Abstract
To elucidate the contribution of LINE-like retrotransposon Zepp elements to the formation and maintenance of chromosomal telomeres, newly formed minichromosomes in irradiated Chlorella vulgaris cells were isolated and structurally characterized. A minichromosome (miniV4) of ∼700 kb in size contained a Zepp cluster taking the place of the telomeric repeats on one terminus, whereas the other end of this chromosome consisted of canonical telomeric repeats. The Zepp copies in this cluster were in a tandem array with their poly(A) tails towards the centromere. Another minichromosome Y32 (∼400 kb in size) was shown to have several copies of Zepp elements on both termini. On the right arm terminus, two copies of Zepp were found in a tandem array with poly(A) tracts facing towards the chromosomal end. The poly(A) tail and the 3′-end of ∼400 bp of the distal copy were replaced by the telomeric repeats. On the 5′-side of the proximal copy was another Zepp element in the reverse orientation. These newly formed telomeric structures are very similar to those previously found in the left arm of chromosome I and the terminus of an unidentified chromosome and support the model of Zepp-mediated restoration and maintenance of Chlorella telomeres.
INTRODUCTION
Telomeres are protein–DNA structures at the ends of eukaryotic chromosomes. They serve two major functions: to distinguish intact from broken chromosomes and to protect chromosomes from degradation and incomplete replication (for a review, see 1). Telomeres also participate in various aspects of functional organization of the nucleus: they interact with each other and with other substructures, like the nuclear scaffold and nuclear envelope, and drive polarized chromosome movements during karyogamy and meiotic prophase I (2). In most organisms, telomeric DNA consists of a tandem array of simple sequence elements, telomeric repeats (1,3,4). In general, the telomeric repeats are synthesized by a specialized reverse transcriptase called telomerase (5). However, there are alternative strategies to maintain and extend telomeric sequences. In Saccharomyces cerevisiae, some recombination mechanisms can restore deficiencies in telomere elongation (6,7). Instead of the canonical telomeric repeats, the Drosophila telomeres are composed of one or more LINE (long interspersed DNA element)-like retrotransposons designated HeT-A and TART, and incomplete DNA replication at the chromosome termini is counterbalanced by frequent transposition of these elements (8–11). Some LINE-like retrotransposable elements have also been shown to integrate or accumulate preferentially in the telomeric or subtelomeric regions, for example, Ty5 of S.cerevisiae (12), TRAS1 and SART1 of Bombyx mori (13,14), Zepp of Chlorella vulgaris (15,16) and GilM and GilT of Giardia lamblia (17).
In the case of C.vulgaris, several copies of Zepp elements accumulate in the telomeric regions (15). Zepps show features characteristic of non-LTR (long terminal repeat), LINE-like elements, including a poly(A) tail, 5′ truncations, two ORFs encoding a gag-like protein (ORF1) and reverse transcriptase (ORF2) and flanking target duplications (16). Zepp is not expressed in the normal growth cycle but can be induced by heat shock treatment. Zepp elements self-integrate: on chromosome I, the Zepp sequence is a target of successive insertions that result in a tandem array of the 3′-regions, with the poly(A) tracts facing toward the centromere. The most distal Zepp copy on this chromosome is inverted so that its poly(A) tail is connected to a very short stretch of the telomeric repeats (15). Another chromosome also had a similar Zepp cluster, but without telomeric repeats that resemble the retrotransposon–telomere structure of the Drosophila telomeres. From these observations, we proposed a telomere-stabilizing role of Zepp elements in certain circumstances, by jumping into telomeric regions and accumulating in a buffer structure for the Chlorella telomere (15).
In recent studies, non-homologous end joining of broken chromosomes in plants has been analyzed in a direct and thorough manner. Linearized plasmid DNA was introduced into tobacco cells and novel joints that formed between the plasmid ends were characterized (18). In another study, double-strand breaks (DSBs) were induced at an I-SceI recognition site within a negative selectable marker gene on a chromosome and new junctions formed on the site were characterized (19). However, these methods are unfortunately not applicable to Chlorella cells because there is no efficient method for DNA-mediated transformation of Chlorella cells.
To elucidate such biological significance of Zepp elements in the formation and maintenance of the Chlorella telomeres, we induced chromosomal breakages in Chlorella cells by irradiation with electron beams and examined newly formed chromosomal ends on minichromosomes. We found that Zepp elements accumulated at one terminus replacing the telomeric repeats on a newly formed minichromosome and also at both ends of another minichromosome, providing supportive evidence for our previous model of Zepp-mediated restoration of the Chlorella telomeres.
MATERIALS AND METHODS
The nucleotide sequence of the Y32 telomeric region described in this work has been deposited in DDBJ/EMBL/GenBank under accession nos AB096693, AB098338, AB098339 and AB098340.
Strains and culture conditions
Chlorella vulgaris C-169 was obtained from the culture collection of the Institute of Molecular and Cellular Biosciences, The University of Tokyo. Cells were cultured photosynthetically in modified Bristol’s medium (MBM) as described previously (20).
Electron beam irradiation
A 5 ml suspension of exponentially growing C.vulgaris C-169 cells (1 × 105 cells/ml) was spread over an MBM plate in a Petri dish (85 mm in diameter) and exposed to an electron beam from a linear accelerator with a 240 KeV tungsten target tube at 20–500 Gy (Mitsubishi Heavy Industries Ltd, Tokyo, Japan). Irradiation at 300 Gy, which permitted the survival of ∼0.01% of cells, was applied to induce extended chromosome rearrangements. After irradiation, plates were incubated in the dark at 25°C. Colonies that appeared on the plates were cultivated in liquid MBM in the light and subjected to contour-clamped homogeneous electric field (CHEF) gel electrophoresis (CHEF-DRII; Bio-Rad, Boston, MA) for karyotyping (21).
Chromosome analysis
Chromosomal DNA molecules of Chlorella cells were separated by CHEF gel electrophoresis as described previously (21). To isolate minichromosomes, CHEF gel electrophoresis was performed in a 1% low melting point agarose (InCert Agarose; Takara Shuzo, Kyoto, Japan) gel in 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA, pH 8.3) with a 3 min switching interval at 5 V/cm for 24 h.
For Southern blotting analysis, probes were labeled with fluorescein using a Gene Image kit (Amersham Pharmacia Biotech, Uppsala, Sweden) and detected with a CDP-Star detection module (Amersham Pharmacia Biotech) according to the manufacturer’s protocol. For detection of the Zepp sequence, an 800 bp HindIII–SphI fragment of the H1.3 clone from the left arm of chromosome I, a 3′ portion immediately adjacent to the poly(A) tail (15), was used as a hybridization probe. The telomeric repeat sequence in pCHt-1 (22) was used as a probe for telomeric repeats.
To detect gradual degradation of DNA fragments derived from the chromosome termini by treatment with Bal31 nuclease, total Chlorella chromosomal DNA (10 µg) or isolated minichromosomal DNA (1 µg) in 500 µl of 1× Bal31 buffer (20 mM Tris–HCl, pH 7.2, 0.6 M NaCl, 12.5 mM MgCl2, 12.5 mM CaCl2 and 1 mM EDTA) was treated with 5 U Bal31 (Takara Shuzo) at 28°C for various time periods. The reaction was terminated by adding EGTA to a final concentration of 50 mM. DNA samples were phenol extracted and precipitated with ethanol before digestion with restriction enzymes.
Cloning of the terminal fragments of minichromosome Y32 (miniY32)
MiniY32 DNA (5 µg) isolated by CHEF gel electrophoresis was treated with T4 DNA polymerase (Toyobo, Osaka, Japan) in 50 mM Tris–HCl, pH 8.0, containing 7 mM MgCl2, 15 mM (NH4)2SO4, 10 mM 2-mercaptoethanol, 0.1 mM EDTA and 0.1 mM dNTPs at 37°C for 1 h. Following digestion with HindIII, the DNA was ligated to the HindIII and HincII sites of pBluescript II SK+ and transformed into Escherichia coli KL10-Gold. The corresponding DNA clones were selected by colony hybridization with the same Zepp probe as described above.
Northern blotting analysis
Total RNA was isolated from Chlorella cells as described previously (15), blotted onto nylon filters and hybridized with 32P-labeled probes. For detection of the Zepp sequence, the same DNA fragment as used in Southern blot hybridization was used as a probe. Hybridization was performed in a solution containing 50% formamide, 5× SSC, 50 mM sodium phosphate (pH 7.0), 0.1% SDS, denatured salmon sperm DNA (50 µg/ml) and 1× Denhardt’s solution. The filter was washed for 15 min at 50°C with 0.2× SSC containing 0.1% SDS. Washed filters were autoradiographed for 24–48 h at –80°C with X-ray film (RX-U; Fuji Film, Tokyo, Japan). In some cases, cells were subjected to electron beam irradiation under the same conditions used for minichromosome induction before RNA isolation.
DNA sequencing and sequence analysis
DNA sequences were determined by the standard chain termination method using an AutoRead sequencing kit and an ALF DNA sequencer (both from Amersham Pharmacia Biotech). DNA sequences were compiled and analyzed with the DNASIS computer program (Hitachi Software, Tokyo, Japan).
RESULTS
Isolation of minichromosomes containing Zepp elements at the telomeric regions
We have recently found that electron beams are very efficient at inducing chromosomal breaks and rearrangements in Chlorella cells (23). In the cells surviving after irradiation, the chromosome separation pattern after CHEF gel electrophoresis changes drastically compared to the wild-type and minichromosomes ranging from 250 to 800 kb in size frequently appear. When chromosomal DNA isolated from such surviving cells was digested with restriction enzymes and subjected to Southern blotting analysis with the 3′-region of the Zepp element (15) as a probe, a few additional bands always appeared compared with the wild-type pattern. The appearance of these extra Zepp-hybridizing bands in surviving cells strongly suggests that Zepp elements may participate in chromosome rearrangements induced by irradiation.
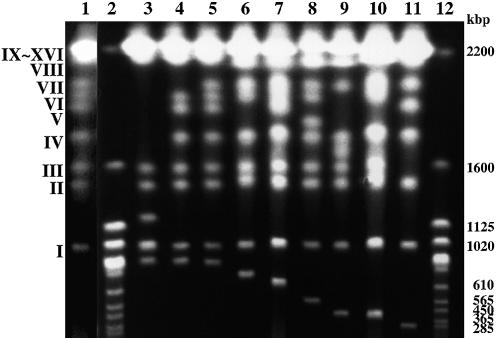
To obtain newly formed chromosome termini that possibly contained Zepp elements, we were interested in isolating minichromosomes from Chlorella survivors after irradiation. Among 242 surviving colonies, 15 clones contained newly formed minichromosomes that could be easily separated by CHEF gel electrophoresis. Nine examples of these are shown in Figure 1, where minichromosomes are recognized as bands beneath chromosome I, the smallest chromosome of C.vulgaris C-169 (980 kb in size) (21). When this chromosome separation pattern was subjected to Southern blotting analysis, all minichromosomes strongly hybridized to the Zepp probe (data not shown).
Figure 1.
Separation of minichromosomes formed in surviving Chlorella cells after electron beam irradiation. Each chromosome was separated by CHEF gel electrophoresis (20). Minichromosomes are seen as bands moving below chromosome I (980 kb). The chromosome numbering was assigned according to previous work (20). Sizes are shown in kilobase pairs (kb). Lane 1, wild-type; lanes 2 and 12, S.cerevisiae as size markers; lane 3, mutant E1; lane 4, mutant T7; lane 5, mutant F6; lane 6, mutant B2; lane 7, mutant V4; lane 8, mutant T8; lane 9, mutant E4; lane 10, mutant Y32; lane 11, mutant P6.
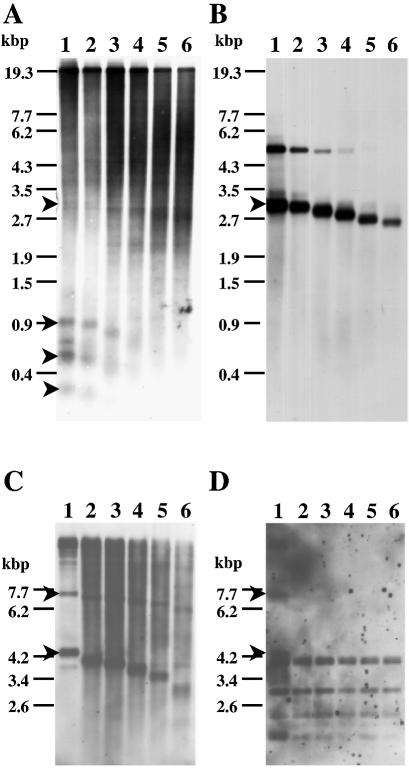
Then, individual minichromosomes were cut out from the gel, treated with Bal31 and various restriction enzymes and subjected to Southern blotting analysis with the Zepp probe. At least two minichromosomes were found to contain Zepp-bearing telomeric regions. In the case of a minichromosome of mutant V4 (miniV4) of ∼700 kb in size, at least three HincII fragments (0.9, 0.6 and 0.3 kb) hybridizing to the Zepp probe were susceptible to Bal31 digestion (Fig. 2A). This result indicates that these fragments were derived from the chromosomal end and contained Zepp sequences. When the same blot was hybridized with telomeric repeats as a probe, two bands (5.7 and 3.3 kb) showed positive signals, only one of which (3.3 kb) decreased in size on Bal31 digestion (Fig. 2B). This band also showed faint hybridization with the Zepp probe (Fig. 2A). Although the hybridization signal of the other band of 5.7 kb was weakened by Bal31 treatment, its size remained exactly the same (Fig. 2B). Such fading of hybridization signals is often observed for DNA fragments treated for long periods with Bal31, but this result may reflect some specific structure of the internal telomeric repeats. The three Zepp-hybridizing terminal bands did not hybridize to the telomeric repeats. Taken together, one terminus of miniV4 should consist of Zepp elements without telomeric repeats whereas the other terminus has canonical telomeric repeats as well as internal Zepp sequences. A few Zepp-hybridizing terminal bands may reflect an unstable state of this terminus. Such a structure is very similar to that previously found in the 2.3 kb HincII fragment of C.vulgaris C-169 (15).
Figure 2.
Detection of terminal fragments of minichromosomes V4 (A and B) and Y32 (C and D). Chromosomal DNA was first treated with Bal31 for various periods and then with HincII (A and B) or HindIII (C and D). Hybridizing bands were detected with an 800 bp 3′-region of Zepp or with the telomeric repeats (pCHt1) as a probe (B and D). Arrowheads indicate hybridizing bands derived from chromosome termini. Lanes 1–6, Bal31 treatment for 0, 30, 60, 120, 180 and 300 s, respectively.
The other case of Zepp-containing telomeres is a minichromosome of mutant Y32 (miniY32, 400 kb). Two Zepp-hybridizing bands (7.0 and 4.2 kb) detected with HindIII (Fig. 2C, lane 1) gradually decreased in size on digestion with Bal31 (Fig. 2C, lanes 2–6). This result clearly indicates that (i) the two bands were from each chromosome terminus and (ii) they contained Zepp sequences. Most importantly, these bands were obviously different from the terminal fragments derived from chromosome I and another chromosome, whose sizes are smaller than 1.5 kb after HindIII digestion (15). In addition, the mutant Y32 contained an intact chromosome I (Fig. 1). Therefore, the 7.0 and 4.2 kb bands were from newly formed termini of miniY32.
When the same blot (Fig. 2C) was probed with Chlorella telomeric repeats as a probe (22), both the 7.0 and 4.2 kb bands showed a hybridization signal (arrowheads in Fig. 2D). However, the signal of both bands soon disappeared on treatment with Bal31, suggesting very short stretches of telomeric repeats at the most distal end of the fragments. Other hybridizing signals seen in Figure 2D are most likely due to the presence of internal regions containing some telomeric repeats on this chromosome.
Isolation and analysis of the nucleotide sequences of the miniV4 and miniY32 telomeric regions
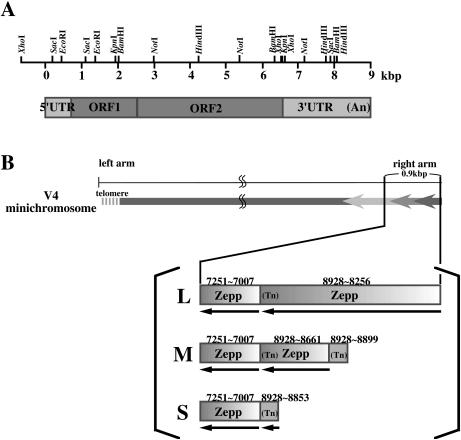
In our previous work, the most distal chromosomal DNA fragments were able to be cloned only after treatment with Klenow enzyme or T4 DNA polymerase (15). Accordingly, both miniV4 and miniY32 DNAs isolated from a CHEF electrophoresis gel were treated with T4 DNA polymerase as described in Materials and Methods before digestion with HincII and HindIII, respectively. The resulting fragments were ligated into the HincII site (for miniV4) and the HincII and HindIII sites (for miniY32) of the vector and introduced into E.coli. Fifteen clones were obtained by colony hybridization with a Zepp probe for the miniV4 fragments. Nucleotide sequences determined for three of them (0.9. 0.6 and 0.3 kb) all showed Zepp sequences. The 935 bp insert (L) (DDBJ/EMBL/GenBank accession no. AB098338) contained two Zepp copies in a tandem array with their poly(A) tails towards the centromere [a 673 bp 3′-end corresponding to positions 8254–8928 of ZA-1 (DDBJ/EMBL/GenBank accession no. AB008896, Fig. 3A) with 17 A residues and a 245 bp 3′-region of another Zepp copy corresponding to ZA-1 positions 7007–7251]. The 583 bp sequence (M) (DDBJ/EMBL/GenBank accession no. AB098339) included three Zepp copies in a similar tandem array (a 30 bp 3′-end from position 8899 to 8928 with A23, a 268 bp 3′-end from positions 8661 to 8928 with A17 and a 245 bp 3′-internal sequence from position 7007 to 7251). The 338 bp insert (S) (DDBJ/EMBL/GenBank accession no. AB098340) contained a 76 bp 3′-end (positions 8853–8928) with A17 and the same 3′-region 245 bp sequence as above. The blunt ends on one side of each clone were commonly at position 7251, corresponding to a HincII site within the Zepp sequence (accession no. AB008896). However, the other ends were not HincII sites and were different from each other, indicating that these clones were not contaminants from other chromosomal regions produced by HincII digestion, but were derived from a chromosomal end. As expected from the results of Figure 2B, there were no canonical telomeric repeats attached to the Zepp sequences, suggesting that these Zepp elements may take the place of the telomeric repeats at this chromosome terminus. These results are summarized in Figure 3B. The right terminus of miniV4 seems to exist in multiple forms, suggesting an unstable state of this terminus.
Figure 3.
Schematic representation of a Zepp element, ZA-1 (A), and the terminal Zepp cluster in miniV4 (B). (A) The full-length Zepp element ZA-1 consists of a 779 bp 5′ untranslated region (5′UTR), ORF1 (1839 bp), ORF2 (3927 bp) and a 2399 bp 3′ untranslated region (3′UTR) with a poly(A) tail of varying size (16). The 800 bp HindIII fragment was used as a probe for Southern blotting analysis. (B) Zepp cluster on the miniV4 terminal region. The 0.9 (L), 0.6 (M) and 0.3 kb (S) fragments detected in Figure 2A contain repeated 3′-regions of Zepp elements in a tandem array with their poly(A) tails towards the centromere. Numbers indicate positions corresponding to the ZA-1 sequence.
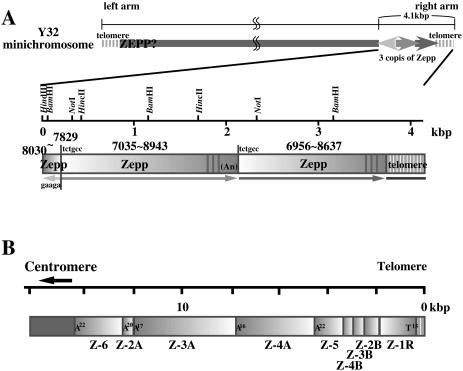
For miniY32, all 24 clones screened by colony hybridization with the Zepp probe contained the same 4.2 kb fragment, but for some reason, no clone with the 7.0 kb fragment was obtained. The nucleotide sequence (4132 bp) of the 4.2 kb fragment was completely determined (DDBJ/EMBL/GenBank accession no. AB096693). A matrix plot analysis showed that an ∼350 bp region at the 3′-end consisted of short tandem repeat sequences and three short stretches of the same repeats were clustered around positions 1250 and 3460. The remaining part consisted of three regions; from the 5′-end, a 220 bp region and two tandem repeats of 1700 bp sequence.
The nucleotide sequence of the 220 bp region was homologous to the 3′-end of the 1700 bp sequence in the reverse orientation. Homology searches through the databases for these sequences revealed that the 220 bp 5′ sequence corresponded to positions 8037–7829 of Zepp (ZA-1) and the 1938 bp sequence adjacent to this region contained a Zepp sequence from position 7035 to 8943 with a poly(A) tail consisting of 17 A residues. Furthermore, another Zepp sequence (positions 6956–8637) followed this poly(A) tract in a tandem array. The short repeats at the 3′-end were found to be the Chlorella telomeric repeat sequence, 5′-TTTAGGG (19), which is consistent with the results from Figure 2D. It looks like a 300–400 bp 3′-end including the poly(A) tract of the most distal copy of Zepp was replaced by the telomeric repeats. It was also found that short stretches of telomeric repeats reiterated three times within a 3′-region of Zepp. A 6 bp sequence, 5′-TCTGCC, immediately adjacent to the poly(A) tract of the second Zepp copy was also found in the upstream flanking region of this copy, corresponding to target duplications. Upstream of this sequence, five nucleotides, 5′-GAAGA, were inserted in the reverse orientation between the most proximal copy and the second copy of Zepp. This sequence may also be a flanking duplication formed by insertion of the proximal copy. Therefore, two copies of Zepp may have integrated at a single position in both orientations. These results are summarized in Figure 4A, compared with the Zepp clusters on the telomeric region of Chlorella chromosome I (Fig. 4B). The Zepp cluster on the telomeric region of miniY32 is very similar to that previously found in the telomeric region of chromosome I. The orientation of the most distal copy of Zepp is the same in both cases, with its 3′-end facing towards the chromosomal end. However, the most distal copy on miniY32 lacked ∼300–400 bp of its 3′-end as well as the poly(A) tract, and instead contained almost the same size of telomeric repeats. LINE-type retrotransposons integrate into a target site from its 3′ poly(A) tail by ‘target-primed reverse transcription’ (24–26). Therefore, this structure suggests that the terminus of miniY32 was broken once and thereafter short telomeric repeats were added.
Figure 4.
Schematic representation of the Zepp cluster on the right terminus of miniY32 (A) compared with the Zepp elements accumulated on the terminus of chromosome I (B). An, poly(A) tracts. Numbers indicate Zepp positions according to the ZA-1 sequence. Possible target site duplication sequences are shown at each Zepp junction.
The other terminus of miniY32 may have a very similar structure because the Bal31 susceptibility of the telomeric repeats and Zepp sequences on the 7.0 kb HindIII fragment was very similar to that of the 4.2 kb fragment (Fig. 2).
Expression of Zepp elements induced by electron beam irradiation
To detect the actual expression of Zepp elements in the irradiated cells, northern blot hybridization was carried out. Total RNA was isolated from wild-type cells 4 and 12 h after electron beam irradiation. As a control, RNA from non-irradiated cells was also analyzed. The results shown in Figure 5 indicate that signals corresponding to the full-length Zepp size (∼9.0 kb) (15) as well as some smaller sizes (∼6.0 kb) appeared in the irradiated cells; the signals were detected at 4 h after irradiation (lane 2) and persisted until at least 12 h after irradiation (lane 3). These signals were absent from non-irradiated cells (lane 1). Non-specific signals of 26S and 18S rRNA and smaller smear bands were also observed, whose nature is not known. These results ensured that the expression of Zepp elements is actually induced by irradiation. Whether this induction is due to DNA breaks caused by irradiation or heat shock effects (15) of irradiation is not known.
Figure 5.
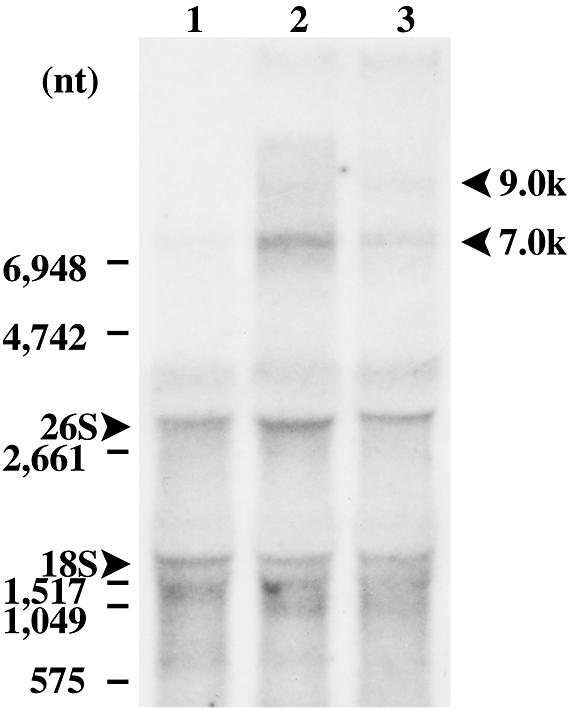
Northern blot hybridization of total RNA isolated from Chlorella cells with the Zepp sequence as probe. Lanes 1–3 contained RNA (10 µg) isolated from non-irradiated cells (control) and cells 4 and 12 h after electron beam irradiation, respectively. The positions of 26S and 18S rRNA are indicated on the left side. The arrows on the right side indicate large Zepp transcripts of ∼9.0 and ∼7.0 kb in size.
DISCUSSION
Participation of Zepp elements in the formation and rearrangements of minichromosomes
On Chlorella chromosome I, Zepp elements form a tandem cluster of six copies immediately next to the small telomeric repeats, in the same orientation relative to the centromere as HeT-A and TART clusters in Drosophila (8–11). A similar Zepp cluster is also exposed on the chromosomal terminus, replacing the telomeric repeats on another chromosome. Based on these observations, we previously proposed a telomere-stabilizing role of Zepp elements in certain circumstances, by jumping into telomeric regions and accumulating in a buffer structure for the telomere (15).
In this work, similar Zepp clusters have been found at the termini of at least two minichromosomes newly formed by irradiation with electron beams. In the case of miniV4, one terminus contained the canonical telomeric repeats (and some Zepp sequences), whereas the other end contained Zepp elements taking the place of the telomeric repeats (Figs 2A and B and 3). The distal Zepp clusters are variable; apparent addition of a new Zepp is recognized by comparing L with M and an extended truncation is obvious between L and S. These structures are different from the similar Zepp clusters on the previously identified terminus of another chromosome (DDBJ/EMBL/BenBank accession no. D89938). Therefore, we have interpreted these Zepp clusters on miniV4 to be a newly formed structure associated with minichromosome formation, probably through the mechanism of Zepp-mediated telomere restoration (15). The Zepp clusters should (tentatively?) restore the broken chromosome terminus. The fate of these terminal structures is very interesting. The latest analysis of subcolonies isolated from mutant V4 by PCR suggested that L was missing but M and S were detected stably in all progeny (data not shown). Structural changes occurring in these cells are now under investigation. At this stage, the Zepp expression pattern in V4 was the same as seen in wild-type C-169, namely no full-length Zepp transcripts were detected without induction.
In the other case, at least one terminus of miniY32 had two Zepp copies inserted in a single site in different orientations, and the most distal Zepp copy lacked 300–400 bp of its 3′-untranslated region (3′-UTR), including a poly(A) tract. Instead, almost the same size of telomeric repeats was added to the 3′-truncated Zepp. LINE-type retrotransposons, including Zepp elements, integrate into the target site from the 3′ poly(A) tail and 3′ truncations never occur under normal conditions. One possible explanation for this structure may be that at a break point on the chromosome, Zepp elements are successively integrated and the 3′-end of the most distal copy was exposed to terminal degradation before serving as a substrate for telomerase. This is consistent with our model (15). In fact, the expression of Zepp RNA was induced under stress conditions, including irradiation (Fig. 5) and heat shock (15).
Alternatively, the newly formed termini of miniY32 may merely be internally located Zepp clusters that are exposed by chromosome breaks and capped by telomerase without any new transposition of Zepp elements. Although this possibility cannot be rigorously excluded, simultaneous occurrence of such Zepp clusters on both ends by chance is highly unlikely. The expected frequency of such a structure is less than 2 × 10–4, provided that 180 copies of Zepp elements with an average size of 4500 bp are distributed randomly on the genome (16). Even if this is the case, Zepp clusters on both ends reflect some functional importance for them, such as stabilizing the telomeres on miniY32, since the telomeric repeats are as short as 300–400 bp on this chromosome. It is also interesting to speculate that Zepp elements at some particular chromosomal locations (other than chromosome termini) may possibly be hypersensitive to Bal31 for some unknown reason. However, internal copies of Zepp elements are totally resistant to Bal31 digestion (15).
Similar terminal Zepp clusters are also possible in other minichromosomes obtained in this work (Fig. 1). When minichromosomal DNAs were subjected to the Bal31 analysis, some (example miniE1, miniT7 and miniF6) showed Zepp-hybridizing, Bal31-susceptible bands in Southern hybridization patterns (data not shown). However, the bands were always large (>9.0 kb) with all restriction enzymes tested and were masked by other Zepp-hybridizing bands. No clone corresponding to these bands has been obtained so far. In the cases of miniB2 (800 kb), miniT8 (580 kb), miniE4 (450 kb) and miniP6 (300 kb), both chromosomal termini consist of canonical telomeric repeats revealed by the appearance of a couple of telomeric repeat-hybridizing, Bal31-susceptible bands in each case. The termini of these minichromosomes seem to be derived from the telomeres of pre-existing chromosomes by translocation or terminal exchanges.
Affinity of Zepp elements for the telomeric region
Zepp is a LINE-like retrotransposon that was first found accumulated at one terminus of Chlorella chromosome I (15). Its characteristic features include a long 3′-UTR of ∼2.4 kb in size (16). Long 3′-UTRs are highly atypical for retroelements and only two other cases have reported such structures so far. Two LINE-like elements, HeT-A and TART, which constitute the telomeres, instead of telomeric repeats, in D.melanogaster, share long 3′-UTR structures (8–11) that are nearly as long as the coding region. This feature prompted speculation that they are required for telomeric chromatin structure and/or terminal transposition (8,9). Another case of similar long 3′-UTRs has recently been found in two retroelements, GilM and GilT, from the parasitic protozoan G.lamblia (17). GilM and GilT are also LINE-like retrotransposons, and are only found in subtelomeric tandem head-to-tail arrays between the single copy genes and terminal arrays of telomeric repeats. However, in this organism, all telomeric repeats are not joined to these elements, since some of them are connected to rDNA or a specific gene. The 3′-UTRs of GilM and GilT are 2.0–2.5 kb in size and share no sequence similarity to each other. These long 3′-UTR structures shared by the telomeric or subtelomeric retrotransposons suggests convergent evolution towards some shared functionality. In this context, it is noteworthy that the Zepp 3′-UTR contains three short stretches of telomeric repeats (two or three copies) at the most terminal region of the element, which suggests either some affinity of this region for telomere binding proteins or it serving as a good substrate for telomerase. Actually, such telomeric repeat sequences on Zepp elements are always in the same orientation as the most terminal telomeric repeats on both chromosome I and miniY32 (Fig. 4A and B). In contrast, the orientation of the Zepp element on the right arm of V4 is reversed, namely the 3′-end faces towards the centromere and there are no canonical telomeric repeats attached to the Zepp repeats. This is also the case for the previously detected terminus on an unidentified chromosome, where three copies of Zepp elements were found in a tandem array in the same orientation as miniV4 (15). Therefore, stabilization through an addition of telomeric repeats by telomerase may require a specific orientation of Zepp elements. Anyway, most Zepp copies found on the telomeric regions of Chlorella chromosomes are 5′ truncated and virtually contain only the 3′-UTR (positions 6545–8943).
As demonstrated in this work, Chlorella Zepp elements provide a good opportunity to understand the dynamic mechanisms involved in telomere formation and maintenance.
REFERENCES
- 1.Zakian V.A. (1995) Telomeres: beginning to understand the end. Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 2.Gilson E., Laroche,T. and Gasser,S.M. (1993) Telomeres and the functional architecture of the nucleus. Trends Cell Biol., 3, 128–134. [DOI] [PubMed] [Google Scholar]
- 3.Zakian V.A. (1989) Structure and function of telomeres. Annu. Rev. Genet., 23, 579–604. [DOI] [PubMed] [Google Scholar]
- 4.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn E.H. (1992) Telomerase. Annu. Rev. Biochem., 61, 13–129. [DOI] [PubMed] [Google Scholar]
- 6.Louis E.J. and Harber,J.E. (1990) The subtelomeric Y′ repeat family in Saccharomyces cerevisiae: an experimental system for repeated sequence evolution. Genetics, 124, 533–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lundblad V. and Blackburn,E.H. (1993) An alternative pathway for yeast telomere maintenance rescue est1– senescence. Cell, 73, 347–360. [DOI] [PubMed] [Google Scholar]
- 8.Biessmann H., Vargeirsdottir,K., Lofsky,A., Chin,C., Ginther,B., Levis,R.W. and Pardue,M.L. (1992) Het-A, a transposable element specifically involved in ‘healing’ broken chromosome ends in Drosophila melanogaster. Mol. Cell. Biol., 12, 3910–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levis R.W., Ganesan,R., Houtchens,K., Taylor,L.A. and Seen,F.-M. (1993) Transposons in place of telomeric repeats at a Drosophila telomere. Cell, 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 10.Seen F.-M. and Levis,R.W. (1994) Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc. Natl Acad. Sci. USA, 91, 12510–12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mason J.M. and Biessmann,H. (1995) The unusual telomeres of Drosophila. Trends Genet., 11, 58–62. [DOI] [PubMed] [Google Scholar]
- 12.Zou S., Ke,N., Kim,J.M., and Voytas,D.F. (1996) The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev., 10, 634–645. [DOI] [PubMed] [Google Scholar]
- 13.Okazaki S., Ishikawa,H. and Fujiwara,H. (1995) TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm, Bombyx mori. Mol. Cell. Biol., 15, 4545–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi H., Okazaki,S. and Fujiwara,H. (1999) A new family of site-specific retrotransposon, SART1, is inserted into telomeric repeats of the silkworm, Bombyx mori. Nucleic Acids Res., 27, 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higashiyama T., Noutoshi,Y., Fujie,M. and Yamada,T. (1997) Zepp, a LINE-like retrotransposon accumulated in the Chlorella telomeric region. EMBO J., 16, 3715–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noutoshi Y., Arai,R., Fujie,M. and Yamada,T. (1998) Structure of the Chlorella Zepp retrotransposon: nested Zepp clusters in the genome. Mol. Gen. Genet., 259, 256–263. [DOI] [PubMed] [Google Scholar]
- 17.Arkhipova I.R. and Morrison,H.G. (2001) Three retrotransposon families in the genome of Giardia lamblia: two telomeric, one dead. Proc. Natl Acad. Sci. USA, 98, 14497–14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorbunova V. and Levy,A.A. (1997) Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res., 25, 4650–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saloman S. and Pucha,H. (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J., 17, 6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamada T. and Sakaguchi,K. (1982) Comparative studies on Chlorella cell walls: induction of protoplast formation. Arch. Microbiol., 132, 10–13. [Google Scholar]
- 21.Higashiyama T. and Yamada,T. (1991) Electrophoretic karyotyping and chromosomal gene mapping of Chlorella. Nucleic Acids Res., 19, 6191–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higashiyama T., Maki,S. and Yamada,T. (1995) Molecular organization of Chlorella vulgaris chromosome I: presence of telomeric repeats that are conserved in higher plants. Mol. Gen. Genet., 246, 29–36. [DOI] [PubMed] [Google Scholar]
- 23.Yamada T., Fujimoto,Y., Yamamoto,Y., Machida,K., Oda,M., Fujie,M., Usami.S. and Nakayama,H. (2003) Minichromosome formation in Chlorella cells irradiated with electron beams. J. Biosci. Bioeng., 95, 601–607. [PubMed] [Google Scholar]
- 24.Luan D.D., Korman,M.H., Jakubczac,J.L. and Eikbush,T.H. (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposon. Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt T. (1999) LINEs, SINEs and repetitive DNA: non-LTR retrotransposons in plant genomes. Plant Mol. Biol., 40, 903–910. [DOI] [PubMed] [Google Scholar]
- 26.Kumar A. and Bennetzen,J.L. (1999) Plant retrotransposon. Annu. Rev. Genet., 33, 479–532. [DOI] [PubMed] [Google Scholar]