Abstract
To obtain a more complete protein profile of the airspace milieu in acute respiratory distress syndrome (ARDS) and to identify new mediators, we analyzed bronchoalveolar lavage fluid (BALF) by shotgun proteomics. Using BALF from three patients, we identified a total of 870 different proteins, a nearly 10-fold increase from previous reports. Among the proteins identified were known markers of lung injury, such as surfactant, proteases, and serum proteins. We also identified several biologically interesting proteins not previously identified in patients with ARDS, including insulin-like growth factor-binding protein-3 (IGFBP-3). Because of the known role of IGFBP-3 in regulating cell survival, we measured IGFBP-3 levels by enzyme-linked immunosorbent assay in ARDS BALF. Normal controls had low levels of IGFBP-3, whereas patients with early ARDS had a significant increase in IGFBP-3. The IGF pathway, acting through the type 1 IGF-receptor, repressed apoptosis of lung fibroblasts but not lung epithelial cells. Furthermore, depletion of IGF in ARDS BALF led to enhanced fibroblast apoptosis. Our data suggest that the IGFBP-3/IGF pathway is involved in the pathogenesis of lung injury, illustrating the power of shotgun proteomics to catalog proteins present in complex biological fluids, such as BALF, from which hypotheses can be developed and tested.
Acute respiratory distress syndrome (ARDS), first described in 1967 by Ashbaugh and colleagues,1 remains an important cause of morbidity and mortality in critically ill patients. ARDS is characterized by an acute pulmonary inflammatory process with epithelial apoptosis and interstitial and intra-alveolar edema, followed by fibroblast proliferation, migration, and fibrosis. The diagnosis of ARDS is based on clinical and radiographical criteria, including acute onset, bilateral infiltrates on chest radiograph, absence of congestive heart failure, and hypoxemia.2 This consensus definition has improved the standardization of clinical research and trials; however, it does not take into account the cause or mechanism of disease.
Much work has focused on the identification of humoral or cellular biological markers of ARDS in hopes that such markers may provide insight into the mechanisms of ARDS and improve the prediction of ARDS in high risk patients and prediction of outcome in ARDS patients.3 To date, no single protein marker identified by traditional laboratory methods has demonstrated the specificity or sensitivity to serve as a reliable predictor of outcome. However, new proteomic methods provide the opportunity to assess the protein profile of a sample that is independent of investigator’s biases and thus has the potential of identifying unsuspected mediators or pathways involved in lung injury.
As a screening strategy to define the bronchoalveolar lavage fluid (BALF) proteome from ARDS patients, we used “shotgun proteomics,” consisting of digestion of proteins in BALF followed by strong-cation exchange fractionation of the peptide mixture and microcapillary-high performance liquid chromatography electrospray ionization tandem mass spectrometry analysis, and then computerized data processing.4 Using strict criteria for matching peptide tandem mass spectra to sequences in a database,5,6 we identified from three patients a total of 897 proteins, of which 79 were identified in all three patients. We selected several of the identified proteins for further testing based on their known functions and potential relevance to lung injury. Expression levels of the candidate proteins were analyzed by enzyme-linked immunosorbent assay (ELISA) in a large sample set of ARDS BALF. Notable among the results, we found insulin-like growth factor (IGF)-binding protein-3 (IGFBP-3) and IGF expression levels correlated with progression of ARDS. Furthermore, we showed that the IGF pathway regulates apoptosis of lung fibroblasts, but not lung epithelial cells, suggesting that the IGF pathway may contribute to the fibroproliferative response in ARDS.
Materials and Methods
Patient Population
The protocol was approved by the Institutional Review Board, University of Washington. Written informed consent was obtained from the patient or responsible relative before patients were entered into the study. Patients with acute lung injury undergoing bronchoscopy for suspected ventilator-associated pneumonia were included in the study as the initial index patients (Table 1).
Table 1.
Clinical Characteristics of ARDS Patients
| Index patients*
|
Retrospective cohort
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | At Risk | ARDS summary | Day 1 | Day 3 | Day 7 | Day 14 | |
| Age | 28 | 49 | 59 | 47 ± 15 | 43 ± 16 | ||||
| Risk factor | |||||||||
| Sepsis | Yes | 3 | 23 | ||||||
| Trauma | Yes | 5 | 25 | ||||||
| Other | Yes | 0 | 13 | ||||||
| Apache score | 16 | 22 | 20 | 19 ± 8 | 22 ± 7 | 22 ± 6 | 21 ± 9 | 23 ± 8 | 19 ± 3 |
| PaO2/FiO2 | 184 | 271 | 290 | 226 ± 83 | 162 ± 61 | 151 ± 55 | 162 ± 58 | 189 ± 71 | 178 ± 43 |
| Static compliance | nd | nd | nd | 40 ± 10 | 37 ± 13 | 41 ± 15 | 37 ± 10 | 28 ± 9 | 35 ± 17 |
| % BAL volume recovered | 38 | 71 | 33 | 57 ± 11 | 50 ± 14 | 54 ± 12 | 50 ± 15 | 43 ± 11 | 44 ± 19 |
| VAP? | No | Yes | No | ||||||
None of the index patients had a diagnosis of pneumonia as a risk factor for acute lung.
In addition, we retrospectively analyzed specimens obtained as part of the University of Washington Specialized Center of Research program in acute lung injury. All patients admitted to the Medical and Surgical Intensive Care Units of Harborview Medical Center between January 1994 and November 1997 were screened prospectively to identify patients who met criteria for predetermined risk criteria for trauma or sepsis “at risk”7 or with established ARDS.7–9 Patients at risk for ARDS did not meet either radiographical or oxygenation criteria for ARDS. Patients were screened prospectively for the onset of ARDS using the following criteria: 1) PaO2/FiO2 < 150 mm Hg or < 200 mm Hg on ≥ 5 cm of H2O positive end-expiratory pressure, 2) diffuse parenchymal infiltrates, 3) pulmonary artery wedge pressure < 18 mm Hg or no clinical evidence of congestive heart failure, and 4) no other obvious explanation for these findings. All patients with ARDS met the criteria of the American-European Consensus Conference definition.2 Day 1 was defined as the first 24 hours after meeting the above criteria for ARDS. The clinical characteristics of the patient groups are shown in Table 1 and have been previously described.7–9 As controls, BALF was obtained from six normal volunteers.
Bronchoalveolar lavage (BAL) was performed as previously described.7–9 Briefly, five separate 30-ml aliquots of 0.89% sterile saline were instilled into the right middle lobe or lingula. The BAL recovery averaged 75 ml (49% return) and was not statistically different between the different ARDS groups by one-way analysis of variance (P > 0.05). BALF was centrifuged immediately after collection, and cell-free supernatants were aliquoted into polypropylene tubes and stored at −70°C. Total protein measurements were made on aliquots of supernatants using a modified Lowry method.10
Peptide Separation and Purification
BALF proteins were concentrated by ice-cold acetone precipitation. BALF containing 2 mg of protein underwent digestion with trypsin (20 μg, sequencing grade; Promega, Madison, WI) overnight at 37°C to allow complete digestion. To prepare for strong-cation exchange chromatography and to reduce the salt concentration, the resulting peptide solutions were diluted eightfold with running buffer (5 mmol/L KH2PO4, 25% acetonitrile, pH 3), and their pH was reduced to ∼2.9 with phosphoric acid (H3PO4). The peptide solutions were passed over a 2.1 × 200 mm, 5-μm particle, 300-Å pore Polysulfoethyl A column (PolyLC; Columbia, MD), washed with running buffer, and then eluted with a 50-minute biphasic gradient of 0 to 25% elution buffer (running buffer plus 350 mmol/L potassium chloride) in 0 to 30 minutes followed by 25 to 100% elution buffer in 30 to 50 minutes. Flow rate was constant at 0.2 ml/minute. Sixteen 2-minute (0.4-ml) fractions were collected. Fractions from strong-cation exchange chromatography were completely dried down in a Speed-Vac (Thermo-Savant, Milford, MA) and redissolved in 0.1% trifluoroacetic acid. To desalt, fractions were loaded onto Oasis mixed-mode cation-exchange cartridges (Waters, Milford, MA), washed with 0.1% tri-fluoroacetic acid, and eluted with 0.1% trifluoroacetic acid, 80% acetonitrile solution. The samples were again dried down and redissolved in 0.2% acetic acid and transferred to autosampler vials for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.8,23 Briefly, an LCQ DECA ion trap mass spectrometer (Thermo Finnigan, San Jose, CA) outfitted with a microelectrospray source (Brechbuehler, Houston, TX) and an HP1100 solvent delivery system (Agilent, Palo Alto, CA) was used to analyze the samples. The samples were automatically delivered by a FAMOS autosampler (LC Packings, San Francisco, CA) to a 100-μm internal diameter, fused silica capillary precolumn packed with 2 cm of 200-Å pore-size Magic C18AQ material (Michrom Bioresources, Auburn, CA) as described elsewhere.11 The samples were washed with solvent A (0.1% formic acid, 5% acetonitrile) on the precolumn and then eluted with a gradient of 10 to 35% solvent B (100% acetonitrile) over 128.5 minutes to a 75-μm × 14-cm fused silica capillary column packed with 100-Å pore-size Magic C18AQ material and then into the mass spectrometer at a constant column-tip flow rate of ∼300 nL/minute. Peptides entering the mass spectrometer were selected for collision induced dissociation by data-dependent methods, and resultant tandem mass spectra were used to obtain protein matches.
Protein Identification Strategies
SEQUEST12 was used to screen tandem mass spectra for matches to peptide sequence by searching against the human International Protein Index database (European Bioinformatics Institute, Cambridge, UK). PeptideProphet and ProteinProphet were used to verify correctness of peptide and protein assignments,5 respectively, and those that displayed Prophet scores of >0.9 were considered identified. To compare different experiments, data were imported into SBEAMS, a relational database management systems that allows comparison across multiple experiments.13 Gene Ontology classifications were used for functional annotations of described genes.14
ELISA
Cytokine measurements were performed in duplicate by ELISA using commercially available kits (IGFBP-3 and heparin-binding EGF-like growth factor (HB-EGF) ELISA; R&D Systems, Minneapolis, MN, and total and free IGF ELISA; Diagnostic Standards Laboratory, Webster, TX). Human serum albumin, β2-microglobulin (Alpha Diagnostic International, Inc., San Antonio, TX), surfactant-D and Clara cell protein (Biovendor LLC, Candler, NC), and fibrinogen (DiaPharma Group, Inc., West Chester, OH) were also detected with commercially available ELISA kits.
Concentrations were extrapolated from simultaneously run standard curves. Differences between experimental conditions and normal controls were assessed with the Mann-Whitney Test using VasserStats software. The Spearman rank order correlation coefficient was determined for ELISA concentrations and total protein concentrations (VasserStats). All tests were two-tailed, and P values of <0.05 were considered significant.
Western Blot Analysis
To detect proteolytic fragments of IGFBP-3, equal volumes of BALF samples were separated by 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to Immobilon, and blocked for 1 hour. Blots were incubated with polyclonal IGFBP-3 antibody (Diagnostic Standards Laboratory), which recognizes the major proteolytic fragments of IGFBP-3, for 1 hour, followed by peroxidase-conjugated secondary antibody for 1 hour, and then developed with ECL. To quantitate relative band intensities, gels were captured with Photoshop (version 7; Adobe Systems Inc., San Jose, CA) and then imported into ImageJ (version 1.30; National Institutes of Health) for analysis.15
Apoptosis Assay
Primary normal human lung fibroblasts (Clonetics, Cambrex Bioscience, Rockland, ME), primary distal lung human epithelial cells (Clonetics) or A549 (American Type Culture Collection, Manassas, VA) were seeded in a 96-well tissue culture plate (2 × 104 cells/well) overnight and serum-starved for 24 hours. Function blocking antibody to the human type 1 IGF receptor (A12), a generous gift from ImClone Systems (New York, NY),16,17 was added to media at indicated concentration for 24 hours. In some experiments, soluble Fas ligand (Alexis Biochemicals, Lausen, Switzerland) or IGFBP-3 (R&D Systems) was added to cells. To assess the contribution of IGF to fibroblast survival in ARDS, BALF was diluted 1:10 with Dulbecco’s modified Eagle’s medium and then incubated with a neutralizing polyclonal antibody (5 ng/ml) to human IGF-I (R&D Systems) or preimmune goat serum for 30 minutes at 4°C before incubation with normal human lung fibroblast. After 48 hours, apoptosis was measured. To prevent detached cells from being aspirated, plates were centrifuged at 200 × g for 10 minutes, and apoptosis was measured using the Cell Death Detection ELISA-plus System (Roche Applied Science, Penzberg, Germany), which detects cytosolic histone-complexed DNA fragments. All experiments were done in triplicate and repeated at least twice. The data are reported as the mean absorbance of triplicate wells mean ± SE or as apoptosis index, defined as the ratio of the mean absorbance of triplicate wells in the experimental condition OD405 nm/control (media alone) OD405 nm.
Results
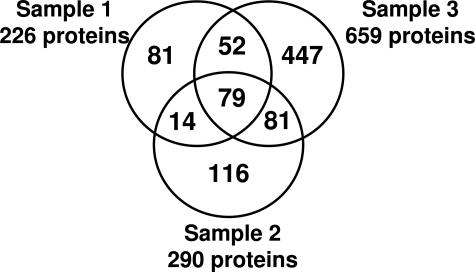
BALF samples from three patients with acute lung injury were analyzed by shotgun proteomics, and a total of 870 unique proteins was identified (downloadable data, including spectra and searchable SEQUEST files, are available at http://www.peptideatlas.org/contributors, authors L.M. Schnapp and S. Donohoe). The numbers of identifications from individual samples were 226, 291, and 659 (Figure 1). Of these, 79 proteins were identified in all three samples (Supplementary Table 1 at http://ajp.amjpathol.org). These identifications represent approximately 10-fold increase in the proteins previously identified in BALF.18
Figure 1.
Summary of protein identifications from three ARDS BALF samples following trypsin digest-cation exchange fractionation.
Our approach identified similar classes of proteins to those previously reported using two-dimensional electrophoresis (2DE).18 Of the 79 proteins common to all three patients, proteins from all cellular compartments were identified, including membrane proteins, cytosolic proteins, nuclear proteins, and cytoskeletal proteins, as well as extracellular and secreted proteins. As expected, we identified albumin in all three samples.19 However, the number of peptides corresponding to albumin varied widely in the patients ranging from 1 to 23 to 79, suggesting that the degree of serum protein leakage varied widely even among these three patients with clinically diagnosed ARDS. We also found extensive coverage of other abundant serum proteins, such as ceruloplasmin (average number of peptides: 13) and fibrinogen α chain (average number of peptides: 117), and other acute phase reactant proteins, such as α1 chymotrypsin (average number of peptides: 31), α2-HS-glycoprotein (average number of peptides: 58), and anti-trypsin inhibitor (average number of peptides: 77). We also identified a number of intracellular proteins, as has been previously reported in lung BAL,18 presumably due to increased cellular turnover and death in lung compartment during lung injury. We confirmed the presence of several serum proteins (albumin, fibrinogen, and β2-microglobulin) and pulmonary proteins (surfactant D and Clara cell protein) by ELISA. Surfactant A2 was also identified, as previously reported by 2DE.18,20 In addition, surfactant B2 (two patients) and surfactant D (one patient) were also identified here but were not found in previous 2DE analysis of BALF. Previous reports speculated that 2DE failed to identify surfactant B2, because of its hydrophobicity, and surfactant D, because of its relative underexpression compared to surfactant A2. Representative spectra are shown in Figure 2.
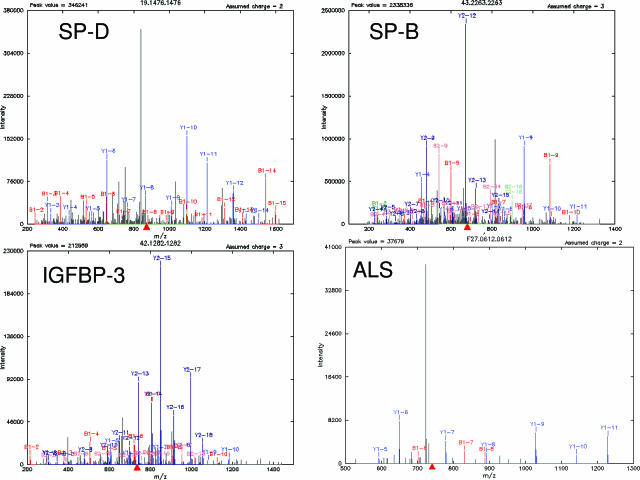
Figure 2.
Representative MS/MS pattern of peptides belonging to surfactant proteins B and D, IGFBP-3, and ALS with various b (red) and y (blue) ions indicated. Green, unable to distinguish between b and y ions. The MS/MS spectra were searched against protein databases leading to identification of indicated proteins.
From the long list of proteins identified by LC-MS/MS shotgun proteomics, we decided to focus on secreted proteins, because they may represent mediators of lung injury. This category included identifications of pre-B-cell colony-enhancing factor in two patients, HB-EGF in one patient and IGFBP-3 and the acid labile subunit (ALS) in two patients, both components of the IGF signaling complex (Figure 2). Pre-B-cell colony-enhancing factor was recently described as an inhibitor of apoptosis that was expressed by neutrophils from septic patients.21 Previous reports showed elevated levels of IGFBP-3 in BALF from patients with idiopathic pulmonary fibrosis (IPF)22,23 and sarcoidosis.24 In contrast, HB-EGF has not been previously associated with acute lung injury.
Validation of Changes in Secreted Proteins in a Large ARDS BALF Sample Set
Proteomics results do not distinguish between reproducible changes and sampling variability during the comparison of data from three different patients. To assess the potential significance of secreted BALF proteins identified by LC-MS/MS shotgun proteomics, we measured expression levels HB-EGF, IGFBP-3, and IGF-I by ELISA in a large BALF sample set that includes patients at different time points in ARDS progression. BALF samples from normal subjects (n = 6), patients at risk for development of ARDS (n = 8), and established ARDS at day 1 (n = 26), day 3 (n = 20), day 7 (n = 10), and day 14 (n = 5) were analyzed.
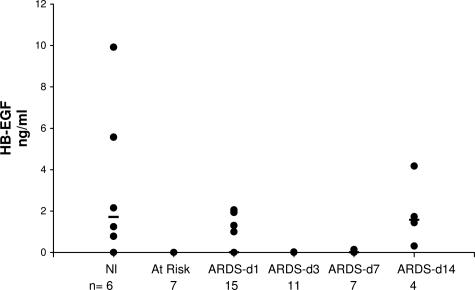
HB-EGF is a potent mitogen and chemotactic factor for fibroblasts.25 Thus, we hypothesized that it might play a role in the fibroproliferative response during acute lung injury. However, ELISA results revealed very low levels of HB-EGF in ARDS BALF and in normal BALF (Figure 3). Furthermore, we did not observe a correlation between HB-EGF levels and progression of lung injury in ARDS. Failure to detect changes in HB-EGF levels in ARDS BALF does not necessarily preclude a role for HB-EGF in lung injury. For instance lack of correlation by ELISA may be due to complex tissue distribution of the multiple forms of HB-EGF26 making it inaccessible in BALF. However, these data illustrate the importance of independently verifying proteins identified by any proteomic screen.
Figure 3.
HB-EGF protein concentrations in BAL from normal volunteers, patients at-risk for ARDS, and patients with established ARDS studied at sequential times. HB-EGF was quantified by human HB-EGF ELISA kit (R&D Systems) per the manufacturer’s direction. The concentration of HB-EGF (black circles) in the samples was determined by interpolation from the standard curve generated with recombinant HB-EGF. All samples were run in duplicate and repeated at least twice. Median (black bars) values are indicated.
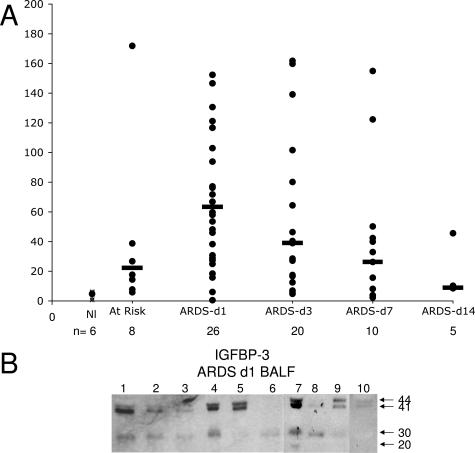
Because of its role in regulating cell survival,27 IGFBP-3 is also a potential candidate protein relevant to the pathogenesis of ARDS. While we found very low levels of IGFBP-3 in BALF from normal controls by ELISA, we detected a marked increase in IGFBP-3 concentration in patients at risk for ARDS and in those with established ARDS (Figure 4A). Because earlier work demonstrated elevated levels of IGFBP-3 in BALF from patients with IPF and sarcoidosis,23,24 we initially speculated that IGFBP-3 would be elevated in late ARDS (day 7 onward), when fibroblast proliferation is a prominent histological feature. In contrast, we found the highest levels early in ARDS (days 1 and 3), with levels decreasing as the disease progressed (Figure 4A). This is consistent with recent evidence that fibroblast activation occurs early in disease.28
Figure 4.
A: IGFBP-3 protein concentrations in BAL from normal volunteers, patients at-risk for ARDS, and patients with established ARDS studied at sequential times. IGFBP-3 was quantified by ELISA kit (R&D Systems) per the manufacturer’s direction. The concentration of IGFBP-3 (black circles) in the samples was determined by interpolation from the standard curve generated with recombinant IGFBP-3. All samples were run in duplicate and repeated at least twice. Median (black bars) values are indicated. All time points were significantly different (P < 0.05) from controls. B: IGFBP-3 immunoblots from 10 different ARDS day 1 BALF. Equal volumes of BALF were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. IGFBP-3 fragments were detected by a polyclonal antibody to IGFBP-3. Intact IGFBP-3 doublet at 41 and 44 kd and the 30- and 20-kd proteolytic fragments are indicated by arrows.
Proteolysis of IGFBP-3 decreases its ability to bind IGF, thereby increasing the bioavailability of the ligand.29 In addition, proteolytic fragments can have independent biological activity.30 However, the IGFBP-3 ELISA does not discriminate between full-length IGFBP-3 and proteolytic fragments. To evaluate the proteolysis of IGFBP-3, we analyzed day 1 and day 3 BALF by Western analysis using an antibody that recognizes the major proteolytic fragments (Figure 4B). The proportion of intact IGFBP-3 to total IGFBP-3 immunoreactivity was determined by densitometry. In at-risk and ARDS day 1 BALF, the majority of IGFBP-3 was present as the intact 41- and 44-kd doublet (at-risk: 53% ± 13; ARDS day 1: 66% ± 14, respectively); the 30-kd fragment was the major proteolytic fragment observed in the samples. Thus, the majority of IGFBP-3 is intact at the time in which concentrations are the highest.
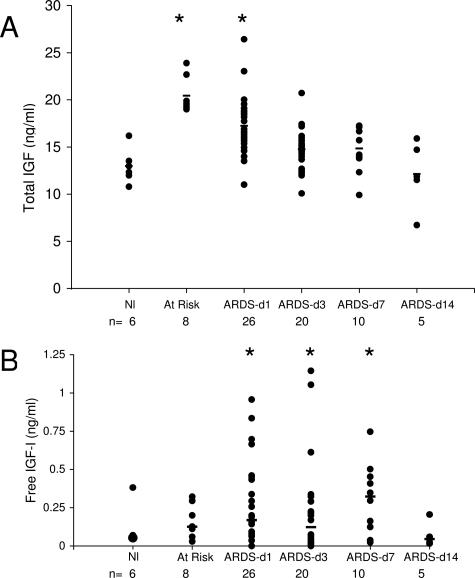
IGFBP-3 is the major binding protein of IGF-I and is bound in a 1:1:1 molar ratio with IGF and the ALS. Although we did not detect IGF in LC-MS/MS proteomics analysis, failure to identify a protein from complex peptide mixtures such as BALF by this screening method does not necessarily indicate absence of the protein.31 However, we subsequently detected IGF in the three index samples by ELISA (free IGF: 0.1, 0.05, and 0.04 ng/ml). In addition, we detected ALS by LC-MS/MS (Figure 2), adding further evidence for a role for the IGF/IGFBP-3 axis in lung injury. Because the ratio of IGF to IGFBP-3 is an important factor in regulating the bioactivity of IGF-I in BALF, we assessed total IGF protein levels by ELISA. The levels of IGF were similar to those observed for IGFBP-3; ie, low levels in normal controls, increased in at-risk and early ARDS patients (day 1 and day 3), and decreased levels in late ARDS patients (Figure 5A). Because bioactivity of IGF is determined by the unbound or free IGF, we also measured free IGF-I in BALF (Figure 5B). As expected, the levels of free IGF were significantly lower than total IGF. Interestingly, the free levels of IGF were elevated later in disease than total IGF. This may be due to changes in other members of the IGFBP family. While it is possible that changes in expression levels simply reflect changes in capillary permeability and serum exudation, this is less likely for several reasons. We found that the majority of change in IGFBP-3 levels could not be accounted for by changes in BALF total protein, with only a fair correlation (r2 = 0.39). Furthermore, there was no correlation between total or free IGF and total protein (r2 = 0.04 and 0.5, respectively, P > 0.05). In addition, measurements of other cytokines and growth factors from the same BALF samples show distinct patterns of expression.32,33
Figure 5.
Total IGF (A) and free IGF (B) protein concentrations (black circles) in BAL from normal volunteers, patients at-risk for ARDS, and patients with established ARDS studied at sequential times. Total and free IGF was quantified by ELISA kit (Diagnostic Standards Laboratory Systems) per the manufacturer’s direction. The concentration of IGF in the samples was determined by interpolation from the standard curve generated with recombinant IGF. All samples were run in duplicate and repeated at least twice. Median (black bars) values are indicated. *, Time points significantly different (P < 0.05) from controls.
IGF/IGF-I Receptor Pathway Regulates Lung Fibroblast but Not Epithelial Cell Apoptosis
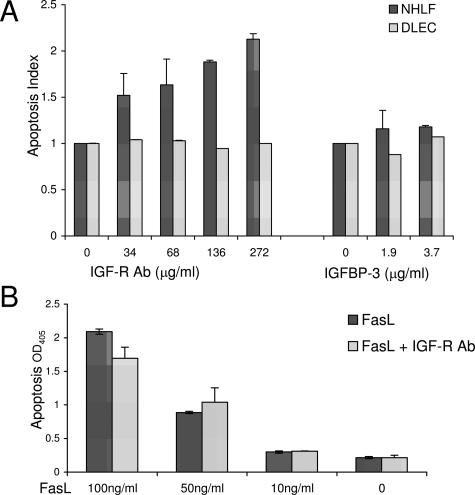
Apoptosis in the lung plays an important role in the development and resolution of acute lung injury. Initial epithelial damage and apoptosis occurs early in ARDS. At the same time, fibroblast activation is thought to occur,28,34 setting the stage for the later fibroproliferative phase of ARDS. In addition, apoptosis of connective tissue cells, such as fibroblasts, may be necessary for injury to resolve. Elevated levels of IGFBP-3 and IGF-I were found in at-risk patients and those with early ARDS, when epithelial damage and death occur. Addition of a blocking antibody to the type 1 IGF receptor (IGF-IR) induced a dose-dependent increase in apoptosis of primary human lung fibroblasts but not primary lung epithelial cells or the human macrophage cell line THP-1 under conditions of serum starvation (Figure 6A and data not shown). Because the Fas pathway is an important contributor to epithelial cell apoptosis in ARDS BALF,35 we asked whether the IGF pathway might influence Fas-mediated apoptosis. However, treatment with IGF-IR antibody did not alter Fas-induced apoptosis of epithelial cells or fibroblasts (Figure 6B and data not shown). Because IGFBP-3 can signal independently of IGF-IR,30,36 we also examined the effect of IGFBP-3 on fibroblast and epithelial cell apoptosis. In contrast to reports showing that IGFBP-3 induced apoptosis in certain cells,37 we did not observe increased apoptosis in fibroblasts or epithelial cells treated with increasing doses of IGFBP-3 (Figure 6). Together, these data suggest that IGF acts through IGF-IR selectively to promote fibroblast survival, with the potential to modify matrix remodeling and repair in acute lung injury.
Figure 6.
Increased apoptosis of primary normal human lung fibroblasts treated with IGF-IR antibody. A: Normal human lung fibroblast or distal lung epithelial cells were serum-starved overnight and then incubated with indicated concentration of IGF-IR antibody or IGFBP-3 overnight. B: A549 cells were serum-starved and incubated with IGF-IR antibody (136 μg/ml) with or without the indicated concentration of Fas ligand. Apoptosis was measured by Cell Death ELISA-plus ELISA (Roche Applied Science) per the manufacturer’s directions. All experiments were done in triplicate and repeated at least twice. Apoptosis index is defined as the ratio of experimental condition OD405 nm:control (media alone) OD405 nm.
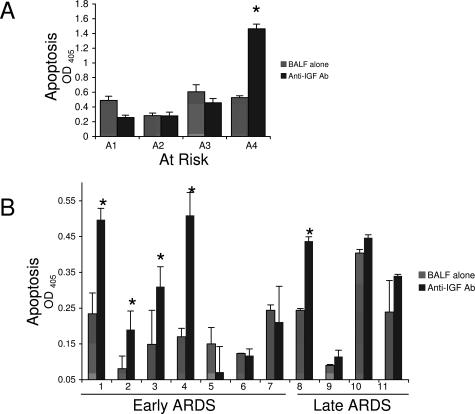
Finally, to determine directly the contribution of the IGF pathway to fibroblast survival during ARDS, we examined fibroblast apoptosis following incubation with ARDS BALF pretreated with a neutralizing antibody to IGF or with preimmune serum (Figure 7). In BALF from early ARDS (<7 days), four of seven samples showed a significant increase in fibroblast apoptosis following IGF neutralization. In BALF from late ARDS (>7 days), one of four samples showed a significant increase in fibroblast apoptosis following IGF neutralization. In BALF from at-risk patients, one of four samples showed a significant increase in fibroblast apoptosis following IGF neutralization (Figure 7A). Thus, the data suggest that in vivo concentrations of IGF contribute to fibroblast survival during many cases of acute lung injury, particularly in early ARDS.
Figure 7.
IGF mediates fibroblast prosurvival signal in ARDS BALF. Fibroblast apoptosis was measured 48 hours following incubation of normal human lung fibroblast with BALF with or without IGF neutralizing antibody (5 ng/ml) from at-risk patients (A) or ARDS patients (B). Apoptosis was measured by Cell Death ELISA-plus ELISA (Roche Applied Science) per the manufacturer’s directions. All experiments were done in triplicate and repeated at least twice. Data are reported as the mean OD415 nm ± SD. *Significant difference (P< 0.05) from BALF alone.
Discussion
Protein Database of BALF
Using shotgun proteomics we identified the largest set of BALF proteins in acute lung injury patients to date. The majority of proteins previously reported from 2DE analysis of BALF from patients with interstitial lung disease and acute lung injury18,20 were also identified in this study by tandem mass spectrometric analysis. Expected markers of lung injury were among those identified such as surfactants A and B, markers of activation and inflammation such as C3a, leukotrienes and markers of matrix remodeling, including collagenases A and B, and proteases. Not surprisingly, many of the proteins identified in BALF from ARDS patients were serum proteins, consistent with the capillary leak syndrome that is characteristic of ARDS.
While even this study underestimates the true protein content of BALF, the number of proteins identified in our study is an order of magnitude greater than previously published reports. Furthermore, we identified proteins in the IGF signaling pathway as well as other secreted proteins that may contribute to disease process. This approach allowed us to identify a new pathway in ARDS and subsequently show changes in expression using a large sample set of ARDS BALF. Bronchoalveolar lavage is a safe method to obtain lung epithelial lining fluid of the airways and alveoli.8 Initial reports of proteomic analysis of normal BALF separated proteins by 2DE, which is based on differences of charge and mass, followed by enzymatic degradation of separated proteins and analysis by mass spectrometry. The initial report of BALF proteomic analysis identified 23 serum proteins in normal BALF, representing 97% of the identified proteins in BALF.38 As the 2DE methodology has improved, the number of proteins identified has increased. A recently published 2DE database of BALF proteins from normal and lung disorders contained 93 proteins.18 However, several aspects of the 2DE method limit the ability to detect particular classes of proteins.39 For example, while the most “abundant” proteins can be readily detected in ARDS BALF by 2DE and MS, low abundance proteins are often not detected without prior fractionation. We chose to avoid prior sample fractionation because of the loss of proteins involved and the limited amount of protein present in our clinical ARDS BALF samples. Therefore, we chose to use cation exchange chromatography of trypsin-digested samples followed by MS/MS analysis to define the ARDS BALF proteome. While this method also has limitations (23), namely the random selection of ions as they enter the tandem mass spectrometer that results in poor overlap in the proteins identified in replicate analyses, it is has the advantage of speed and ease. Because a primary LC-MS/MS proteomic screen is unable to sample all of the proteins present,31 additional methods, including isolation of subpopulations of proteins and further refinement of the methodologies, should increase the yield of protein identifications. In addition, one can estimate relative abundance by comparing sequence coverage and number of peptides identified.40 Therefore, LC-MS/MS analysis is an excellent screening tool to initially characterize a sample of mostly unknown protein composition, but this must be followed by corroboration of interesting results using independent methods.
Potential Role of the IGF/IGFBP-3 Pathway
Following initial LC-MS/MS proteomic analysis of BALF, we focused on further characterization of IGF pathway components. Because of the known role of the IGF pathway in regulation of cell survival,27,29 we tested whether it might play a role in the development and resolution of acute lung injury. IGF-binding proteins are a family of six related proteins that bind IGF-I and -II with high affinity. IGFBP-3, the major circulating IGFBP, binds IGF-I in conjunction with an acid-labile glycoprotein subunit (ALS) to form a circulating complex. Because of the high affinity of IGFBP-3 for IGF-I, it has a major role in controlling the bioavailability of IGFs. IGF-I bound to IGFBP-3 does not interact with IGF-IR and thus fails to induce a prosurvival signal. IGF-I bound to IGFBP-3 has a longer half-life and may act as a stable reservoir of IGF-I. We found that the concentrations of IGFBP-3 and IGF were highest in at risk and early ARDS, and then decreased as disease progressed. IGFBP-3 is subject to cleavage by a number of proteases, including plasmin, matrix metalloproteases, and cathepsins. Because IGFBP-3 fragments have lower affinity for IGF than does the intact binding protein, proteolysis of IGFBP-3 is the major mechanism for release of IGF and increasing IGF bioavailability. Increased IGFBP-3 proteolysis was observed in BALF from other lung diseases. For example, BALF from sarcoid patients contained elevated IGFBP-3, most in the form of 30-kd (proteolyzed) fragment.24 However, we found the majority of IGFBP-3 was intact, not proteolyzed, despite the presumed proteolytic environment during ARDS.
Results from in vitro studies showed that the IGF pathway regulates survival of fibroblasts, not epithelial cells. In addition, we showed that neutralization of IGF in ARDS BALF increased fibroblast apoptosis. This was most pronounced in early ARDS. This suggests that IGF detected in BALF is biologically active and may play a role in regulating fibroblast survival in ARDS. We speculate that the lack of effect of IGF neutralization in some samples is due to additional pathways that regulate fibroblast survival. For example, interleukin-1β in lung edema fluid induces fibroblast proliferation.28 In addition, surfactant A41 and fibronectin-derived peptides42 also affect fibroblast apoptosis. While histological evidence of fibroproliferation is observed 5 to 7 days after the onset of ARDS, there is increasing evidence that fibroblasts are activated very early in ARDS. For example, procollagen III peptide, a marker of collagen synthesis, is elevated in BALF early in ARDS and remains elevated for 7 days or longer.34,43 Pulmonary edema fluid obtained from acute lung injury patients within 4 hours of intubation has an increased mitogenic effect on human lung fibroblasts.28 In concert, the data suggest that fibroblast activation is already occurring at the time acute lung injury is clinically apparent. Because IGF is elevated early in ARDS and regulates fibroblast survival, we speculate that the IGF pathway may contribute to early fibroblast activation in ARDS.
Dysregulation of cell survival and proliferation is a feature of many lung diseases, including ARDS and IPF. However, less is known about the role of the IGF pathway in lung disease. IGF-I was originally described in the lung as alveolar macrophage-derived growth factor. IGF-I mRNA was elevated in bleomycin-induced pulmonary fibrosis in mice.44 IGF-I was implicated as a major fibroblast mitogen in BALF from patients with sarcoidosis24 and systemic sclerosis,45 and macrophage-derived IGF-I inhibited apoptosis of a fibroblast cell line.46 In eight patients with fibroproliferative ARDS (day 7), biopsies showed increased IGF-I immunostaining, which correlated with increased cell proliferation.47 Elevated levels of IGFBP-3 were detected in BALF from patients with IPF22,23 and sarcoidosis.24 A recent study showed IGFBP-3 increased collagen and fibronectin synthesis by fibroblasts.48 Furthermore, fibroblasts derived from patients with IPF had increased expression of IGFBP-3 compared to normal controls.48 One of the interesting features about ARDS is that the lung injury (and fibrosis) resolves in the majority of patients. One possibility is that apoptosis of fibrogenic cells (ie, fibroblasts) is essential for the resolution of lung injury.49–51 Because the IGF/IGFBP-3 pathway is a key determinant of cell survival, dampening this pathway may be necessary both for normal scarring to resolve and to prevent a prolonged fibrogenic response (ie, fibrosis).
In summary, we used LC-MS/MS proteomic analysis as an initial screening method to sample the BALF proteome in ARDS. As such it provided an excellent means to define the ARDS BALF proteome and allowed us to hypothesize roles for select identified proteins. Proteins from this list suspected of playing a role in disease progression were selected for further investigation in a large clinical sample set. The results showed that the expression of IGF and IGFBP-3 changed as ARDS progressed. Further, we identified a role of IGF pathway in mediating fibroblast survival in vitro and in vivo. Finally, we speculate that the IGF pathway, through regulation by IGFBP-3, controls fibroblast survival, which contributes to the fibroproliferative response in acute lung injury.
Supplementary Material
Acknowledgments
We thank Gus Matute-Bello and Naoki Hagimoto for assistance with apoptosis assays, Jeremy Kahn for statistical advice, Matthew Rosengart for assistance with samples, Steve Plymate for helpful discussions, and Elaine Raines for critical review of the manuscript.
Footnotes
Address reprint requests to Lynn M. Schnapp, M.D., Pulmonary and Critical Care Medicine, University of Washington, Harborview Medical Center, 325 9th Ave., Box 359640, Seattle, WA 98104. E-mail lschnapp@u.washington.edu.
Supported by the National Institutes of Health (grants RO1 HL73028 and SCCOR P50 HL073996 to L.M.S.), Mass Spectrometry Core for WWAMI (Washington, Wyoming, Alaska, Montana, and Idaho) Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (grant 1U54AI57141-01), Proteomics Core for Center for Ecogenetics and Environmental Health (grant 5P30ES007033-10 to D.R.G.), and by the National Heart, Lung, and Blood Institute, National Institutes of Health (under contract N01-HV-28179 to the Seattle Proteome Center).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Ashbaugh DG, Bigelow DB, Petty TL, Levine BE. Acute respiratory distress in adults. Lancet. 1967;2:319–323. [Google Scholar]
- Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, LeGall JR, Morris A, Spragg R. Report of the American-European Consensus conference on acute respiratory distress syndrome: definitions, mechanisms, relevant outcomes, and clinical trial coordination. J Crit Care. 1994;9:72–81. doi: 10.1016/0883-9441(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Pittet JF, Mackersie RC, Martin TR, Matthay MA. Biological markers of acute lung injury: prognostic and pathogenetic significance. Am J Respir Crit Care Med. 1997;155:1187–1205. doi: 10.1164/ajrccm.155.4.9105054. [DOI] [PubMed] [Google Scholar]
- Griffin TJ, Aebersold R. Advances in proteome analysis by mass spectrometry. J Biol Chem. 2001;276:45497–45500. doi: 10.1074/jbc.R100014200. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Matute-Bello G, Liles WC, Radella F, 2nd, Steinberg KP, Ruzinski JT, Jonas M, Chi EY, Hudson LD, Martin TR. Neutrophil apoptosis in the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1997;156:1969–1977. doi: 10.1164/ajrccm.156.6.96-12081. [DOI] [PubMed] [Google Scholar]
- Steinberg KP, Milberg JA, Martin TR, Maunder RJ, Cockrill BA, Hudson LD. Evolution of bronchoalveolar cell populations in the adult respiratory distress syndrome. Am J Respir Crit Care Med. 1994;150:113–122. doi: 10.1164/ajrccm.150.1.8025736. [DOI] [PubMed] [Google Scholar]
- Greene KE, Wright JR, Steinberg KP, Ruzinski JT, Caldwell E, Wong WB, Hull W, Whitsett JA, Akino T, Kuroki Y, Nagae H, Hudson LD, Martin TR. Serial changes in surfactant-associated proteins in lung and serum before and after onset of ARDS. Am J Respir Crit Care Med. 1999;160:1843–1850. doi: 10.1164/ajrccm.160.6.9901117. [DOI] [PubMed] [Google Scholar]
- Lowry O, Rosebrough N, Farr A, Randall R. Protein measurement with the Folin-Phenol reagents. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Yi EC, Lee H, Aebersold R, Goodlett DR. A microcapillary trap cartridge-microcapillary high-performance liquid chromatography electrospray ionization emitter device capable of peptide tandem mass spectrometry at the attomole level on an ion trap mass spectrometer with automated routine operation. Rapid Commun Mass Spectrom. 2003;17:2093–2098. doi: 10.1002/rcm.1150. [DOI] [PubMed] [Google Scholar]
- Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nature Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- Baliga NS, Pan M, Goo YA, Yi EC, Goodlett DR, Dimitrov K, Shannon P, Aebersold R, Ng WV, Hood L. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc Natl Acad Sci USA. 2002;99:14913–14918. doi: 10.1073/pnas.192558999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS: ImageJ. http://rsb.info.nih.gov/ij/. Bethesda, MD, US National Institutes of Health, 1997–2005 [Google Scholar]
- Burtrum D, Zhu Z, Lu D, Anderson DM, Prewett M, Pereira DS, Bassi R, Abdullah R, Hooper AT, Koo H, Jimenez X, Johnson D, Apblett R, Kussie P, Bohlen P, Witte L, Hicklin DJ, Ludwig DL. A fully human monoclonal antibody to the insulin-like growth factor I receptor blocks ligand-dependent signaling and inhibits human tumor growth in vivo. Cancer Res. 2003;63:8912–8921. [PubMed] [Google Scholar]
- Wu JD, Odman A, Higgins LM, Haugk K, Vessella R, Ludwig DL, Plymate SR. In vivo effects of the human type I insulin-like growth factor receptor antibody A12 on androgen-dependent and androgen-independent xenograft human prostate tumors. Clin Cancer Res. 2005;11:3065–3074. doi: 10.1158/1078-0432.CCR-04-1586. [DOI] [PubMed] [Google Scholar]
- Noel-Georis I, Bernard A, Falmagne P, Wattiez R. Database of bronchoalveolar lavage fluid proteins. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;771:221–236. doi: 10.1016/s1570-0232(02)00114-9. [DOI] [PubMed] [Google Scholar]
- Hirsch J, Hansen KC, Burlingame AL, Matthay MA. Proteomics: current techniques and potential applications to lung disease. Am J Physiol. 2004;287:L1–L23. doi: 10.1152/ajplung.00301.2003. [DOI] [PubMed] [Google Scholar]
- Bowler RP, Duda B, Chan ED, Enghild JJ, Ware LB, Matthay MA, Duncan MW. Proteomic analysis of pulmonary edema fluid and plasma in patients with acute lung injury. Am J Physiol. 2004;286:L1095–L1104. doi: 10.1152/ajplung.00304.2003. [DOI] [PubMed] [Google Scholar]
- Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC. Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest. 2004;113:1318–1327. doi: 10.1172/JCI19930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston C, Jagirdar J, Lee TC, Hur T, Hintz RL, Rom WN. Enhanced insulin-like growth factor molecules in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1995;151:1597–1603. doi: 10.1164/ajrccm.151.5.7537587. [DOI] [PubMed] [Google Scholar]
- Pala L, Giannini S, Rosi E, Cresci B, Scano G, Mohan S, Duranti R, Rotella CM. Direct measurement of IGF-I and IGFBP-3 in bronchoalveolar lavage fluid from idiopathic pulmonary fibrosis. J Endocrinol Invest. 2001;24:856–864. doi: 10.1007/BF03343942. [DOI] [PubMed] [Google Scholar]
- Allen JT, Bloor CA, Knight RA, Spiteri MA. Expression of insulin-like growth factor binding proteins in bronchoalveolar lavage fluid of patients with pulmonary sarcoidosis. Am J Respir Cell Mol Biol. 1998;19:250–258. doi: 10.1165/ajrcmb.19.2.3080. [DOI] [PubMed] [Google Scholar]
- Raab G, Klagsbrun M. Heparin-binding EGF-like growth factor. Biochim Biophys Acta. 1997;1333:F179–F199. doi: 10.1016/s0304-419x(97)00024-3. [DOI] [PubMed] [Google Scholar]
- Iwamoto R, Mekada E. Heparin-binding EGF-like growth factor: a juxtacrine growth factor. Cytokine Growth Factor Rev. 2000;11:335–344. doi: 10.1016/s1359-6101(00)00013-7. [DOI] [PubMed] [Google Scholar]
- LeRoith D, Roberts CT., Jr The insulin-like growth factor system and cancer. Cancer Lett. 2003;195:127–137. doi: 10.1016/s0304-3835(03)00159-9. [DOI] [PubMed] [Google Scholar]
- Olman MA, White KE, Ware LB, Simmons WL, Benveniste EN, Zhu S, Pugin J, Matthay MA. Pulmonary edema fluid from patients with early lung injury stimulates fibroblast proliferation through IL-1 beta-induced IL-6 expression. J Immunol. 2004;172:2668–2677. doi: 10.4049/jimmunol.172.4.2668. [DOI] [PubMed] [Google Scholar]
- Clemmons DR. Insulin-like growth factor binding proteins and their role in controlling IGF actions. Cytokine Growth Factor Rev. 1997;8:45–62. doi: 10.1016/s1359-6101(96)00053-6. [DOI] [PubMed] [Google Scholar]
- Lalou C, Lassarre C, Binoux M. A proteolytic fragment of insulin-like growth factor (IGF) binding protein-3 that fails to bind IGFs inhibits the mitogenic effects of IGF-I and insulin. Endocrinology. 1996;137:3206–3212. doi: 10.1210/endo.137.8.8754741. [DOI] [PubMed] [Google Scholar]
- Yi EC, Marelli M, Lee H, Purvine SO, Aebersold R, Aitchison JD, Goodlett DR. Approaching complete peroxisome characterization by gas-phase fractionation. Electrophoresis. 2002;23:3205–3216. doi: 10.1002/1522-2683(200209)23:18<3205::AID-ELPS3205>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Goodman RB, Strieter RM, Martin DP, Steinberg KP, Milberg JA, Maunder RJ, Kunkel SL, Walz A, Hudson LD, Martin TR. Inflammatory cytokines in patients with persistence of the acute respiratory distress syndrome. Am J Respir Crit Care Med. 1996;154:602–611. doi: 10.1164/ajrccm.154.3.8810593. [DOI] [PubMed] [Google Scholar]
- Park WY, Goodman RB, Steinberg KP, Ruzinski JT, Radella F, 2nd, Park DR, Pugin J, Skerrett SJ, Hudson LD, Martin TR. Cytokine balance in the lungs of patients with acute respiratory distress syndrome. Am J Respir Crit Care Med. 2001;164:1896–1903. doi: 10.1164/ajrccm.164.10.2104013. [DOI] [PubMed] [Google Scholar]
- Chesnutt AN, Matthay MA, Tibayan FA, Clark JG. Early detection of type III procollagen peptide in acute lung injury. Pathogenetic and prognostic significance. Am J Respir Crit Care Med. 1997;156:840–845. doi: 10.1164/ajrccm.156.3.9701124. [DOI] [PubMed] [Google Scholar]
- Matute-Bello G, Liles WC, Steinberg KP, Kiener PA, Mongovin S, Chi EY, Jonas M, Martin TR. Soluble Fas ligand induces epithelial cell apoptosis in humans with acute lung injury (ARDS). J Immunol. 1999;163:2217–2225. [PubMed] [Google Scholar]
- Franklin SL, Ferry RJ, Jr, Cohen P. Rapid insulin-like growth factor (IGF)-independent effects of IGF binding protein-3 on endothelial cell survival. J Clin Endocrinol Metab. 2003;88:900–907. doi: 10.1210/jc.2002-020472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim ML, Levitt Katz LE, Davis J, Dotzler WC, Cohen P, Ferry RJ., Jr Insulin-like growth factor binding protein-3 is a novel mediator of apoptosis in insulin-secreting cells. Growth Horm IGF Res. 2004;14:216–225. doi: 10.1016/j.ghir.2003.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DY, Hook GE. Pulmonary alveolar proteinosis: analysis of airway and alveolar proteins. Am Rev Respir Dis. 1979;119:979–990. doi: 10.1164/arrd.1979.119.6.979. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Corthals GL, Zhang Y, Rochon Y, Aebersold R. Evaluation of two-dimensional gel electrophoresis-based proteome analysis technology. Proc Natl Acad Sci USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Vazquez de, Lara L, Becerril C, Montano M, Ramos C, Maldonado V, Melendez J, Phelps DS, Pardo A, Selman M. Surfactant components modulate fibroblast apoptosis and type I collagen and collagenase-1 expression. Am J Physiol. 2000;279:L950–L957. doi: 10.1152/ajplung.2000.279.5.L950. [DOI] [PubMed] [Google Scholar]
- Hadden HL, Henke CA. Induction of lung fibroblast apoptosis by soluble fibronectin peptides. Am J Respir Crit Care Med. 2000;162:1553–1560. doi: 10.1164/ajrccm.162.4.2001015. [DOI] [PubMed] [Google Scholar]
- Clark JG, Milberg JA, Steinberg KP, Hudson LD. Type III procollagen peptide in the adult respiratory distress syndrome. Association of increased peptide levels in bronchoalveolar lavage fluid with increased risk for death. Ann Intern Med. 1995;122:17–23. doi: 10.7326/0003-4819-122-1-199501010-00003. [DOI] [PubMed] [Google Scholar]
- Maeda A, Hiyama K, Yamakido H, Ishioka S, Yamakido M. Increased expression of platelet-derived growth factor A and insulin-like growth factor-I in BAL cells during the development of bleomycin-induced pulmonary fibrosis in mice. Chest. 1996;109:780–786. doi: 10.1378/chest.109.3.780. [DOI] [PubMed] [Google Scholar]
- Harrison NK, Cambrey AD, Myers AR, Southcott AM, Black CM, du Bois RM, Laurent GJ, McAnulty RJ. Insulin-like growth factor-I is partially responsible for fibroblast proliferation induced by broncho-alveolar lavage fluid from patients with systemic sclerosis. Clin Sci (Lond) 1994;86:141–148. doi: 10.1042/cs0860141. [DOI] [PubMed] [Google Scholar]
- Wynes MW, Frankel SK, Riches DW. IL-4-induced macrophage-derived IGF-I protects myofibroblasts from apoptosis following growth factor withdrawal. J Leukoc Biol. 2004;76:1019–1027. doi: 10.1189/jlb.0504288. [DOI] [PubMed] [Google Scholar]
- Krein PM, Sabatini PJ, Tinmouth W, Green FH, Winston BW. Localization of insulin-like growth factor-I in lung tissues of patients with fibroproliferative acute respiratory distress syndrome. Am J Respir Crit Care Med. 2003;167:83–90. doi: 10.1164/rccm.2201012. [DOI] [PubMed] [Google Scholar]
- Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol. 2005;166:399–407. doi: 10.1016/S0002-9440(10)62263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan SH. The myofibroblast in pulmonary fibrosis. Chest. 2002;122:286S–289S. doi: 10.1378/chest.122.6_suppl.286s. [DOI] [PubMed] [Google Scholar]
- Uhal BD. Apoptosis in lung fibrosis and repair. Chest. 2002;122:293S–298S. doi: 10.1378/chest.122.6_suppl.293s. [DOI] [PubMed] [Google Scholar]
- Iredale JP, Benyon RC, Pickering J, McCullen M, Northrop M, Pawley S, Hovell C, Arthur MJ. Mechanisms of spontaneous resolution of rat liver fibrosis. Hepatic stellate cell apoptosis and reduced hepatic expression of metalloproteinase inhibitors. J Clin Invest. 1998;102:538–549. doi: 10.1172/JCI1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.