Abstract
The prokaryotic DNA polymerase III β homodimeric clamp links the replication complex to DNA during polynucleotide synthesis. This clamp is loaded onto DNA and unloaded by the clamp loader complex, the δ subunit of which by itself can bind to and open the clamp. β Clamps from diverse bacteria were examined using contrast hierarchical alignment and interaction network (CHAIN) analysis, a statistical approach that categorizes and measures the evolutionary constraints imposed on protein sequences by natural selection. Some constraints are subtle inasmuch as they are unique to certain bacteria. Examination of corresponding molecular interactions within structures of the Escherichia coli β dimeric and δ–β complexes reveals that N320, Y323 and R176, which are subject to very strong constraints, form a substructure that may serve as a platform for leveraging and directing δ-induced conformational changes. N320 may play a prominent role, as it is strategically situated between this substructure and regions linked to δ binding and opening of β’s dimeric interface. R176 appears to act as a relay between the δ binding site and the clamp’s central hole. Other residues subject to strong constraints are likewise associated with structurally important features. For example, two pairs of interacting residues, R269/E304 and K74/E300, form salt bridges at the dimeric interface, while the C-terminal residues M362, P363, M364 and R365 appear to play key roles in δ binding. Q149 and K198 appear to sense DNA within the clamp’s central hole while other residues may relay this information to the δ binding site. Mutagenesis experiments designed to explore possible mechanisms are proposed.
INTRODUCTION
In order for prokaryotic DNA polymerase III to track along the rapidly moving replication fork during DNA synthesis, it requires a topological link to the DNA. This is provided by an accessory subunit, the β sliding clamp (reviewed in 1). The β clamp forms a stable homodimeric ring (2) (Fig. 1A) that surrounds the DNA and thereby allows the catalytic α subunit, with which it interacts, to move quickly along the DNA for tens of thousands of base pairs without falling off (3,4). The β ring forms via head-to-tail interactions between two semicircular monomers.
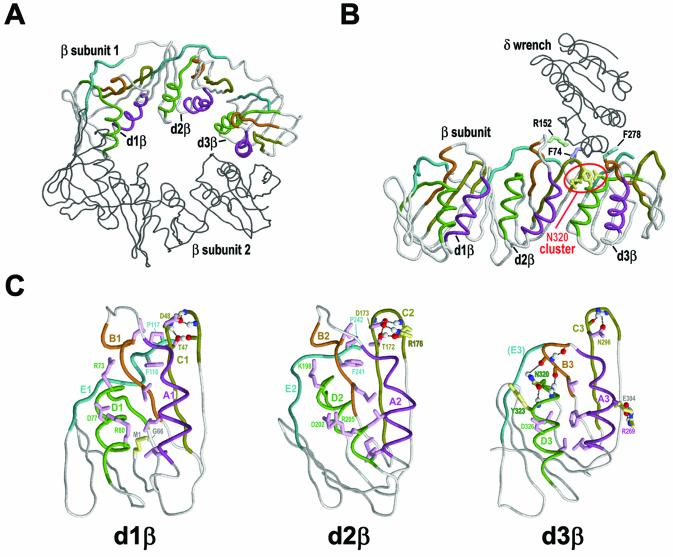
Figure 1.
Structural features of β clamps. The three domains of the β subunits are designated here as d1β, d2β and d3β, respectively. The backbone traces of the β subunits are shown with motif and satellite regions colored as for the corresponding alignments in Figure 2. Figures were generated using RasMol (29). (A) Locations of β domains and conserved motifs within the β homodimer (PDB: 2POL). Motif regions are indicated for one of the two β subunits (2). (B) Locations of β domains and conserved motifs within the δ–β complex (PDB: 1JQL) (15). Several residues discussed in the text are shown in order to help orient the other figures relative to the entire δ–β complex. See text for details. (C) Structural features of the three β domains. Motif regions and key residues corresponding to each β domain’s shared and specific constraints are indicated. Residues specifically discussed in the text are labeled. Hydrogen bonds are depicted as dotted lines. Color scheme: side chains of residues corresponding to β domain-shared and β domain-specific constraints (magenta and yellow, respectively); side chains of conserved residues in δ (grayish blue); main chain traces and residue designations (colored by motif regions as in Fig. 2); oxygen, nitrogen and hydrogen atoms establishing hydrogen bonds (red, blue and white, respectively).
Each β monomer consists of three distinct, yet structurally very similar, domains (Fig. 1C), so that the β homodimeric ring contains a total of six domains where each of two β sheets in each domain hydrogen bonds to a β sheet in an adjacent domain. The ring’s outer surface thus forms six β sheets, one spanning each adjacent pair of domains, that together support 12 α helices lining the inside of the ring (2). Some of these α helices contain basic residues that favor association with acidic polynucleotide phosphates without establishing specific contacts, so that the clamp may freely slide along the DNA.
The β clamp is structurally related to functionally analogous eukaryotic and archaeal PCNA (5–7), and bacteriophage T4 (gp45) (8) sliding clamps, though these differ inasmuch as they form homotrimers from subunits consisting of two domains each. The structural similarity between the β clamp and these other clamps is essentially undetectable at the sequence level.
The β ring must be opened both for assembly onto DNA prior to polynucleotide synthesis and for removal from DNA after replication is completed. Loading and unloading of the β clamp is mediated by the clamp loader complex (9–11), the minimum components of which are three copies of the γ subunit and one copy each of the δ and δ′ subunits (12,13). The δ subunit is a key clamp loader component inasmuch as δ alone is sufficient to bind to and open the clamp (10). Biochemical analysis suggests that the binding energy of δ is harnessed to force open one of the dimeric interfaces of the clamp, such that the opened β ring retains its dimeric structure (14). The crystal structure of the δ–β complex (15) (Fig. 1B) has provided substantial insight into this process (reviewed in 1,16,17). Based on this structure, it was proposed (15) that the δ subunit, which is likened to a ‘wrench’, induces conformational changes at the β clamp dimeric interface thereby triggering an inherent spring-loaded structural mechanism that opens the β ring.
Here I examine evolutionary clues to the β clamp’s inherent structural mechanisms using a statistical approach called contrast hierarchical (CH) alignment and interaction network (CHAIN) analysis (18). CHAIN analysis aligns a set of related sequences, classifies the aligned sequences into distinct categories, and pinpoints those residues within each category subject to the strongest selective constraints. Here I examine three categories of selective constraints: (i) constraints shared by all three β domains (designated here as d1β, d2β and d3β), (ii) constraints specific to individual domains (from all bacterial groups) and (iii) constraints specific to individual domains from the γ proteobacteria, a group that includes the Escherichia coli β clamp. Other procedures then search for various types of molecular interactions associated with key residues within available E.coli β clamp structures. This identifies structural features critical to the clamp’s biological function and that thus presumably play important mechanistic roles. Clues regarding these mechanisms may be gleaned from the structural locations and chemical properties of key residues.
MATERIALS AND METHODS
CHAIN analysis of β clamp domains
CHAIN analysis, which is described in detail elsewhere (18), generates two types of output: a ‘CH alignment’ and structural displays of the corresponding molecular interactions. Figures 2–4 show three β clamp CH alignments, each of which is split into three sub-alignments (designated A–C) corresponding to the three β clamp domains. In constructing each of these, CHAIN analysis multiply aligns three sets of related sequences: (i) a ‘foreground set’, (ii) a ‘display set’, which is a subset of the foreground set, and (iii) a ‘background set’, which is a superset of the foreground set.
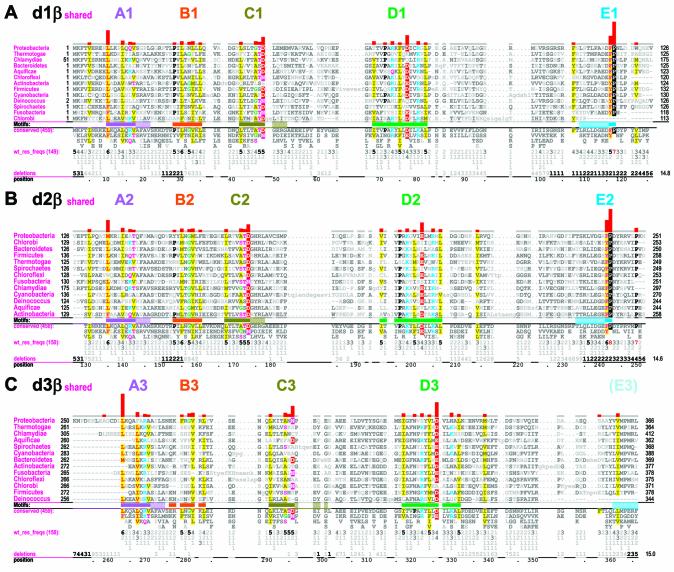
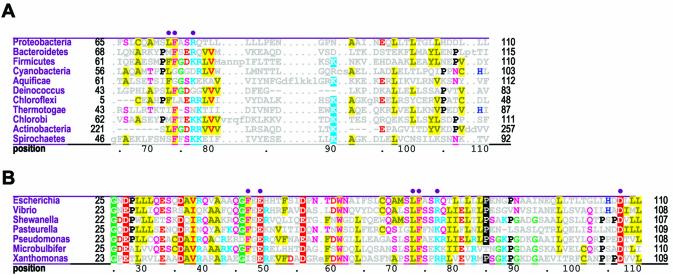
Figure 2.
β Domain-shared constraints. CH alignments for each of the three β clamp domains are shown (see Materials and Methods). The displayed sequences in each alignment represent β clamps from major prokaryotic taxa, which are indicated in the leftmost column. The first sequence (proteobacteria) is the E.coli β clamp, which serves as the query in this and the other CHAIN analyses described here. The bars directly below the aligned sequences indicate the motif regions discussed in the text and are colored to match the main-chain traces for these regions in Figures 1, 6 and 7; solid bars indicate the main motifs while adjacent hashed bars indicate associated satellite regions. The full foreground sequence alignment is not shown directly, but rather is merely represented by the conserved residue patterns below the alignment. The corresponding weighted residue frequencies (‘wt_res_freqs’) are given in integer tenths below conserved residues. For example, a ‘5’ in integer tenths indicates that the corresponding residue directly above it occurs in 50–60% of the (weighted) sequences. Deletion frequencies are similarly given in integer tenths (black; range 10–100%) or hundredths (gray; range 1–9%) as indicated. Histograms above the alignments display the relative strengths of the inferred selective constraints acting at each position (using a quasi-logarithmic scale); aligned residues corresponding to the most constrained positions are highlighted for emphasis. (A) β Domain 1-shared constraints. (B) β Domain 2-shared constraints. (C) β Domain 3-shared constraints.
Figure 4.
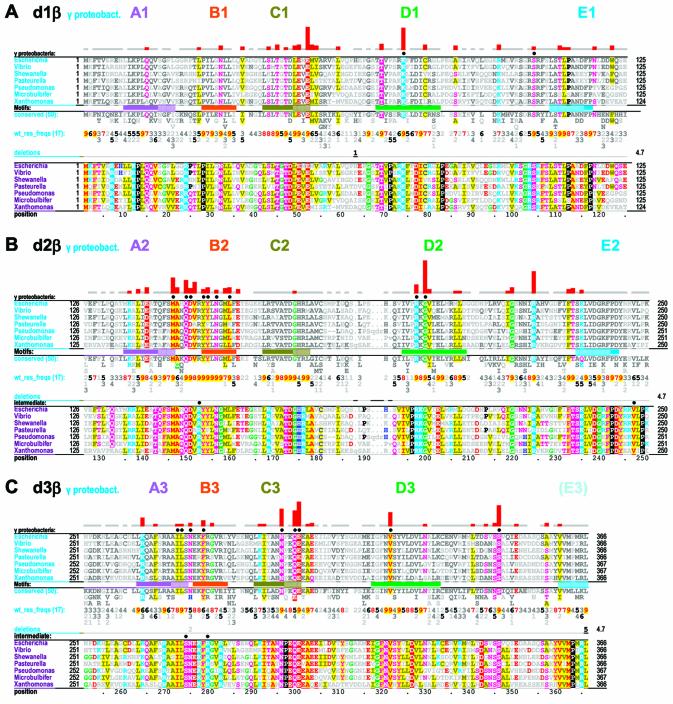
γ Proteobacterial-specific constraints. Two alignments are shown for each domain. The first is a CH alignment of the β domain from families within the γ proteobacterial species (indicated in the leftmost column). The second highlights all conserved residues within the same display set without attempting to categorize them (termed γ proteobacterial-total constraints). The second alignment thus helps identify intermediate constraint categories corresponding to conserved γ proteobacterial residues that are inconsistently conserved within the three categories specifically examined here (see Materials and Methods). The solid circles directly above alignments indicate residues specifically mentioned in the text or shown in Figures 6 and 7. See the legend to Figure 2 for further descriptions. (A) γ Proteobacterial-specific and -total constraints within β domain 1. (B) γ Proteobacterial-specific and -total constraints within β domain 2. (C) γ Proteobacterial-specific and -total constraints within β domain 3.
The foreground set corresponds to those sequences whose selective constraints are being measured. Note that the foreground sequence alignment is not explicitly displayed as such, but rather is merely represented by the conserved patterns and residue frequencies below the aligned sequences actually displayed (Figs 2–4). The display set corresponds to the aligned sequences of interest within the foreground set and thus only these are explicitly shown. In Figures 2 and 3 the display set corresponds to 13 representative β clamps from distinct major prokaryotic taxa, while in Figure 4 the display set corresponds to seven representative β clamps from families within one of these taxa, the γ subdivision of the proteobacteria.
Figure 3.
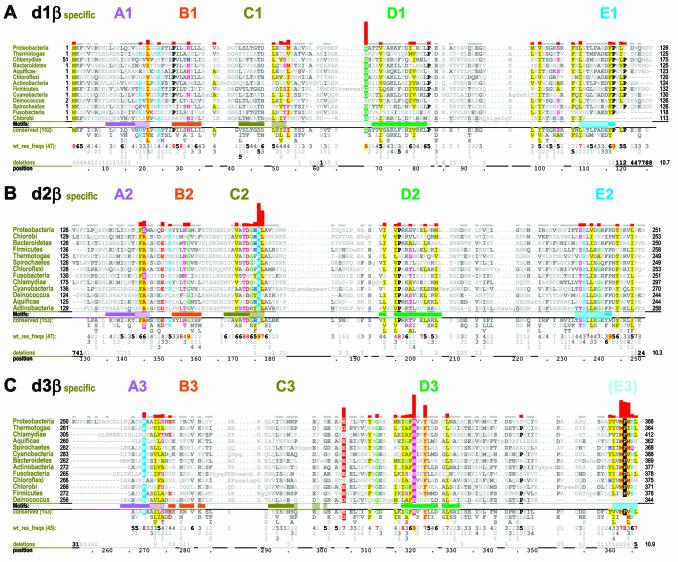
Domain-specific selective constraints. CH alignments of β clamp domains from major prokaryotic taxonomic groups are shown. See legend to Figure 2 for further descriptions. (A) β Domain 1-specific constraints. (B) β Domain 2-specific constraints. (C) β Domain 3-specific constraints.
CHAIN analysis measures selective constraints by determining the degree to which conserved residue positions in the foreground alignment contrast with the residues observed at corresponding positions in an alignment of the background set, which contains a broader category of sequences than the foreground set. More specifically, constraints are measured in terms of the difficulty of obtaining the amino acids observed at a particular position in the foreground alignment by randomly drawing amino acids from the distribution observed at that position in the background alignment. Foreground positions with compositions that closely resemble the background will thus be found to have little or no category-specific constraints, while positions with compositions incompatible (or that ‘contrast’) with the background will be found to have strong category-specific constraints. CH alignments display these constraints both quantitatively in histograms (using quasi-logarithmic scaling) and qualitatively through highlighting of aligned residues (Figs 2–4).
Here I examine three constraint categories, termed ‘β domain-shared’, ‘β domain-specific’ and ‘γ proteobacteria-specific’ constraints. β Domain-shared constraints act upon more than one domain. In this case, subsequences corresponding to all three β domains constitute the foreground set and the overall frequency of amino acids generally observed in proteins serves as an implicit background set at each position (Fig. 2). β Domain-specific constraints act on only one domain. In this case, subsequences corresponding to a specific β domain constitute the foreground set and available sequences for all three domains (which thus corresponds to the foreground in the previous category) constitute the background set (Fig. 3). γ Proteobacterial-specific constraints specifically act on β domains within γ proteobacterial clamps. In this case, subsequences corresponding to a particular γ proteobacterial β domain constitute the foreground set and subsequences from all bacteria for that domain (which again corresponds to the foreground set in the previous category) constitute the background set (Fig. 4). CHAIN analysis also allows for another category of ‘intermediate’ constraints, which here corresponds to residue positions that are highly conserved in γ proteobacteria but that are inconsistently conserved across these three foreground sequence sets and thus belong to a category outside of those specifically examined here (see Fig. 4). For a complete description of CHAIN analysis see Neuwald et al. (18).
CHAIN analysis procedures specific to this study
Several CHAIN analysis procedures were specifically adapted to this study. In particular, the background alignment for measuring β domain-shared constraints was obtained by initializing the CHAIN analysis iterative multiple alignment procedure with a structural alignment of all three E.coli β domains. This structural alignment was obtained using the CE program (19). For analysis of β domain-specific constraints, the foreground set was constructed from 153 full-length β clamp sequences from 13 bacterial taxa (i.e. sequence fragments were eliminated manually). In contrast, I used a standard CHAIN analysis optimization procedure, called ‘Bayesian partitioning with pattern selection’ (18), to automatically define the γ proteobacterial foreground set. Also, for analysis of both the δ and β subunits of DNA polymerase III, display sets consist only of orthologous proteins from bacterial species for which the entire genome has been sequenced. Note that δ subunit display sequences were obtained from the same organisms as the corresponding β clamp sequences. Orthologs were identified using the COG database (20) or, when unavailable by this means, through searches of a given organism’s proteome against a multiple alignment profile composed of previously validated δ or β orthologs.
Sequences displayed in alignments
Sequence identifiers for the β clamp display set representing the 13 major prokaryotic taxa (Figs 2 and 3) are: Proteobacteria, 16131569 (PDB: 2POLA); Thermotogae, 15643032; Chlamydiae, 15604794; Bacteroidetes, 23135730; Aquificae, 15606912; Chloroflexi, 22972815; Actinobacteria, 23016960; Firmicutes, 16799081; Cyanobacteria, 16329463; Deinococcus, 15805043; Spirochaetes, 15638997; Fuso bacteria, 19703871; and Chlorobi, 21672842. Sequence identifiers for the β clamp display set representing seven genera (six families) of γ proteobacteria (Fig. 4) are: Escherichia, 16131569 (PDB: 2POLA); Vibrio, 27360551; Microbulbifer, 23027046; Shewanella, 24371609; Pasteurella, 15603025; Pseudomonas, 11348446; and Xanthomonas, 21229480.
Sequence identifiers for the δ subunit display set representing 11 major prokaryotic taxa (Fig. 5A) are: Proteobacteria, 16128623; Bacteroidetes, 23138213; Firmicutes, 16800584; Cyanobacteria, 22299116; Aquificae, 15606373; Deino coccus, 15806263; Chloroflexi, 22974008; Thermotogae, 15642955; Chlorobi, 21674760; Actinobacteria, 23018560; and Spirochaetes, 15594800. Sequence identifiers for the δ subunit display set representing seven genera (six families) of γ proteobacteria are: Escherichia, 16974835 (PDB: 1JQJC); Vibrio, 27359869; Shewanella, 24372755; Pasteurella, 15603081; Pseudomonas, 9950180; Microbulbifer, 23029615; and Xanthomonas, 21232048.
Figure 5.
Conserved residues within the δ subunit of the clamp loader complex. Residues discussed in the text or shown in Figures 6 and 7 are indicated by the solid circles above the alignments. (A) Alignment of δ subunits from major prokaryotic taxa. (B) Alignment of δ subunits from γ proteobacterial families.
RESULTS AND DISCUSSION
β Domain category-specific constraints
β Domain-shared constraints (Fig. 2) are for the most part associated with four sequence motifs (designated A1–D1, A2–D2 and A3–D3 in β domains 1–3, respectively). These motifs, though often subtly conserved, span the major prokaryotic taxa and presumably reflect basic structural features required for the β domain’s general function. A prominent fifth motif (designated E1 and E2 in Fig. 2A and B, respectively) occurs within the two interdomain linkers connecting domains 1–2 and domains 2–3.
β Domain-specific constraints (Fig. 3) often overlap with or are adjacent to one of the five motif regions and I will thus refer to these as satellite regions of the main motifs. In Figures 2–4 these satellite regions are designated using hashed extensions of the solid colored bars designating the main motif regions. A cluster of domain 3-specific constraints near the β clamp’s C-terminus is designated the E3 region, because it structurally corresponds to the E1 and E2 satellite region.
In general, both β domain-shared and β domain-specific constraints seldom involve residues that directly interact either with the other β subunit within the homodimeric clamp (2) or with the δ subunit within the δ–β complex (15). Certain previously noted salt bridge contacts (2,14), for example, are very poorly conserved within these two categories. Nevertheless, the hydrophobic nature of several key residues establishing contact between subunits, such as L78-β, F106-β, L108-β, I272-β and L273-β (2), is conserved. Mutation of two of these hydrophobic residues, namely I272-β and L273-β, to alanine disrupts the β dimeric structure (14). In contrast, many residue positions subject to the strongest constraints in these two categories are substantially buried within the β structure and thus are unlikely to directly participate in protein–protein interactions. This is consistent with the notion that distinct modes of interaction between δ and β have evolved within different bacterial taxa.
These observations motivated an analysis of γ proteobacterial-specific constraints (Fig. 4), which indeed reveals that certain residues establishing salt bridges and hydrogen bonds at the dimeric interface (2,14) are well conserved within this group (see below). Many other γ proteobacterial-specific constraints are also evident. Despite the fact that these organisms are rather closely related, sufficient evolutionary time has elapsed subsequent to their divergence for quantification of selective constraints inasmuch as several aligned positions (e.g. 62, 181 and 287) have substantially diverged, while others remain highly conserved. This analysis also indicates that underlying mechanisms associated with certain structural features of the E.coli β clamp may not generalize to bacteria outside of the γ proteobacteria.
Conserved residues within the δ subunit
Before considering the structural roles of conserved β domain residues, we will first consider conserved residues in δ that interact with the β clamp. Based on the structure of the δ–β complex (15) there are two key residues in δ that mediate binding to β, L73-δ and F74-δ. These two residues are moderately conserved across major prokaryotic taxa (Fig. 5A), but most of the remaining residues in δ that contact β are either very weakly conserved or non-conserved. Within the γ proteobacteria, however, these two key residues are well conserved (Fig. 5B), as are various other residues in δ that directly or indirectly interact with the β clamp (see below). This again suggests that specific taxa have evolved distinct modes of interaction between δ and β and that interacting δ and β residues have coevolved within the γ proteobacteria to function in conjunction with each other.
β Domain-shared constraints
Analysis of β domain-shared constraints not only provides insight into the basic structure of the β domain, but also helps interpret the role of each domain’s unique features inasmuch as these are built upon this common structure. This section thus provides a conceptual framework for subsequent discussions of domain-specific and γ proteobacterial-specific constraints.
Constraints shared by all three β domains. A likely cause for the β domain-shared constraints is the need to maintain the common β domain structure itself (shown for each of the three E.coli domains in Fig. 1C). Examination of these structures reveals that, indeed, most of the more weakly conserved positions within the four β domain motifs correspond to hydrophobic residues projecting into the core of each domain. Similarly, a structural role seems likely for a highly conserved aspartate or asparagine within motif C (D48-β, D173-β and N296 in Fig. 2A–C, respectively) and for a conserved threonine, serine or asparagine at the previous sequence position (T47-β, T172-β and N295-β, respectively). For both of these positions the residue side chains typically form hydrogen bonds with the backbone of a sharp turn between two β strands (Fig. 1C), presumably thereby stabilizing the turn.
There are other conserved positions shared by all three domains, however, that appear to be unrelated to maintaining the β domain’s structural fold. For example, basic residues (K and R) are favored at two positions in motif D (Fig. 2). These residues lie within helices facing the interior of the ring and, as previously suggested (2), are likely involved in generating a positive electrostatic potential facilitating non-specific interactions with the backbone phosphates of the encircled DNA. Though inconsistently conserved, these positive charges may nevertheless maintain a sufficiently strong positive potential due to the redundancy provided by the six β domains. Located directly between these two basic residue positions within motif D is a conserved aspartate or glutamate (D77-β, E202-β and D326-β in Fig. 2A–C, respectively) that appears to play a non-structural role—perhaps functioning as an acidic counterbalance to the adjacent basic residue positions. The residue at this position within domain 3 (D326-β) also interacts with the highly conserved domain 2-specific residue R176-β (see below).
A motif shared by interdomain linkers. The residues exhibiting the strongest constraints shared by more than one β domain corresponds to motif E, which is located within the two linkers connecting adjacent domains and which contains the pattern [FY]-P (Fig. 2A and B). In considering a possible function for this [FY]-P pattern, note that the proline side chain restricts the interdomain linker’s backbone flexibility, which is further restricted by the packing of this proline against the adjacent phenylalanine or tyrosine. Also, upon binding of δ to the β clamp, F74-δ contacts the E2 motif proline P242-β. Thus, one role for the [FY]-P motif may be to stabilize the δ binding pocket while, at the same time, perhaps limiting δ-induced conformation changes in regions preceding the [FY]-P motif. This raises the question, however, as to why the [FY]-P motif within the domain 1 to 2 linker (F116-β and P117-β) is also conserved. A straightforward explanation is that this [FY]-P motif performs a similar role, given that this region is predicted to interact with the clamp loader γ subunit (13), which is structurally related to the δ subunit (13).
β Domain-specific constraints
The strongest β domain 1-specific constraint (Fig. 3A) corresponds to a single, strikingly conserved glycine (G66-β) located near the D1 motif. Within the available crystal structures of β clamps, this glycine contacts the initial methionine (M1-β), which is, of course, 100% ‘conserved’. This methionine’s side chain sulfur atom is located near the peptide bond between G66-β and A67-β and its β carbon makes a van der Waal’s contact with the α carbon of G66-β. Perhaps due to glycine’s inherent backbone flexibility, these interactions are also associated with bending of the β strand containing G66-β and hydrogen bonding of this strand with the clamp’s N-terminus. Moreover, within the available structures, G66-β fails to form non-covalent interactions with any other residues—except for backbone hydrogen bonds to adjacent residues within the clamp’s β sheet. Thus, although its function is unclear, the contact between M1-β and G66-β appears to be more than a coincidence.
The domain 1-specific constraints acting upon I78-β, F106-β and L108-β, though relatively weak, are worth mentioning because these residues establish hydrophobic contacts at the dimeric interface between β subunits (see Fig. 6E inset). These constraints are less striking because similar residues establish contacts at domain interfaces within the β subunit and thus these constraints are not strictly domain specific.
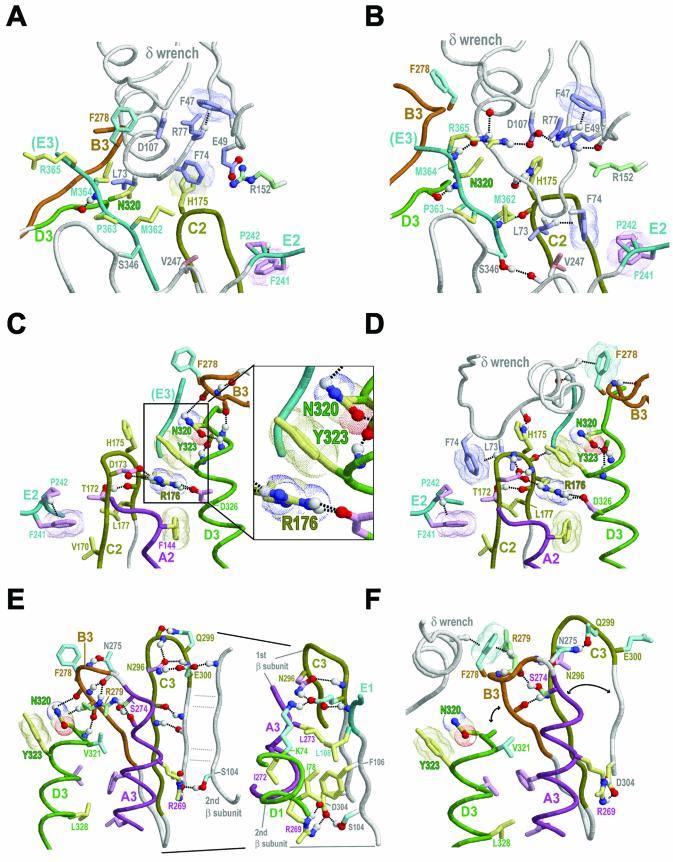
Figure 6.
(Opposite) Structural analysis of category-specific constraints within the β dimeric and δ–β complexes. Representations and coloring are as described in Figure 1 with the following additional color schemes: residue side chains corresponding to γ proteobacterial-specific and -intermediate constraints (cyan and green, respectively); hydrogen bonding carbons (colored as their corresponding side chains). Amino-aromatic and van der Waals interactions are depicted as dot clouds. Dotted lines into clouds depict CH-π interactions (30). See text for details. (A) Theoretical interaction between the unbound δ and β structures showing potential steric clashes. This figure was generated by superimposing the structure of δ from the γ clamp loader complex (13) upon the structure of δ from the δ–β complex (15) and the structure of β from the dimeric complex (2) upon the structure of β from the δ–β complex. This results in three major steric clashes (atom distances <1 Å): (i) one between L73-δ and the β domain 3-specific conserved residues N320-β and M364-β; (ii) one between δ’s β interaction helix and residues within the B3 loop (of which only F278-β is shown); and (iii) one between E49-δ and R152-β. (B) The same view as in (A) for the actual δ–β complex. Conformational changes in both δ and β resolve the steric clashes observed in (A). (C) The δ binding region (domains 2–3) of the β dimer viewed from the center of the β ring. This view is from the opposite direction relative to that shown in (A). The B3 loop and N320-β both serve as N-caps (27,28) for the D3 helix. The inset shows a more detailed view of the N320 cluster (see text). (D) The same view as in (C) for the δ–β complex. Here, only N320-β serves as an N-cap (27,28) for the D3 helix. The N320-β capping hydrogen bonds form new alliances in the δ–β complex relative to the β dimer. (E) The dimeric interface between domain 3 of one β subunit and domain 1 of the other subunit. This view is roughly from the center of the β ring. The inset shows additional details regarding conserved residues at the dimeric interface, including K74-β within domain 1, which electrostatically interacts with, and hydrogen bonds to, E300-β within domain 3. Both residues are subject to strong γ proteobacterial-specific constraints (Fig. 4). (F) The same view as in (E) for the δ–β complex.
The strongest domain 2-specific constraints are acting on two sequence adjacent residue positions corresponding to R176-β and L177-β within motif C2 (Fig. 3B). Notably, R176-β contacts two residues that are highly conserved within domain 3, Y323-β (Fig. 3C) and D326-β (Fig. 2C). (D326-β is subject to a strong domain-shared constraint; see above.) Within the δ–β complex, L177-β contacts the two key δ residues L73-δ and F74-δ (Fig. 6D).
Several residues are subject to strong domain 3-specific constraints (Fig. 3C), which is perhaps to be expected considering that domain 3 harbors both a dimeric interface and sites of interaction with the δ wrench. The most notable of these residues are N320-β, Y323-β and three residues near the C-terminus, M362-β, P363-β and M364-β. Two other domain 3-specific conserved residue positions correspond to E304-β and R269-β, which in the E.coli β clamp structure form a salt bridge between the A3 helix and the β strand adjacent to (but not across) the clamp’s dimeric interface (see Fig. 6E). Notably, R269-β is also the most buried interface residue (116 Å2). The need to maintain this salt bridge, which presumably plays a critical role at the dimeric interface, may explain the selective constraints acting on these residues. I272-β and L273-β, which are near R269-β within the A3 helix and (as mentioned above) are buried at the homodimeric interface (Fig. 6E inset), are likewise subject to domain 3-specific constraints.
γ Proteobacterial-specific constraints
Certain γ proteobacterial-specific conserved residues (Fig. 4) establish contacts across the β clamp dimeric interface. For example, the residues K74-β and E300-β, which are subject to strong γ proteobacterial-specific constraints, correspond to a salt bridge across the dimeric interface (Fig. 6E inset). Likewise, the residues at positions corresponding to I272-β and L273-β, which presumably play stabilizing roles at the dimeric interface (Fig. 6E inset), are nearly always isoleucine and leucine, respectively, within the γ proteobacteria (Fig. 4C), but are replaced by other hydrophobic residues in other bacteria (Fig. 3C).
Other residues subject to γ proteobacterial-specific constraints presumably perform other roles (Fig. 4). Although a full understanding of these roles clearly requires additional structural and biochemical analysis, here I will consider only those residues for which possible roles may readily be suggested. For example, S346-β, which is subject to a strong γ proteobacterial-specific constraint, both contacts L73-δ and hydrogen bonds to the backbone oxygen of V247-β, which extensively contacts both L73-δ and F74-δ (Fig. 6B). Although the position corresponding to V247-β is poorly conserved across diverse bacteria, it is essentially always hydrophobic and, within γ proteobacteria closely related to E.coli, essentially always valine (Fig. 4). Similar observations regarding γ proteobacterial-specific residues will be presented—in conjunction with further consideration of residues specific to other categories—in the following sections.
N320-β appears to play a strategic role in δ–β binding
To explore the biological implications of this analysis, we first consider the relationship between various residues subject to category-specific constraints and conformational changes required for binding of δ to the β dimer. To better understand these conformational changes, it is helpful to first consider the major steric clashes that occur between the unbound δ and β structures (as shown in Fig. 6A) when these are superimposed upon their corresponding bound structures, because such clashes clearly need to be resolved upon binding of β by δ. This reveals three major clashes: (i) a L73-δ clash with N320-β and, to a lesser extent, with M364-β; (ii) a severe clash between δ’s β interaction helix and three residues in the B3 motif region (positions 276–279) that are conserved within the γ proteobacteria (Fig. 4C); and (iii) a clash between R152-β and E49-δ, two other residues conserved within the γ proteobacteria (Figs 4B and 5B). These clashes are resolved within the δ–β complex (Fig. 6B) via conformational changes within both δ and β that (considering δ’s biological function) presumably correspond in some way to the clamp opening mechanism. In the following sections I explore how a comparison of these conformational changes with category-specific constraints provides evolutionary clues regarding this mechanism.
The steric clash between N320-β and L73-δ is particularly intriguing in this regard inasmuch as N320-β corresponds to the residue subject to the strongest domain 3-specific constraint (Fig. 3B), while, as mentioned above, L73-δ is a key residue mediating binding of δ to β (15). Furthermore, N320-β is structurally associated with Y323-β, R176-β and M364-β, which are likewise subject to some of the strongest domain-specific constraints, and adjoins to the B3 motif region, which (as is discussed below) undergoes significant conformational changes upon interaction with δ. Some of the most striking evolutionary clues thus point to N320-β as playing a strategic role and N320-β thus serves as an appropriate starting point for discussion.
R176-β may be a relay between the δ binding site and the β clamp’s central hole
One explanation for the strong selective constraint acting on N320-β is its structural interaction with Y323-β, a conserved residue that in turn interacts with R176-β. R176-β occupies the aligned position subject to the strongest β domain 2-specific constraint (Fig. 3B). These three residues thus constitute the most distinctive β clamp structural feature, which I term the ‘N320 cluster’ (Fig. 6C inset). Within this cluster the aromatic ring of Y323-β forms an amino-aromatic interaction with the side chain amino group of N320-β and, on the other side of the ring, a similar cation-π interaction with the guanidinium group of R176-β (21–24). Observations discussed in this and the next section suggest that R176-β acts as a relay between the δ binding site and regions near the β clamp’s central hole.
The R176-β guanidinium group is flipped by 180° in the δ–β complex relative to its conformation in either subunit of the β homodimer. This flip, which the B factors associated with these structures suggest is real, provides a mechanism for exchanging one set of precise molecular interactions for another because the guanidinium group possesses both a positive charge that can participate in electrostatic interactions and a large, rigid, planar array containing five hydrogen bond donors (25). In both the β dimeric and δ–β structures this guanidinium group hydrogen bonds with the side chain of D326-β (Fig. 6C and D), which corresponds to the aspartate or glutamate within motif D that is generally conserved in all β domains (see Fig. 2 and discussion above). The R176-β guanidinium group also hydrogen bonds with D173-β, which corresponds to an aspartate or asparagine in motif C that is conserved in all β domains (Fig. 2) and that—in conjunction with a sequence adjacent conserved threonine or serine (T172-β)—typically stabilizes the tight turn between the two β strands associated with motif C (Fig. 6C and D). All of these residues are well conserved in β clamps, suggesting that the corresponding interactions are associated with critical functions.
To further explore the role of R176-β, note that L177-β, which is at the base of the δ binding pocket (Fig. 6C and D), and H175-β, which is highly buried (106 Å2) upon binding of δ to β (15), are both sequence adjacent to R176-β. Although the aligned position corresponding to H175-β is weakly conserved across diverse bacteria (Fig. 3B), the residue side chains at this position are nearly always five or six atom aromatic rings that, in general, can serve as surface protrusions favorable for binding to the corresponding δ subunit. A lysine that sometimes occurs at this position is likewise compatible with a similar ‘knob-in-hole’ binding mechanism, such as is seen, for example, for a lysine residue in Ran GTPase that inserts into the hole of the β propeller domain of Ran’s exchange factor, RCC1 (26). Note also that T172-β, the above-mentioned conserved threonine that hydrogen bonds to the backbone oxygen of R176-β, also directly contacts L73-δ and F74-δ upon binding of δ (Fig. 6D). Thus, sequence adjacent residues on either side of R176-β, all of which also interact with δ, appear to provide a mechanism for coupling binding of δ to precise repositioning of R176-β.
Possible links between the δ binding site and DNA sensing residues
Links between δ binding sites and the clamp’s central hole. CHAIN analysis suggests that there are at least two structural links connecting β’s δ binding site with residues near the clamp’s central hole, through which DNA is thread. One of these links involves R176-β while the other involves another arginine, R152-β; both links also involve other conserved residues.
For the first link, the δ-induced conformational changes in R176-β, as discussed in the previous section, may be relayed to F144-β and S145-β—two residues that pack up against R176-β (Fig. 7A and B) and that correspond to the domain 2-specific conserved pattern [FY]-[SA] associated with motif A2 (Fig. 3B). Notably, R176-β’s positively charged guanidinium group is positioned to electrostatically interact with the negative dipole moment at the A2 helix C-terminal region, in which F144-β and S145-β are located. The N320-β cluster, which includes R176-β (Fig. 6C inset), may serve as a stable platform for leveraging and directing δ-induced conformational changes toward this region or, conversely, for directing conformational changes in this region back toward δ. Note that the loop connected to this end of the A2 helix (Fig. 7A and B) leads to the B2 region and is located within the β clamp’s central hole.
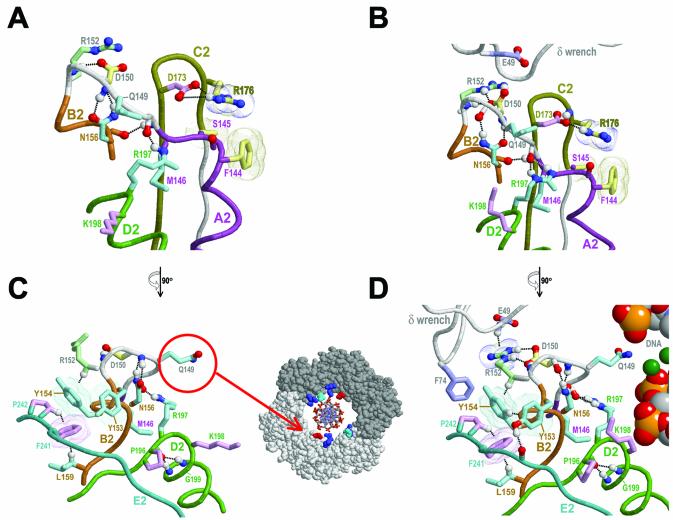
Figure 7.
Links between bound δ and conserved residues near the central hole of the β clamp. See legend to Figure 6 for a description of representations; see text for details. (A) The structural features near the linker region between the A2 and B2 motifs within the β dimer. (B) The same view as in (A) for the δ–β complex. (C) Side view of the β dimer region shown in (A). The inset shows how the conserved asparagine Q149-β (red) protrudes into the central hole of the β ring roughly to the same degree as certain basic residues (blue and cyan) proposed to interact with DNA (modeled). Q149-β, which is in domain 2, structurally corresponds to one of these basic residues within domain 1, R24-β (cyan). (D) Side view of the δ–β complex in (B) with modeled DNA in the center of the β ring.
Near the B2 end of this loop is a second structural link to bound δ involving the less conserved domain 2-specific residue R152-β (Fig. 3B) that, nevertheless, is well conserved among the γ proteobacteria (Fig. 4B). As mentioned above, R152-β and E49-δ would sterically clash were it not for δ-induced conformational changes (see Fig. 6A), and within the δ–β complex these two residues electrostatically interact (Fig. 7B and D). Note also that a cluster of γ proteobacteria-specific conserved residues occurs within the region from F144-β up to and including the B2 motif (Fig. 4B) and that a number of these residues establish alternative molecular interactions within the β dimer versus the δ–β complex (Fig. 7A and C versus B and D, respectively).
Possible DNA sensing mechanisms. Two structural features associated with these γ proteobacterial constraints point to a possible mechanism linking the sensing of encircled DNA to the δ binding site. The first feature consists of a glutamine (Q149-β) that is located in the loop connecting motifs A2 and B2 and an adjacent aspartate (D150-β) that interacts with R152-β (Fig. 7C and D). Notably, in both the β dimeric and δ–β structures, Q149-β protrudes into the β clamp’s central hole (Fig. 7C and D). Indeed, the only other residues that protrude into the hole to this extent correspond to weakly conserved basic residues predicted to non-specifically associate with encircled DNA (Fig. 7C inset). Furthermore, Q149-β, which is within domain 2, structurally corresponds to one of these basic residues within domain 1, namely R24-β.
This suggests that Q149-β is likely to sense the presence of DNA within the β clamp’s central hole to a similar extent as these basic residues and, indeed, perhaps more so, as interaction of a non-basic residue with DNA backbone phosphates would be less favorable and thus more likely to induce a conformational change in Q149-β. Indeed, the residue corresponding to Q149-β in non-proteobacterial species is very often acidic (Fig. 3B), in which case any interaction with backbone phosphates would be repulsive. This influence of DNA on the conformation of Q149-β may be relayed back to the δ binding pocket via D150-β, which is sequence adjacent to Q149-β and which, in the δ–β complex, hydrogen bonds with R152-β, a residue directly contacting δ (Fig. 7D).
A second possible structural feature for sensing of encircled DNA centers on three residue positions (197–199) near the N-terminal end of the motif D2 helix (Fig. 7). Two of these residues, R197-β and G199-β, are subject to γ proteobacteria-specific constraints (Fig. 4B), while a third residue, K198-β, is one of two conserved basic residues within the D2 helix (Fig. 2B) that are predicted to associate with backbone phosphates of encircled DNA (see above). Surprisingly, within the available δ–β clamp crystal structures, which are not associated with DNA, K198-β protrudes less into the central hole than does Q149-β (not shown). Thus, it is easy to envision that, upon association with DNA, K198-β would reposition itself closer to the clamp’s central hole. Moreover, because K198-β lies between R197-β and G199-β, its repositioning—in combination with the backbone flexibility of G199-β and the inherent affinity of R197-β itself for DNA—may distort or disrupt both the N-terminal end of the D2 helix and a hydrogen bond between R197-β and the Q149-β loop (Fig. 7). R197-β also packs up against M146-β (Fig. 7), a γ proteobacteria-specific conserved residue within the Q149-loop that is adjacent to S145-β (Fig. 4B). Thus, any DNA-associated repositioning of R197-β and Q149-β may be propagated back to R176-β, the proposed relay to the δ binding pocket, via the F144-S145 region, which packs up against R176-β. Other γ proteobacterial-specific residues within motif B2 (Fig. 4B) may similarly relay conformational changes back to other δ-interacting regions, such as the F241-P242 motif (Fig. 7C and D).
These proposed structural links between the clamp’s central hole and δ binding regions might operate in either direction. That is, either DNA sensing residues could help trigger the release of δ and closure of the clamp around DNA or binding of δ to a DNA loaded clamp could trigger conformational changes in DNA sensing residues that may assist in unloading of the clamp. Of course both possibilities may apply, in which case the ATP/ADP-bound states of the clamp loader complex (of which δ is but one component) would likely determine directionality. It should be stressed, however, that specific aspects of such a mechanism are unlikely to apply to organisms outside of the γ proteobacteria.
Links between N320-β, the C-terminal E3 region and δ binding sites
Returning our focus again to the strategic residue N320-β, we next examine its relationship to the C-terminal E3 region, which contains several residues subject to strong domain-specific constraints and which binds to δ (Fig. 3C). N320-β directly contacts one of these residues, M364-β, and, as discussed above, both N320-β and M364-β would sterically clash with L73-δ (Fig. 6A) were it not for δ-induced conformational changes. These changes include both unwinding of the C-terminal end of δ’s β-interacting helix and repositioning of L73-δ and F74-δ into the β clamp’s δ binding pocket (Fig. 6B).
Notably, M364-β is sequence adjacent to R365-β, the most buried residue upon binding of β to δ (155 Å2). Within the δ–β complex, R365-β’s positively charged side chain interacts with the negative dipole moment of the restructured C-terminus of δ’s β-interacting helix (Fig. 6B). R365-β also hydrogen bonds to an aspartate (D107-δ) that is highly conserved among γ proteobacterial δ subunits (Fig. 5B) and that is linked—via similarly conserved δ residues (Fig. 6B)—to δ’s site of interaction with R152-β (as discussed above).
Likewise, the two residues directly preceding M364-β, namely M362-β and P363-β, also contact δ within the δ–β complex (as does M364-β itself) and are subject to strong domain 3-specific constraints (as are M364-β and N320-β) (Fig. 3C). Notably, binding of δ to β places L73-δ on the other side of M362-β (Fig. 6B) relative to the δ–β model based on the unbound structures (Fig. 6A). The M362-β residue position nearly always contains a methionine, the most flexible of the long hydrophobic residues (25). Thus, this methionine may facilitate transient conformational changes allowing it to reposition itself around and pack up against L73-δ. P363-β adds rigidity to the backbone conformation of the E3 region, which may help precisely position the nearby R365-β, M364-β and M362-β residues and thereby facilitate their interactions with δ.
Links from N320-β and the B3 loop to the β dimeric interface
In the unbound β structure, N320-β and adjacent backbone atoms interact both with the backbone atoms of the B3 loop and with the side chain of R279-β (Fig. 6E), a residue within the B3 loop that is highly conserved within γ proteobacteria families closely related to E.coli. The B3 loop also appears to act as an N-cap (27,28) for the D3 helix. In contrast, the corresponding regions within domains 1 and 2 form short helices (Fig. 1C) that do not act as N-caps in either the bound or unbound structures.
Upon binding, δ’s β-interacting helix packs up against and disrupts the B3 loop’s interaction with N320-β (Fig. 6E and F), and R279-β is repositioned behind F278-β, a γ proteobacteria-specific residue (Fig. 4C) that extensively contacts δ’s β-interacting helix (Fig. 6F). This brings R279-β spatially close to R365-β (not shown), perhaps thereby also facilitating R365-β’s association with δ, as discussed in the previous section. This movement of the B3 region is also associated with rearrangements between strands within the β sheet spanning the dimeric interface between domains 3 and 1. In particular, hydrogen bonds between the B3 strand and an adjacent C3 strand and between the B3 loop and N296-β are formed (Fig. 6E and F), while other hydrogen bonds between the C3 strand and an adjacent strand at the dimeric interface are disrupted. Other γ proteobacteria-specific conserved residues, such as S274-β, N275-β and Q299-β, partake in these rearrangements by establishing alternative hydrogen bonds within the bound versus the unbound forms (Fig. 6E and F).
The B3 loop conformational change also appears coupled to the adjacent A3 helix inasmuch as the A3 and B3 regions are covalently attached via a one-residue long backbone linker corresponding to N275-β, one of the γ proteobacterial conserved residues. Notably, this linker is considerably shorter (by five residues) than the corresponding domain 1 and 2 linkers. (Incidentally, unlike the domain 3 linker, these other linkers protrude into the clamp’s central hole and contain residues proposed to interact with DNA, namely R24-β and Q149-β.) The section of the A3 helix directly adjacent to this one-residue linker contains the conserved residues L273-β, I272-β and R269-β, all of which make substantial contacts at the dimeric interface (Fig. 6E inset). Recall too that R269-β forms a salt bridge with D304-β, a conserved residue near the end of the β strand that lies at the dimeric interface. Thus, the B3 region’s short covalent attachment to the A3 helix, its alternative interactions with the C3 region, and the associated alternative interactions between γ proteobacterial conserved residues appear to constitute a mechanism linking δ’s β-interacting helix to conformational changes at the dimeric interface and, presumably, to opening of the clamp.
Nevertheless, some of these conformational changes may be artifacts inasmuch as the δ–β structure actually corresponds to a mutant form of the β clamp, in which both I272-β and L273-β have been changed to alanines (15). Given their locations, however, the only secondary structural features within the open clamp that the I272A and L273A mutations seem likely to disrupt are the hydrogen bonds between β strands on either side of the C3 loop. The possible maintenance of these strand interactions in the open wild-type clamp suggests a more straightforward clamp opening mechanism in which δ-induced conformational changes are propagated further, thereby leading to disruption of strand interactions directly at the dimeric interface.
Hydrogen bonds involving side chains of conserved interface residues (Fig. 6E inset) may facilitate such a mechanism inasmuch as these interactions are more easily disrupted by structural perturbations than are hydrophobic interactions (due to the tighter geometric restrictions of hydrogen bonds) and are more readily replaced by solvent interactions (due to their hydrophilic nature).
CONCLUSIONS
The category-specific selective constraints identified here point to key residues and structural features associated with the β clamp’s biological functions. Although CHAIN analysis provides no direct information on what those functions may in fact be, it is clear from experimental analyses that they include: formation of a clamp-like structure, binding to δ and to other clamp loader subunits, δ-induced opening of the clamp, and the release of δ and closure of the clamp upon association with DNA. Insights may thus be obtained by considering how well these functions are explained by the structures and chemical properties of those residues subject to strong category-specific constraints. In this regard, the analysis here provides mechanistic clues concerning (i) binding of δ to β, (ii) δ-mediated opening of the clamp’s dimeric interface and (iii) linkage between δ-associated regions of the clamp and its central hole, through which DNA is thread. It also points to a prominent structural feature, a stacked arrangement of the N320-β, Y323-β and R176-β residues, that appears to be associated with all three of these functions. Further analysis involving weaker selective constraints would likely provide additional clues regarding residues with supporting roles, but, for the sake of clarity, these are not discussed here.
This analysis suggests various mutagenesis studies designed to experimentally probe specific aspects of underlying mechanisms. Indeed, conservative replacements of key residues in one β domain can be proposed based on homology to corresponding residues within the other β domains. For example, the location of Q149-β within domain 2 and its relationship to other conserved residues suggests that it plays a role in sensing the presence of DNA within the clamp’s central hole. The structurally equivalent residue within domain 1 corresponds to a conserved basic residue, R24-β, that also projects into the central hole. The conservative mutation Q149R thus might be expected to disfavor any DNA-induced conformational change at this position by favoring continued protrusion of the mutant lysine residue into the central hole after loading of the clamp onto DNA. Furthermore, biochemical analysis of this mutant could directly explore Q149-β’s possible role in DNA-induced release of the clamp loader complex. Other conservative mutations at key positions in β may likewise be suggested, such as: a D304N mutation to probe the role of the salt bridge between R269-β and D304-β; a N320D mutation to eliminate N320-β’s amino-aromatic interaction with Y323-β; and replacement of the short linker between the A3 and B3 regions with the corresponding linker between the A1 and B1 or the A2 and B2 regions.
Such mutagenesis studies would aim at deciphering the molecular meaning of the patterns of selective constraints revealed by CHAIN analysis. Indeed, these constraints reflect an underlying molecular logic that, in the light of comprehensive experimental data, may well require chemical and structural descriptions as detailed and precise as a complex mathematical proof to fully understand. Thus, the analysis here merely serves as an initial crude step toward such a molecular description.
Acknowledgments
ACKNOWLEDGEMENTS
I thank Natarajan Kannan for critical reading of the manuscript and helpful suggestions. This work was supported by NIH grant LM06747 to A.F.N.
REFERENCES
- 1.Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2002) Clamp loaders and sliding clamps. Curr. Opin. Struct. Biol., 12, 217–224. [DOI] [PubMed] [Google Scholar]
- 2.Kong X.P., Onrust,R., O’Donnell,M. and Kuriyan,J. (1992) Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell, 69, 425–437. [DOI] [PubMed] [Google Scholar]
- 3.Stukenberg P.T., Studwell-Vaughan,P.S. and O’Donnell,M. (1991) Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J. Biol. Chem., 266, 11328–11334. [PubMed] [Google Scholar]
- 4.Naktinis V., Turner,J. and O’Donnell,M. (1996) A molecular switch in a replication machine defined by an internal competition for protein rings. Cell, 84, 137–145. [DOI] [PubMed] [Google Scholar]
- 5.Krishna T.S., Kong,X.P., Gary,S., Burgers,P.M. and Kuriyan,J. (1994) Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell, 79, 1233–1243. [DOI] [PubMed] [Google Scholar]
- 6.Kelman Z. and O’Donnell,M. (1995) Structural and functional similarities of prokaryotic and eukaryotic DNA polymerase sliding clamps. Nucleic Acids Res., 23, 3613–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumiya S., Ishino,Y. and Morikawa,K. (2001) Crystal structure of an archaeal DNA sliding clamp: proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci., 10, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moarefi I., Jeruzalmi,D., Turner,J., O’Donnell,M. and Kuriyan,J. (2000) Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol., 296, 1215–1223. [DOI] [PubMed] [Google Scholar]
- 9.Yao N., Turner,J., Kelman,Z., Stukenberg,P.T., Dean,F., Shechter,D., Pan,Z.Q., Hurwitz,J. and O’Donnell,M. (1996) Clamp loading, unloading and intrinsic stability of the PCNA, beta and gp45 sliding clamps of human, E. coli and T4 replicases. Genes Cells, 1, 101–113. [DOI] [PubMed] [Google Scholar]
- 10.Leu F.P., Hingorani,M.M., Turner,J. and O’Donnell,M. (2000) The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J. Biol. Chem., 275, 34609–34618. [DOI] [PubMed] [Google Scholar]
- 11.Marians K.J. (1992) Prokaryotic DNA replication. Annu. Rev. Biochem., 61, 673–719. [DOI] [PubMed] [Google Scholar]
- 12.Onrust R., Finkelstein,J., Turner,J., Naktinis,V. and O’Donnell,M. (1995) Assembly of a chromosomal replication machine: two DNA polymerases, a clamp loader and sliding clamps in one holoenzyme particle. III. Interface between two polymerases and the clamp loader. J. Biol. Chem., 270, 13366–13377. [DOI] [PubMed] [Google Scholar]
- 13.Jeruzalmi D., O’Donnell,M. and Kuriyan,J. (2001) Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell, 106, 429–441. [DOI] [PubMed] [Google Scholar]
- 14.Stewart J., Hingorani,M.M., Kelman,Z. and O’Donnell,M. (2001) Mechanism of β clamp opening by the δ subunit of Escherichia coli DNA polymerase III holoenzyme. J. Biol. Chem., 276, 19182–19189. [DOI] [PubMed] [Google Scholar]
- 15.Jeruzalmi D., Yurieva,O., Zhao,Y., Young,M., Stewart,J., Hingorani,M., O’Donnell,M. and Kuriyan,J. (2001) Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell, 106, 417–428. [PubMed] [Google Scholar]
- 16.O’Donnell M., Jeruzalmi,D. and Kuriyan,J. (2001) Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol., 11, R935–R946. [DOI] [PubMed] [Google Scholar]
- 17.Davey M.J., Jeruzalmi,D., Kuriyan,J. and O’Donnell,M. (2002) Motors and switches: AAA+ machines within the replisome. Nature Rev. Mol. Cell. Biol., 3, 826–835. [DOI] [PubMed] [Google Scholar]
- 18.Neuwald A.F., Kannan,N., Poleksic,A., Hata,N. and Liu,J.S. (2003) Ran’s C-terminal, basic patch and nucleotide exchange mechanisms in light of a canonical structure for Rab, Rho, Ras and Ran GTPases. Genome Res., 13, 673–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shindyalov I.N. and Bourne,P.E. (1998) Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 20.Tatusov R.L., Koonin,E.V. and Lipman,D.J. (1997) A genomic perspective on protein families. Science, 278, 631–637. [DOI] [PubMed] [Google Scholar]
- 21.Burley S.K. and Petsko,G.A. (1986) Amino-aromatic interactions in proteins. FEBS Lett., 203, 139–143. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell J.B., Nandi,C.L., McDonald,I.K., Thornton,J.M. and Price,S.L. (1994) Amino/aromatic interactions in proteins: is the evidence stacked against hydrogen bonding? J. Mol. Biol., 239, 315–331. [DOI] [PubMed] [Google Scholar]
- 23.Flocco M.M. and Mowbray,S.L. (1994) Planar stacking interactions of arginine and aromatic side-chains in proteins. J. Mol. Biol., 235, 709–717. [DOI] [PubMed] [Google Scholar]
- 24.Gallivan J.P. and Dougherty,D.A. (1999) Cation-pi interactions in structural biology. Proc. Natl Acad. Sci. USA, 96, 9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richardson J.S. and Richardson,D.C. (1989) Principles and patterns of protein conformation. In Fasman,G.D. (ed.), Prediction of Protein Structure and the Principles of Protein Conformation. Plenum, New York, pp. 1–98. [Google Scholar]
- 26.Renault L., Kuhlmann,J., Henkel,A. and Wittinghofer,A. (2001) Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1). Cell, 105, 245–255. [DOI] [PubMed] [Google Scholar]
- 27.Presta L.G. and Rose,G.D. (1988) Helix signals in proteins. Science, 240, 1632–1641. [DOI] [PubMed] [Google Scholar]
- 28.Richardson J.S. and Richardson,D.C. (1988) Amino acid preferences for specific locations at the ends of α helices. Science, 240, 1648–1652. [DOI] [PubMed] [Google Scholar]
- 29.Sayle R.A. and Milner-White,E.J. (1995) RASMOL: biomolecular graphics for all. Trends Biochem. Sci., 20, 374. [DOI] [PubMed] [Google Scholar]
- 30.Weiss M.S., Brandl,M., Suhnel,J., Pal,D. and Hilgenfeld,R. (2001) More hydrogen bonds for the (structural) biologist. Trends Biochem. Sci., 26, 521–523. [DOI] [PubMed] [Google Scholar]