Abstract
Large deletions in WNK1 are associated with inherited arterial hypertension. WNK1 encodes two types of protein: a kidney-specific isoform (KS-WNK1) lacking kinase activity and a ubiquitously expressed full-length isoform (L-WNK1) with serine threonine kinase activity. Disease is thought to result from hypermorphic mutations increasing the production of one or both isoforms. However, the pattern of L-WNK1 expression remains poorly characterized. We generated transgenic mice bearing a murine WNK1 BAC containing the nlacZ reporter gene for monitoring L-WNK1 expression during development and adulthood. We observed previously unsuspected early expression in the vessels and primitive heart during embryogenesis, consistent with the early death of WNK1−/− mice. The generalized cardiovascular expression observed in adulthood may also suggest a possible kidney-independent role in blood pressure regulation. The second unsuspected site of L-WNK1 expression was the granular layer and Purkinje cells of the cerebellum, suggesting a role in local ion balance or cell trafficking. In the kidney, discordance between endogenous L-WNK1 and transgene expression suggests that either cis-regulatory elements important for physiological renal expression lie outside the BAC sequence or that illegitimate interactions occur between promoters. Despite this limitation, this transgenic model is a potentially valuable tool for the analysis of spatial and temporal aspects of WNK1 expression and regulation.
Familial hyperkalemic hypertension (FHHt, no. OMIM145260) is a hereditary form of hypertension caused by mutations affecting two members (WNK1 and WNK4) of a new family of serine threonine kinases, the “with no lysine (K),” or WNK, kinases.1,2 WNK4 is found primarily in the distal nephron, and a number of recent experiments have shown this kinase to be important in the regulation of ion handling.3–5 However, the distribution of WNK1 is more complex due mostly to the production of different isoforms. A long ubiquitous isoform (L-WNK1) containing the kinase domain is produced under the control of proximal promoters whereas a shorter isoform is produced specifically in the distal nephron (KS-WNK1).6,7 This shorter isoform has no kinase activity and is generated from an alternative promoter upstream from exon 4a. It is unclear whether the FHHt-causing deletions in intron 1 of WNK1 affect the qualitative and quantitative production of these two isoforms. The typical renal symptoms of FHHt indicate a probable change in expression pattern, at least in the distal nephron. We previously observed an increase in WNK1 mRNA levels in leukocytes of affected patients, suggestive of hypermorphic mutations.1 This mechanism is consistent with recent in vitro findings that L-WNK1 inhibits WNK4, which itself inhibits the sodium co-transporter NCC, the potassium transporter ROMK1, and chloride transtubular transfer.8 It is also consistent with the inhibition of L-WNK1 by KS-WNK19 and with the effects of L-WNK1 on the epithelial sodium transporter ENaC.10,11
The ubiquitous nature of WNK1 probably renders the whole situation much more complex. Indeed, L-WNK1 is produced in several epithelia, the heart, muscle, and brain. However, its distribution remains poorly characterized. Expression studies in adults have mostly involved multitissue Northern blots6,7,12–15 or the application of immunohistochemistry techniques to epithelia.15 In other organs, such as the heart, it has proved difficult to obtain consistent results with techniques such as immunohistochemistry and Northern blotting due to alternative splicing of WNK1, hindering studies into the possible multiple functions of WNK1. Finally, WNK1 expression during development has not yet been studied despite its probable importance, as reflected by the early death of WNK1−/− embryos.16
As a first step toward an analysis of the consequences of FHHt-causing WNK1 mutations, we designed an in vivo model for monitoring L-WNK1 expression during development and adulthood. We generated transgenic mice bearing the murine WNK1(mWNK1) locus modified to include a reporter gene. Species-specific variation in transcription factor binding sites may be responsible at some level for species-specific differences in gene expression and regulation. We constructed a mouse transgene to prevent these interferences between cis- and trans regulatory elements and, thus, potential discordance between the transgene and the endogenous gene expression. Moreover, we had previously shown that the organization, structure, and expression of the WNK1 gene are similar in mice and humans.7 The construct used was a bacterial artificial chromosome (BAC RP24-212e14) containing 47.4 kb upstream from the transcription start site and 11.2 kb downstream from the last mWNK1 exon, into which we incorporated in exon 2 the nuclear lacZ (nlacZ) gene as a reporter. Furthermore we introduced a stop codon in the KS-WNK1-specific exon 4a to prevent KS-WNK1 overexpression. We used this BAC reporter assay to analyze L-WNK1 expression in a physiological model, during embryogenesis and adulthood, with high sensitivity and single-cell resolution. This analysis provided unexpected findings, concerning WNK1 expression in the cardiovascular and central nervous systems in particular. This transgenic model is of potential value for the analysis of spatial and temporal aspects of WNK1 expression and regulation.
Materials and Methods
Generation of the Transgene
BAC Identification
BAC RP24-212e14 spanning the mWNK1 locus was identified on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). The RPCI-24 library was constructed from C57BL/6 genomic DNA using the pTARBAC1 vector (CHORI). The chosen BAC included the complete mWNK1 gene sequence. It began at nucleotide −47440 relative to the mWNK1 transcription start site (sequenced BAC end: gi 13218338) and ended 11.4 kb after the last mWNK1 exon (sequenced BAC end: gi 13218335).
BAC Modifications
Escherichia coli strains EL-250 and EL-350, electroporation conditions with BAC DNA and targeting cassettes, selection of recombinant clones, and excision of the selection cassette were described by Lee and colleagues17 and Liu and colleagues.18 The BAC construct was checked at each step by DNA digestion and direct sequencing of the targeted regions, to ensure that the structure of the original BAC was conserved and that the clones included the homologous recombination events.
LoxP Site Deletion from the pTARBAC1 Vector of the RP24-212e14 Clone
We deleted the LoxP site from the backbone to prevent recombination between this site and the loxP sequences inserted in the BAC. We constructed a targeting cassette containing an ampicillin (amp) resistance gene replacing the LoxP sequence based on pTamp.17 In the targeted BAC, the LoxP site was replaced by the amp gene.
Exon 4a-Targeting Cassette
The exon 4a-targeting cassette was generated by inserting a translation stop codon just downstream from the start codon in exon 4a, followed by the SV40 polyadenylation signal (SV40pA). The kanamycin (kan) resistance gene was incorporated into the cassette to facilitate positive selection for homologous recombination in E. coli. LoxP sites flanking the kan gene made it possible to remove this gene by arabinose induction of the Cre recombinase carried in EL350 before the generation of transgenic mice, thereby excluding the possibility of promoter interference effects. Two DNA fragments cloned by polymerase chain reaction (PCR) were incorporated at the ends of the targeting cassette for bacterial homologous recombination at the exon 4a locus (Figure 1B). The 346-bp Ex4a-5′ sequence was cloned by PCR using forward primer 5′-CAAAAGTCAAGGAGGCAGAGC-3′ and reverse primer 5′-CATCAGAAAAGGCAACAGACTT-3′, and the 359-bp Ex4a-3′ sequence was cloned using forward primer 5′-TAAATTCTCATTGCTGCTGCTGT-3′ and reverse primer: 5′-GTTTGTTTGCCAACTAATCTGCT-3′.
Figure 1.
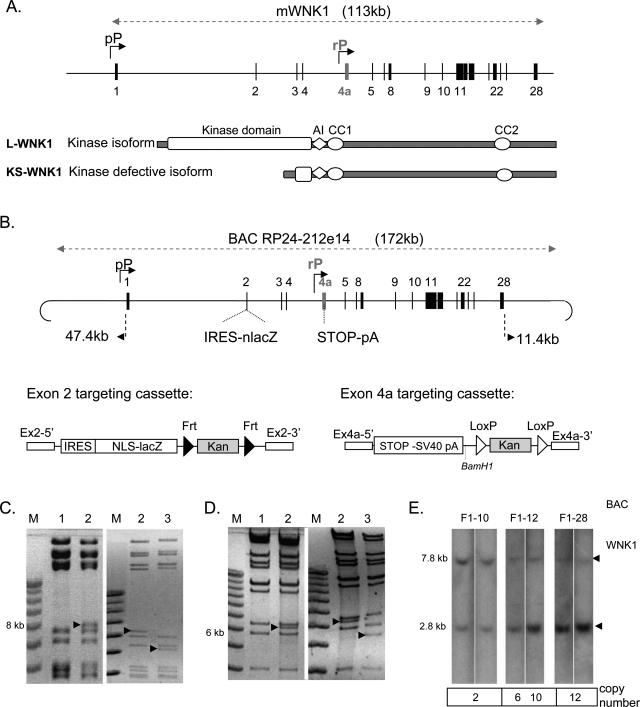
Strategy for studying the production of WNK1 kinase domain-containing isoforms. A: Genomic structure of the mouse WNK1 gene. The genomic segment encompassing WNK1 is represented as a horizontal line, and exons are indicated by numbered vertical lines. The two alternative known promoters for mWNK1 are shown (bent arrows). The proximal promoters (pP) control transcription for the kinase domain-containing isoform (L-WNK1) and a renal promoter (rP) drives expression for the isoform lacking the kinase domain (KS-WNK1). AI, autoinhibitory domain; CC, coiled coil domain. B: Schematic diagram of the targeting cassettes used to generate the modified RP24-212e14 BAC and the resulting reporter BAC with the nlacZ gene inserted into exon 2 and a translation stop codon inserted into exon 4a, abolishing KS-WNK1 production. The size of the genomic sequences included in the BAC, on the 5′ and 3′ sides of WNK1, is indicated. The exon 2-targeting cassette was generated by inserting the nuclear lacZ gene, with the SV40 polyadenylation signal (nlacZ), downstream from an IRES sequence (internal ribosomal entry site). The kanamycin resistance gene (Kan) is flanked by two FRT sites (black arrows), which are recognized by the Flp recombinase. At each end of the cassette, sequences homologous to the 5′ (Ex2-5′) and 3′ (Ex2-3′) sequences of exon 2 facilitate recombination with the BAC. The exon 4a-targeting cassette was generated by inserting a transcription stop signal and the SV40 polyadenylation signal (STOP-SV40polyA), the kan gene flanked by two loxP sites (white arrows) and sequences homologous to exon 4a at each end (Ex4-5′ and Ex4a-3′). C: Exon 2 modification in the BAC was confirmed by EcoRV digestion. The exon 2 band displayed an appropriate increase in size and a decrease in size consistent with the deletion of the kan gene after recombinase treatment. D: Exon 4a modification in the BAC was confirmed by BamHI digestion. The fingerprint displayed an appropriate 6-kb band, confirming stop codon insertion and a decrease in size after recombinase treatment consistent with kan removal. Diagnostic changes are indicated by arrows. Lane 1: Parental BAC; lane 2: recombinant BAC with kanamycin resistance; lane 3: recombinant BAC with kan removed by Flp or Cre induction. E: The BAC transgene copy number was quantified by Southern blotting of DNA from F1 mice digested with PstI and probed with an ex4a probe. The expected size of the endogenous WNK1 gene is 7.8 kb and that for the transgene is 2.8 kb.
Exon 2-Targeting Cassette
The exon 2-targeting cassette was generated by inserting the gene encoding β-galactosidase (lacZ) together with a nuclear localization signal (n) and the SV40pA signal downstream from an internal ribosome entry site sequence (IRES). Flp recognition target (FRT) sites flanking the kan gene made it possible to remove this gene by arabinose induction of Flp recombinase before the generation of transgenic mice. Two DNA fragments cloned by PCR were incorporated at the ends of the targeting cassette for bacterial homologous recombination at the exon 2 locus (Figure 1B). The 375-bp Ex2-5′ sequence was cloned by PCR using forward primer 5′-GTTT-TTCGAGACAGGGTTTCTCT-3′ and reverse primer 5′-TTCTTTGAATCTCTGCCTTTCAG-3′, and the 380-bp Ex2-3′ sequence was cloned using forward primer 5′-GGAATCCACAGTAAAAGGGAAAA-3′ and reverse primer 5′-CCATGTCTAGTGCTTCCTCTGTT-3′. In the targeted BAC, IRES-nlacZ was inserted 42 bases downstream from the first base of exon2.
Generation and Identification of L-WNK1-nlacZ Transgenic Mice
Transgenic mice were generated by microinjection of the circular BAC DNA into fertilized (C57BL/6JxSJL) F2 eggs at a concentration of 1 ng/μl using standard techniques.19 Injected eggs were reimplanted the day after the injection into pseudopregnant CD1(ICR) foster mothers. Genomic DNA was prepared from mouse tails and analyzed by Southern blot or PCR analysis. For Southern blot analysis, 15 μg of DNA was digested by PstI and subjected to electrophoresis. The Ex4a 5′ sequence was used as a probe and allowed the detection of a 7.8-kb fragment in the endogenous mWNK1 gene and a 2.8-kb fragment in the BAC transgene. Transgene copy number for each transgenic line was determined by analysis of hybridization signals with the Molecular Imager FX and Quantity one program (Bio-Rad, Marnes la Coquette, France). For PCR analysis, the primers used were forward primer 5′-GTTTTTCGAGACAGGGTTTCTCT-3′ and reverse primer 5′-AACGCACACCGGCCTTATTCC-AAG-3′.
Analysis of Transgene Expression
Heterozygous males were crossed with nontransgenic females (C57BL/6N). Embryos were staged taking embryonic day (E) 0.5 as the day of the vaginal plug.
X-gal Staining
Embryos and adult tissues were dissected in phosphate-buffered saline (PBS), fixed by incubation in 4% paraformaldehyde for 10 to 20 minutes at 4°C (depending on the embryonic stage or the size of the tissue), rinsed twice in PBS, and stained overnight at room temperature with 0.4 mg/ml 5-bromo-4-chloro-3-indolyl-b-d-galactopyranoside (X-Gal; Invitrogen, Cergy Pontoise, France) in PBS, 5 mmol/L K3Fe, 5 mmol/L K4Fe, 2 mmol/L MgCl2, 0.2% Nonidet P-40. They were then postfixed in paraformaldehyde. For cryostat sectioning, embryos and adult tissues were transferred to 15% sucrose in PBS at room temperature after paraformaldehyde fixation and then embedded in 7% gelatin and 15% sucrose in PBS before freezing by immersion in isopentane at −140°C. Cryosections (10 μm) were cut from these tissues and rinsed in PBS before X-gal staining overnight at room temperature, as previously described, with postfixing and counterstaining with eosin.
Immunohistochemistry
We cut 200-μm kidney sections with a vibratome after perfusion of anesthetized transgenic mice with 4% paraformaldehyde via the right atrium. These sections were stained overnight in X-gal solution at room temperature before paraffin-embedding. Sections (8 to 10 μm) were deparaffinized, rehydrated, and endogenous peroxidase inactivated with 3% H2O2 in PBS. They were then blocked with the avidin-biotin blocking kit (Vector Laboratories, Burlingame, CA) and 5% normal goat serum in PBS and incubated with a polyclonal anti-NCC (thiazide-sensitive Na-Cl co-transporter) antibody (dilution 1/2000),20 anti-NKCC2 (BSC-1) (renal Na+, K+, 2Cl− co-transporter) antibody (dilution 1/400),21 or anti-β-ENaC (epithelial sodium channel β subunit) antibody (dilution 1/500)22 for 2 hours in PBS, 1% bovine serum albumin, and 0.025% Tween. Sections were then washed twice in PBS and incubated for 30 minutes with biotinylated goat anti-rabbit IgG (Vector Laboratories). This secondary antibody was amplified and detected with the Vectastain ABC kit and the DAB peroxidase substrate kit (Vector Laboratories).
In Situ Hybridization
The L- and KS-WNK1 isoforms were detected with RNA probes labeled with digoxigenin-labeled uridine triphosphate as recommended by the manufacturer (Roche Diagnostics, Meylan, France) and binding to exon 4a (KS-WNK1), exons 1 to 4 (L-WNK1) or exons 25 to 27 (L- and KS-WNK1). Whole-mount in situ hybridization on embryos (E9.5 to E13.5) and adult tissues was performed as previously described.23 In situ hybridization on microtome sections (10 μm) was performed as previously described.24
Alternative Splicing
Total RNA from mouse tissues were reverse-transcribed by using the MMLV RT (Gibco BRL-Life Technologies, Invitrogen) and random hexamers (Roche Diagnostics). Exon 4b splice variant was detected with primers binding to exons 2 and 4b or to exons 4a and 4b.
Real-Time Quantitative PCR on Microdissected Tubules
Adult mouse kidneys were microdissected at the plate-forme d’explorations fonctionnelles rénales in vivoand ex vivo, Institut des Cordeliers (Paris, France) as previously described.25 Real-time quantitative RT-PCR (QRT-PCR) assays were performed in a LightCycler (Roche Diagnostics) with the intercalation of SYBR Green (Roche Di-agnostics) as a fluorescence reporter. Primers were designed to discriminate between the long and kidney-specific isoforms. Forward primer from exon 1 (5′-GACAGTCTACAAAGGTCTGGACAC-3′) and reverse primer from exon 2 (5′-GACCCTTTAACATTTCAGCCTCTTC-3′) were used to amplify L-WNK1, and primers from exon 4a (5′-TTGTCATCATAAATTCTCATTGCTG-3′) and exon 5 (5′-AGGAATTGCTACTTTGTCAAAACTG-3′) were used to amplify KS-WNK1. Forward (5′-TTATGGAAGGTGCAGCCACTA-3′) and reverse (5′-CTGTTGCACCAGTAGCCATATCT-3′) primers were used to amplify exon 4b from both L- and KS-WNK1 transcripts. Forward primer from exon 4a (5′-TTATTGTAAATTCTCATTGCTGCTG-3′) and reverse primer from exon 4b (5′-ATATCTTGGCAGGCCAGTCTTTAT-3′) were used to amplify exon 4b from KS-WNK1 transcripts, and forward primer from exon 4 (5′-GAATATCCATACTCAGAATGCCAAA-3′) and reverse primer from exon 4b (5′-CCTGGTGATGACTCTTTTCCACTA-3′) were used to amplify exon 4b from L-WNK1 transcripts. Primers from the lacZ gene (lacZ forward 5′-TACACCAACGTAACCTATCCCATTA-3′ and lacZ reverse 5′-GAGTTAACGCCATCAAAAATAATTC-3′) were used to analyze the lacZ mRNA distribution along the nephron. For all of the samples, the amount of PCR product was calculated as a percentage of a RNA standard (whole kidney RNA) per mm of tubule length (arbitrary unit). Data were then corrected for PCR efficiency as previously described.26
Nucleotide Sequence Accession Number
The nucleotide sequence of exon 4b of the mouse WNK1 gene has been deposited in the GenBank database under GenBank accession no. DQ402040.
Results
Generation of L-WNK1-nlacZ Genomic Reporter Mice
The ubiquitous kinase domain-containing isoform (L-WNK1) is produced under the control of proximal promoters upstream from and into exon 1 (pP), whereas the kidney-specific isoform with no kinase activity (KS-WNK1) is produced under the control of an alternative promoter (rP) in intron 4 (Figure 1A). We analyzed L-WNK1 expression by means of a genomic reporter assay, using a BAC bearing the mWNK1 locus (Figure 1B). We constructed two targeting cassettes—one to introduce the nlacZ reporter gene into mWNK1 exon 2 (Figure 1B) and the other to insert a stop codon into exon 4a, thereby preventing production of the KS-WNK1 isoform (Figure 1B). The homologous recombinant events were checked by means of several restriction enzyme digestions (Figure 1, C and D). Homologous recombination sites and selection cassette excision were checked by BAC sequencing.
In the resulting construct, the nlacZ gene was fused to the exon 2 coding sequence, resulting in β-galactosidase expression under control of the proximal promoters of the BAC clone. Internal translation initiation from the IRES sequence located just upstream from the nlacZ coding sequence led to production of an efficient β-galactosidase enzyme rather than a WNK1-β-galactosidase fusion protein. We obtained four transgenic founders from this construct, and transmission of the BAC transgene to the F1 generation was confirmed by genomic Southern blotting (Figure 1E). Three of the four lines expressed the transgene. Transgenic lines 10 and 28 carried 2 and 12 copies of the transgene, respectively, at a single chromosomal location. Transgenic line 12 carried 6 and 10 copies at two chromosomal locations and was separated into two lines, each with transgene insertion at a single chromosomal location. The four independent transgenic lines had almost identical L-WNK1-nlacZ expression profiles, with only slight differences in expression intensities.
WNK1 Is Strongly Expressed in the Cardiovascular System
Cardiac Expression
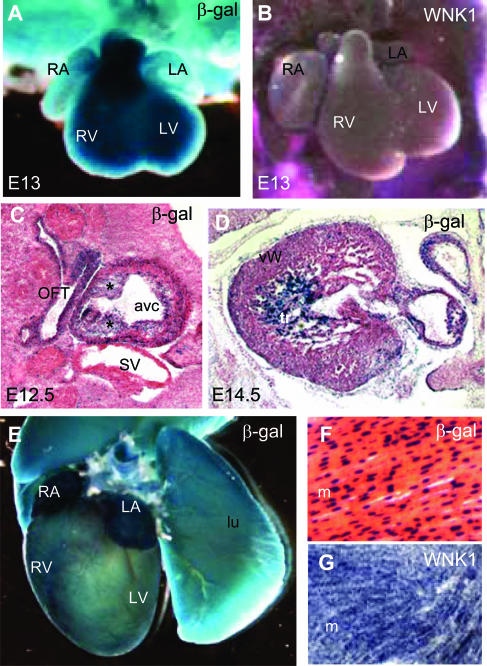
The L-WNK1-nlacZ transgene was strongly expressed in the heart during early development. Whole-mount observation as early as day 7.5 of development revealed ubiquitous low-level nlacZ expression in the embryo. Marked changes in X-gal staining occurred during rapid organogenesis, between stages E8 and E9. During this period, reporter expression increased in the developing primitive heart (see Supplementary Figure 1 at http://ajp.amjpathol.org). The pattern of L-WNK1-nlacZ expression in E13 heart (Figure 2A) was similar to that of endogenous WNK1 (Figure 2B). We stained embryo cryosections to locate nlacZ expression more precisely in the primitive heart. At E12.5, expression was strongest in the outflow tract and atrioventricular canal (Figure 2C). At E14.5, expression was strongest in the trabeculations of the ventricular wall (Figure 2D).
Figure 2.
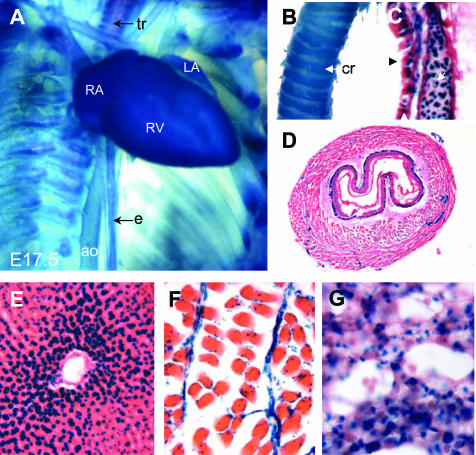
L-WNK1 expression in the developing and adult heart. A and E: Whole-mount X-gal staining. B and G:In situHybridization with the L-WNK1 mRNA probe. C, D, and F: X-gal staining on frozen sections counterstained with eosin. A: At E13, the heart is undergoing remodeling and L-WNK1-nlacZ transgene was strongly expressed in the right atrium (RA), left atrium (LA), right ventricle (RV), and left ventricle (LV). B: L-WNK1 mRNA was detected in all the cardiac chambers. C: We detected at E12.5 strong β-galactosidase activity in precursors of the cardiac valves (asterisks) in the atrioventricular canal (avc). OFT, outflow tract; SV, sinus venosus. D: At E14.5, the transgene is strongly expressed in trabeculae (tr) of the ventricular wall (vw). E and F: Adult heart, showing ubiquitous expression of the transgene (lu, lung), confirmed by in situ hybridization analysis of myocardium (m) transverse sections (G). Original magnifications: ×10 (A, B); ×40 (C, D); ×2.5 (E); ×250 (F, G).
L-WNK1 cardiac expression was maintained in adults. In toto X. gal staining demonstrated sustained expression of the transgene (Figure 2E). Histological analysis showed that the transgene was expressed homogeneously and that its product was present in cardiomyocytes (Figure 2F). Endogenous cardiac L-WNK1 expression was confirmed by in situ hybridization (Figure 2G).
Vascular Expression
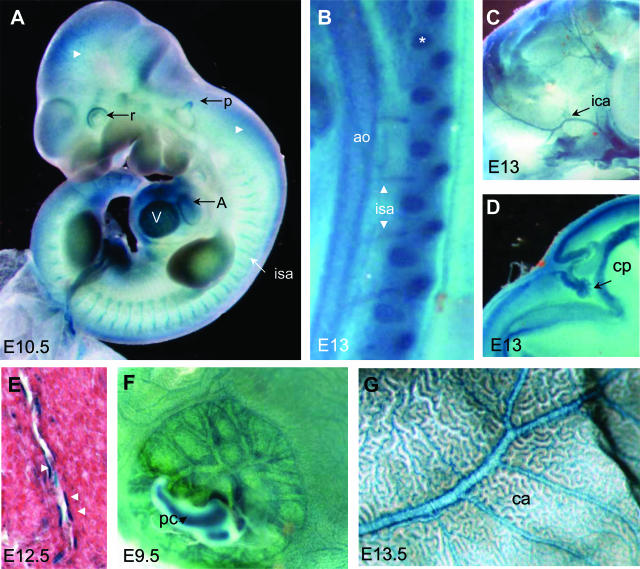
At E10.5 the transgene was expressed throughout the developing vasculature (Figure 3A). β-Galactosidase activity was clearly detected in the intersegmental arteries and the capillaries of perineural tissues. At E13, β-gal activity was observed in the aorta and intersegmental arteries (Figure 3B), internal carotid arteries (Figure 3C), and vessels of the choroid plexus (Figure 3D). We stained cryosectioned E12.5 embryos to locate nlacZ expression precisely. The transgene was expressed ubiquitously at a low level, but was much more strongly expressed in the endothelial cells of arteries and veins (Figure 3E). The transgene was also expressed in the extraembryonic vasculature. It was detected in placenta (Figure 3F) and yolk sac vessels throughout development and throughout its capillary bed (Figure 3G).
Figure 3.
Embryonic and extraembryonic expression of the L-WNK1-nlacZ transgene in the vascular system. A–D and F–G: Whole-mount X-gal staining. E: X-gal staining on frozen sections counterstained with eosin. A: E10.5 embryo X-gal stained for a short time, showing marked expression in the primitive heart (A, atrium; V, ventricle), in the intersegmental arteries (isa), peripheral vessels of the neural tube (white arrows), pinna of the ear (p), and in the retina (r). B: At E13, nlacZ was still strongly expressed in intersegmental arteries, aorta (ao), and cartilage (asterisks). C and D: Whole-mount staining also clearly demonstrated β-galactosidase activity in the internal carotid artery (ica; C) and choroid plexus (cp; D). E: Small vessels also displayed strong expression in endothelial cells (black arrows). F and G: Extraembryonic tissues (F): placental cord (pc) and vessels of placenta and yolk sac (G). E13.5 yolk sac showing staining in the arterioles and throughout the capillary bed (ca; G). Original magnifications: ×5 (A); ×10 (B–D, F–G); ×250 (E).
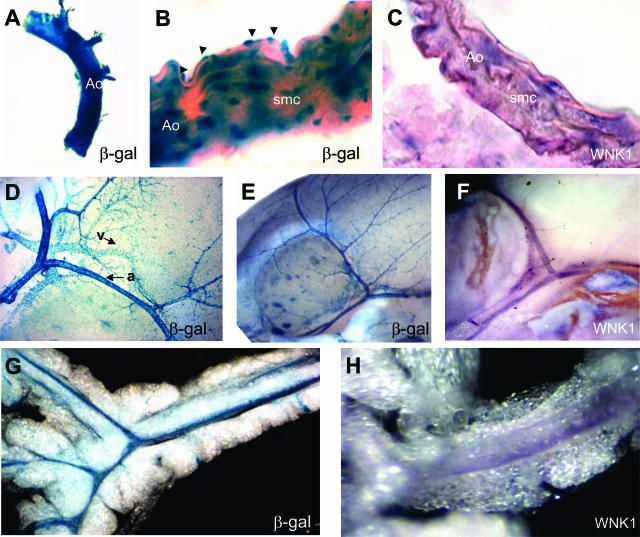
As development progressed, nlacZ expression was evident in many blood vessels throughout the body, in large and small arteries, capillaries, and veins. β-Galactosidase activity was detected in the adult abdominal aorta (Figure 4, A and B), vessels in the brain (Figure 4, D and E), the diaphragm, kidneys, skeletal muscle, and mesentery (Figure 4G). X-gal staining was detected in the small arteries and their connecting arterioles at the brain surface, and in the veins and venules (Figure 4, D and E).
Figure 4.
L-WNK1 expression in the adult vascular system. A, D, E, and G: Whole-mount X-gal staining. B: X-gal staining of frozen sections counterstained with eosin. In situ (C) and in toto (F, H) hybridization with the L-WNK1 mRNA probe. A–C: Transgene and L-WNK1 expression in the aorta. The staining of aorta cryosections shows expression in both endothelial (arrows) and smooth muscle cells (smc). Strong expression is also evident in small arteries (a) and veins (v) at the surface of the brain (D, E), and of the mesentery (G). Endogenous L-WNK1 expression was confirmed in the brain (F) and mesenteric vessels (H). Original magnifications: ×2.5 (A); ×250 (B); ×100 (C); ×10 (D); ×5 (E, F, G); ×10 (H).
We stained cryosections of adult aorta with X-gal (Figure 4B) to identify the expressing cell types in the blood vessels. Both endothelial cells and smooth muscle cells were stained. The corresponding endogenous expression was assessed by in situ hybridization with an L-WNK1 RNA probe. Expression was confirmed in the aorta (Figure 4C), brain vessels (Figure 4F), and mesenteric vessels (Figure 4H). In the aorta, endogenous WNK1 expression was detected in the vascular endothelium and smooth muscle cell layers (Figure 4C).
Expression in the Central Nervous System
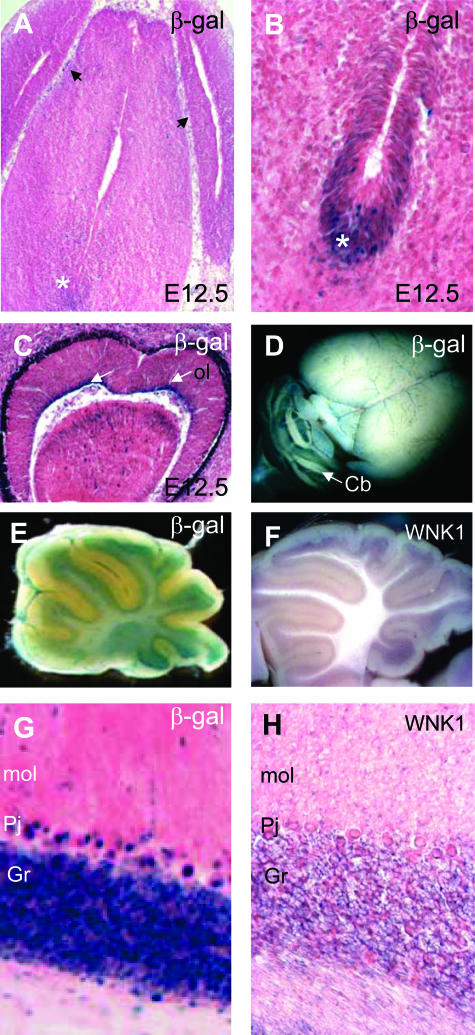
In embryos, almost no L-WNK1-nlacZ transgene expression was observed in the central nervous system, except in the capillaries surrounding the neural tube (Figure 5A) and some neurons of the floor plate (Figure 5B). The transgene was also expressed in epithelial cells of the optic layer (Figure 5C). In adult mice, the transgene was expressed in the cerebellum (Figure 5D). The most intense region of X-gal staining was the granular layer and cerebellar Purkinje cells, with only weak staining observed in the molecular layer and white matter (Figure 5, E and G). Expression levels in these layers appeared constant from 3 days after birth into adulthood (see Supplementary Figure 2 at http://ajp.amjpathol.org). In toto hybridization (Figure 5F) and in situ hybridization on cryosections (Figure 5H) gave similar results.
Figure 5.
L-WNK1 production in the central nervous system. A–C and G: X-gal staining on frozen sections counterstained with eosin. D and E: Whole-mount X-gal staining. L-WNK1 in toto (F) and in situ (H) hybridization. A: At E12.5, capillaries surrounding the neural tube (black arrows) were positively stained whereas most neurons were not. B: At 12.5, cells of the floor plate (asterisks) showed strong positive staining. C: Expression in the epithelial cells of the optic layer (ol). In adults, L-WNK1 expression was restricted to the cerebellum (Cb; D–H), the granular layer (g), cerebellar Purkinje cells (Pj), and to a lesser extent, the molecular layer (mol). Original magnifications: ×40 (A, C); ×100 (B, G, H); ×2.5 (D); ×5 (E, F).
Expression in Other Tissues
During embryogenesis, β-galactosidase activity was detected in the esophagus and in the cartilaginous rings of the trachea (Figure 6A). In adults, β-galactosidase activity was detected in the chondrocytes of the cartilaginous rings and in the epithelia of the esophagus and trachea (Figure 6, B–D). In adult mice, β-galactosidase activity was ubiquitous. Muscle, lung, liver, and intestine also presented strong β-galactosidase activity, consistent with the results of Northern blots. Histological analysis showed positive staining of the hepatocytes (Figure 6E) and skeletal muscle cells (Figure 6F). However in muscle, the strongest expression of the L-WNK1-nlacZ transgene was detected in vascular cells. In the lung, β-galactosidase activity was observed in the vessels and capillaries and also in pneumocytes (Figure 6G).
Figure 6.
Expression in the esophagus, trachea, liver, muscle, and lungs of embryos and adults. A and B: Whole-mount X-gal staining. D–G: X-gal staining on frozen sections counterstained with eosin. A: Expression in the esophagus (e), trachea (tr), aorta (ao), right atrium (RA), right ventricle (RV), left atrium (LA), in E17.5 embryo. In adult, L-WNK1-nlacZ transgene expression was detected in the cartilaginous rings of the trachea (cr; B), the tracheal epithelium (black arrow) and the chondrocytes (white arrow; C) and in the epithelium of the esophagus (D). Staining for β-galactosidase activity in liver (E), skeletal muscle (F), and lung (G). Original magnifications: ×2.5 (A); ×5 (B); ×40 (D);×100 (C, E, F, G).
Renal WNK1 Expression
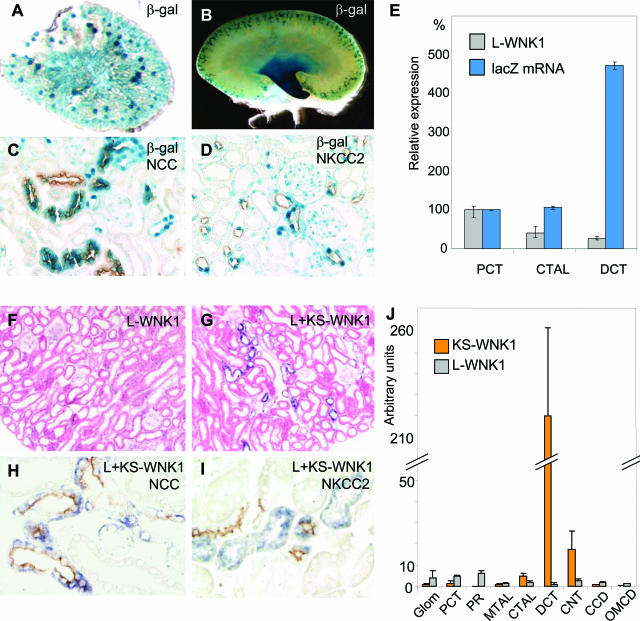
The transgene was expressed in the kidney vasculature of embryos and neonates. In toto staining of E17.5 kidney showed sustained expression of the transgene. At birth, expression was strongest in the glomerular capillaries as shown by X-gal staining of vibratome sections (Figure 7A). Cross sections of adult kidneys from all four transgenic lines showed strong β-galactosidase activity in cortical tubules and the papilla and uniformly low levels of expression in other cells, including those of the glomerular capillaries (Figure 7, B–D). We investigated the possible nephron segment-specific expression of the L-WNK1-nlacZ transgene by staining kidney sections from transgenic mice with anti-NKCC2, anti-NCC, and anti-β-ENaC antibodies. Anti-NCC staining partly coincided with β-galactosidase staining. However, a few tubules were β-galactosidase-positive and NCC-negative whereas others were NCC-positive and β-galactosidase-negative (Figure 7C). β-Galactosidase staining did not show the same distribution as anti-β-ENaC antibody staining (not shown), but β-galactosidase activity was co-localized with anti-NKCC2 antibody staining in the cortex (Figure 7D). The expression of the L-WNK1-nlacZ transgene was confirmed by real-time RT-PCR on microdissected tubules that showed a fivefold greater abundance of nlacZ mRNA in the distal than in the proximal convoluted tubule (Figure 7E).
Figure 7.
L-WNK1-nlacZ transgene expression in kidney. A: X-gal staining of a 100-μm vibratome transverse kidney section from an embryo. B: Whole-mount X-gal staining on kidney from adult mouse. C and D: Double-staining with X-gal and immunolabeling. E: Relative expression of nlacZ mRNA and L-WNK1 mRNA in proximal convoluted tubule, CTAL, and DCT. QRT-PCR results are expressed as percentage of nlacZ and L-WNK1 expression fixed at 100% in proximal convoluted tubule. Histograms represent means, and bars indicate the minima and maxima for six experiments (L-WNK1) or two experiments (nlacZ). Renal WNK1 expression. F: In situ hybridization with a probe specific for L-WNK1, with eosin counterstaining. G:In situ hybridization with a probe recognizing both L- and KS-WNK1, with eosin counterstaining. H: Immunolocalization of NCC and in situ hybridization of WNK1 (both L and KS isoforms). I: Immunolocalization of NKCC2 and in situ hybridization of WNK1 (both L and KS isoforms). J: Relative abundance of L- and KS-WNK1 mRNA along the length of the mouse nephron, as determined by QRT-PCR. Experiments were performed on glomerulus (Glom), proximal convoluted tubule (PCT), pars recta (PR), medullar thick ascending limb of Henle’s loop (MTAL), CTAL, DCT, connecting tubule (CNT), cortical collecting duct (CCD), and outer medullar collecting duct (OMCD) RNA extracts. Results (arbitrary U per mm tubule length) are expressed as means ± SE from four to six animals. Original magnifications, ×5 (A); ×10 (B); ×250 (C, D, H, I); ×100 (F, G).
Together, these results show that the L-WNK1-nlacZ transgene expression was strong in the distal part of the cortical thick ascending limb of Henle’s loop (CTAL) and the proximal part of the distal convoluted tubule (DCT1). This strong expression contrasted with the uniform low level of L-WNK1 expression previously reported7,14 and confirmed in this study by in situ hybridization with the exon 1-4 RNA probe (Figure 7F). Furthermore, in situ hybridization with a 3′ WNK1 RNA probe binding to both the L- and KS-WNK1 isoforms gave a strong hybridization signal restricted to the cortical kidney (Figure 7G), with no staining of the papilla (not shown). This specific cortical expression of the endogenous WNK1 gene co-localized with NCC (Figure 7H), but not with NKCC2 (Figure 7I), consistent with low levels of expression of the L-WNK1 isoform and expression of the KS-WNK1 isoform predominantly in the DCT.
The quantitative discordance between the production of β-galactosidase and the endogenous L-WNK1 mRNA may reflect the greater sensitivity of histochemical detection of β-galactosidase activity than of mRNA detection by in situ hybridization. We used real-time RT-PCR on isolated microdissected tubules to analyze in more detail endogenous WNK1 expression in the nephron. We assessed the relative abundance and renal distribution of the L- and KS-WNK1 isoforms, using primers amplifying either exons 1 and 2 or exons 4a and 5 (Figure 7J). A striking difference in distribution and abundance was shown in the mouse adult kidney, consistent with in situ hybridization results. Primers amplifying sequences corresponding to the L-WNK1 isoform revealed uniform, low-level expression along the nephron. In contrast, the KS-WNK1 isoform was present primarily in the DCT and, to a lesser extent, in the CTAL and the connecting tubule (CNT).
We analyzed this discordance further by investigating whether the specific pattern of transgene expression in the kidney could be accounted for by the use of other promoters. 5′RACE-PCR experiments on mouse kidney WNK1 cDNA, using a primer specific to exon 2, identified no sequences other than the proximal promoters (not shown). The amplification of sequences from exon 2 to 3 by QRT-PCR on microdissected tubules gave a similar profile along the length of the nephron as did the amplification of sequences from exons 1 to 2 (not shown). Thus, expression of the transgene could not be accounted for by the use of an alternative promoter.
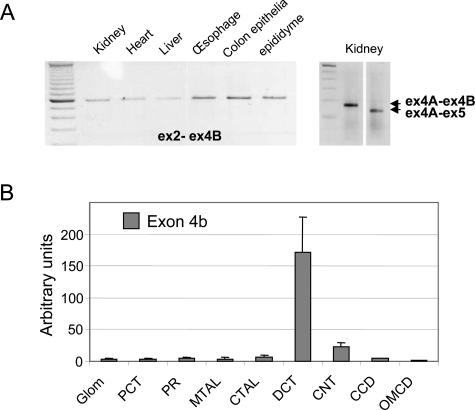
Finally, because the reporter gene was located on exon 2 in the BAC construct, we investigated whether cell-specific alternative splicing of the first four exons of L-WNK1 could occur. Alignment of an EST from 13-day mouse embryos (gi 26095199) with the L-WNK1 sequence revealed the presence of a 129-bp exon located between exons 4 and 5 (exon 4b) previously shown to be expressed in the renal cortex.14 Splicing between exons 2 and 4b (L-WNK1) was confirmed by RT-PCR on all tested mouse tissues (Figure 8A, left). In the kidney, RT-PCR also generated an amplicon between exons 4a and 4b (KS-WNK1) (Figure 8A, right). Nevertheless, the major product amplified from exons 4a to 5 was a 120-bp molecule corresponding to a transcript in which exon 4b was removed by splicing (Figure 8A, right). We then investigated the expression profile of the exon 4b-containing isoforms by QRT-PCR on mouse microdissected nephron (Figure 8B) and relative abundance of the L- and KS- exon 4b-containing isoforms by QRT-PCR on kidney. Exon 4b was present in both the L- and KS-WNK1 isoforms. However, it accounted for only ∼1/30th of the total amounts of both isoforms. Interestingly, this new exon contains an in-frame stop codon when spliced with exon 4 (in the L-WNK1 isoform) or with exon 4a (in the KS-WNK1 isoform). Exon 4b splicing may thus regulate WNK1 production. However, it is not conserved between mice and humans.
Figure 8.
A novel exon 4b alternatively spliced in L- and KS-WNK1. A: Agarose gel electrophoresis of RT-PCR products from mouse tissues. Amplified exons (ex) to which the primers bound and the corresponding amplified fragments are indicated by arrows. B: Quantification of WNK1 exon 4b-containing transcripts in mouse kidney by QRT-PCR on microdissected nephrons.
Discussion
We have demonstrated here the feasibility of a BAC transgenic approach for the detailed analysis of L-WNK1 expression during embryogenesis and adulthood. This strategy gives sensitive detection of a reporter gene with single cell resolution, which is often difficult to achieve for endogenous genes. For example, in vessels the histochemical detection of the lacZ gene product has proven to be more sensitive than the detection of Dll4 mRNA by in situ hybridization.27 Using this genomic reporter assay, we confirmed that the L-WNK1 isoform is produced in many organs in addition to the polarized epithelia in which it was first detected,15 including the cardiovascular system, in which strong, early expression was observed. The 172-kb genomic fragment containing the mWNK1 locus drove a pattern of expression in adult mice similar to that of the endogenous gene, analyzed by in situ hybridization, in all parts of the body. In the kidney, difference between the distribution of L-WNK1 mRNA along the nephron and β-galactosidase activity or lacZ mRNA could be observed. These results were replicated in four different transgenic lines and indicate that this BAC—containing the 47 kb upstream from the mWNK1 gene associated with the entire 34-kb intron 1 sequence and the first 11 kb of the 3′ flanking region—contains the regulatory elements required to confer a spatiotemporal expression pattern very similar to that of the endogenous gene. It is therefore a potentially useful, sensitive tool for analyzing the regulation of L-WNK1 expression in response to several physiological stimuli in vivo and for studying the consequences of changes in the spatial, temporal, and quantitative pattern of expression after the deletion of intron 1 of WNK1, as in FHHt-disease causing mutations.
One of our main findings was the expression of L-WNK1 in the cardiovascular system early in embryogenesis. Expression was detected as early as day 8.5 in the developing vasculature and primitive heart. It was strongest during the initial stages of heart development, consistent with a peak of expression during active periods of differentiation. Expression was observed throughout the myocardium but was particularly strong in the trabeculated walls of the ventricle. Cardiac trabeculations originate in the cardiogenic mesoderm and were initially thought to deliver oxygen and nutrients to the ventricles in the absence of the coronary vascular system. It has also been suggested that they act as contractile elements, maintaining ventricular wall stiffness, and serve to distribute the electrical impulse for contraction throughout the ventricles.28 In the adult transgenic mouse, we observed diffuse L-WNK1 expression, consistent with previous Northern blot results showing stronger expression in the heart than in other tissues. Despite high levels of WNK1 mRNA, convincing staining of heart with an anti-WNK1 antibody was not seen in a previous study.15 The antibody used recognizes a peptide encoded in exon 12. It is known that there is alternative splicing of WNK1.7,13,29 The major WNK1 form in the heart corresponds to a L-WNK1 transcript in which exon 12 was removed by splicing,7 which thus explains why it is not recognized by the antibody. Our results show that expression was not restricted to a particular subtype of cells. Further investigation is required to determine whether changes in WNK1 expression affect cell contractility or cardiac function. FHHt patients are not known to display any cardiac abnormality other than arterial hypertension, but it has yet to be determined whether the disease-causing mutations selectively modify expression of the L-WNK1 and/or the KS-WNK1 isoform. Mice lacking L-WNK1 were recently generated by means of a gene-trap strategy in embryonic stem cells.16 Embryos homozygous for the mutation died early in utero, before 13 days of development. Because cardiovascular defects are the most frequent cause of early death in utero in engineered mice,30 our findings suggest that L-WNK1 may be essential for embryonic cardiovascular development. WNK1 has been shown to activate the ERK5 mitogen-activated protein kinase cascade,31 a pathway implicated in multiple physiological functions, such as cell survival, proliferation, and differentiation; it is also essential for cardiac development, endothelial cell growth, and maintenance of vascular integrity during adulthood.32 In adult vessels, L-WNK1 was detected in endothelial cells and in vascular smooth muscle cells. Further investigation is required to determine whether this expression pattern could partly account for the low blood pressure of L-WNK1 +/−mice.
We also unexpectedly detected L-WNK1 in the cerebellum, in both embryos and adults, but almost no L-WNK1 was detected in the cerebral cortex. This molecule was essentially restricted to the granular layer and cerebellar Purkinje cells. In INS-1 pancreatic β-cell line, WNK1 has been co-localized with synaptotagmin 2 (Syt 2),33 a protein involved in the regulation of membrane trafficking and vesicle fusion via a calcium-sensing mechanism.34 According to Lee and colleagues,33 WNK1 selectively binds to and phosphorylates Syt 2, whereas WNK4 does not. This interaction may lead to the retention or insertion of proteins in the plasma membrane, thereby locally regulating ion balance. Our data demonstrate the strong expression of L-WNK1 in cerebellar Purkinje cells in which Syt 2 transcripts are very abundant from postnatal day 15 until adulthood, at the time of major synaptogenesis,35 suggesting that this molecule plays a specific role in these cells. In the cerebellum, Purkinje cells form synapses with parallel fibers that form strategic elements of its neuronal circuitry.36 During movement, tracking position and velocity are signaled by simple spike discharges from Purkinje cells.37 L-WNK1 may regulate ion handling and thus activity in Purkinje cells. Integrated studies of the morphological and functional consequences of WNK1 inactivation should therefore also focus in particular on movements controlled by the cerebellum.
The main clinical and biological features of FHHt and the detection of WNK4 and KS-WNK1 in the renal cortex have led to many studies investigating the pathophysiological effects of WNK1 and WNK4 on ion handling in the distal nephron. It has been demonstrated that WNK4 regulates diverse ion flux pathways by modulating NCC and ROMK1 activity and by regulating paracellular permeability.4 However, much less is known about the respective roles of L-WNK1 and KS-WNK1. Expression studies in Xenopus oocytes have suggested that L-WNK1 antagonizes the inhibitory effect of WNK4 on NCC,38 and that this inhibition depends on the catalytic activity of WNK1.39 In HEK cells, it has been demonstrated that L-WNK1 inhibits ROMK1, whereas KS-WNK1 reverses this inhibition.40 L-WNK1 has also been shown to bind the serum- and glucocorticoid-inducible protein kinase SGK1, activating this protein by increasing its phosphorylation, leading to activation of the epithelial sodium channel in Xenopus oocytes.10 It might be difficult to see how L-WNK1 could play a prominent role in the distal nephron because its mRNA is present in much smaller amounts than KS-WNK1 in this tissue7 and because KS-WNK1 may itself act as a dominant-negative regulator of L-WNK1.9,40 Nevertheless, a recent study demonstrates that L-WNK1 is up-regulated and KS-WNK1 is down-regulated in the kidney of rats fed with a low potassium diet. An experimental model making it possible to follow the production of each isoform separately would thus be very informative. However, although we used a BAC encompassing more than 170 kb of the mWNK1 locus, our analyses of transgene and endogenous L-WNK1 expression, characterized by in situ hybridization and quantitative RT-PCR on microdissected tubules, were not entirely concordant. BAC transgene expression, like endogenous L-WNK1 expression, was diffuse along the length of the nephron and in the glomerular capillaries but was clearly much stronger in the distal part of the CTAL and the proximal part of the DCT than endogenous L-WNK1 expression. The same profile was obtained for all four transgenic lines analyzed. Because this BAC provided the regulatory elements and the essential chromatin environment for correctly regulated expression in other tissues, the high levels of expression observed in the DCT and the distal part of the CTAL may be due to the absence of additional cis-acting elements involved in L-WNK1 expression in the kidney. We searched for conserved nongenic sequences upstream and downstream from the BAC ends and identified several sequences for future analysis as potential regulatory elements. However, it is also possible that transgene expression reflects a modification to L-WNK1 expression in response to stress conditions. An increase in the level of renal L-WNK1 transcripts has been observed in animals on a low-potassium diet.40 It remains unclear whether this up-regulation takes place specifically in the CTAL and DCT. Strong L-WNK1 expression in the DCT may inhibit ROMK1 and modulate potassium secretion. It may also abolish the inhibitory effect of WNK4 on NCC activity,38 thereby activating Na+ reabsorption.
Alternatively, given that the renal expression of the transgene resembles that of KS-WNK1, the discordance between endogenous L-WNK1 and transgene expression in the kidney may be due to an illegitimate interaction between the proximal promoters of L-WNK1 and the renal enhancer located upstream from exon 4a. This renal enhancer is highly active in renal epithelial cells.7 This illegitimate interaction may result from a regulation mechanism that is different in the context of the transgene. There may be an insulator between the L-WNK1 promoters and the renal enhancer. This insulator may be inactivated by methylation,41 a phenomenon known to occur in transgenes.42 Alternatively, the renal promoter and the next proximal promoter may be too close together, with no barrier element, in the case of head to tail insertion of multicopies of the transgene. This appears likely because our four transgenic lines had integrated 2 to 12 copies of the BAC transgene at the same chromosomal location.
Finally, this study demonstrated the physiological expression of an additional WNK1 isoform in mouse that is not present in human. Both L-WNK1 and KS-WNK1 may contain an alternatively spliced exon, exon 4b, and these alternatively spliced forms have been detected in the renal cortex.14 Exon 4b is 129 bp long and contains a premature stop codon, leading to a predicted protein of 459 residues, truncated after catalytic kinase subdomain VII. The corresponding L-WNK1 isoform therefore has no kinase activity. The inclusion of exon 4b just after exon 4a should lead to the production of a very short (41 residues) KS-WNK1 isoform. RT-PCR experiments showed that exon 4b-containing isoforms are produced in only small amounts. Exon 4b is not removed by splicing in only 3% of KS-WNK1 and L-WNK1 transcripts. We have demonstrated the occurrence of a similar rare alternative splicing event in the human L-WNK1 isoform.7 The use of an alternative splicing site in intron 3, 83 bp upstream from exon 4, leads to the introduction of a premature stop codon and the production of an enzyme with no catalytic activity. Thus, for both mouse and human WNK1 genes, expression may be regulated by both classical transcriptional means and by alternative splicing leading to the production of truncated isoforms. This mechanism may modulate WNK1 activity, depending on the physiological conditions.
In conclusion, we describe here an in vivo reporter assay based on the use of the sensitive and stable nlacZ reporter, for studies of the expression of WNK1 kinase domain-containing isoforms in transgenic mice. The pattern of transgene expression in embryos and adult mice was indistinguishable from that of the endogenous WNK1 gene in all parts of the body except the kidney, making this a sensitive model for studying the transcriptional regulation of WNK1. Using this model, we showed that kinase-containing WNK1 isoforms are produced early in development, particularly in the cardiovascular system, suggesting a possible unsuspected role in vascular development and blood pressure regulation. We also showed that the regulation of WNK1 kinase domain-containing isoforms might involve regulatory cis-acting elements outside the BAC sequence. These elements may repress L-WNK1 production in the distal nephron.
Supplementary Material
Acknowledgments
We thank Dominique Eladari for helpful technical advice; Julie Sappa for critical reading and editing of the manuscript; Dr. M. Knepper, National Institutes of Health, Bethesda, Maryland, for providing the NKCC2 antibody; Dr. J. Loffing, University of Lausanne, Lausanne, Switzerland, for providing anti-NCC and anti-β-ENaC antibodies; Dr. N.G. Copeland, National Cancer Institute, Frederick, Maryland, for providing E. coli strains EL 250, EL 350, PTamp, and PL452 plasmids.
Footnotes
Address reprint requests to Dr. Xavier Jeunemaitre, INSERM U772, College de France, 11, place Marcelin Berthelot, 75005 Paris, France. E-mail: xavier.jeunemaitre@college-de-france.fr.
Supported by grants from INSERM, the Fondation de France, the Agence Nationale pour la Recherche, the Ministry of Education and Research (fellowship to C.D.), and the University Pierre et Marie Curie (to C.D.).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Wilson F, Disse-Nicodème S, Choate K, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford D, Lipkin G, Achard J-M, Feely M, Dussol B, Berland Y, Unwin R, Simon D, Farfel Z, Jeunemaitre X, Lifton R. Mutations in WNK kinases reveal a novel mechanism of human hypertension. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- Hadchouel J, Delaloy C, Faure S, Achard JM, Jeunemaitre X. Familial hyperkalemic hypertension. J Am Soc Nephrol. 2006;98:421–428. doi: 10.1681/ASN.2005030314. [DOI] [PubMed] [Google Scholar]
- Yamauchi K, Rai T, Kobayashi K, Sohara E, Suzuki T, Itoh T, Suda S, Hayama A, Sasaki S, Uchida S. Disease-causing mutant WNK4 increases paracellular chloride permeability and phosphorylates claudins. Proc Natl Acad Sci USA. 2004;101:4690–4694. doi: 10.1073/pnas.0306924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Wilson FH, Lalioti M, Toka H, Qin H, Lifton RP. WNK kinases: molecular regulators of integrated epithelial ion transport. Curr Opin Nephrol Hypertens. 2004;13:557–562. doi: 10.1097/00041552-200409000-00012. [DOI] [PubMed] [Google Scholar]
- Wilson FH, Kahle KT, Sabath E, Lalioti MD, Rapson AK, Hoover RS, Hebert SC, Gamba G, Lifton RP. Molecular pathogenesis of inherited hypertension with hyperkalemia: the Na-Cl cotransporter is inhibited by wild-type but not mutant WNK4. Proc Natl Acad Sci USA. 2003;100:680–684. doi: 10.1073/pnas.242735399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- Delaloy C, Lu J, Houot AM, Disse-Nicodeme S, Gasc JM, Corvol P, Jeunemaitre X. Multiple promoters in the WNK1 gene: one controls expression of a kidney-specific kinase-defective isoform. Mol Cell Biol. 2003;23:9208–9221. doi: 10.1128/MCB.23.24.9208-9221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle KT, Wilson FH, Lifton RP. Regulation of diverse ion transport pathways by WNK4 kinase: a novel molecular switch. Trends Endocrinol Metab. 2005;16:98–103. doi: 10.1016/j.tem.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Subramanya AR, Yang CL, Zhu X, Ellison DH. Dominant-negative regulation of WNK1 by its kidney-specific kinase-defective isoform. Am J Physiol Renal Physiol. 2006;290:F619–F624. doi: 10.1152/ajprenal.00280.2005. [DOI] [PubMed] [Google Scholar]
- Xu BE, Stippec S, Chu PY, Lazrak A, Li XJ, Lee BH, English JM, Ortega B, Huang CL, Cobb MH. WNK1 activates SGK1 to regulate the epithelial sodium channel. Proc Natl Acad Sci USA. 2005;102:10315–10320. doi: 10.1073/pnas.0504422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naray-Fejes-Toth A, Snyder PM, Fejes-Toth G. The kidney-specific WNK1 isoform is induced by aldosterone and stimulates epithelial sodium channel-mediated Na+ transport. Proc Natl Acad Sci USA. 2004;101:17434–17439. doi: 10.1073/pnas.0408146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP. Human hypertension caused by mutations in WNK kinases. Science. 2001;293:1107–1112. doi: 10.1126/science.1062844. [DOI] [PubMed] [Google Scholar]
- Verissimo F, Jordan P. WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene. 2001;20:5562–5569. doi: 10.1038/sj.onc.1204726. [DOI] [PubMed] [Google Scholar]
- O’Reilly M, Marshall E, Speirs HJ, Brown RW. WNK1, a gene within a novel blood pressure control pathway, tissue-specifically generates radically different isoforms with and without a kinase domain. J Am Soc Nephrol. 2003;14:2447–2456. doi: 10.1097/01.asn.0000089830.97681.3b. [DOI] [PubMed] [Google Scholar]
- Choate KA, Kahle KT, Wilson FH, Nelson-Williams C, Lifton RP. WNK1, a kinase mutated in inherited hypertension with hyperkalemia, localizes to diverse Cl−-transporting epithelia. Proc Natl Acad Sci USA. 2003;100:663–668. doi: 10.1073/pnas.242728499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, BeltrandelRio H, Buxton EC, Edwards J, Finch RA, Friddle CJ, Gupta A, Hansen G, Hu Y, Huang W, Jaing C, Key BW, Jr, Kipp P, Kohlhauff B, Ma ZQ, Markesich D, Payne R, Potter DG, Qian N, Shaw J, Schrick J, Shi ZZ, Sparks MJ, Van Sligtenhorst I, Vogel P, Walke W, Xu N, Zhu Q, Person C, Sands AT. Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci USA. 2003;100:14109–14114. doi: 10.1073/pnas.2336103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press,; 1994 [Google Scholar]
- Nijenhuis T, Hoenderop JG, Loffing J, van der Kemp AW, van Os CH, Bindels RJ. Thiazide-induced hypocalciuria is accompanied by a decreased expression of Ca2+ transport proteins in kidney. Kidney Int. 2003;64:555–564. doi: 10.1046/j.1523-1755.2003.00128.x. [DOI] [PubMed] [Google Scholar]
- Kim GH, Ecelbarger CA, Mitchell C, Packer RK, Wade JB, Knepper MA. Vasopressin increases Na-K-2Cl cotransporter expression in thick ascending limb of Henle’s loop. Am J Physiol. 1999;276:F96–F103. doi: 10.1152/ajprenal.1999.276.1.F96. [DOI] [PubMed] [Google Scholar]
- Loffing J, Pietri L, Aregger F, Bloch-Faure M, Ziegler U, Meneton P, Rossier BC, Kaissling B. Differential subcellular localization of ENaC subunits in mouse kidney in response to high- and low-Na diets. Am J Physiol. 2000;279:F252–F258. doi: 10.1152/ajprenal.2000.279.2.F252. [DOI] [PubMed] [Google Scholar]
- Eichmann A, Yuan L, Breant C, Alitalo K, Koskinen PJ. Developmental expression of pim kinases suggests functions also outside of the hematopoietic system. Oncogene. 2000;19:1215–1224. doi: 10.1038/sj.onc.1203355. [DOI] [PubMed] [Google Scholar]
- Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- Virlon B, Cheval L, Buhler JM, Billon E, Doucet A, Elalouf JM. Serial microanalysis of renal transcriptomes. Proc Natl Acad Sci USA. 1999;96:15286–15291. doi: 10.1073/pnas.96.26.15286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissant A, Lourdel S, Baillet S, Paulais M, Marvao P, Teulon J, Imbert-Teboul M. Heterogeneous distribution of chloride channels along the distal convoluted tubule probed by single-cell RT-PCR and patch clamp. Am J Physiol. 2004;287:F1233–F1243. doi: 10.1152/ajprenal.00155.2004. [DOI] [PubMed] [Google Scholar]
- Benedito R, Duarte A. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr Patterns. 2005;5:750–755. doi: 10.1016/j.modgep.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544–556. doi: 10.1038/nrg843. [DOI] [PubMed] [Google Scholar]
- Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Kruzynska-Frejtag A, Kneer PL, Machnicki M, Koushik SV. What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis. 2003;35:1–21. doi: 10.1002/gene.10152. [DOI] [PubMed] [Google Scholar]
- Xu BE, Stippec S, Lenertz L, Lee BH, Zhang W, Lee YK, Cobb MH. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem. 2004;279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Lee JD. Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J Mol Med. 2004;82:800–808. doi: 10.1007/s00109-004-0602-8. [DOI] [PubMed] [Google Scholar]
- Lee BH, Min X, Heise CJ, Xu BE, Chen S, Shu H, Luby-Phelps K, Goldsmith EJ, Cobb MH. WNK1 phosphorylates synaptotagmin 2 and modulates its membrane binding. Mol Cell. 2004;15:741–751. doi: 10.1016/j.molcel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Chapman ER. Synaptotagmin: a Ca(2+) sensor that triggers exocytosis? Nat Rev Mol Cell Biol. 2002;3:498–508. doi: 10.1038/nrm855. [DOI] [PubMed] [Google Scholar]
- Berton F, Iborra C, Boudier JA, Seagar MJ, Marqueze B. Developmental regulation of synaptotagmin I, II, III, and IV mRNAs in the rat CNS. J Neurosci. 1997;17:1206–1216. doi: 10.1523/JNEUROSCI.17-04-01206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CM, Miyamoto H, Huang RH: The mouse cerebellum from 1 to 34 months: parallel fibers. Neurobiol Aging 2005, [Epub ahead of Print] [DOI] [PubMed] [Google Scholar]
- Roitman AV, Pasalar S, Johnson MT, Ebner TJ. Position, direction of movement, and speed tuning of cerebellar Purkinje cells during circular manual tracking in monkey. J Neurosci. 2005;25:9244–9257. doi: 10.1523/JNEUROSCI.1886-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Angell J, Mitchell R, Ellison DH. WNK kinases regulate thiazide-sensitive Na-Cl cotransport. J Clin Invest. 2003;111:1039–1045. doi: 10.1172/JCI17443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CL, Zhu X, Wang Z, Subramanya AR, Ellison DH. Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. J Clin Invest. 2005;115:1379–1387. doi: 10.1172/JCI22452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazrak A, Liu Z, Huang CL. Antagonistic regulation of ROMK by long and kidney-specific WNK1 isoforms. Proc Natl Acad Sci USA. 2006;103:1615–1620. doi: 10.1073/pnas.0510609103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Sasaki H. Imprinting and looping: epigenetic marks control interactions between regulatory elements. Bioessays. 2005;27:1–4. doi: 10.1002/bies.20171. [DOI] [PubMed] [Google Scholar]
- Schumacher A, Koetsier PA, Hertz J, Doerfler W. Epigenetic and genotype-specific effects on the stability of de novo imposed methylation patterns in transgenic mice. J Biol Chem. 2000;275:37915–37921. doi: 10.1074/jbc.M004839200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.