Abstract
Short-range transcriptional repressors are locally acting factors that play important roles in developmental gene expression in Drosophila. To effect repression, Knirps and other short-range repressors bind the CtBP corepressor, but these repressors also function via CtBP-independent pathways. Possible mechanistic differences between CtBP-dependent and -independent repression activities are poorly understood. The distinct activities might provide qualitatively different activities necessary in different promoter contexts, or they might combine to give quantitatively different effects. We analyze separately the CtBP-dependent and CtBP-independent domains of Knirps previously characterized in the embryo to determine possible functional distinctions of the two repression activities. Both domains are active in cell culture and are dependent on the same residues required for activity in the embryo. The domains have similar properties with respect to distance-dependent repression and resistance to inhibition by the deacetylase inhibitor trichostatin A. In tests of repressor–activator specificity, the extent of repression was related not to the chemical nature of the activation domain but to the total activation potential. This result indicates that the balance of competing activation and repression signals is decisive in determining the effectiveness of repressors on genetic switches, suggesting that multiple repression activities are utilized to provide quantitatively, rather than qualitatively, distinct outputs.
INTRODUCTION
Transcriptional repressors play important and diverse roles in the regulation of gene expression in development. In Drosophila, early embryonic gene patterning requires the activity of a number of transcriptional repressors that have been characterized as short-range repressors, including the proteins encoded by the knirps, giant, Krüppel and snail genes (1,2). These proteins repress transcription when bound to sites within ∼100 bp of activators or the transcriptional initiation site, thus they have much more limited ranges of action than most transcriptional activators, which can function over long distances and from many different orientations (1,2). Short-range repression plays a vital role in precise control of complex transcriptional control regions, where individual enhancers in a locus can be shut down by the localized activity of the repressors, without impeding the activity of other nearby enhancers (3).
The knirps gene encodes a DNA binding transcription factor expressed in broad regions in the blastoderm embryo and in specific tissues later in embryogenesis (4). Knirps protein represses a number of target genes in the embryo, including the pair-rule gene even-skipped (eve), which is expressed in a series of seven transverse stripes under the control of modular enhancers (5). Knirps regulation is important for proper function of the eve stripe 3/7 and stripe 4/6 enhancers, and Knirps binding sites are found within these enhancers (6,7). Introduction of discrete Knirps binding sites within heterologous enhancers or near basal elements is sufficient to confer repression by the endogenous Knirps protein in transgenic embryo assays, and using these assays, the distance dependence of Knirps was mapped with respect to adjacent activator sites or the start site of transcription (8,9).
Critical to the normal activity of Knirps and other short-range repressor proteins is the ability to interact with the CtBP corepressor, an evolutionarily conserved protein implicated in transcriptional repression in Drosophila and vertebrates (10,11). CtBP is homologous to the family of α-hydroxyacid dehydrogenases, binds to NAD and NADH via a Rossman fold domain, and contains evolutionarily conserved residues in the presumptive active site. Recent reports suggest that CtBP possesses a weak dehydrogenase activity, in contrast to earlier findings (11–15). This corepressor interacts with the DNA binding repressors by contacting a small motif similar to P-DLS-K/R. Binding of CtBP to its protein targets has been suggested to be regulated by the redox state of the nucleus through the ratio of NAD/NADH (12). All proteins that have been characterized as short-range repressors in Drosophila have been found to functionally interact with CtBP, but CtBP also interacts with proteins whose range of action has not been defined, such as Brinker and Su(H), and it also appears to interact with the long-range repressor Hairy in an antagonistic mode (16–19). In vertebrates, CtBP interacts with the C-terminus of the adenovirus E1A protein, downregulating the transformation activity of E1A, and cellular factors such as the Ikaros and Zeb proteins, where it appears to contribute to their repression activity (reviewed in 10,11). The mechanism by which CtBP proteins mediate repression is not currently understood, but CtBP proteins have been reported to interact with histone deacetylase (HDAC) activities and proteins in several studies (14,20–24). Affinity purification of human CtBP has identified additional cofactors, including histone methyl transferases and the CoREST factor (15).
The CtBP cofactor is not the sole mediator of repression activity by Knirps, Giant and Krüppel. These proteins also repress transcription through CtBP-independent activities, and in the case of Knirps, the CtBP-dependent and CtBP-independent repression activities have been mapped to overlapping, but distinct portions of the protein (9,25). The biological significance of the multiple repression activities of short-range repressors is not understood, but different transcriptional regulatory elements appear to respond differently to the two activities. In the embryo, some enhancers require both CtBP-dependent and CtBP-independent activities for effective repression to occur, while other enhancers can be repressed solely by CtBP-independent means (9,25,26). For example, repression of the Krüppel gene in anterior regions requires both Giant and CtBP, while repression in central regions of the embryo requires only Giant (25). Similarly, the eve stripe 3/7 enhancer, which responds to low levels of Knirps, is repressed in the absence of CtBP, while the eve stripe 4/6 enhancer, which responds only to high levels of Knirps, requires CtBP (9; P.Struffi, M.Corado, M.Kulkarni and D.N.Arnosti, submitted). Distinct repression activities might employ qualitatively different mechanisms, allowing repression to occur in different settings, or the activities might be mechanistically similar, allowing for quantitatively greater repression when both activities are employed.
To better understand the functional characteristics of CtBP-dependent and CtBP-independent repression activities of Knirps, we have employed cell culture assays. Such assays are commonly used to measure repression activity, but it is not generally known whether transcription factors work the same way in transient transfections as they do in a native chromatin context. Thus, we previously correlated the activity of repressors in the embryo, where they are most likely to exhibit their normal activity, with their activity in cell culture, where we can more easily manipulate the system (9,27,28). In embryo studies, we have identified amino acid residues required for CtBP-dependent and CtBP-independent activity of Knirps, and the exact ranges of action (generally <100 bp from an activator or promoter). Using this information, we have compared here the separate repression functions of Knirps in parallel to determine possible qualitative or quantitative differences. Our results suggest that the two repression domains exhibit strikingly similar properties, suggesting their combination in vivo might be called upon in distinct developmental settings to provide quantitatively sufficient levels of repression by Knirps.
MATERIALS AND METHODS
Cloning of pAc2T and p5G2T reporters
The CMV basal promoter sequence from –50 to +75 was amplified from pUHC13-3 (29) using a 5′ oligonucleotide bearing BamHI, NotI and SphI sites and a 3′ oligonucleotide bearing an XhoI site and inserted into pBluescript cut with BamHI and XhoI to generate pBSCMV. Two Tet operator sites (2× TRE) were inserted into the SphI site of pBSCMV by ligating the following oligonucleotide 5′-GGTCGACTCCCTATCAGTGATAGAGAAAAGTGTCCCTATCAGTGATAGAGAGCATG-3′ (Tet binding sites underlined) and its complement into the plasmid to make p2T. The luciferase reporter plasmid pUHC13–3 (29) was digested with XhoI and BamHI to remove the original 7× TRE and CMV promoter, and an XhoI/BamHI fragment from p2T containing 2× TRE and the CMV basal promoter (–50 to +75) was inserted, creating p2TLuc. To generate pAc2T, a 2.2 kb of Drosophila Actin 5C enhancer fragment was amplified from pAX (29) with PCR primers 5′-AAGGAAAAAAGCGGCCGCATCATGAATGGCATCAACTCTG-3′ and 5′-AAGGAAAAAAGCGGCCGCTATGGGTATGAATTGTTCGCC-3′ (NotI sites underlined) digested with NotI, and then ligated into the NotI site of p2TLuc to obtain pAc2T. To make p5G2T, the five Gal4 binding sites from pUAS-lacZ (30) were PCR amplified with oligonucleotides bearing NotI sites and cloned into the NotI site of p2TLuc. To move the 2× TRE element from –50 in pAc2T to –75, –105 or –160, 25 or 55 bp spacers (9) were inserted into the SphI site between the basal promoter and TRE elements to obtain pAc2T-75luc, pAc2T-105luc and pAc2T-160luc.
Tet and Gal4 repressors and activators
To generate genes for Tet repressors, the Escherichia coli Tet-repressor DNA binding domain (amino acids 1–207) was amplified by PCR from pRevTet off VP16 (Clontech) using a 5′ oligonucleotide containing a HindIII site and a Kozak sequence 5′ of an initiation codon (underlined), 5′-GGGGCCAAGCTTGGATCCCAACCAAAATGTCAAGATTAGATAAAAGTAAAGTGATT-3′ and a 3′ oligonucleotide having in-frame KpnI and XbaI sites for insertion of repressor coding sequences 5′-GGGCCTCTAGAGCACGGGGATCCGGACCCACTTTCACATTTAAGTTGTTTTTCTAA-3′. The amplification product was digested with HindIII and XbaI and ligated with the previously digested pAX vector (27), producing pAXTet, which expresses the cDNA under control of the Actin 5C promoter. KpnI and XbaI fragments encoding transcriptional repressors were obtained from Drosophila transformation vectors encoding Gal4 chimeras (9,28), and ligated with pAxTet to obtain pTet-Knirps N, Nmut, C, Cmut, pTet-Giant and pTet-CtBP. Gal4-Groucho was obtained from A. Courey (31) and Tet-VP16 was described in Chen et al. (27). pAXGal4-VP16, pAXGal4-AD and pAXGal4-Sp1 were created by cloning BamHI/XbaI fragments encoding Gal4 (1–92) activators (32) into pAX (27) to express that gene under the actin 5C enhancer. Gal4-Bicoid Q and Gal4-Bicoid 3Q were obtained from N. Dostatni (33).
Cell culture and transient transfections
Drosophila S2 cells (American Type Culture Collection) were grown at 22–24°C in Schneider Drosophila medium (Gibco BRL) containing 10% fetal bovine serum and penicillin/streptomycin. For reporter assays, Drosophila S2 cells were plated in 12-well plates at a density of 1 × 106 cells per well and transfected by the calcium phosphate method. Transfection cocktails contained 0.5 µg of either pAc2T or p5G2T, 0.25 µg of pTet repressor, variable amounts of pAcGal4-fused activators and 0.1 µg of pAc/lacZ internal control for each well. pBluescript was used to equalize the total amount of DNA transfected. After 5 h, 1 ml of medium with or without 1 µg/ml doxycycline was added to the cells, which were then incubated for 24 h. The medium was changed once more, with or without 1 µg/ml doxycycline or addition of trichostatin A (TSA) (Sigma) in 10% DMSO or vehicle alone, then cells were incubated for 20 h. Cells were harvested and lysed with reporter assay lysis buffer (Promega). Luciferase assays were performed following the manufacturer’s instructions (Promega) and analyzed with a Berthold Lumat 9501. Each experiment was performed in duplicate at least three times. Luciferase activity was corrected for the transfection efficiency using β-gal activity as an internal control and to a normalized Bradford protein assay (Bio-Rad).
Gel shift analysis
Drosophila S2 cells were plated in 100-mm plates at a density of 1 × 107 cells per plate and transfected with 20 µg of each chimeric construct. After 48 h, the nuclear extracts were prepared and analyzed by gel shift assay as previously described (27).
RESULTS
Short-range repressors and reporters
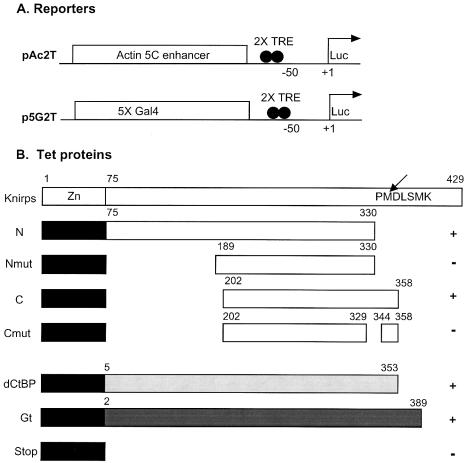
To assay independently the CtBP-dependent and -independent repression activities of Knirps, we fused CtBP-independent (Knirps N) and CtBP-dependent (Knirps C) portions of Knirps to the Tet-repressor DNA binding domain (Fig. 1). Previous studies in embryos indicated that these domains can repress when fused to a heterologous DNA binding protein (9,25,28). The Tet DNA binding domain used here binds to DNA in the absence of the drug doxycycline (27). Addition of the drug releases the repressor from the reporter gene and abolishes repression, providing a way to verify that effects on reporter gene expression are direct. As additional controls, mutant proteins that were inactive in embryos were tested, as well as the TetR DNA binding domain itself (‘Stop’). The Knirps repressor proteins were tested in parallel with the entire Giant repression domain and the Drosophila CtBP corepressor, both of which are short-range repressors in embryos and have been tested in cell culture assays (25,27,34). The repressors were targeted to promoters of genes activated by the actin 5C enhancer or a synthetic enhancer activated by Gal4 activator fusions (Fig. 1A).
Figure 1.
Reporters and short-range repressors tested. (A) Assays utilized luciferase reporter genes activated by the actin 5C enhancer or a cluster of Gal4 binding sites. Two Tet operator binding sites are located at –50 bp with respect to the transcriptional initiation site. (B) A schematic diagram of the Knirps repressor, with CtBP binding motif (arrow, PMDLSMK) between residues 331 and 337 as indicated. N-terminal CtBP-independent repression domain (75–330) and C-terminal CtBP-dependent repression domains were fused to the Tet-repressor DNA binding domain (black box). Inactive mutant proteins Nmut (189–330) and Cmut (202–329, 344–358) were also assayed as controls. The entire Giant repression domain (Gt) and the CtBP corepressor protein were also fused to the Tet DNA binding domain. Pluses and minuses (right) indicate the previously determined activity of the repression domains in embryo assays (9,25).
Expression of Tet-repressor chimeras
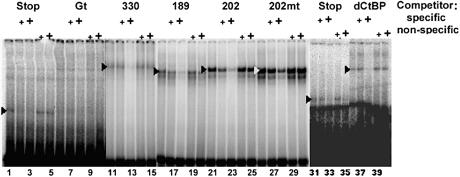
To compare protein expression levels of transfected genes, nuclear extracts from cells expressing the recombinant proteins were tested by gel electrophoretic mobility shift assays (Fig. 2). Expression levels of Tet-repressor proteins differed but, importantly, the expression of the wild-type Knirps fragments (Knirps N and C, lanes 11 and 21) were very similar to the corresponding mutant forms (Knirps Nmut and Cmut, lanes 16 and 26), indicating that the inactivity of the mutants was not due to lack of expression or DNA binding activity. The specificity of binding was established by the ability to compete the complexes with excess specific, but not with non-specific, DNA probes. The TetR DNA binding domain and the CtBP repressor were expressed at lower, but detectable levels. The Giant protein was not detected at all, but was functional, as shown in the following experiments.
Figure 2.
Measurement of Tet-repressor binding activity. Nuclear extracts of S2 Schneider cells transfected with genes for Tet-repressor constructs were assayed by gel electrophoretic mobility shift. Specificity of binding was tested by the addition of 10- or 100-fold molar excess specific or non-specific unlabeled oligonucleotide. Positions of complexes formed by Stop (lanes 1–5, 31–35), Knirps N-terminal CtBP-independent repression domain (N, lanes 11–15), an inactive mutant (Nmut, lanes 16–20), Knirps C-terminal CtBP-dependent repression domain (C, lanes 21–25), an inactive mutant (Cmut, lanes 26–30) and CtBP (dCtBP, lanes 36–40) are marked with arrowheads. Levels of Giant protein were undetectable, even at longer exposures. The inactive Knirps mutant proteins (Nmut and Cmut) were expressed at equal or slightly higher levels than the corresponding active proteins, demonstrating that the inactivity of the mutant proteins is not simply due to poor expression or binding. The Giant and CtBP proteins, which were more active than the Knirps repressors in some assays, were expressed at lower levels than the Knirps N and Knirps C proteins, indicating that their potency is not simply due to higher expression levels.
Both CtBP-independent and CtBP-dependent Knirps repression domains are active in cell culture assays
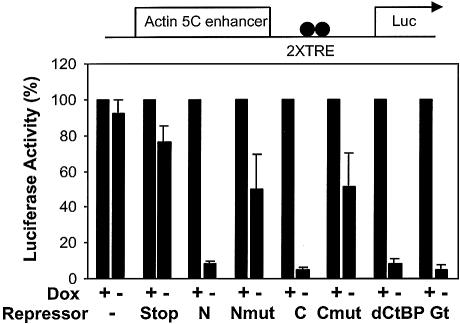
Repression activity of the Tet-fusion proteins was assayed on a reporter activated by the actin 5C enhancer, with two promoter-proximal Tet R binding elements. Tet-Knirps fusion proteins containing intact CtBP-dependent (C) and CtBP-independent (N) repression domains mediated ∼10-fold repression (Fig. 3). A deletion of 13 amino acids that removes the CtBP binding motif from Knirps C reduced repression activity from ∼10- to only ∼2-fold, indicating that the bulk of the repression activity is dependent on CtBP binding. A mutation in the N-terminal, CtBP-independent repression domain also reduced repression activity from ∼10- to ∼2-fold. Repression in all cases was abolished by treatment with doxycycline, indicating that the repression activity was dependent on DNA binding by the repressor proteins. Repression levels by CtBP and Giant in these assays were comparable with those achieved with Knirps N or C.
Figure 3.
Both CtBP-dependent and CtBP-independent Knirps repression domains function as repressors in transient assays. An actin 5C activated luciferase reporter construct was cotransfected into Schneider cells with Tet-repressor constructs. Ten-fold or greater repression was noted in the presence of either Knirps N or C repression domains, similar to that exhibited by the CtBP and Giant proteins. Considerably weaker (2-fold) effects were noted with mutant repression domains previously found to be inactive in the embryo (9). Activity of the reporter gene in the presence of doxycycline (Dox) (conditions under which the repressor proteins do not bind DNA) was used to normalize the reporter readout. (The absolute differences between luciferase activities in the presence of doxycycline were <50%.) Transfection efficiency was corrected for by normalizing to the signal from a cotransfected lacZ reporter and total protein (correction for transfection efficiency typically changed the uncorrected values <5%). Experiments in this and subsequent figures were performed in triplicate, with duplicate readings of each extract; standard deviations are shown.
Both Knirps repression domains exhibit distance-dependent repression
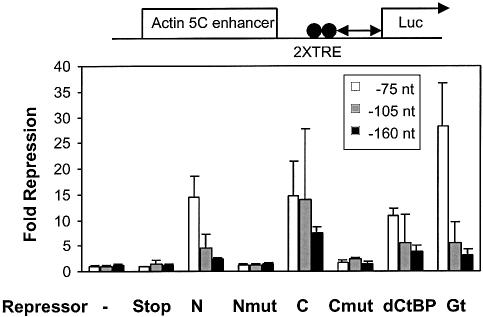
In embryos, Knirps and Giant can repress when bound near the basal promoter, but repression is highly distance dependent, showing marked attenuation if the proteins are over ∼100 bp from the initiation site (9,28). The Actin 5C reporter plasmid was modified to move the Tet binding sites from –50 to –75, –105 or –160 bp with respect to the transcriptional initiation site, and the panel of repressor proteins was assayed in transient transfection assays (Fig. 4). (In these assays, activity of repressors was identical with binding sites at –50 or –75 bp.) All repressors that were active on the reporter with binding sites at –75 bp showed a marked drop-off in repression activity on templates with distal binding sites at –160 bp. For Knirps N and Giant, there was a significant loss of repression at –105 bp. This strong distance dependence closely mirrors the effects seen on chromatinized, integrated trangene reporter genes in embryos, and indicates for the first time that the distance effects of Drosophila short-range repressors is equally apparent in cell culture assays. These assays also demonstrate that the overall distance dependence of the intact Knirps repressor is mirrored by similar short-range activities of the Knirps N and Knirps C repression domains. As expected, on all templates, the Knirps mutant proteins exhibited a low and unchanged activity.
Figure 4.
Knirps, Giant and CtBP repressors show strong distance dependence in transfection assays. Actin 5C enhancer reporters containing Tet operator sites at –75 (open bars), –105 (shaded bars) or –160 (black bars) were cotransfected with Tet-repressor constructs. The Tet DNA binding domain and inactive Knirps mutant had little or no activity on all templates, while the Knirps N and C repression domains, as well as Giant and CtBP, showed a sharp distance dependence, with fold repression close to background levels at –160 bp, similar to the distance effects noted with endogenous repressor proteins in transgenic embryos (9,28). Fold repression was calculated by dividing the signal obtained in the presence of doxycycline (no repression) by the signal obtained in the absence of doxycycline (repression).
Effect of histone deacetylase inhibitor trichostatin A
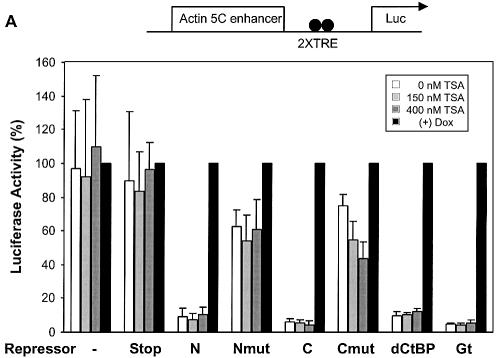
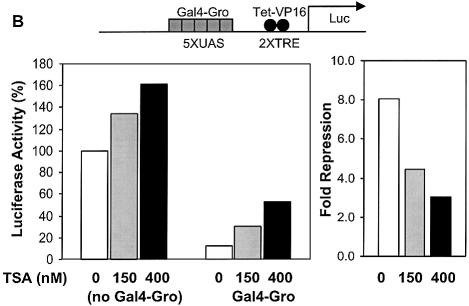
A number of transcriptional repressors have been found to interact with HDACs, enzymes that may facilitate repression by deacetylating histone tails and forming a more compact chromatin structure (31,35). However, it is not known if HDAC activities play a role in short-range repression. Therefore, we tested the effect of the HDAC inhibitor TSA on repression mediated by Knirps, Giant and CtBP. Repression levels measured on the actin 5C reporter were not significantly affected by the addition of 150 or 400 nm TSA, suggesting that these repressors act in a TSA insensitive manner (Fig. 5A). To ascertain that these levels of the drug were effective under these conditions, we tested in parallel other repressors proven to be sensitive to TSA. Because Tet-Rpd3 and Tet-Gro fusions proved unstable, we measured repression by a Gal4-Groucho repressor, a protein that has been demonstrated to interact with the Rpd3 HDAC (31). As expected, the repression effectiveness of this protein was TSA sensitive, showing a dose-dependent loss of repression activity, from 8-fold in the absence of TSA to 3-fold in the presence of 400 nM TSA, results similar to previous findings (Fig. 5B) (31). Similar results were noted with TSA inhibition of Gal4-Rpd3 repression activity (data not shown).
Figure 5.
Repression by Knirps, Giant and CtBP resistant to HDAC inhibitor TSA. (A) Addition of 0 (open bars), 150 (light-gray bars) or 400 nM TSA (dark-gray bars) had no effect on repression of reporter activity by active Tet repressors. Weak repression activity by Knirps Cmut appeared to slightly increase at the highest levels of added TSA. Activities were normalized to those obtained in the presence of doxycycline (no repression). (B) TSA effect on Groucho repression as a control for TSA treatment effectiveness. Cells grown and transfected under identical conditions as in (A) were cotransfected with a reporter containing binding sites for a Gal4-Groucho corepressor and the VP16 activator, in the presence of 0, 150 or 400 nM TSA (left). In the absence of TSA, reporter activity is repressed 8-fold by Gal4-Groucho (right). Repression effectiveness was decreased to 3-fold by the addition of TSA. A non-specific stimulation of this reporter by TSA was noted previously (31). Results shown are representative of two experiments.
Repressor–activator specificity
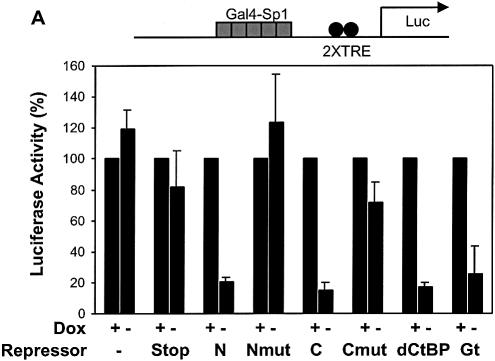
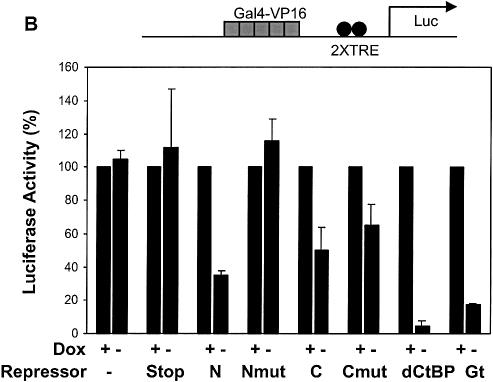
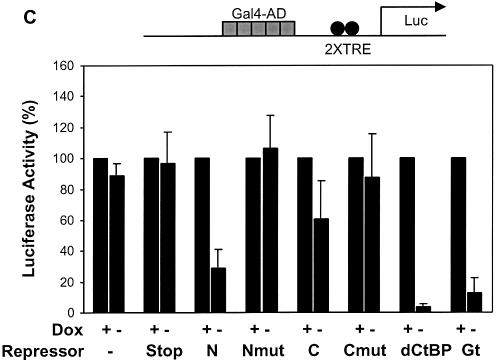
Some CtBP-interacting repressors have been reported to exhibit activator specificity (36); therefore, we tested whether the repression domains would show specificity against distinct activation domains. Gal4-activator proteins were targeted to a reporter containing five Gal4 binding sites upstream of two Tet operator sequences. On this reporter, activation by the relatively weak Sp1 activation domain was strongly inhibited by both Knirps N and Knirps C, as well as Giant and CtBP (Fig. 6A). The mutant repressor proteins showed little or no repression activity in this assay, demonstrating the specificity of the repression activity. In contrast, when the same reporter was activated by the stronger Gal4 or VP16 activation domains (>1000-fold higher activity on this reporter/µg transfected plasmid), the Knirps C repressor domain was not significantly more effective than the mutant (≤2-fold repression) (Fig. 6B and C). Repression by the Knirps N domain was ∼3-fold, better than the mutant control, but not as potent as the Giant and CtBP repression domains (6- and 20-fold, respectively). The weak level of repression exhibited by both Knirps N and C against the strong transcriptional activators distinguishes these repressors from Giant and CtBP. As discussed below, it seems unlikely that this difference in effectiveness is a reflection of mechanistic differences between CtBP and Knirps, at least in the case of the Knirps C domain, which works by interacting with CtBP. It is possible that a higher local concentration of CtBP in the case of Tet-CtBP permits repression even in the presence of potent activators.
Figure 6.
Inverse correlation between repression and activator strength. A reporter activated by a cluster of five Gal4 binding sites was cotransfected with plasmids expressing Gal4-activator and Tet-repressor proteins. (A) Gal4-Sp1-activated transcription was strongly inhibited (≥5-fold repression) by both Knirps N and C domains, while mutants were inactive. CtBP and Giant exhibited repression activity similar to the Knirps repression domains. (B and C) Repression of Gal4 VP16 and Gal4 AD activators. Knirps C did not exhibit activity greater than the mutant form of the protein, while the Knirps N showed only weak activity. CtBP and Giant mediated 5–20-fold repression. Signals were normalized to those obtained in the presence of doxycyline (no repression).
Activation quantity, not type of activator domain, affects repression
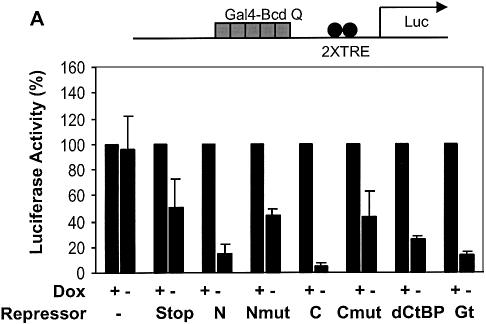
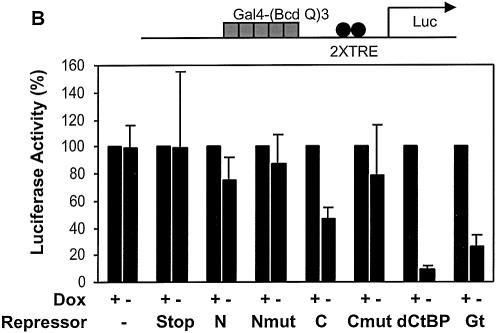
Unlike the case with the Sp1 activator, Gal4 and VP16 were poorly repressed by Knirps N, and resistant to repression by Knirps C, perhaps reflecting a qualitatively unique activation mechanism. Alternatively, the differences in repressibility might be due to quantitative effects, and not associated with a particular activation pathway or the chemical nature of the activation domain. To distinguish between these two possibilities, we assayed transcription driven by two versions of the same activator, the glutamine-rich activation domain of Bicoid (33). A single copy of this activation domain fused to the Gal4 DNA binding domain works as a weak activator, comparable with the Sp1 activation domain. Both Knirps N and Knirps C repressed reporters activated by the Bcd Q domain, with mutant repression domains showing only weak effects, similar to those seen with the actin 5C enhancer (Fig. 7A). In contrast to the weak activity of BcdQ, three copies of the Bcd Q activation domain fused together provided a strong activation signal, >100-fold that seen with the monomeric form (33). This activator was not significantly repressed by Knirps N and only weakly by Knirps C (Fig. 7B). Similar to the situation with the Gal4 and VP16 activators, Giant and CtBP were more effective than Knirps, mediating ∼3-fold and ∼7-fold repression, respectively. Thus, at least in the case of the Bicoid activation domain, the ability of Knirps repression domains to function effectively seems related to the overall strength of activation output, rather than the chemical nature of the activation domain.
Figure 7.
Activator strength, rather than the nature of the activation domain, dictates repression effectiveness. (A) The weak Gal4-Bicoid activator containing one copy of the glutamine-rich domain was effectively repressed by both Knirps N and C domains (7–15-fold), significantly above that seen with mutant repression domains (∼2-fold). Giant and CtBP both repressed expression, although CtBP was reproducibly weaker in this context than in others tested. (B) A strong Gal4-Bicoid activator containing three copies of the same glutamine-rich domain was not effectively repressed by either Knirps N or C domains, relative to the mutant proteins. CtBP was relatively more effective against this activator, mediating ∼10-fold repression, while Giant mediated ∼4-fold repression.
DISCUSSION
Recent work has established that short-range transcriptional repressors act through CtBP-dependent and CtBP-independent pathways, but the biological relevance of this dual mode of action is not well understood. Furthermore, the biochemical mechanisms employed by these activities are unknown. As an initial step in understanding the activity of short-range repressors, we have characterized the individual repression activities of Knirps, the first case where the separate activities of a bona fide short-range repressor have been studied in parallel. We demonstrate that both CtBP-dependent and CtBP-independent activities can function in transfection assays (Fig. 3). The similar distance dependence exhibited by Knirps N and Knirps C might indicate that similar mechanisms are used, despite the differential requirement for the CtBP corepressor. The distance-dependent repression seen in transfection assays is strikingly similar to that observed in embryo assays with endogenous proteins, underscoring the parallels between the two systems (Fig. 4) (9,28). The chromatin state of transiently transfected DNA is not well defined, but transiently transfected genes are generally found to be more susceptible to activation than are the endogenous counterparts present in the chromosomes, suggesting a looser chromatin configuration. Clearly, the activities of short-range repressors do not require the type of chromatin configuration present in the chromosome, a point which is supported by previous studies with intact repression domains. From these results, it is also clear that the promoter proximal repression activity is not just due to steric hindrance or occlusion of the basal machinery, for Knirps C and Cmut are of virtually identical size, yet show dramatic differences in activity.
HDAC enzymes have been implicated in the activity of many transcriptional repressors (31,35). Although CtBP itself has not been reported to possess HDAC activity, the protein can be coimmunoprecipitated with HDACs or deacetylase activity. In some studies, CtBP-mediated repression was reported to be inhibited by the HDAC inhibitor TSA (37), but other studies found CtBP-mediated repression to be TSA insensitive (21,22). Using conditions where we could observe TSA effects on the known HDAC-dependent repressor Groucho, we observed a complete insensitivity of Giant, Knirps and CtBP to TSA, strongly suggesting that repression by these proteins does not involve a TSA-sensitive HDAC activity (Fig. 5A and B). Consistent with this finding, a mutation in the HDAC rpd3 gene does not perturb repression mediated by short-range repressors (38). However, other HDACs, resistant to TSA, might function as cofactors. Indeed, TSA-insensitive class II HDACs have been found to bind to vertebrate CtBP (24).
We previously identified two factors that were critical for the effective function of short-range repressors: the distance between activators and repressors, and the concentration of activators (9,25,28). An issue that had not been previously explored was whether the nature of the activation domain might play an important role in responding to a repressor. Some CtBP-dependent mammalian repressor domains do exhibit repressor activator specificity; for example, the C-terminus of the ZEB repressor that contains a CtBP binding motif is specific for the muscle-specific MEF2 transcriptional activator (36). Drosophila repressors have been tested against a range of endogenous transcriptional activators, but no evidence of activator specificity has been identified. Our results indicate susceptibility to repression appears to be a function of the total transcriptional output rather than the chemical nature of the activation domain. This point is made especially effectively in the comparison of two versions of the Bicoid glutamine-rich activation domain. Activation mediated by proteins carrying one copy of the Bicoid glutamine-rich activation domain is effectively blocked by Knirps, Giant and CtBP, while a stronger activator consisting of three copies of the same domain is resistant to Knirps repression (Fig. 7A and B). Interestingly, this same activation domain is repressed by a tethered CtBP molecule, despite the weak activity of the Knirps C domain that is wholly dependent on its CtBP binding motif. Perhaps tethering CtBP directly via a DNA binding domain raises the local concentration of the factor more effectively than tethering it via the Knirps C domain. The effectiveness of Giant, another CtBP-dependent repressor, against the more potent activators, also suggests that the quantitative level of CtBP repression can vary, depending on how the cofactor is tethered to the promoter. Thus, rather than showing activator specificity, these short-range repressors appear to counterbalance activation potential, and mediate repression when the activation potential is not too great, whether from a high level of activator protein, or from the presence of a potent activation domain. A similar situation is observed in the embryo, where the susceptibility of Gal4 activators to repression by Giant and Knirps is dependent on the number and affinity of the activator binding sites (M. Kulkarni, unpublished observations).
Previously, the observation that distinct activities of Knirps or Giant were required to repress different enhancers in vivo raised the possibility that their CtBP-dependent and CtBP-independent repression activities had distinct mechanisms of repression that were suitable in different contexts (9,25,26). The similarity of function of the two Knirps repression domains, as well as their differential responses to quantitatively different levels of transcriptional activation, suggests instead that combining these activities provides quantitatively, rather than qualitatively, different levels of repression. In the case of repression of the Krüppel gene by Giant, in anterior regions, high levels of the Bicoid activator would require combined short-range repressor activities, while in central regions, where activator concentrations are lower, the CtBP-independent repression alone would suffice. Similarly, a single repression activity by Knirps suffices to repress eve stripe 3/7, which can respond to very low levels of Knirps, perhaps because the large number of Knirps binding sites loads a large amount of the repressor on this element (6,7). Thus, proper regulatory decisions by developmentally regulated cis elements are proposed to reflect quantitative balances between activator and repressor outputs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Kulkarni, M. Sutrias-Grau and two anonymous reviewers for helpful comments, A. Courey and N. Dostatni for plasmids, and E. Fernandez-Villatoro for technical assistance. This work was supported by NIH Grant GM56976 to D.N.A.
REFERENCES
- 1.Gray S. and Levine,M. (1996). Transcriptional repression in development. Curr. Opin. Cell Biol., 8, 358–364. [DOI] [PubMed] [Google Scholar]
- 2.Courey A.J. and Jia,S. (2001) Transcriptional repression: the long and the short of it. Genes Dev., 15, 2786–2796. [DOI] [PubMed] [Google Scholar]
- 3.Small S., Arnosti,D.N. and Levine,M. (1993) Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development, 119, 767–772. [PubMed] [Google Scholar]
- 4.Chen C.K., Kuhnlein,R.P., Eulenberg,K.G., Vincent,S., Affolter,M., and Schuh,R. (1998) The transcription factors KNIRPS and KNIRPS RELATED control cell migration and branch morphogenesis during Drosophila tracheal development. Development, 125, 4959–4968. [DOI] [PubMed] [Google Scholar]
- 5.Fujioka M., Emi-Sarker,Y., Yusibova,G.L., Goto,T. and Jaynes,J.B. (1999) Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers and multi-stripe positioning by gap gene repressor gradients. Development, 126, 2527–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small S., Blair,A. and Levine,M. (1996) Regulation of two pair-rule stripes by a single enhancer in the Drosophila embryo. Dev. Biol., 175, 314–324. [DOI] [PubMed] [Google Scholar]
- 7.Berman B.P, Nibu,Y., Pfeiffer,B.D., Tomancak,P., Celniker,S.E., Levine,M., Rubin,G.M. and Eisen,M.B. (2002) Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl Acad. Sci. USA, 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnosti D.N., Gray,S., Barolo,S., Zhou,J. and Levine,M. (1996) The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J., 15, 3659–3666. [PMC free article] [PubMed] [Google Scholar]
- 9.Keller S.A., Mao,Y., Struffi,P., Margulies,C., Yurk,C.E., Anderson,A.R., Amey,R., Moore,S., Ebels,J.M., Foley,K., Corado,M. and Arnosti,D.N. (2000) dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein. Mol. Cell. Biol., 20, 7247–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinnadurai G. (2002) CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell, 9, 213–224. [DOI] [PubMed] [Google Scholar]
- 11.Turner J. and Crossley,M. (2001) The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bioessays, 23, 683–690. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q., Piston,D.W. and Goodman,R.H. (2002) Regulation of corepressor function by nuclear NADH. Science, 295, 1895–1897. [DOI] [PubMed] [Google Scholar]
- 13.Kumar V., Carlson,J.E., Ohgi,K.A., Edwards,T.A., Rose,D.W., Escalante,C.R., Rosenfeld,M.G. and Aggarwal,A.K. (2002) Transcription corepressor CtBP is an NAD(+)-regulated dehydrogenase. Mol. Cell, 10, 857–869. [DOI] [PubMed] [Google Scholar]
- 14.Balasubramanian P., Zhao,L.J. and Chinnadurai,G. (2003) Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett., 537, 157–160. [DOI] [PubMed] [Google Scholar]
- 15.Shi Y., Sawada,J., Sui,G., Affar,E.B., Whetstine,J.R., Lan,F., Ogawa,H., Luke,M.P.-S., Nakatani,Y. and Shi,Y. (2003) Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature, 422, 735–738. [DOI] [PubMed] [Google Scholar]
- 16.Hasson P., Muller,B., Basler,K. and Paroush,Z. (2001) Brinker requires two corepressors for maximal and versatile repression in Dpp signalling. EMBO J., 20, 5725–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morel V., Lecourtois,M., Massiani,O., Maier,D., Preiss,A. and Schweisguth,F. (2001) Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol., 11, 789–792. [DOI] [PubMed] [Google Scholar]
- 18.Poortinga G., Watanabe,M. and Parkhurst,S.M. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H. and Levine,M. (1999) Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sundqvist A., Sollerbrant,K. and Svensson,C. (1998) The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein-histone deacetylase complex. FEBS Lett., 429, 183–188. [DOI] [PubMed] [Google Scholar]
- 21.Sundqvist A., Bajak,E., Kurup,S.D., Sollerbrant,K. and Svensson,C. (2001) Functional knockout of the corepressor CtBP by the second exon of adenovirus E1a relieves repression of transcription. Exp. Cell Res., 268, 284–293. [DOI] [PubMed] [Google Scholar]
- 22.Koipally J. and Georgopoulos,K. (2000) Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem., 275, 19594–19602. [DOI] [PubMed] [Google Scholar]
- 23.Melhuish T.A. and Wotton,D. (2000) The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J. Biol. Chem., 275, 39762–39766. [DOI] [PubMed] [Google Scholar]
- 24.Zhang C.L., McKinsey,T.A., Lu,J.R. and Olson,E.N. (2001) Association of COOH-terminal-binding protein (CtBP) and MEF2-interacting transcription repressor (MITR) contributes to transcriptional repression of the MEF2 transcription factor. J. Biol. Chem., 276, 35–39. [DOI] [PubMed] [Google Scholar]
- 25.Strunk B., Struffi,P., Wright,K., Pabst,B., Thomas,J., Qin,L. and Arnosti,D.N. (2001) Role of CtBP in transcriptional repression by the Drosophila giant protein. Dev. Biol., 239, 229–240. [DOI] [PubMed] [Google Scholar]
- 26.La Rosee-Borggreve A., Hader,T., Wainwright,D., Sauer,F. and Jäckle,H. (1999) hairy stripe 7 element mediates activation and repression in response to different domains and levels of Kruppel in the Drosophila embryo. Mech. Dev., 89, 133–140. [DOI] [PubMed] [Google Scholar]
- 27.Ryu J.R., Olson,L.K. and Arnosti,D.N. (2001) Cell-type specificity of short-range transcriptional repressors. Proc. Natl Acad. Sci. USA, 98, 12960–12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewitt G.F., Strunk,B.S., Margulies,C., Priputin,T., Wang,X.-D., Amey,R., Pabst,B.A., Kosman,D., Reinitz,J. and Arnosti,D.N. (1999) Transcriptional repression by the Drosophila giant protein: cis element positioning provides an alternative means of interpreting an effector gradient. Development, 126, 1201–1210. [DOI] [PubMed] [Google Scholar]
- 29.Gossen M., Freundlieb,S., Bender,G., Muller,G., Hillen,W. and Bujard,H. (1995) Transcriptional activation by tetracyclines in mammalian cells. Science, 268, 1766–1769. [DOI] [PubMed] [Google Scholar]
- 30.Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 31.Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seipel K., Georgiev,O. and Schaffner,W. (1992) Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J., 11, 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janody F., Sturny,R., Schaeffer,V., Azou,Y. and Dostatni,N. (2001) Two distinct domains of Bicoid mediate its transcriptional downregulation by the Torso pathway. Development, 128, 2281–2290. [DOI] [PubMed] [Google Scholar]
- 34.Phippen T.M., Sweigart,A.L., Moniwa,M., Krumm,A., Davie,J.R. and Parkhurst,S.M. (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J. Biol. Chem., 275, 37628–37637. [DOI] [PubMed] [Google Scholar]
- 35.Ng H.H. and Bird,A. (2000) Histone deacetylases: silencers for hire. Trends Biochem. Sci., 25, 121–126. [DOI] [PubMed] [Google Scholar]
- 36.Postigo A.A. and Dean,D.C. (1999) Independent repressor domains in ZEB regulate muscle and T-cell differentiation. Mol. Cell. Biol., 19, 7961–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Criqui-Filipe P., Ducret,C., Maira,S.M. and Wasylyk,B. (1999) Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J., 18, 3392–3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mannervik M. and Levine,M. (1999) The Rpd3 histone deacetylase is required for segmentation of the Drosophila. Proc. Natl Acad. Sci. USA, 96, 6797–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]