Abstract
Lysosomal acid lipase (LAL) hydrolyzes cholesteryl esters and triglycerides to generate free fatty acids and cholesterol in the cell. The downstream metabolites of these compounds serve as hormonal ligands for nuclear receptors and transcription factors. Genetic ablation of the lal gene in the mouse caused malformation of macrophages and inflammation-triggered multiple pathogenic phenotypes in multiple organs. To assess the relationship between macrophages and lal−/− pathogenic phenotypes, a macrophage-specific doxycycline-inducible transgenic system was generated to induce human LAL (hLAL) expression in the lal−/− genetic background under control of the 7.2-kb c-fms promoter/intron2 regulatory sequence. Doxycycline-induced hLAL expression in macrophages significantly ameliorated aberrant gene expression, inflammatory cell (neutrophil) influx, and pathogenesis in multiple organs. These studies strongly support that neutral lipid metabolism in macrophages contributes to organ inflammation and pathogenesis.
Macrophages produce and secrete cytokines, chemokines, and growth factors that influence gene expression, cell proliferation/differentiation, and apoptosis in organ tissues through paracrine and autocrine mechanisms, making them vitally important to the physiological functions of multiple organs. Many diseases are tightly associated with malformation and malfunction of macrophages within organs. Design and establishment of a system to specifically express “genes of interest” in macrophages in a temporal/spatial fashion will greatly assist in understanding the molecular mechanisms of macrophage differentiation/maturation and the functional roles of macrophages in disease formation. The tetracycline-inducible transgenic mouse system has proven to be very effective in inducing genes of interest in a temporal and spatial manner. In this system, “activator” transgenic mice bear the reverse tetracycline-responsive transactivator (rtta) fusion protein under control of a cell type-specific promoter. The rtta expression is restricted to specific cell types in transgenic mice. In a separate transgenic mouse line, the gene of interest is under control of the tet operator DNA-binding sequence that is linked to a minimal promoter. After crossbreeding, expression of the gene of interest can be precisely controlled by addition or removal of tetracycline or doxycycline in double transgenic mice. This strategy has been proven to be very effective in many organ systems, including the lung.1,2
To design a tetracycline-controlled macrophage-specific system, the key is to select a macrophage-specific promoter. It has been reported previously that the c-fms gene, which encodes the receptor for macrophage colony-stimulating factor (CSF-1), is selectively expressed in macrophage and trophoblast cell lineages. In a transgenic mouse study, 7.2-kb of the 5′-flanking regulatory sequence and the downstream intron 2 of the c-fms gene have been defined to direct specific expression of the green fluorescent protein (GFP) reporter gene in the same locations as the endogenous gene, therefore providing an ideal DNA sequence to direct gene expression in mononuclear phagocyte lineage during embryonic development and in bone marrow, peripheral blood, and all adult tissues.3
Cholesteryl esters and triglycerides are important components in neutral lipids, which can be hydrolyzed by lysosomal acid lipase (LAL) in the lysosome of cells to generate free cholesterol and free fatty acids. After LAL cleaves these lipids, they exit the lysosome and enter the cytosol. An lal−/− mouse model has been generated by gene targeting.4 lal−/− mice developed pathogenic phenotypes in multiple organs, including lung, liver, spleen, adrenal glands, and small intestine.4–6 These pathogenic phenotypes are caused by abnormal gene expression. For example, Affymetrix GeneChip microarray analysis of lal−/− mice showed aberrant gene expression for inflammatory cytokines/chemokines, endopeptidases (eg, matrix metalloproteinases), apoptosis inhibitors, transcription factors, and oncogenes in the lung.5,7 These gene profile changes correlated well with pulmonary phenotype progression in the lal−/− lung in an age-dependent manner. It is known that many metabolic derivatives of free cholesterols and free fatty acids serve as hormonal ligands for nuclear receptors and transcription factors that have profound and diverse functions in gene regulation, cell proliferation, differentiation, and apoptosis. In a rescue study, treatment of peroxisome proliferator-activated receptor γ (PPARγ) ligands (downstream metabolites of LAL) significantly improved lal−/− pulmonary inflammation and aberrant gene expression, suggesting that inflammation-triggered pathogenesis during LAL deficiency is partially caused by inactivation of PPARγ because of a lack of ligand production. Because abnormal appearance of macrophages is the major manifestation of lal−/− mice within multiple organs, we hypothesize that a deficiency of LAL greatly affects the normal functions of macrophages and leads to various pathogenic phenotypes.
To test this hypothesis, doxycycline-controlled c-fms 7.2-kb promoter/intron2-rtTA and (tetO)7-CMV-hLAL transgenic mouse lines were generated. After crossbreeding the double transgenic lines into lal−/− mice, expression of hLAL in macrophages significantly ameliorated tissue inflammation, pathogenic phenotypes, and aberrant gene expression in several organs, supporting a concept that cholesteryl ester and triglyceride metabolism is essential for the normal biological function of macrophages.
Materials and Methods
Animal Care
All scientific protocols involving the use of animals in this study have been approved by the Institution Animal Care and Usage Committee and followed guidelines established by the Panel on Euthanasia of the American Veterinary Medical Association. Protocols involving the use of recombinant DNA or biohazardous materials followed guidelines established by the National Institutes of Health. Animals were housed in an Institution Animal Care and Usage Committee-approved facility. Animals were regularly screened for common respiratory pathogens and murine viral hepatitis. Experiments involving animal sacrifice used CO2 narcosis to minimize animal discomfort.
Generation of Transgenic Mice
To generate c-fms-rtTA transgenic mice, the cDNA fragment of rtTA was amplified by polymerase chain reaction (PCR) using a downstream primer (5′-AAGGAAAAAAGCGGCCGCGTACATTGAGCAACTGAC-3′) and an up-stream primer (AAGGAGGGCCCGCCACCATGTCTAGATTAGATAAA-3′). The PCR product was digested with ApaI/NotI and subcloned into the p7.2ΔMCS construct containing the 7.2-kb c-fms promoter/intron2 fragment. The c-fms-rtTA expression cassette containing the 7.2-kb c-fms promoter/intron2 fragment, the rtTA cDNA, and the SV40 polyadenylation signaling sequence was microinjected into eggs of FVB/N mice by the Transgenic Core Facility at the University of Cincinnati, College of Medicine. Founder lines were identified by a pair of primers covering the 7.2-kb c-fms promoter/intron2 sequence (5′-TGATTGAAGGGTCCAGACTCATTC-3′) and an rtTA cDNA coding region sequence (5′-AGTGTAGGCTGCTCTACACCAAGC-3′).
To generate the (tetO)7-CMV-hLAL transgenic mouse line, hLAL cDNA was amplified by PCR using a downstream primer (5′-AAGGAAAAAGCGGCCGCTTATCACTTGTCATCGTCGTCCTTGTAGTCCTGATATTTCCTCATTAG-3′) that contains a Flag sequence and an upstream primer (5′-CTAGACGCGTGCCACCATGAAAATGCGGTTCTTGG-3′). The PCR product was digested with MluI/NotI and subcloned downstream of the CMV minimal promoter linked to seven Tet-responsive elements (7× TRE) at the MluI and NotI sites in the pTRE2 vector (BD Bioscience-Clontec, Mountain View, CA). The expression cassette containing the CMV promoter, the hLAL cDNA, and the β-globin polyadenylation signaling sequence was excised and purified for microinjection into FVB/N mice. Founder lines were identified by a pair of primers covering the pTRE2 vector sequence (5′-ACGCCATCCACGCTGTTTTG-3′) and the hLAL cDNA sequence (5′-AGACAACTGGTTTGGGACCTTTG-3′).
Isolation of Macrophages from Bronchoalveolar Lavage Fluid (BALF) for Antibody Array Analysis
BALF was collected by perfusing each lung of wild-type or lal−/− mice with 1-ml aliquots of 0.9% sodium chloride and withdrawing back the fluids three times. BALFs from eight mice were combined and centrifuged for 5 minutes at 1000 rpm and 4°C to collect macrophage pellets. Purified macrophages were lysed for protein preparation. After determination of protein concentrations, expression levels of cytokines/chemokines were determined with the Mouse Inflammation Antibody Array System from RayBio Cytokine Antibody Array service (RayBiotech, Inc., Norcross, GA). Fold changes of cytokines and chemokines between lal−/− and wild-type macrophages were analyzed by the software service from the same company. The intensity of each signal was scanned and normalized with backgrounds.
Isolation of Total RNA from Whole Lung Tissue for Reverse Transcriptase (RT)-PCR Assay
Whole lung tissues were dissected from doxycycline-treated (4-month) or untreated c-fms-rtTA/(tetO)7-CMV-hLAL double mice or c-fms-rtTA/(tetO)7-CMV-hLAL/lal−/− triple mice after mice were anesthetized by intraperitoneal injection. Whole lung tissues were homogenized in RLT lysis buffer for total RNA purification as recommended by the manufacturer (Qiagen, Valencia, CA). Total RNAs were purified using the Qiagen total RNA purification kit as recommended by the manufacturer (Qiagen).
For semiquantitative RT-PCR assay, total RNAs were used to detect mRNA expression by a SuperScript One-Step RT-PCR Kit (Invitrogen, Carlsbad, CA). Primers for RT-PCR amplification of Api6, MafB, MMP-9, MMP-12, Spi-C, and GAPDH genes were described previously.7 Primers for tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, IL-6, CCL7, and CXCL1 were purchased from SuperArray Bioscience (Frederick, MD). Primers for RT-PCR amplification of hLAL were an upstream primer corresponding to the hLAL cDNA sequence (5′-CACATTCTCCTGCTGGAACTTCTG-3′) and a downstream primer corresponding to the pTRE vector sequence (5′-CCTGAAAACTTTGCCCCCTC-3′).
Tissue Lipid Extraction and Determination of Cholesteryl Ester and Triglyceride Concentrations
Total tissue lipids were extracted from the liver and small intestine by the Folch method.4 Concentrations of cholesteryl esters and triglycerides were determined as previously described.6,8
Liver Extraction and Lysosomal Acid Lipase Activity Assays
Liver tissues were homogenized, sonicated, and extracted in tissue lysis buffer (100 mmol/L NaPO4, pH 6.8, 1 mmol/L ethylenediamine tetraacetic acid, 10 mmol/L dithiothreitol, 0.5% NP-40, and 0.02% sodium azide) followed by centrifugation at 4°C, 10,000 × g for 15 minutes. Aliquots of the supernatant were stored at −20°C until assayed. Protein concentration of the liver extract was determined by the BCA assay following the protocol from manufacturer (Pierce, Rockford, IL). hLAL activities were estimated with the fluorogenic substrate 4-methyl-umbelliferyl-oleate (4-MUO). The assay was performed in a microtiter plate at 37°C at pH 5.5 using a 4-methylumbelliferone (4-MU) standard curve. In brief, 4-MUO in hexane (100 mg/ml) was diluted 1 to 100 in 4% Triton X-100. The assay used 50 μl of the diluted substrate, 25 μl of diluted liver extract, and 125 μl of assay buffer (0.2 mol/L Na2Ac and 0.01% Tween 80, pH 5.5) at 37°C for 30 minutes. The reaction was stopped by adding 100 μl of 0.75 mol/L Tris, pH 8.0. The resulting fluorescence signal was detected at excitation 360 nm and emission 460 nm. The relative fluorescence units from each sample were directly compared with that of the standard curve. One unit of activity is defined as producing 1 μmol/L 4-MU per minute. All assays were conducted in duplicate, and the results were the mean of three experiments. Assays were linear within the time frame used, and less than 10% of substrates were cleaved.
Oil Red-O Staining
Frozen tissue sections were prepared from liver and intestine after a standard cryostat procedure. Tissue section slides were stained with Oil Red-O solution (0.5% in propylene glycol) in a 60°C oven for 10 minutes and placed in 85% propylene glycol for 1 minute; slides were counterstained in hematoxylin.
Immunohistochemistry
Tissues from liver, intestine, and lung were collected after mice were anesthetized. For lung tissue preparation, lungs were inflation-fixed with 4% paraformaldehyde in phosphate-buffered saline overnight at 4°C. All tissues were washed with phosphate-buffered saline and dehydrated by a series of increasing ethanol concentrations, followed by paraffin embedding. Sections were incubated with rat anti-mouse Ly6G antibody (1:500; BD Biosciences PharMingen) or rabbit anti-mouse macrophage antibody (1:200; Accurate Chemical & Scientific Co., Westbury, NY) as the primary antibody. Tissue sections were washed and treated with biotinylated secondary antibodies. To visualize signals, interactions were detected with the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA) according to the manufacturer’s instructions.
Results
Inflammatory Cytokine/Chemokine Expression in Macrophages of Wild-Type and lal−/− Mice
Abnormal appearance and accumulation of macrophages in multiple organs suggests that they play a major role in contributing to pathogenesis in lal−/− mice. These pathogenic phenotypes are associated with inflammation. To establish that macrophages are a primary source for mediating inflammation in lal−/− mice, wild-type and lal−/− macrophages were isolated from BALF. Macrophages were lysed by using cell lysis buffer (from RayBiotech). The expression levels of various cytokines and chemokines in macrophages were measured by inflammatory antibody array analysis (Table 1). Expression levels of many cytokines and chemokines were changed (most were up-regulated) in lal−/− macrophages in comparison with wild-type macrophages. This strongly suggests that the LAL activity in macrophages controls production of pro-inflammatory cytokines and chemokines, and malfunction of macrophages has a primary effect in lal−/− pathogenesis in various organs.
Table 1.
Aberrant Expression of Cytokines/Chemokines in lal−/− Bronchoalveolar Macrophages in Antibody Array Analysis
| Name | WT BALF macrophage | KO BALF macrophage |
|---|---|---|
| POS | 27,135.08 | 27,135.08 |
| NEG | 0.00 | 0.00 |
| BLC | 0.00 | 1055.47 |
| CD30L | 70.50 | 868.94 |
| Eotaxin | 2288.00 | 1493.28 |
| Eotaxin-2 | 3233.32 | 1010.42 |
| FAS ligand | 3998.55 | 1174.42 |
| Fractalkine | 4590.73 | 3100.23 |
| G-CSF | 5598.22 | 5100.65 |
| GM-CSF | 1557.38 | 5563.10 |
| IFN-γ | 1734.26 | 1627.01 |
| IL-1α | 2075.22 | 2556.13 |
| IL-1β | 0.00 | 364.96 |
| IL-2 | 0.00 | 93.26 |
| IL-3 | 0.00 | 0.00 |
| IL-4 | 0.00 | 685.93 |
| IL-6 | 0.00 | 0.00 |
| IL-9 | 0.00 | 0.00 |
| IL-10 | 0.00 | 0.00 |
| IL-12-p40/p70 | 1111.31 | 322.02 |
| IL-12-p70 | 3417.25 | 4985.92 |
| IL-13 | 0.00 | 6295.13 |
| IL-17 | 0.00 | 2587.81 |
| I-TAC | 0.00 | 1488.35 |
| KC | 0.00 | 343.14 |
| Leptin | 0.00 | 371.30 |
| LIX | 0.00 | 1621.38 |
| Lymphotactin | 0.00 | 0.00 |
| MCP-1 | 0.00 | 0.00 |
| M-CSF | 0.00 | 56.66 |
| MIG | 0.00 | 0.00 |
| MIP-1-α | 0.00 | 963.26 |
| MIP-1-γ | 3893.44 | 10,003.87 |
| RANTES | 2591.14 | 7414.30 |
| SDF-1 | 2509.11 | 4039.91 |
| TCA-3 | 1496.49 | 4414.37 |
| TECK | 0.00 | 1594.64 |
| TIMP-1 | 0.00 | 1476.38 |
| TIMP-2 | 0.00 | 0.00 |
| TNF-α | 0.00 | 0.00 |
| sTNF RI | 0.00 | 0.00 |
| sTNF RII | 0.00 | 0.00 |
POS, positive control; NEG, negative control; BLC, B-lymphocyte chemoattractant; G-CSF, granulocyte colony stimulating factor; GM-CSF, granulocyte-macrophage colony stimulating factor; IFN, interferon; I-TAC, interferon-inducible T-cell α-chemoattractant; KC, keratinocyte cytokine; LIX, LPS-induced CXC chemokine; M-CSF, macrophage colony stimulating factor; MIG, monokine induced by γ-interferon; MIP, macrophage inflammatory protein; RANTES, regulated upon activation, normal T-cell expressed, and presumably secreted; SDF, stromal cell-derived factor; TCA, T-cell activation; TECK, thymus-expressed chemokine; TIMP, tissue inhibitor of metalloproteinases.
Generation of c-fms-rtTA Transgenic Mice
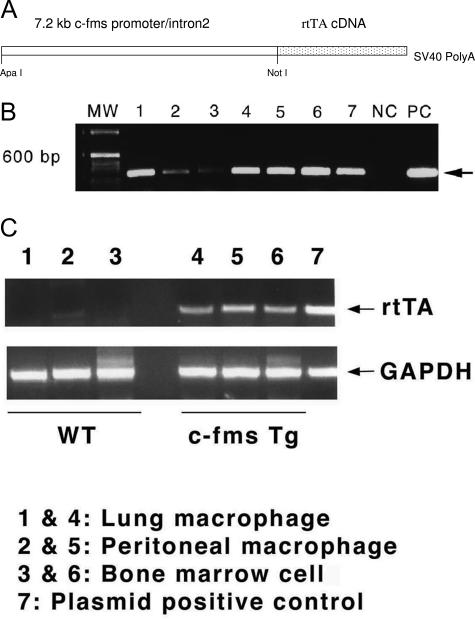
To test the primary role of macrophages in lal−/− pathogenesis, we designed a system to specifically express hLAL in macrophages. This is achieved by subcloning the cDNA fragment of rtTA into the p7.2ΔMCS construct that contains the 7.2-kb c-fms promoter/intron2 fragment (Figure 1A). The c-fms-rtTA expression cassette DNA containing the 7.2-kb c-fms promoter/intron2 fragment, the rtTA cDNA, and the SV40 polyadenylation signaling sequence was isolated and microinjected into eggs of FVB/N mice. Seven positive clones were identified after screening by PCR using a pair of primers covering the 7.2-kb c-fms promoter/intron2 sequence and the rtTA cDNA sequence (Figure 1B). The clones are designated as c-fms-rtTA transgenic founder lines. Among these founders, only two founders showed positive transgene F1 generations. Founder 77 was chosen for further characterization of rtTA expression in the macrophage lineage compartment. Alveolar macrophages, peritoneal macrophages, and bone marrow cells were isolated from this founder line for mRNA purification. A pair of primers corresponding to the rtTA cDNA sequence was used for detection of rtTA expression by RT-PCR. Both macrophages and bone marrow cells showed rtTA expression (Figure 1C). As a control, no expression was detected in isolated AT II cells (data not shown).
Figure 1.
Generation of c-fms rtTA transgenic mice. A: Illustration of c-fms-rtTA transgene design. B: PCR genotyping of c-fms-rtTA transgenic founders. MW, molecular weight marker; NC, negative control from tail DNAs of a negative pup; PC, positive control using the c-fms-rtTA construct as a DNA template. C: Expression of rtTA mRNA in peritoneal macrophages, lung macrophages, and bone marrow cells of c-fms-rtTA transgenic mice.
Generation of (tetO)7-CMV-hLAL Transgenic Mice
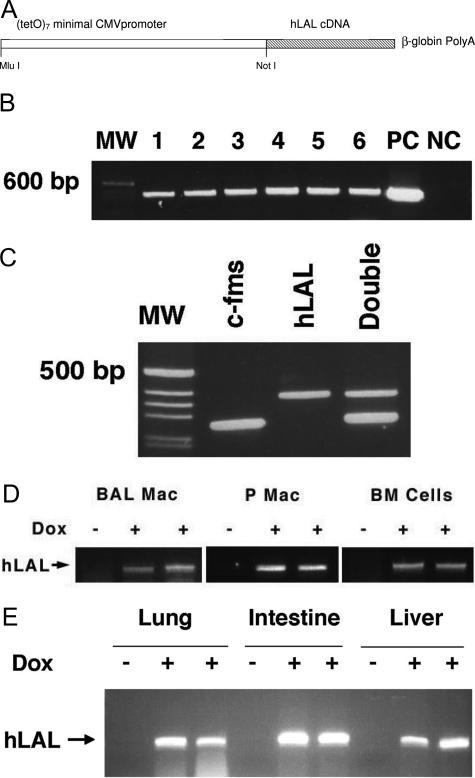
In parallel, the cDNA fragment of hLAL was subcloned into the pTRE construct that contains seven copies of the tet operator DNA binding sequence linked to a minimal CMV promoter and β-globin polyA signals (Figure 2A). The expression cassette of the (tetO)7-CMV-hLAL construct was microinjected into eggs of FVB/N mice. Six positive clones were obtained by PCR using a pair of primers covering the hLAL cDNA and pTRE sequences (Figure 2B). These clones were designated as (tetO)7-CMV-hLAL founder lines. All founders showed positive F1 generations. Founder 556 was crossbred with the c-fms Founder 77 line to obtain double transgenic mice (Figure 2C). After treatment of c-fms-rtTA/(tetO)7-CMV-hLAL double transgenic mice with doxycycline-treated food for 1 month, macrophages (from peritoneal and lung lavage fluid) and bone marrow cells showed induced hLAL mRNA expression (Figure 2D). Increased hLAL mRNA expression level was also observed in liver, lung, and intestinal tissues that contain macrophages (Figure 2E).
Figure 2.
Generation of (tetO)7-CMV-LAL transgenic mice. A: Illustration of (tetO)7-CMV-hLAL transgene design. B: PCR genotyping of (tetO)7-CMV-hLAL transgenic founders. MW, molecular weight marker; NC, negative control from tail DNAs of a negative pup; PC, positive control using (tetO)7-CMV-hLAL construct as a DNA template. C: PCR genotyping of c-fms-rtTA/(tetO)7-CMV-hLAL double transgenic mice. MW, molecular weight marker. D: Expression of hLAL mRNA in bronchio-alveolar macrophages (BAL Mac), peritoneal macrophages (P Mac), and bone marrow (BM) cells. E: Expression of hLAL mRNA in lung, intestine, and liver of c-fms-rtTA/(tetO)7-CMV-hLAL double transgenic mice after doxycycline (Dox) treatment (+). Untreated (−) double transgenic mice were the control.
Macrophage-Specific Expression of hLAL Reduced Neutral Lipid Accumulation in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
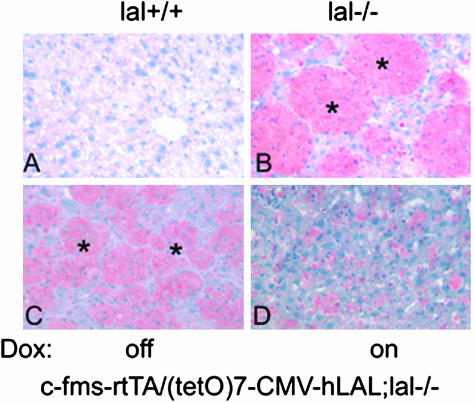
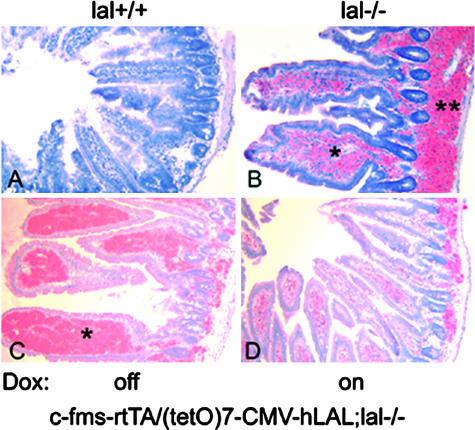
LAL deficiency in mice caused massive neutral lipid accumulation in Kupffer cells in the liver, lamina propria (inside of epithelia villia), and the base of villia in the intestine and in macrophages and alveolar type II cells in the lung.4–6,8 To test whether deficiency of LAL in macrophages is responsible for lipid accumulation, c-fms-rtTA/(tetO)7-CMV-hLAL double transgenic mice were introduced into the lal−/− background by crossbreeding to generate c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice. Without doxycycline treatment, neutral lipid accumulation was still observed in the liver and the intestine of c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice, similar to lal−/− mice (Figures 3 and 4; identified in the asterisk area). However, expression of hLAL in macrophages induced by doxycycline treatment significantly reduced neutral lipid accumulation in the same liver and intestinal areas of triple lal−/− mice by Oil-Red-O staining (Figures 3 and 4). This clearly indicates the effectiveness of doxycycline-induced hLAL activity in degrading neutral lipids in triple lal−/− mice.
Figure 3.
Macrophage-specific expression of hLAL reduced neutral lipid accumulation in the liver of c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Frozen sections of liver were stained with Oil Red-O. Massive neutral lipid accumulation (red color) was detected in Kupffer cells (macrophages) in livers of lal−/− (B) and c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice (C) without doxycycline treatment as indicated by asterisks. D: Doxycycline treatment significantly reduced Oil Red-O staining in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice. Wild-type lal+/+ mice (A) were the negative control for Oil Red-O staining.
Figure 4.
Macrophage-specific expression of hLAL reduced neutral lipid accumulation in the intestine of c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Frozen sections of small intestine were stained with Oil Red-O. Massive neutral lipid accumulation (red color) was detected in the lamina propria and the base of villi in the intestine of lal−/− (B) and c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice (C) without doxycycline treatment as indicated by asterisks. D: Doxycycline treatment significantly reduced Oil Red-O staining in the c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice. Wild-type lal+/+ mice (A) were the negative control for Oil Red-O staining.
Macrophage-Specific Expression of hLAL Restored the LAL Activity in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
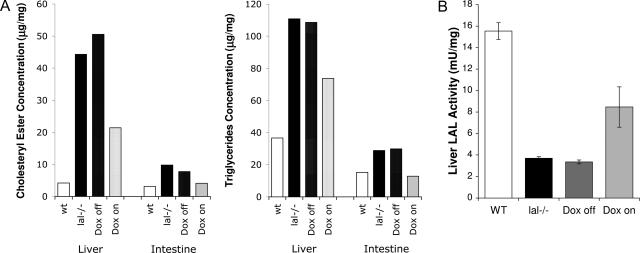
To monitor changes of neutral lipid degradation at the biochemical level in addition to Oil-Red-O staining, concentrations of cholesteryl esters and triglycerides were measured in doxycycline-treated and untreated triple mice. In agreement with Figures 3 and 4, concentrations of cholesteryl esters and triglycerides were significantly higher in single lal−/− and triple lal−/− mice without doxycycline treatment. Expression of hLAL in macrophages by doxycycline treatment significantly decreased concentrations of cholesteryl esters and triglycerides (Figure 5A). A direct measurement of the LAL enzymatic activity using the fluorogenic substrate 4-MUO was also performed. Proteins were isolated from the livers of doxycycline-treated and untreated c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice. Wild-type and lal−/− mice were used as controls. Although doxycycline-untreated triple mice showed a similar LAL activity with that in lal−/− mice, treatment of doxycycline partially restored the LAL activity (Figure 5B). Taken together, the neutral lipid metabolism was partially recovered by expression of hLAL in macrophages of c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice.
Figure 5.
Macrophage-specific expression of hLAL restored the LAL activity in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice. A: Concentrations of cholesteryl esters (left) and triglycerides (right) in the liver and small intestine of wild-type mice, lal−/− mice, untreated (Dox off) triple lal−/− mice, and doxycycline-treated (Dox on) triple lal−/− mice. B: The liver LAL activity in wild-type mice, lal−/− mice, untreated (Dox off) triple lal−/− mice, and doxycycline-treated (Dox on) triple lal−/− mice were measured using the fluorogenic substrate, 4-MUO in vitro.
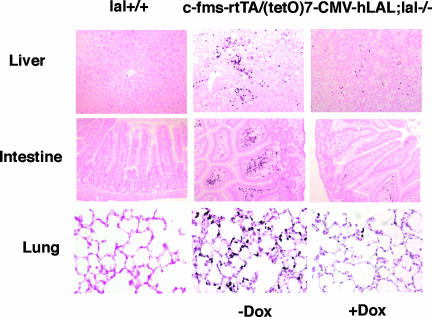
Macrophage-Specific Expression of hLAL Reduced Neutrophil Infiltration in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
Neutrophil infiltration is a major inflammatory manifestation in lal−/− mice.5,7 Triple lal−/− mice were treated with doxycycline for 3 months to induce hLAL expression in macrophages. The liver, intestine, and lung were examined for assessment of neutrophil infiltration because these organs displayed dramatically pathogenic phenotypes during LAL deficiency.4–6,8 As demonstrated in Figure 6, all three organs from triple lal−/− mice without doxycycline treatment exhibited significant neutrophil infiltration versus wild-type mice. After doxycycline induction in triple lal−/− mice, expression of LAL in macrophages significantly attenuated neutrophil influx into liver, intestine, and lung tissues. These results indicate that LAL activity in macrophages plays an essential role in controlling tissue inflammation in various organs during LAL deficiency.
Figure 6.
Macrophage-specific expression of hLAL reduced neutrophil infiltration in c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Paraffin embedded liver, intestine, and lung tissue sections from wild-type (lal+/+) and untreated (−Dox) or doxycycline-treated (+Dox) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice were immunostained with Ly6G antibody. Small black dots in various sections represent stained neutrophils.
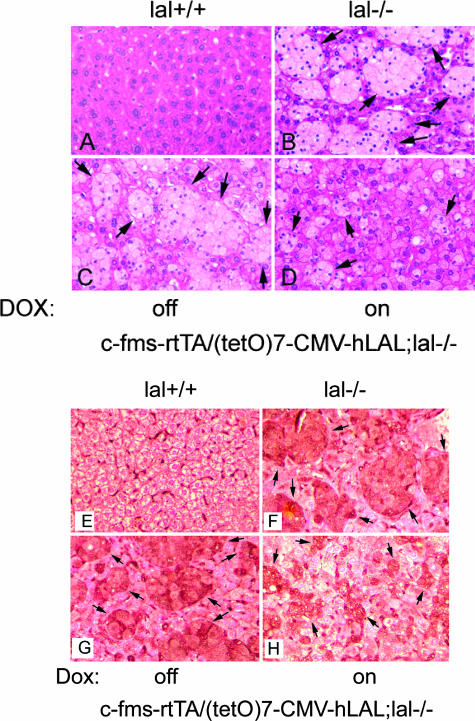
Macrophage-Specific Expression of hLAL Reduced Liver Pathogenesis in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
We reported previously that LAL deficiency in mice caused severely pathogenic phenotypes and enlargement in the size of the liver in association with foamy macrophage formation (Kupffer cells).4,6,8 To specifically address the functional role of macrophages in liver pathogenic processes, c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice with or without doxycycline treatment were examined. At the histological level, triple lal−/− mice without doxycycline treatment displayed massive foamy Kupffer cell formation (Figure 7C), similar to lal−/− mice (Figure 7B). After 3 months of doxycycline treatment beginning from 1 month of age, the size of foamy Kupffer cells was significantly reduced (Figure 7D). It appears that lipid-filled hepatocytes remained unchanged in treated triple lal−/− mice, indicating cell type-specific hLAL rescue in the liver. This is confirmed by immunohistochemical staining using macrophage-specific antibody, in which positively stained and lipid-stored macrophages were significantly reduced in the livers of treated triple lal−/− mice (Figure 7, E–H). These results suggest that LAL activity in macrophages contribute to lal−/− pathogenesis in the liver.
Figure 7.
Macrophage-specific expression of hLAL reduced liver pathogenesis in c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Histological (A–D) and immunohistochemical (E–H) comparison of liver sections from untreated (C and G) and doxycycline-treated (D and H) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Wild-type (A and E) and lal−/− (B and F) mice were used as negative and positive controls, respectively, for foamy Kupffer cell formation. Kupffer cells are indicated by arrows.
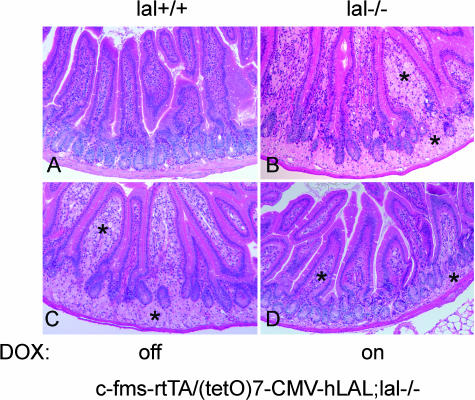
Macrophage-Specific Expression of hLAL Reduced Intestine Pathogenesis in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
To address the functional role of macrophages in the intestinal pathogenic process, a histological study of intestines from c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice was performed. Similar to lal−/− mice (Figure 8B), macrophage accumulation was observed in the lamina propria (inside of epithelia villia) and the base of villia in doxycycline-untreated triple lal−/− mice (Figure 8C, identified in the asterisk area). Doxycycline treatment significantly reduced both the size and the number of macrophages in triple lal−/− mice (Figure 8D).
Figure 8.
Macrophage-specific expression of hLAL reduced intestine pathogenesis in c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Histological comparison of intestine sections from untreated (C) and doxycycline-treated (D) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Wild-type (A) and lal−/− (B) mice were negative and positive controls.
Macrophage-Specific Expression of hLAL Reduced Lung Pathogenesis in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
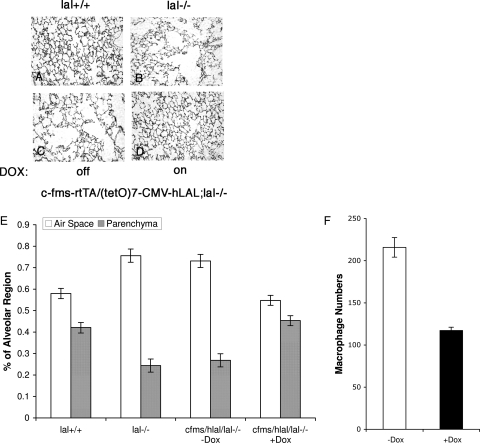
In the lung, alveolar space enlargement (emphysema) was closely associated with macrophage proliferation in lal−/− mice.5 Extracellular membrane degrading enzymes such as MMP-9 and MMP-12 were highly expressed in the lal−/− lung during pulmonary pathogenesis. To assess the functional role of macrophages in this pulmonary pathogenic process, histological analyses of c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice with or without doxycycline treatment were performed. In untreated triple mice (Figure 9C), the alveolar space area was significantly larger than that seen in the wild-type lung (Figure 9A), similar to that seen in the single lal−/− lung (Figure 9B). Expression of hLAL in macrophages for 5 months significantly reversed emphysema in doxycycline-treated triple lal−/− mice (Figure 9D). A quantitative analysis revealed a reduced ratio between the alveolar airspace area and the parenchyma area in the lal−/− lung after macrophage hLAL expression (Figure 9E). In the immunohistochemical staining study using a macrophage-specific antibody, positively stained macrophages were significantly reduced in the lung of doxycycline-treated versus untreated triple lal−/− mice (Figure 9F). Therefore, LAL activity in macrophages is essential for maintaining normal alveolar structure and function in the lung.
Figure 9.
Macrophage-specific expression of hLAL reduced pulmonary pathogenesis in c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. A–D: Histological comparison of lung sections from untreated (C) and doxycycline-treated (D) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Wild-type (A) and lal−/− (B) mice were controls. E: Morphometric analysis of alveolar space in wild-type (lal+/+) mice, lal−/− mice, and untreated (−Dox) and doxycycline-treated (+Dox) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. Differences between ratios of airspace/parenchyma were analyzed by analysis of variance. P < 0.05. Values are means ± SD; n = 3 (mice). F: Immunohistochemical staining comparison of lung sections from untreated and doxycycline-treated c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice using an antibody specifically against macrophage surface antigens. Positively stained cells from various microscopic fields of each sample were counted. Differences between untreated (−Dox) and doxycycline-treated (+Dox) samples were analyzed by analysis of variance. P < 0.05. Values are means ± SD; n = 3 (mice).
Macrophage-Specific Expression of hLAL Reduced Aberrant Gene Expression in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− Triple Mice
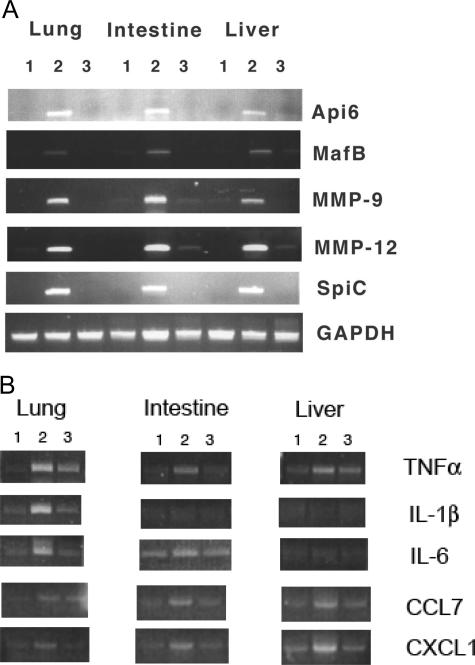
Pathogenic phenotypes are caused by aberrant gene expression in lal−/− mice. Gene profile changes have previously been assessed in the lal−/− lung by Affymetrix GeneChip microarray analysis. This analysis identified aberrant expression of many genes during LAL deficiency. These gene products include cytokines/chemokines, MMPs, apoptosis inhibitors, transcription factors, and oncogenes.5,7 Using these molecules as biomarkers to assess how expression of hLAL in macrophages corrects aberrant gene expression in the lung, liver, and intestine, mRNA expression levels were examined by RT-PCR. As demonstrated in Figure 10A, Api6, MMP-9, MMP-12, MafB, and Spi-C were highly overexpressed in the lung, liver, and intestine of c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice, in a manner similar to single lal−/− mice.7 Expression of hLAL by doxycycline treatment in macrophages significantly reduced the expression levels of these molecules in triple lal−/− mice. This indicates that these molecules contribute to pathogenic progression in multiple organs during LAL deficiency. A group of cytokines and chemokines were also up-regulated in lungs of lal−/− mice as we previously reported.5 A set of cytokines/chemokines in this group (TNF-α, CCL7, and CXCL1) showed significant reduction in triple lal−/− mice after doxycycline treatment in the lung, liver, and intestine. Another set of cytokines/chemokines in this group (IL-1β and IL-6) only showed changes in the lung but not in the intestine and liver, after doxycycline treatment (Figure 10B). This observation suggests that pathogenesis in different organs share common genes and pathways with certain distinctions during LAL deficiency.
Figure 10.
Macrophage-specific expression of hLAL reduced aberrant gene expression in c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. A: RT-PCR analysis of Api-6, MMP-9, MMP-12, Spi-C, MafB, and GAPDH in lung, intestine, and liver of wild-type (lane 1), untreated (lane 2), and doxycycline-treated (lane 3) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice. B: RT-PCR analysis of TNF-α, IL-1β, IL-6, CCL7, and CXCL1 in lung, intestine, and liver of wild-type (lane 1), untreated (lane 2), and doxycycline-treated (lane 3) c-fms-rtTA/(tetO)7-CMV-LAL;lal−/− triple mice.
Discussion
It is intriguing that many seemingly unrelated disease phenotypes of various organs coexist in lal−/− mice. Some common cellular and molecular mechanisms must exist to link these pathogenic processes together. Based on previous observations, abnormal macrophages were observed in multiple organs in lal−/− mice, including the liver, intestine, and lung.4–8 Therefore, macrophages play a central role in pathogenic progression during LAL deficiency. Aberrant expression levels of multiple inflammatory cytokines and chemokines were monitored in association with lal−/− macrophages (Table 1). These inflammatory molecules can change the local microenvironment by binding to the residential cells to affect gene expression (Figure 10) and cause pathogenic phenotypes in various organs. Establishment of an in vivo macrophage-specific controllable system is essential to elucidate and dissect molecular, cellular, and physiological mechanisms in lal−/− mice. In this report, we generated a c-fms 7.2-kb promoter/intron2-rtTA transgenic line, in which rtTA expression is restricted to macrophages as previously reported.3 In the c-fms 7.2-kb promoter/intron2-directed GFP transgenic system, GFP has been detected in Kupffer cells of the liver, lamina propria of the small intestine, and bronchio-alveolar macrophages of the lung. This expression pattern overlaps with abnormal macrophage malformation in lal−/− mice as determined by Oil Red-O staining of neutral lipids.4–6,8 Macrophages in these areas appear foamy and lipid filled. Therefore, the c-fms 7.2-kb promoter/intron2 DNA sequence is ideal for directing LAL expression in vivo to correct macrophage-mediated pathogenic phenotypes in lal−/− mice. After crossbreeding these mice with the (tetO)7-CMV-hLAL transgenic mouse line, the induced expression level of hLAL was readily detectable by RT-PCR in macrophages and in bone marrow cells (Figure 2D), as well as in liver, intestine, and lung tissues that contain macrophages (Figure 2E). The LAL enzymatic activity was restored with correction of neutral lipid metabolism (Figure 5).
After crossbreeding c-fms-rtTA/(tetO)7-CMV-hLAL double transgenic mice with lal−/− mice, expression of hLAL in lal−/− macrophages by doxycyline treatment significantly reduced neutral lipid accumulation in the liver and intestine as assessed by Oil Red-O staining (Figures 3 and 4) in association with attenuated inflammatory neutrophil infiltration. Neutrophil infiltration has been observed in the liver, intestine, and lung of lal−/− mice (Figure 6), suggesting that inflammation is a major trigger for pathogenic progression in various lal−/− organs. In general, inflammation starts with stimulation of vascular endothelial cells by pro-inflammatory cytokines and chemokines generated by either regional macrophages or residential cells.9 Cytokines and chemokines regulate leukocyte trafficking, homing, and migration. The process begins with activating intracellular signaling cascades in endothelial cells, which influences expression of an array of inflammation-associated genes, including a variety of adhesion molecules. Increased expression of adhesion molecules facilitates migration and adhesion of phagocytic neutrophils to vascular endothelium at vessel sites adjacent to inflammatory sites. Neutrophils migrate through vessel walls to infiltrate inflammatory tissues. Activated neutrophils produce both free radicals (eg, superoxide) and proteinases (eg, neutrophil elastase and MMPs) causing tissue damage. Correction of neutrophil infiltration and pathogeneses in multiple organs by restoring LAL production in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice suggests that macrophages play an important role in controlling inflammation-triggered pathogenesis through production and secretion of cytokines and chemokines. This has been confirmed by RT-PCR analysis in the lung, liver, and intestine (Figure 10B). Although migrating macrophages are a major source and site for production of cytokines and chemokines, partial correction of pathogenic phenotypes in c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice suggests that other residential cell types in these organs also contribute to aberrant production of inflammatory cytokines and chemokines leading to tissue pathogenesis. This has been confirmed by the observation that TNF-α and IL-6 were up-regulated in the lung but not in macrophages (Table 1). Interactions between macrophages and residential cells are important for tissue pathogenic development in lal−/− mice.
Based on previous and current observations, it appears that neutral lipid metabolism is essential for controlling malformation and malfunction of macrophages caused by aberrant gene expression in lal−/− mice. Deficiency of LAL in macrophages blocks the normal synthesis of various hormonal ligands for nuclear receptors such as PPARγ and other transcription factors such as nuclear factor E2 p45-related factor 2 (Nrf2).10–13 Treatment with PPARγ ligands significantly attenuated lal−/− pulmonary inflammation and aberrant gene expression in the lungs.7 In addition to PPARγ, there is emerging evidence indicating that other lipid mediators such as Nrf2 contribute to cyclooxygenase product function in inflammation.13 Expression of hLAL in macrophages of c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice restored the production of ligands for nuclear receptors and lipid-mediating transcription factors that suppress synthesis and secretion of pro-inflammatory cytokines and chemokines. As a consequence, pathogenic phenotypes were ameliorated in various lal−/− organs. Recently, biochemical pathways were identified that use polyunsaturated fatty acids to generate resolvins and protectins that can expedite inflammatory resolution.14 These mediators might be regulated and enhanced by hLAL. Expression of hLAL in macrophages may restore these pathways.
In addition to the cytokines, chemokines, and lipid-mediated nuclear receptors/transcription factors mentioned above, other common molecules also contribute to lal−/− pathogenic processes in various organs. While studying the relationship between aberrant gene expression and pathogenic progression in the lal−/− lung, a set of genes was identified with aberrant expression, including Api6, MMP-9, MMP-12, MafB, and Spi-C.7 To see whether aberrant expression of these genes occurs in other organs, RT-PCR analysis was performed. Interestingly, Api6, MMP-9, MMP-12, MafB, and Spi-C were all up-regulated in the liver and intestine in addition to the lung. In c-fms-rtTA/(tetO)7-CMV-hLAL;lal−/− triple mice, doxycycline treatment significantly reduced expression of these molecules in all tested organs (Figure 10A). Api6 has been shown to be an apoptotic inhibitor with a molecular weight of 37 kd (331 amino acids) and belongs to the scavenger receptor cysteine-rich superfamily.15,16 Transcription factor MafB belongs to the maf proto-oncogene family and is expressed in a wide variety of tissues and encodes a 311-amino acid protein containing a typical bZip motif in its carboxy-terminal region.17,18 Spi-C belongs to the Ets family of transcription factors.19 Ets proteins comprise a large family of transcription factors involved in a variety of cellular processes. The MMPs are a group of zinc-dependent endopeptidases including collagenases, gelatinases, and stromelysins.20 These molecules cause a profound impact on tissue structure by degrading components of the extracellular matrix.21,22 Although it is unclear how the LAL activity in macrophages affects pathogenic expression of these molecules at this time, it is obvious that these molecules are crucial to the pathogenic progress in various lal−/− organs. Study of these molecules will provide significant insight into the molecular mechanisms and linkages between various pathogenic phenotypes in lal−/− mice.
Footnotes
Address reprint requests to Cong Yan, Ph.D., The Center for Immunology, Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, 635 Barnhill Dr., Indianapolis, IN 46202, or Hong Du, Ph.D., Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave., Cincinnati, OH 45229-3039. E-mail:coyan@iupui.edu and Hong,Du@cchmc.org.
Supported by National Institute of Health grants HL-061803 and HL-067862 (to C.Y. and H.D.).
References
- Yang L, Naltner A, Yan C. Overexpression of dominant negative retinoic acid receptor alpha causes alveolar abnormality in transgenic neonatal lungs. Endocrinology. 2003;144:3004–3011. doi: 10.1210/en.2002-0121. [DOI] [PubMed] [Google Scholar]
- Lian X, Qin Y, Hossain SA, Yang L, White A, Xu H, Shipley JM, Li T, Senior RM, Du H, Yan C. Overexpression of Stat3C in pulmonary epithelium protects against hyperoxic lung injury. J Immunol. 2005;174:7250–7256. doi: 10.4049/jimmunol.174.11.7250. [DOI] [PubMed] [Google Scholar]
- Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR, Hume DA. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- Du H, Duanmu M, Witte D, Grabowski GA. Targeted disruption of the mouse lysosomal acid lipase gene: long-term survival with massive cholesteryl ester and triglyceride storage. Hum Mol Genet. 1998;7:1347–1354. doi: 10.1093/hmg/7.9.1347. [DOI] [PubMed] [Google Scholar]
- Lian X, Yan C, Yang L, Xu Y, Du H. Lysosomal acid lipase deficiency causes respiratory inflammation and destruction in the lung. Am J Physiol Lung Cell Mol Physiol. 2004;286:L801–L807. doi: 10.1152/ajplung.00335.2003. [DOI] [PubMed] [Google Scholar]
- Du H, Heur M, Duanmu M, Grabowski GA, Hui DY, Witte DP, Mishra J. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res. 2001;42:489–500. [PubMed] [Google Scholar]
- Lian X, Yan C, Qin Y, Knox L, Li T, Du H. Neutral lipids and peroxisome proliferator-activated receptor-{gamma} control pulmonary gene expression and inflammation-triggered pathogenesis in lysosomal acid lipase knockout mice. Am J Pathol. 2005;167:813–821. doi: 10.1016/s0002-9440(10)62053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Schiavi S, Levine M, Mishra J, Heur M, Grabowski GA. Enzyme therapy for lysosomal acid lipase deficiency in the mouse. Hum Mol Genet. 2001;10:1639–1648. doi: 10.1093/hmg/10.16.1639. [DOI] [PubMed] [Google Scholar]
- Winyard PG. Key stages in the acute inflammatory response and their relevance as therapeutic targets. Methods Mol Biol. 2003;225:3–6. [Google Scholar]
- Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–240. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- Willson TM, Lambert MH, Kliewer SA. Peroxisome proliferator-activated receptor gamma and metabolic disease. Annu Rev Biochem. 2001;70:341–367. doi: 10.1146/annurev.biochem.70.1.341. [DOI] [PubMed] [Google Scholar]
- Uhlig S. Who tidies up the lung?: listen to cox-2 and nrf2. Am J Respir Crit Care Med. 2005;171:1198–1199. doi: 10.1164/rccm.2503003. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Savill J. Resolution of inflammation: the beginning programs the end. Nat Immunol. 2005;6:1191–1197. doi: 10.1038/ni1276. [DOI] [PubMed] [Google Scholar]
- Gebe JA, Llewellyn M, Hoggatt H, Aruffo A. Molecular cloning, genomic organization and cell-binding characteristics of mouse Spalpha. Immunology. 2000;99:78–86. doi: 10.1046/j.1365-2567.2000.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Hirokami Y, Matsuhashi N, Takatsuka H, Naito M. Increased susceptibility of thymocytes to apoptosis in mice lacking AIM, a novel murine macrophage-derived soluble factor belonging to the scavenger receptor cysteine-rich domain superfamily. J Exp Med. 1999;189:413–422. doi: 10.1084/jem.189.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- Kataoka K, Fujiwara KT, Noda M, Nishizawa M. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol Cell Biol. 1994;14:7581–7591. doi: 10.1128/mcb.14.11.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Martensson A, Liberg D, Leanderson T. Spi-C, a novel Ets protein that is temporally regulated during B lymphocyte development. J Biol Chem. 1999;274:10259–10267. doi: 10.1074/jbc.274.15.10259. [DOI] [PubMed] [Google Scholar]
- Moraes TJ, Chow CW, Downey GP. Proteases and lung injury. Crit Care Med. 2003;31:S189–S194. doi: 10.1097/01.CCM.0000057842.90746.1E. [DOI] [PubMed] [Google Scholar]
- Shapiro SD, Senior RM. Matrix metalloproteinases, matrix degradation and more. Am J Respir Cell Mol Biol. 1999;20:1100–1102. doi: 10.1165/ajrcmb.20.6.f151. [DOI] [PubMed] [Google Scholar]
- Visse R, Naguse H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]