZ gene was then introduced through genetic crossing. These strains were then assayed for target gene silencing. A β-galactosidase assay showed no reduction in target enzyme activity when transcription of the inverted repeat was activated (data not shown). However, RNA analysis indicated that the 6.2 kb panhandle transcript was being generated in this strain (Fig. 2B). Initially, this indicated that the panhandle RNA did not inhibit target gene activity in this system. However, since we have shown that the expression of both antisense RNA and additional sense RNA has a dose-dependent effect on gene silencing, efforts were made here to elevate the steady-state level of the panhandle RNA. To this end, an episomal plasmid containing the lacZ panhandle cassette was made and introduced into the lacZ expressing strain. β-Galactosidase activity was reduced by ∼60% in this strain, while addition of thiamine to the culture medium returned β-galactosidase activity to control levels (Fig. 2C), indicating that lacZ inhibition was dependent on expression of the panhandle gene. Northern analysis showed that the episomally based panhandle gene was expressed at a similar level to the antisense lacZ gene and was approximately 10-fold higher than the strain containing a single copy of the panhandle gene (Fig. 2B).
Figure 2.
lacZ panhandle-mediated gene silencing. (A) The lacZ panhandle construct contains the full-length lacZ gene (with an internal frameshift mutation) which is followed by the inverted 5′ 2.5 kb lacZ fragment. Intramolecular hybridisation generates an RNA with 2.5 kb RNA duplex and a 1 kb loop. The nmt1 promoter and terminator sequences are indicated by P and T, respectively. (B) The relative steady-state level of the panhandle lacZ RNA (6.2 kb) expressed from single copy and multi-copy genes is shown in comparison to episomally expressed antisense lacZ RNA (3.7 kb). The northern blot was probed with the nmt1 fragment to identify the nmt1-driven episomal transcripts in addition to the endogenous nmt1 transcript. The lacZ signals were normalised to the endogenous nmt1 transcript (1.3 kb) and quantitated by phosphorimager analysis. (C) The target strain was transformed with the episomally expressed lacZ panhandle and analysed for β-galactosidase activity. The appropriate plasmids were co-introduced to complement auxotrophy. The panhandle strain was assayed in the presence of thiamine (grey). At least three independent colonies were assayed in triplicate for each strain.
To investigate whether expression of the panhandle RNA was impacting on the cellular phenotype of these transformants, cultures were grown to mid-logarithmic phase and then viewed under the light microscope. It was observed that there was no difference in the growth rate of the panhandle-containing strain compared to control cells lacking this construct. Furthermore, there was no difference in the general morphology of the cells (data not shown). We also performed experiments to show that the panhandle lacZ was forming dsRNA in vivo. To this end we generated a panhandle construct containing a functional lacZ gene and a control construct with the 3′ inverted portion of panhandle as a direct repeat. Both were introduced into a yeast strain lacking the lacZ target gene. While the control strain showed high levels of β-galactosidase activity, virtually no activity was detectable in the panhandle-expressing strain (data not shown). This indicated that the lacZ sequence was unable to be translated, probably due to panhandle formation. Together, these data indicated that a construct capable of forming an intramolecular RNA duplex could inhibit target gene activity in a dose-dependent manner.
Co-expression of antisense and sense RNA enhances inhibition of a c-myc target
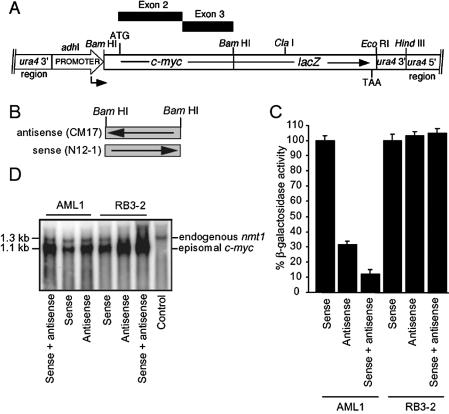
To test the ability of dsRNA to specifically interfere with other target sequences in fission yeast, complementary c-myc sequences were co-expressed in a strain containing an integrated c-myc–lacZ fusion cassette (Fig. 3A) (33). The target sequence is capable of generating β-galactosidase, and as the target sequence is fused to the 5′ end of the lacZ gene, gene silencing of the c-myc sequence will also result in down-regulation of lacZ expression. A 792 bp antisense c-myc fragment from exon 2 of the human c-myc gene (named CM-17) was previously found to suppress β-galactosidase activity within the c-myc–lacZ fusion target strain (AML1) by 47% (33). Both the sense and antisense c-myc genes were under control of the conditional nmt1 promoters. The region of the c-myc–lacZ target to which this fragment is homologous is shown in Figure 3B. The CM-17 fragment was subcloned into the BamHI site of pREP4 in the sense orientation to generate pN12-1. The antisense c-myc vector (pCM-17) and the sense c-myc vector were then transformed into AML1 both independently and together. When β-galactosidase activity was normalised for plasmid segregation it was found that co-expression of the antisense and sense constructs enhanced c-myc suppression by an additional 20% compared with the antisense c-myc vector alone, while expression of pN12-1 alone showed no inhibition of enzyme activity (Fig. 3C). This enhancement of gene silencing was consistently observed with this human gene target. Transformation of RB3-2 (the strain expressing only the lacZ target) with the antisense and sense c-myc constructs resulted in no down-regulation of β-galactosidase activity, indicating that the action of dsRNA is sequence-specific in the fission yeast model (Fig. 3C). Northern analysis demonstrated that the c-myc constructs were being expressed in all strains analysed (Fig. 3D). The RNA was probed with the nmt1 fragment which hybridises to both the endogenous nmt1 sequence (1.3 kb) and the episomally expressed c-myc sequences. Normalisation of the c-myc RNA to the endogenous nmt1 transcript indicated that both antisense and sense constructs were being expressed in the co-transformed strains as the steady-state level was approximately equivalent to the sum of that seen in the strains transformed with the sense or antisense constructs only (Fig. 3D).
Figure 3.
dsRNA-mediated suppression of a c-myc target. (A) The AML1 strain contains a c-myc–lacZ fusion cassette integrated at the ura4 locus. Exons 2 and 3 of the human c-myc gene were employed as a target. The relative position of the c-myc antisense fragment is shown. The transcription initiation site is indicated by the bent arrow. The straight arrow represents the normal direction of transcription for a particular DNA fragment. (B) The region of the c-myc target from which the antisense fragment was derived is shown. The fragments are aligned to the illustration in (A). (C) The target strains AML1 and RB3-2 were transformed with the sense construct pN12-1 (sense), the antisense construct pCM-17 (antisense) or both (antisense + sense). Transformants were grown in the absence of thiamine and assayed for β-galactosidase activity. Strains were transformed with appropriate control plasmids to complement auxotrophy. (D) RNA was probed with the nmt1 fragment. The control strain did not contain the episomally expressed c-myc sequence (1.1 kb) but shows the endogenous nmt1 transcript at 1.3 kb.
Detection of small interfering lacZ RNA
Given our observations that antisense RNA and panhandle RNA suppressed expression of the homologous lacZ gene, we decided to examine the relevant transformants for the presence or absence of lacZ-specific siRNA, a hallmark of gene regulation by RNAi (22). Using a hydrolysed lacZ-specific sense RNA probe, small RNAs were clearly identified in the pandhandle-expressing yeast transformants (Fig. 4). Small RNAs were also observed in the antisense RNA-expressing strain, albeit to a much lesser extent. These same RNAs were not detected in transformants containing vector controls or expressing only lacZ sense RNA. These results indicate that antisense RNA- and panhandle RNA-mediated regulation of lacZ gene expression in S.pombe involves the production of small RNAs of the size range expected for a mechanism involving RNAi. However, no correlation between the level of siRNAs present in the transformants and the degree of gene silencing was found in this study. Nevertheless, this is the first demonstration of single gene regulation through RNAi in any yeast species and further supports the utility of fission yeast as a model for better understanding the biological role(s) of this evolutionarily conserved mechanism of controlling gene expression (28).
Figure 4.
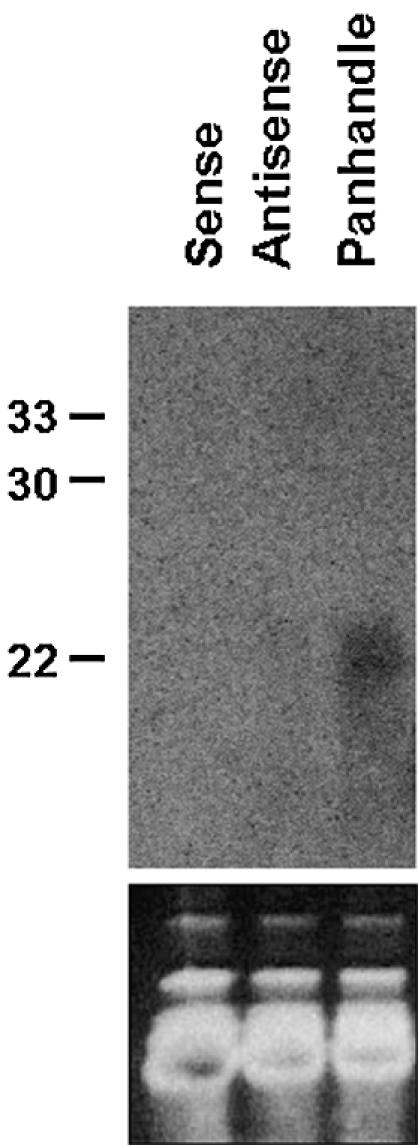
Detection of siRNA in dsRNA-expressing strains. Total RNAs spanning 20–30 bases in size were fractionated on a 15% 8 M urea TBE gel, transferred to nylon membrane and probed with a radiolabelled lacZ sense RNA that had been sheared to an approximate length of 50 nt. DNA oligos were also run as size controls. The ethidium bromide stained gel, which shows the total RNA spanning 20–30 bases, indicates equal RNA loading.
Co-expression of aes2 with panhandle RNA enhances gene silencing
Recent studies on RNA-mediated gene silencing, including our current work, suggest that antisense RNA, co-suppression and dsRNA-mediated interference may share similar mechanisms (40). We therefore explored whether overexpression of a host-encoded factor previously shown to enhance antisense RNA-mediated gene silencing (34) could also enhance dsRNA-mediated regulation. To this end, the effect of co-expressing the antisense enhancing sequence, aes2, with a lacZ panhandle construct (pM53-1) in a yeast strain containing the lacZ target gene (RB3-2) was tested. The aes2 factor was isolated from a S.pombe cDNA library and encodes part of the elongation factor EF-Tu. It was previously shown to enhance antisense RNA-mediated inhibition of the lacZ reporter gene by an additional 15% (34) (Fig 5). The panhandle vector pM53-1 was either co-transformed with the control plasmid pREP2 or with the aes2 plasmid into the target strain RB3-2. Under these conditions, the aes2 transformant displayed an additional 35% suppression of β-galactosidase activity when compared to the transformant expressing only the panhandle lacZ RNA (Fig. 5). This result indicated that expression of the aes2 gene could specifically enhance both antisense RNA- and, to a greater degree, panhandle RNA-mediated gene inhibition. In addition to the presence of siRNA in both the antisense- and panhandle-expressing strains this data suggests that both antisense- and dsRNA-mediated gene silencing may share a common mechanism.
Figure 5.
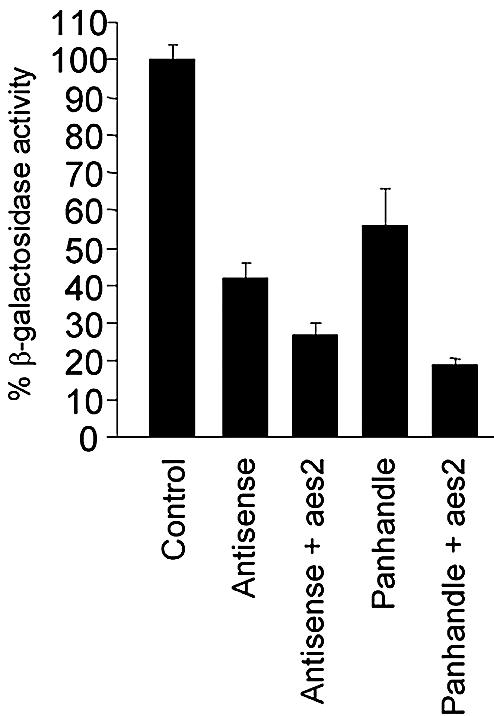
Co-expression of lacZ panhandle construct and aes2 factor. β-Galactosidase activity was determined in co-transformants. The control strain was RB3-2 co-transformed with pREP2 and pREP4 to complement auxotrophy. The panhandle strain was RB3-2 transformed with pM53-1 and pREP2. Five independent colonies were assayed in triplicate for each strain.
DISCUSSION
RNA-mediated gene silencing encompasses a variety of phenomena including antisense-mediated gene suppression, co-suppression and dsRNA-mediated gene interference (RNAi in animals), all of which have been implicated in the cellular control of gene expression and defence against viral infection (40). Recent evidence suggests that all of these gene silencing categories may operate through similar mechanisms, with the generation of dsRNA being the key factor (18). We have presented evidence to suggest that dsRNA (i) is a key mediator in antisense RNA-mediated gene silencing in fission yeast, (ii) acts in a dose-dependent manner and (iii) acts through an RNAi-like mechanism.
If antisense RNA was acting by hybridising to its target mRNA and directly inhibiting translation, it would follow that excess non-coding sense RNA would act as a competitor of the antisense RNA:target mRNA interaction. This competitive reaction would titrate out the available antisense RNA allowing for coding sense mRNA to be translated, with a consequent reduction in target inhibition. However, additional expression of both genomic and episomal non-encoding lacZ sense RNA failed to increase the level of β-galactosidase activity, but instead stimulated gene suppression. We have previously shown that the level of suppression within a given yeast strain was dependent upon the absolute level of antisense RNA (30). Similarly, results presented here suggest that gene silencing is also dependent on the dose of sense RNA. The concentration of sense RNA was increased by first expressing the sense transgene in single copy (stably integrated) and then in multiple copies (episomally maintained) in lacZ-expressing strains. Episomal co-expression of the complementary lacZ genes resulted in a further enhancement of lacZ gene silencing compared with their co-expression in single copy. This suggested that dsRNA was functioning in a dose-dependent manner. In addition, we have observed that increasing the steady-state level of target lacZ mRNA enhances the level of antisense RNA-mediated gene silencing (G.M. Arndt and D. Atkins, unpublished data). Consistent with gene silencing in other organisms, the increase in gene inhibition was associated with a concomitant reduction in lacZ mRNA, although a complete reduction was not seen. The specificity of dsRNA-mediated gene interference was also demonstrated by targeting a c-myc–lacZ fusion gene with co-expressed sense and antisense c-myc constructs. In this case, enhanced gene silencing was observed when additional c-myc RNA was generated while there was no effect of c-myc dsRNA on the lacZ target gene alone.
Expression of a lacZ panhandle construct also inhibited target gene expression, consistent with the prediction that dsRNA was the integral factor in antisense RNA-mediated gene silencing. A construct which generated a transcript with a 1 kb loop and 2.5 kb of intramolecular complementarity inhibited the lacZ gene by ∼60%. When one-tenth of the level of panhandle RNA was expressed no gene silencing was observed, again demonstrating that dsRNA-mediated gene silencing was dose-dependent in this system. The ability of a sequence with the potential to form an intramolecular RNA duplex to inhibit target gene expression is consistent with the use of similar constructs in plants (3), C.elegans (41) and trypanosomes (42). Furthermore, short lacZ-specific RNAs (∼21–23 nt long) were identified in strains expressing the panhandle construct, and to a lesser extent in the antisense RNA-expressing strain. The presence of siRNA is one of the hallmarks of the RNAi mechanism of gene silencing and therefore demonstrates for the first time that fission yeast employs the RNAi pathway for gene regulation.
The inability of the panhandle construct to inhibit gene expression to a greater extent than the antisense construct in this study may be due to the following reasons. First, it has recently been demonstrated that panhandle constructs are less effective at inhibiting target genes than hairpin RNA produced from direct inverted repeats (43). Secondly, it is well known that long inverted repeats can lead to genomic instability in prokaryotes and lower eukaryotes (44). It has also been shown that, even in systems where dsRNA acts in sub-stoichiometric amounts, the gene silencing effect is more robust when higher doses of dsRNA are introduced (1). However, we found that the panhandle strain had the highest levels of the siRNA species while the level of target gene suppression was not as robust as that seen in the antisense-expressing strain. Therefore, although increasing the potential formation of dsRNA correlated with the level of gene silencing, there is no correlation between the level of siRNA and gene inhibition in this system. This suggests that the nature of the siRNA population impacts on the observed level of gene silencing. For example, the antisense strains have the ability to form dsRNA of the entire length of the lacZ gene (3.5kb), however, the panhandle transcript only forms dsRNA of the 5′ 2.5 kb lacZ sequence. Therefore, while the panhandle generates more siRNA it could be that the short dsRNAs from the 3′ end of the lacZ gene are more efficient at silencing gene expression.
We have shown that overexpression of a cofactor of antisense RNA-mediated gene silencing (aes2) can also enhance dsRNA-mediated gene silencing. If antisense RNA acts through a dsRNA intermediate in this system then a factor that enhances antisense RNA-mediated gene silencing should also stimulate dsRNA-mediated gene inhibition. To test this hypothesis we co-expressed the previously identified aes2 factor (elongation factor EF-Tu lacking the mitochondrial localization domain) with the panhandle lacZ construct to investigate if it could also stimulate dsRNA-mediated gene silencing. Surprisingly, it reduced β-galactosidase activity by an additional 35% in the presence of the panhandle RNA. These data demonstrate that both antisense RNA- and dsRNA-mediated gene silencing pathways can be modulated by overexpression of the same host-encoded factor. The notion that dsRNA and antisense RNA pathways may share a common mechanism is further supported by our previous observation that the ATP-dependent RNA helicase, ded1, also enhances antisense RNA-mediated gene silencing (34). Both ded1 and EF-Tu act on dsRNA. The first acts in translation initiation by unwinding RNA duplexes while EF-Tu transports tRNA to the ribosome by first recognising the RNA duplex in tRNA stems. Interestingly, the level of gene silencing was enhanced by aes2 to a much greater extent in the panhandle strain than in the antisense strain. This coincided with the presence of a higher level of short lacZ-specific RNAs in the panhandle strain, again suggesting that the nature of the dsRNA may influence the level of gene silencing. The precise molecular role of aes2, and other aes factors (34), is unknown at present and will require further analysis using the fission yeast system described.
In conclusion, we have shown here that co-expression of complementary transcripts can enhance antisense RNA-mediated gene silencing, that panhandle RNA can inhibit target gene expression and that the antisense RNA- and dsRNA-gene silencing mechanisms may be linked through a common RNAi-like mechanism. Finally, the genetic tractability of fission yeast, including the ease of targeted integration, amenability to antisense RNA- and dsRNA-mediated gene silencing and knowledge of its genomic sequence, should allow for its continued use as an experimental model for the elucidation of gene silencing mechanisms, including investigation of components of the RNAi machinery that are essential for cell survival.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge Nham Tran and Hong-Ping Zhang for performing the short dsRNA experiments.
REFERENCES
- 1.Hammond S.M., Caudy,A.A. and Hannon,G.J. (2001) Post-transcriptional gene silencing by double-stranded RNA. Nature Rev. Genet., 2, 110–119. [DOI] [PubMed] [Google Scholar]
- 2.Fire A., Xu,S., Montgomery,M., Kostas,S., Driver,S. and Mello,C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 3.Waterhouse P., Graham,M. and Wang,M. (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl Acad. Sci. USA, 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misquitta L. and Paterson,B. (1999) Targeted disruption of gene function in Drosophila by RNA interference (RNA-i): a role for nautilus in embryonic somatic muscle formation. Proc. Natl Acad. Sci. USA, 96, 1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanchez-Alvarado A. and Newmark,P. (1999) Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl Acad. Sci. USA, 96, 5049–5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lohmann J., Endl,I. and Bosch,T. (1999) Silencing of developmental genes in Hydra. Dev. Biol., 214, 211–214. [DOI] [PubMed] [Google Scholar]
- 8.Wargelius A., Ellingsen,S. and Fjose,A. (1999) Double-stranded RNA induces specific developmental defects in Zebrafish embryos. Biochem. Biophys. Res. Commun., 263, 156–161. [DOI] [PubMed] [Google Scholar]
- 9.Li Y.-X., Farrell,M., Liu,R., Moharty,N. and Kirby,M. (2000) Double-stranded RNA injection produces null phenotypes in Zebrafish. Dev. Biol., 217, 394–405. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 11.Paddison P.J., Caudy,A.A. and Hannon,G.J. (2002) Stable suppression of gene expression by RNAi in mammalian cells. Proc. Natl Acad. Sci. USA, 99, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacque J.M., Triques,K. and Stevenson,M. (2002) Modulation of HIV-1 replication by RNA interference Nature, 418, 435–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruiz F., Vayssie,L., Klotz,C., Sperling,L. and Madaddu,L. (1998) Homology-dependent gene silencing in Paramecium. Mol. Biol. Cell, 9, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cogoni C. and Macino,G. (1997) Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci., 2, 438–443. [Google Scholar]
- 15.Voinnet O. and Baulcombe,D. (1997) Systemic signalling in gene silencing. Nature, 389, 553. [DOI] [PubMed] [Google Scholar]
- 16.Scherczinger C. and Knecht,D. (1993) Co-suppression of Dictyostelium discoideum myosin II heavy-chain gene expression by a sense orientation transcript. Antisense Res. Dev., 3, 207–217. [DOI] [PubMed] [Google Scholar]
- 17.Bahramian M. and Zarbl,H. (1999) Transcriptional and posttranscriptional silencing of rodent a1(I) collagen by a homologous trancriptionally self-silenced transgene. Mol. Cell. Biol., 19, 274–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery M. and Fire,A. (1998) Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet., 14, 255–258. [DOI] [PubMed] [Google Scholar]
- 19.Dougherty W. and Parks,T. (1995) Transgenes and gene suppression: telling us something new? Curr. Opin. Cell Biol., 7, 399–405. [DOI] [PubMed] [Google Scholar]
- 20.Cogoni C. and Macino,G. (1999) Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr. Opin. Microbiol., 2, 657–662. [DOI] [PubMed] [Google Scholar]
- 21.Zamore P., Tuschl,T., Sharp,P. and Bartel,D. (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell, 101, 25–33. [DOI] [PubMed] [Google Scholar]
- 22.Elbashir S.M., Lendeckel,W. and Tuschl,T. (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev., 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 24.Knight S.W. and Bass,B.L. (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science, 293, 2269–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voinnet O., Vain,P., Angell,S. and Baulcombe,D. (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- 26.Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 27.Reinhart B.J. and Bartel,D.P. (2002) Small RNAs correspond to centromere heterochromatic repeats. Science, 297, 1831. [DOI] [PubMed] [Google Scholar]
- 28.Hall I.M., Shankaranarayana,G.D., Noma,K., Ayoub,N., Cohen,A. and Grewal,S.I. (2002) Establishment and maintenance of a heterochromatin domain. Science, 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- 29.De Backer M.D., Raponi,M. and Arndt,G.M. (2002) RNA-mediated gene silencing in non-pathogenic and pathogenic fungi. Curr. Opin. Microbiol., 5, 323–329. [DOI] [PubMed] [Google Scholar]
- 30.Raponi M., Atkins,D., Dawes,I. and Arndt,G. (2000) The influence of antisense gene location on target gene regulation in the fission yeast Schizosaccharomyces pombe. Antisense Nucleic Acid Drug Dev., 10, 29–34. [DOI] [PubMed] [Google Scholar]
- 31.Arndt G.M., Atkins,D., Patrikakis,M. and Izant,J. (1995) Gene regulation by antisense RNA in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 248, 293–300. [DOI] [PubMed] [Google Scholar]
- 32.Clarke M., Patrikakis,M. and Atkins,D. (2000) Comparative analysis of artificial antisense RNA regulation in fission yeast and human cells. Biochem. Biophys. Res. Commun., 268, 8–13. [DOI] [PubMed] [Google Scholar]
- 33.Arndt G.M., Patrikakis,M. and Atkins,D. (2000) A rapid genetic screening system for identifying gene-specific suppression constructs for use in human cells. Nucleic Acids Res., 28, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raponi M. and Arndt,G.M. (2002) Dominant genetic screen for cofactors that enhance antisense RNA-mediated gene silencing in fission yeast. Nucleic Acids Res., 30, 2546–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno S., Klar,A. and Nurse,P. (1991) Molecular and genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 36.Prentice H. (1992) High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res., 20, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–1390. [DOI] [PubMed] [Google Scholar]
- 38.Catalanotto C., Azzalin,G., Macino,G. and Cogoni,C. (2002) Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev., 16, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heyer W.-D., Sipiczki,M. and Kohli,J. (1986) Replicating plasmids in Scizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol. Cell. Biol., 6, 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fire A. (1999) RNA-triggered gene silencing. Trends Genet., 15, 358–363. [DOI] [PubMed] [Google Scholar]
- 41.Tavernarakis N., Wang,S., Dorovkov,M., Ryazanov,A. and Driscoll,M. (2000) Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nature Genet., 24, 180–183. [DOI] [PubMed] [Google Scholar]
- 42.Shi H., Djikeng,A., Mark,T., Wirtz,E., Tschudi,C. and Ullu,E. (2000) Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA, 6, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith N., Singh,S., Wang,B.-B., Stoutjesdijk,P., Green,A. and Waterhouse,P. (2000) Total silencing by intron-spliced hairpin RNAs. Nature, 407, 320–321. [DOI] [PubMed] [Google Scholar]
- 44.Gordenin D. and Resnick,M. (1998) Yeast ARMS (DNA at-risk motifs) can reveal sources of genome instability. Mutat. Res., 400, 45–58. [DOI] [PubMed] [Google Scholar]