Abstract
Multiply damaged sites (MDS) are defined as greater than/equal to two lesions within 10–15 bp and are generated in DNA by ionizing radiation. In vitro repair of closely opposed base damages ≥2 bp apart results in a double strand break (DSB). This work extends the in vitro studies by utilizing clusters of uracil DNA damage as model lesions to determine whether MDS are converted to DSBs in bacteria. Lesions were positioned within the firefly luciferase coding region, transformed into bacteria (wild-type, uracil DNA glycosylase-deficient, ung–, or exonuclease III and endonuclease IV-deficient, xth–nfo–) and luciferase activity measured following repair. DSB formation was expected to decrease activity. Two closely opposed uracils separated by ≤7 bp decreased luciferase activity in wild-type and xth–nfo–, but not ung– bacteria. Growth of bacteria to obtain plasmid-containing colonies demonstrated that the plasmid was destroyed following the mis-repair of two uracils positioned 7 bp apart. This study indicates a DSB is formed when uracil DNA glycosylase initiates repair of two closely opposed uracils ≤7 bp apart, even in the absence of the major apurinic endonucleases. This work supports the in vitro studies and demonstrates that DNA repair is not always advantageous to cells.
INTRODUCTION
DNA damaging agents such as ionizing radiation (1) and bleomycin sulfate (2) introduce clustered lesions or multiply damaged sites (MDS) into DNA. Even though other chemicals such as hydrogen peroxide and metabolically produced reactive oxygen species introduce similar types of DNA damage, fewer lesions are required to produce a lethal event by ionizing radiation and bleomycin sulfate (3). This is believed to be due to the lack of repairability of the MDS. MDS contain oxidized purines, pyrimidines, apurinic/apyrimidinic (AP) sites or single strand breaks (SSBs), on the same (4) or opposing DNA strands (5–8). We and others (1,9,10) have postulated that attempts to repair MDS can convert non-lethal or mutagenic base damage into double strand breaks (DSBs). Oxidative DNA damage is predominantly repaired by base excision repair (reviewed in 11), which involves the removal of the damaged base and deoxyribose, resulting in a SSB-repair intermediate. Initiation of repair at two opposed base damages could therefore produce a DSB prior to the completion of repair, producing a lethal lesion. In fact, allowing time for DNA repair following irradiation of mammalian (12,13) or bacterial (14,15) cells results in an increase in the production of DSBs, and bacterial mutants deficient in three DNA glycosylases have significantly fewer DSBs introduced during the first 8 min following X-irradiation (15).
To further understand how repair of MDS may be detrimental to the cell, it is necessary to work with defined lesions, where the type of damage and distance separating the lesions are known. Extensive work using a variety of defined MDS has been performed in vitro with purified prokaryote and eukaryote repair enzymes (reviewed in 16–18). In general, if two lesions are situated in opposite strands and >2 bp apart, the DNA glycosylases and AP endonucleases initiate repair and introduce two closely opposed SSBs. Reconstitution of the Escherichia coli repair pathway demonstrated that a MDS consisting of an 8-oxo-7,8 dihydroguanine (8-oxodG) and a SSB is converted to a DSB, even when all the enzymes are present for complete repair (19). Very little work has been performed with defined MDS in cells. This work aims to extend the in vitro MDS studies by examining MDS repair in wild-type and DNA repair-deficient bacterial cells, to determine whether MDS are converted to DSBs under physiological conditions. As a model of MDS, we have chosen to study clustered uracil DNA damages, which are also repaired by base excision repair. In E.coli, there are two enzymes that initiate repair of uracil DNA damage (U): uracil DNA glycosylase (Ung) and double strand uracil DNA glycosylase (Dug), which is also known as mismatched uracil glycosylase (Mug). Ung can remove U from single- and double-stranded DNA and is able to excise the U if it is base-paired with an adenine (A) or a guanine (G) (20). Mug is predominantly active on 3,N4-ethenocytosine·G, U·G or U·T pairs in double-stranded DNA and is less efficient at removing U from a U·A base pair (21,22). In this study, we have only examined U·A base pairs and therefore it is likely that we are studying the initiation of repair by Ung. Ung is a monofunctional DNA glycosylase and excises the U leaving an AP site. Exonuclease III and endonuclease IV are the major AP endonucleases that cleave at the AP site, resulting in a SSB with 5′ deoxyribose phosphate and 3′ hydroxyl termini. The 5′deoxyribose phosphate is removed by Rec J and the 5′ phosphate and 3′ hydroxyl termini are utilized by DNA polymerase I and DNA ligase to insert the missing nucleotide and complete repair. The first intermediate in the repair of two closely opposed Us is therefore an MDS consisting of two closely opposed AP sites. Even though the majority of AP sites generated by ionizing radiation are oxidized (23,24), it is possible that an MDS consisting of two AP sites similar to this Ung-repair intermediate could be introduced by ionizing radiation. Radical attack at the base can result in the breakage of the N-glycosylic bond and the formation of an AP site (24).
Previously, Dianov et al. (10) demonstrated that replication of a plasmid containing two uracils situated 12 bp apart in opposite strands, enhances the frequency of recombination only if Ung is present in the bacteria. The proposed explanation was that the two uracils are converted to a DSB by base excision repair and the DSB stimulates recombination. Our work extends this study by examining repair of two uracils situated at different distances apart (0–33 bp) and orientated 5′ or 3′ to each other. We have produced a new assay that utilizes the firefly luciferase reporter in bacteria to determine whether closely opposed base damages are converted to DSBs in cells, in the absence of DNA replication. Repair was studied in wild-type, uracil DNA glycosylase-deficient (ung–) and exonuclease III- and endonuclease IV-deficient (xth–nfo–) bacteria. As expected, two closely opposed uracils separated by ≤7 bp decreased luciferase activity in wild-type and xth–nfo–, but not ung– bacteria. However, uracil residues separated by 13–33 bp did not decrease activity in wild-type cells.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were purchased from Operon Technologies Inc. (Alameda, CA), contained 5′ phosphate termini and were purified following polyacrylamide gel electrophoresis. Oligonucleotides used to generate double-stranded DNA molecules containing no damage, a single uracil or two closely opposed uracil residues are shown in Tables 1 and 2. A double-stranded oligonucleotide inserted into the XhoI site of pBestluc was generated by annealing two oligonucleotides: 5′ TCGAGTCAAGCGGTCAACTATGAAGAACTG 3′ and 5′ TCGACACTTCTTCATAGTTGACCGCTTGAC 3′. The insertion was checked by sequencing using a primer 5′ CATAAAGGCCAAGAAGGGC 3′. Primers used for PCR did not contain 5′ phosphate modifications and were not purified by polyacrylamide gel electrophoresis. Two primer sets were used (Fig. 1): primer 1 (5′ ATGTGGATTTCGAGTCGTCT 3′) and primer 2 (5′ ATATCGATTCCAATTCAGCG 3′), and primer 3 (5′ TGGATGGCTACATTCTG 3′) and primer 4 (5′ GTCATCGTCGGGAAGACCTGCCACGCC 3′).
Table 1. Double-stranded oligonucleotides containing no damage or a single uracil.
| Name | Sequence |
|---|---|
| NT position (Top strand) | 1 5 10 15 20 25 30 35 40 |
| Undamaged | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U2 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAAUTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U5 | 5′ TAAAUACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U13 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCUATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U14 | 5′ TAAATACAAAGGAUATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U15 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTAUAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U16 | 5′ TAAATACAAAGGATAUCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U21 | 5′ TAAATACAAAGGATATCAGGUGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |
| U36 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATUGGAAT 3′ |
| 3′ TAATTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ |
The position of the uracil (U) lesions is numbered relative to the nucleotide (NT) on the top strand of the oligonucleotide.
Table 2. Double-stranded oligonucleotides containing two uracil lesions.
| Name | Position of two Us relative to each other | Sequence | |
|---|---|---|---|
| 5′ or 3′ | Base pairs apart | ||
| U – 34 | 5′ | 33 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATUGGAAT 3′ |
| 3′ TAAUTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 21 | 5′ | 20 | 5′ TAAATACAAAGGATATCAGGTGGCCCCCGCTGAATUGGAAT 3′ |
| 3′ TAATTTATGTTTCCTAUAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 19 | 5′ | 18 | 5′ TAAATACAAAGGATATCAGGUGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAAUTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 14 | 5′ | 13 | 5′ TAAATACAAAGGATAUCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAAUTTATGTTTCCTATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 8 | 5′ | 7 | 5′ TAAATACAAAGGATATCAGGUGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCUATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 6 | 5′ | 5 | 5′ TAAATACAAAGGATATCAGGUGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTAUAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U – 1 | 5′ | 0 | 5′ TAAATACAAAGGATAUCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTAUAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U + 1 | 3′ | 0 | 5′ TAAATACAAAGGAUATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCTAUAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
| U + 8 | 3′ | 7 | 5′ TAAAUACAAAGGATATCAGGTGGCCCCCGCTGAATTGGAAT 3′ |
| 3′ TAATTTATGTTTCCUATAGTCCACCGGGGGCGACTTAACCTTAGC 5′ | |||
When the two uracils are positioned 5′ to each other, the name is designated with a minus and when they are 3′ to each other, the name is designated with a plus. The number in the name designates the nucleotide position of the second uracil relative to the first.
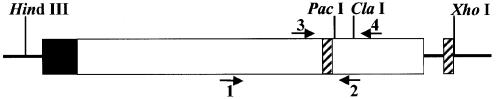
Figure 1.
p3′luc is a derivative of pBestluc, and is a high copy vector that expresses firefly luciferase (clear box) from the tac promoter (shaded box) in bacteria. This vector has unique PacI and ClaI restriction sites situated in the 3′ end of the luciferase coding region. A double-stranded oligonucleotide was inserted at the XhoI site to establish a direct repeat sequence (hatched box). The arrows indicate the position of DNA primers used for PCR analysis.
Bacteria
Wild-type (BW35) and kanamycin-resistant exonuclease III- and endonuclease IV-deficient (xth–nfo–) bacteria were obtained from Dr Susan S. Wallace (University of Vermont, Burlington, VT). Tetracycline-resistant uracil DNA glycosylase-deficient (ung–) bacteria (BD2008, a lambda-minus derivative of BD2007) (25) were obtained from Dr Bernard Weiss (Emory University, Atlanta, GA). All bacterial strains were isogenic. Xth–nfo– and ung– bacteria were grown at 40 µg/ml kanamycin and 10 µg/ml tetracycline, respectively, either on solid growth medium or in liquid culture during the preparation of electrocompetent bacteria. Electrocompetent bacteria were generated according to Seidman et al. (26).
Plasmids
pPROLar.A122 (Dr R. Henry, University of Arkansas-Fayetteville, AR) and pACYC184 (New England Biolabs, Beverly, MA) are low copy vectors with p15A origins of replication that encode resistance to kanamycin (50 µg/ml) and chloramphenicol (34 µg/ml), respectively. pBestluc (Promega, Madison, WI) is a high copy vector with a pUC origin of replication that encodes resistance to ampicilin or carbenicillin (50 µg/ml). pBestluc expresses the firefly luciferase open reading frame from a tac promoter. At the 3′ end of the open reading frame is a unique XhoI site, into which we inserted a double-stranded oligonucleotide to generate p3′luc (Fig. 1). The oligonucleotide was designed to destroy the XhoI site at the 5′ terminus of the oligonucleotide, but leave the XhoI site intact at the 3′ terminus of the insertion site. This oligonucleotide contained 24 bp that was identical to sequence situated upstream of a unique PacI site in the luciferase coding region; 421 bp upstream from the insertion site. p3′luc therefore contains a direct repeat sequence surrounding the unique PacI and ClaI restriction sites.
Insertion of undamaged or uracil-containing oligonucleotides into p3′luc
In order to generate double-stranded oligonucleotides, 6 pmol of each complementary strand of DNA was mixed in 10 mM Tris–HCl (pH 8), 50 mM NaCl in 15 µl vol and heated to 85°C for 5 min, prior to cooling the DNA to room temperature during a 1 h time period. p3′luc was linearized using PacI and ClaI restriction enzymes, subjected to electrophoresis through a 0.7% agarose gel and the linear DNA isolated from the gel using the Qiaquick Gel Extraction kit (Qiagen Inc., Valencia, CA). The concentration of the linear DNA was quantified using an Agilent 2100 Bioanalyzer (Agilent Technologies, Wilmington, DE). A 30 µl ligation reaction containing 1.5 µg (∼600 fmol) of linear p3′luc, 1.8 pmol double-stranded oligonucleotide, 1 mM rATP, 50 mM Tris–HCl (pH 7.5), 7 mM MgCl2, 1 mM dithiothreitol and 6 U T4 DNA ligase (Stratagene, La Jolla, CA) was incubated overnight at 4°C. Salts were then removed from the DNA using the Qiaquick Nucleotide Removal kit (Qiagen Inc.) and the DNA eluted using 30 µl water (pH 8.4).
DNA repair assay
Electrocompetent bacteria were co-transformed with 4 µl (∼200 ng) of the ligation reaction and either 5 or 0.1 ng of pPROLar.A122 or pACYC184 at 2.5 kV, 200 Ω, 25 µF. Initial studies utilized 5 ng of the low copy plasmid; however, we later found 0.1 ng of this second plasmid to be sufficient to normalize the efficiencies of the different transformations. Following transformation, the bacteria were incubated at 37°C and 250 r.p.m. in a rotating incubator for 4 h in 1.5 ml Luria–Bertani medium (LB) containing 50 or 100 µM novobiocin (Sigma-Aldrich, St Louis, MO) and 1 mM IPTG (Sigma-Aldrich). The effectiveness of the novobiocin varied for different lot numbers of the chemical, therefore the optical density at 600 nm was checked for one of the transformations at 0, 1 and 4 h to monitor the growth of the culture. Growth was inhibited for at least 1 h during the incubation and drastically reduced during the remainder of the experiment. After 4 h, the culture was grown on triplicate plates of chloramphenicol (34 µg/ml) or kanamycin (50 µg/ml) containing solid medium to determine the level of transformation of each sample. The type of antibiotic depended upon which low copy plasmid was used in the transformation. An additional antibiotic was added to the solid medium if the bacterial strain being studied was resistant to a particular antibiotic, e.g. ung– are resistant to 10 µg/ml tetracycline. To measure luciferase activity, cell-free extracts were prepared from 1 ml of bacterial culture according to manufacturers’ recommendations (Promega). Briefly, cells were collected by centrifugation, resuspended in 50 µl 0.1 M K2HPO4, 2 mM EDTA and frozen in –80°C ethanol. After defrosting the cells, 150 µl of lysis reagent (1.25 mg/ml lysozyme, 2.5 mg/ml BSA and 1× cell culture lysis reagent; Promega) was added and the solution incubated for 10 min at room temperature prior to centrifugation. Cell-free extract (20 µl) was mixed with luciferase assay reagent (100 µl; Promega) in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). The relative light units per kanamycin or chloramphenicol-resistant colony were calculated. This activity for each transformation was expressed as a percentage of the activity of the undamaged sequence.
Analysis of p3′luc DNA following DNA repair
Electrocompetent bacteria were co-transformed with 1 µl (∼50 ng) of the ligation reaction and 0.1 ng pACYC184 at 2.5 kV, 200 Ω, 25 µF. Following transformation, the bacteria were incubated at 37°C and 250 r.p.m. in a rotating incubator for 4 h in 1.5 ml LB medium containing 100 µM novobiocin (Sigma-Aldrich) and 1 mM IPTG (Sigma-Aldrich). After 4 h, the culture was grown overnight on triplicate plates of chloramphenicol (34 µg/ml) or ampicillin (50 µg/ml) containing solid medium. The number of colonies was counted and a ratio calculated of the number of ampicillin-resistant (AmpR)/the number of chloramphenicol-resistant (CmR) colonies. Individual AmpR colonies were used to inoculate a 3 ml LB–ampicillin culture and plasmid DNA was isolated after overnight growth. PCR reactions containing plasmid DNA were performed using 125 nM each of primers 1 and 2 or primers 3 and 4 (Fig. 1) in a 50 µl reaction containing 10 mM Tris–HCl (pH 9), 50 mM KCl, 0.1% Triton X-100, 2 mM MgCl2, 200 µM each of dGTP, dCTP, dATP and dTTP and 2 U Taq DNA polymerase (Promega). The reactions were performed using a Perkin Elmer 9600 GeneAmp Gene System for 30 cycles using an annealing temperature of 47°C. Products were visualized following electrophoresis through an agarose gel.
RESULTS
We wanted to design an assay in which we could determine whether an MDS is converted to a DSB during DNA repair in bacteria. Since the MDS are generated by ligating double-stranded oligonucleotides into a plasmid, it was essential to design a sensitive assay to detect the repair of a small amount of ligation product. Previously, Dianov et al. (10) examined the effect of two uracils, situated 12 bp apart in opposing strands, on recombination in bacteria. This latter assay allows not only DNA repair but also replication. Replication of DNA containing two base damages in opposing strands results in each of the base damages being annealed to an undamaged complementary strand and loss of the MDS. Repair would have to occur prior to replication to study the products of MDS repair. This could decrease the sensitivity of the assay. We have utilized a reporter gene to detect the introduction of DSBs. p3′luc is a bacterial expression vector that expresses firefly luciferase. There are two unique restriction sites (PacI and ClaI) situated 45 bp apart in the 3′ end of the open reading frame (Fig. 1). We have shown previously that this encodes a part of the protein critical for activity (27). Deletion of sequence or breakage of the plasmid between the PacI–ClaI sites results in the loss of activity. We therefore inserted oligonucleotides containing the MDS between these restriction sites. Conversion of the MDS to a DSB was expected to cause a decrease in luciferase activity. We also designed the vector to contain a 24 bp direct repeat sequence encompassing the MDS insertion site (Fig. 1). If a DSB was generated by base excision repair, we would expect the repeat sequence to stimulate repair of the plasmid by recombination as found by Dianov et al. (10), resulting in deletion of the 421 bp situated between the repeat sequences.
This assay involves the ligation of a double-stranded oligonucleotide into the PacI and ClaI sites, transformation of the ligation reaction into bacteria and measurement of luciferase activity after 4 h incubation in the presence of novobiocin. Novobiocin inhibits DNA gyrase, does not induce the SOS DNA-repair pathway (28) and has been shown previously to inhibit bacterial and plasmid replication (29,30). Initial experiments demonstrated that we could inhibit bacterial growth with ≥50 µM novobiocin and that 4 h was the optimal time to measure luciferase activity (data not shown). Since in vitro studies predicted that the MDS would be converted to a DSB and possibly destroy the p3′luc, we decided to co-transform the bacteria with a second plasmid encoding a different antibiotic resistance to p3′luc. This was used to normalize the luciferase activity for the efficiency of transformation of each sample.
Effect of a single uracil on luciferase activity
Each oligonucleotide containing a single uracil was annealed to an undamaged oligonucleotide (Table 1), ligated into p3′luc and tested to see whether a single uracil altered luciferase activity compared with undamaged sequence. Each uracil used to form MDS (Table 2) was tested as a single lesion. This was particularly important for the lesions on the transcribed strand, as removal of the uracil generating an AP site or a nucleotide gap could generate a deleted transcript (31) or decrease the efficiency of transcription (32). The uracil residues were base paired with an adenine to prevent the production of mutated transcripts (33) and mismatch repair. To prevent a decrease in the efficiency of promoter clearance the lesions were positioned at the 3′ end of the coding region, away from the promoter. As can be seen from Figures 2A, 3A and 4A, a single uracil lesion did not alter the luciferase activity measured from extracts of wild-type, ung– or xth–nfo– bacteria. No difference was seen between a uracil positioned on the transcribed strand or the non-transcribed strand. A control ligation, which did not contain oligonucleotides, was also performed for each experiment. A variation was seen with different batches of purified PacI, ClaI linearized p3′luc. Luciferase activity for the control ligation was 0.5–10% of the undamaged sequence.
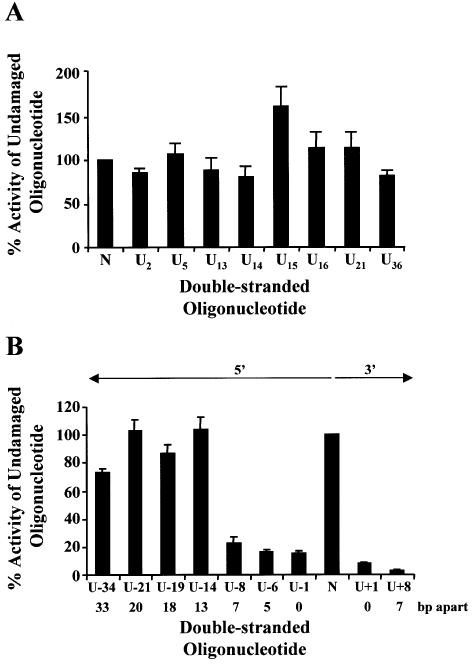
Figure 2.
Repair of uracil DNA damage in wild-type bacteria (BW35) Double-stranded oligonucleotides containing a single uracil (A), two closely opposed uracils (B) or no damage (represented as N on the graphs) were ligated into p3′luc and the ligation reaction co-transformed with either pPROLar.A122 or pACYC184 into wild-type bacteria. After a 4 h incubation, the luciferase activity was measured in a cell-free extract and the activity normalized to the number of kanamycin- or chloramphenicol-resistant colonies obtained after overnight growth on solid medium. These results were used to determine the percentage of activity measured for each sample compared with the undamaged control sequence. Triplicate transformations were performed for each ligation reaction and at least two ligations were examined. The average and the standard error are shown graphically. For substrates containing two uracils (B), the number of base pairs (bp) separating the two uracils, as well as the orientation (5′ or 3′) of the uracils with respect to each other, is indicated.
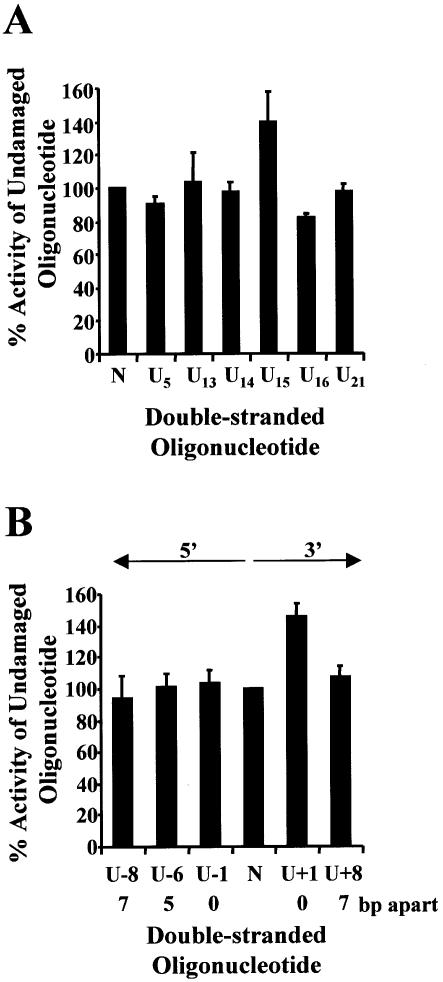
Figure 3.
Repair of uracil DNA damage in uracil DNA glycosylase- deficient bacteria (BD2008). Double-stranded oligonucleotides containing a single uracil (A), two closely opposed uracils (B) or no damage (represented as N on the graphs) were ligated into p3′luc and the ligation reaction co-transformed with pACYC184 or pPROLar.A122 into ung– bacteria. After a 4 h incubation, the luciferase activity was measured in a cell-free extract and the activity normalized to the number of kanamycin- or chloramphenicol-resistant colonies obtained after overnight growth on solid medium. These results were used to determine the percentage of activity measured for each sample compared with the undamaged control sequence. Triplicate transformations were performed for each ligation reaction and at least two ligations were examined. The average and the standard error are shown graphically. For substrates containing two uracils (B), the number of base pairs (bp) separating the two uracils, as well as the orientation (5′ or 3′) of the uracils with respect to each other, is indicated.
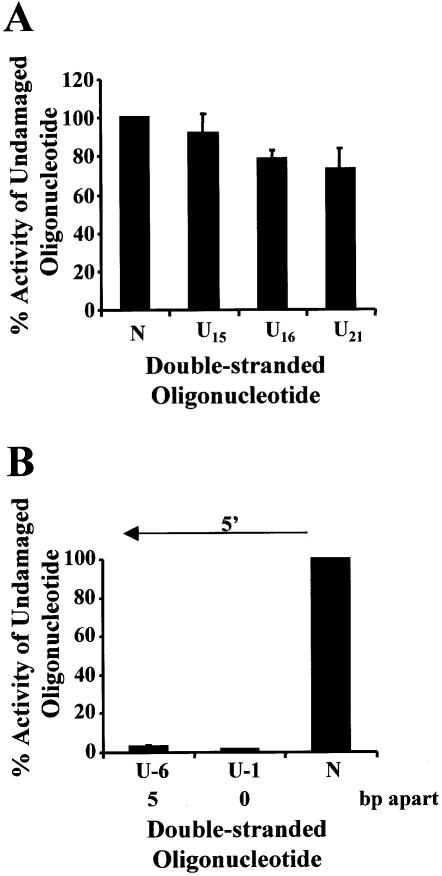
Figure 4.
Repair of uracil DNA damage in exonuclease III- and endonuclease IV-deficient bacteria (xth–nfo–). Double-stranded oligonucleotides containing a single uracil (A), two closely opposed uracils (B) or no damage (represented as N on the graphs) were ligated into p3′luc and the ligation reaction co-transformed with pACYC184 into xth–nfo– bacteria. After a 4 h incubation, the luciferase activity was measured in a cell-free extract and the activity normalized to the number of chloramphenicol-resistant colonies obtained after overnight growth on solid medium. These results were used to determine the percentage of activity measured for each sample compared with the undamaged control sequence. Triplicate transformations were performed for each ligation reaction and at least three ligations were examined. The average and the standard error are shown graphically. For substrates containing two uracils (B), the number of base pairs (bp) separating the two uracils, as well as the orientation (5′) of the uracils with respect to each other, is indicated.
Effect of clustered uracils on luciferase activity
When uracil lesions were positioned in opposing DNA strands, either 5′ or 3′ to each other, and separated by ≤7 bp, a 5–30 fold decrease in luciferase activity was detected in wild-type bacteria (Fig. 2B). The same ligation reactions were then transformed into ung– bacteria. In the absence of uracil DNA glycosylase, the clustered uracil lesions did not decrease luciferase activity (Fig. 3B). However, when two uracils were positioned in opposing strands, 5′ to each other and separated by ≤5 bp, luciferase activity decreased in xth–nfo– bacteria (Fig. 4B).
To try to define the size of MDS in our system, the opposing uracil lesions were positioned at different distances apart. When the two uracils were positioned 5′ to each other and 13–33 bp apart, luciferase activity did not decrease in comparison with the undamaged sequence in wild-type bacteria (Fig. 2B).
Analysis of plasmid remaining after DNA repair
Production of a DSB by repair, could result in the destruction of the linear plasmid by nucleases within the cell, or repair by recombination and deletion of the sequence. Both of these scenarios would result in the loss of luciferase activity. Degradation of the plasmid would also decrease the number of AmpR colonies that could be grown from the culture following repair. We therefore wanted to determine whether repair of a single uracil (U5) or two closely opposed uracils (U + 8) decreased the number of AmpR colonies in the culture in comparison with undamaged sequence. By co-transforming the ligation reactions with pACYC184 and determining the number of CmR colonies, we were able to normalize the number of AmpR colonies for variations in transformation efficiency. As can be seen from Table 3, the ratio of the AmpR/CmR colonies was significantly decreased for the p3′luc containing a MDS with two uracils situated 3′ to each other in opposite strands and 7 bp apart (U + 8) in comparison with either a single uracil (U5) or undamaged sequence. There was no significant difference between the single uracil and the undamaged sequence, or between the MDS and the control ligation containing no insert. To determine whether mis-repair of the sequence had occurred, plasmid DNA was isolated from AmpR colonies and subjected to PCR analysis. Two sets of primers were used in the analysis: primers 1 and 2, which were expected to generate a 602 bp product, and primers 3 and 4, which amplify a 189 bp product (Fig. 1). AmpR colonies that contained deletions were found in ligations of the undamaged sequence and the single uracil (U5), as well as the control ligation and the MDS (U + 8; Table 3). All of the deletions were detected using primers 3 and 4, and were similar in size (∼45–50 bp) when subjected to gel electrophoresis through a 2% agarose gel (data not shown). Two of the plasmids carrying deletions from the U + 8 ligation were sequenced and found to be deleted in the PacI–ClaI region and a few base pairs on either side of these sites. As can be seen from Table 3, ligations of U + 8 and ‘no insert’ had a high percentage of colonies carrying this small deletion (72 and 85%, respectively), while a lower percentage was found in colonies from the undamaged and U5 ligations (13 and 21%, respectively).
Table 3. Analysis of plasmid remaining after DNA repair.
| Sample | AmpR/CmR | Number of colonies analyzed by PCR | Number containing a deletion |
|---|---|---|---|
| Undamaged | 7.2 ± 1.5 | 55 | 7 |
| U5 | 4.5 ± 0.6 | 56 | 12 |
| U + 8 | 1.0 ± 0.3a | 54 | 39 |
| No insert | 1.2 ± 0.2a | 20 | 17 |
Following transformation of wild-type bacteria with the ligation reaction and pACYC184 and a 4 h incubation in the presence of novobiocin, bacteria were grown on solid medium containing either chloramphenicol or ampicillin. AmpR and CmR colonies were counted after overnight growth and a ratio calculated for AmpR/CmR colonies. Two ligation reactions and six transformations were performed to generate the data, except for the no insert control where one ligation and three transformations were performed. The average and standard error are shown.
aStatistical difference compared with the undamaged sequence and the U5 sequence, which contains a single uracil (P < 0.05). Plasmid was isolated from AmpR colonies and analyzed by PCR for deletion of sequence.
DISCUSSION
We have designed an assay utilizing the firefly luciferase reporter to detect the introduction of a DSB at an MDS in bacteria. We have utilized closely opposed uracil DNA damages as a model for MDS. Uracil DNA damage and oxidative DNA damage are predominantly repaired by base excision repair. In fact, removal of uracil by Ung results in the introduction of an AP site, which is similar to oxidative damage found in MDS (5–8). In this system, a DSB introduced at the MDS, or mis-repair resulting in deletion of sequence, causes a decrease in luciferase activity. To ensure that a single lesion did not alter activity, each single uracil was tested for its effect on luciferase activity. Single uracil residues situated in either the transcribed or non-transcribed strand (Table 1) did not decrease luciferase activity in wild-type, ung– or xth–nfo– bacteria (Figs 2A, 3A and 4A). This indicates that a single uracil did not decrease the efficiency of transcription, and that conversion of the uracil to an AP site or nucleotide gap did not produce a substantial amount of deleted transcript or aberrant protein in our system. Since the uracil residues were base paired with an adenine, we did not expect mutated transcripts to be generated, even if the uracil was converted to an AP site. Escherichia coli RNA polymerase has been found to insert an adenine opposite an AP site during transcription (32).
Previous in vitro studies examining the repair of MDS with purified prokaryote or eukaryote repair enzymes or cell-free extracts have demonstrated that, in general, two damages situated in opposing DNA strands and immediately 5′ or 3′ to each other do not result in a DSB; the initiation of repair at one of the lesions inhibits the repair of the second lesion (reviewed in 16–18). Recently, we have shown that this inhibition of repair can enhance the mutation frequency of 8-oxodG if it is positioned immediately 5′ and opposed to a second 8-oxodG (27). However, in the work described here, two uracils situated in opposing DNA strands and immediately 5′ or 3′ to each other (U – 1 and U + 1; Table 2) did decrease luciferase activity to 15 and 8% of the undamaged sequence, respectively, in wild-type bacteria (Fig. 2B). This suggests that a DSB was produced during the attempt to repair these MDS. Similar MDS in ung– bacteria did not result in a decrease in luciferase activity (Fig. 3B). This indicates that repair had to be initiated by Ung to generate the DSB. The binding and/or catalytic activity of Ung was therefore not disrupted by a SSB or AP site (uracil repair-intermediates) situated immediately 5′ or 3′ to uracil, unlike the DNA glycosylases studied previously in vitro: formamidopyrimidine DNA glycosylase (Fpg) binding was disrupted by the presence of a SSB (19), endonuclease III and Fpg showed a decrease in catalytic activity at dihydrothymine situated in close opposition to an AP site (34) and a reduced kcat was measured for Fpg cleavage at an 8-oxodG closely opposed to an AP site or SSB (34). This may be explained by the fact that Ung can remove uracil from single-stranded DNA, whereas the other DNA glycosylases are specific for lesions in double-stranded DNA and do require protein–DNA contacts on the opposite strand to the lesion.
Following the action of Ung, MDS consisting of two AP sites may exist. Exonuclease III and endonuclease IV are the two major AP endonucleases in bacteria. Cleavage of bistranded AP sites by exonuclease III in vitro is reduced when the second AP site is immediately 5′ or 3′ to the first AP site (35). Inhibition is greater if the lesions are 5′ to each other and appears to be due to the generation of a SSB at one of the AP sites. It would have been expected therefore that the U + 1, but not the U – 1, lesion would have decreased luciferase activity. Interestingly, a decrease in luciferase activity was also found if the U – 1 and U – 6 MDS were transformed into xth–nfo– bacteria (Fig. 4B). This suggests that breakage at the AP sites occurred via a mechanism or pathway distinct from the AP endonucleases, and that this system may act as a ‘back-up’ in wild-type cells to repair AP sites when exonuclease III and endonuclease IV are unable to cleave the damage. Two possible alternatives are the AP lyase activities in the cell and the nucleotide excision repair (NER) pathway, which has also been shown to cleave at an AP site (36,37). However, as discussed above, the DNA glycosylases with associated AP lyase activity were also inhibited in vitro by a second lesion situated 5′ or 3′ to the target lesion. This would indicate that NER might play a significant role in MDS repair. However, it is also possible that when all the components of the base excision repair pathway are present in the cell, the AP lyases are able to cleave more efficiently. Recently, it was shown that endonuclease IV can enhance the activity of endonuclease III in vitro, and even promote the formation of an endonuclease III–DNA complex (38), and inhibition of Fpg at an 8-oxodG situated immediately 3′ to a SSB is decreased when endonuclease IV, DNA polymerase I and DNA ligase are added (19). Work in vitro using mammalian cell nuclear extract also demonstrated that an AP site can be cleaved even when it is situated immediately 5′ or 3′ to a second AP site (39). Although it is beyond the scope of this work, further studies are planned using bacteria mutated in the AP endonucleases and NER proteins to try to determine what pathway is involved in cleaving these specific MDS.
In agreement with the in vitro studies, MDS containing two uracils in opposite DNA strands and situated 5–7 bp apart 5′ to each other, or 7 bp apart 3′ to each other, resulted in a decrease in luciferase activity in wild-type bacteria (Fig. 2B), suggesting the formation of a DSB. Similar lesions did not cause a decrease in luciferase activity in ung– bacteria (Fig. 3B), indicating that repair had to be initiated by Ung to cause a decrease in activity. The decrease in activity could be due to degradation of the linear plasmid after the formation of a DSB, or deletion of sequence following recombination. The p3′luc assay system was designed to contain a direct repeat encompassing the MDS (Fig. 1), which should stimulate recombination if a DSB is introduced. After repair, cultures transformed with ligations of undamaged sequence, a single uracil (U5) or the U + 8 MDS, were grown to obtain AmpR (containing p3′luc) or CmR (containing pACYC184) colonies. Degradation of p3′luc during repair was expected to decrease the AmpR/CmR colonies in the culture for wild-type bacteria. The U + 8 MDS resulted in an AmpR/CmR colony ratio significantly lower (approximately seven times) than the undamaged sequence and the sequence containing a single uracil, and similar to the ligation that did not contain a double-stranded oligonucleotide insert (Table 3). This indicates that a large proportion of the p3′luc containing the MDS was destroyed during repair. It is unlikely that the low number of AmpR colonies obtained from the U + 8 sample was caused by a low efficiency of ligation, since the luciferase activity measured in ung– bacteria was equivalent for the ligations of the undamaged and MDS-containing oligonucleotides (Fig. 3). PCR was used to determine whether deletions had been introduced into the surviving p3′luc. Although a large number of colonies from the U + 8 sample did contain deletions (72%), the deletion was very small (∼45 bp) and was situated between the PacI–ClaI restriction sites. In fact, when two samples were sequenced they were found to be deleted only in the PacI–ClaI region, which is the site of insertion of the double-stranded oligonucleotide in the DNA repair assay. A similar percentage of deleted sequences (85%) of the same size and at the same position were found in the no insert control AmpR colonies and a lower percentage of the same deletion (13–21%) was also found in plasmid obtained from the AmpR colonies from the undamaged and single uracil samples (Table 3). The plasmid with the deletion cannot be related to the uracil damage or MDS repair by recombination as it was found in the undamaged sample. Since plasmid containing the same deletion was present in the ‘no insert’ control, it is likely this circular molecule was generated during the ligation reaction by the mis-joining of the PacI–ClaI linear termini and is therefore a background ligation product. The lack of large deletions or recombination products in the AmpR colonies could be explained by the inhibition of recombination during the assay. Novobiocin is added to inhibit DNA gyrase and DNA replication. However, DNA gyrase has been implicated in recombination. DNA gyrase is required for the synapsis of a pair of Mu DNA ends during recombination in the replicative Mu transposition pathway (40), and site-specific recombination catalyzed by the E.coli Tn3 resolvase (41) and the bacteriophage lambda Int system (42) require DNA supercoiling, which is provided by DNA gyrase.
The majority of the in vitro studies did not examine MDS that were >5 bp apart and a variety of enzymes were able to initiate repair once the second lesion was ≥2 bp apart (reviewed in 16–18). Dianov et al. (10) did demonstrate that two uracils situated in opposing strands and 12 bp apart did increase recombination frequency in wild-type bacteria. We therefore wanted to try and define the size of MDS in our system. MDS were tested that consisted of two uracils ranging from 13 to 33 bp apart (Fig. 2). None of these lesions decreased luciferase activity in wild-type cells. The two uracil lesions were repaired completely if they were ≥13 bp apart, defining the size of the MDS, in our plasmid system in bacteria, as two uracils separated by <13 bp. Since hydrogen bonding between the bases of the two strands will be an important factor in maintaining DNA structure during complete repair of the two SSBs, the minimum distance between two opposing SSBs that results in a DSB will likely be altered by the GC content of the DNA sequence and therefore may vary with sequence context. Future studies will determine whether the size of a MDS is altered by the types of damage that form the clustered lesion.
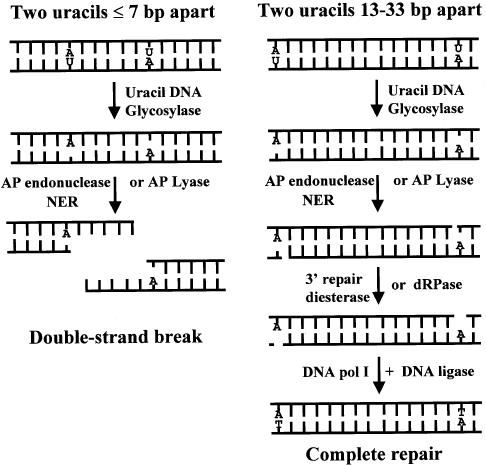
In summary, if two uracils are positioned in opposite DNA strands and situated ≤7 bp apart in bacteria, uracil DNA glycosylase initiates base excision repair and the AP sites are converted to SSB-repair intermediates by the AP endonucleases (exonuclease III and endonuclease IV), AP lyases and/or NER in the cell (Fig. 5). The two SSB-repair intermediates form a double-strand break and the DNA can be degraded by endogenous nucleases. In the absence of Ung a double-strand break is not formed. If the two uracils are 13–33 bp apart, complete repair occurs. This work therefore supports the in vitro studies using defined MDS and indicates that clustered damage introduced by ionizing radiation can be converted to lethal DSBs by the initiation of repair in bacteria.
Figure 5.
Closely opposed uracils (≤7 bp apart) are converted to a double-strand break in bacteria. When two uracils (U) are in opposing DNA strands and situated either 5′ or 3′ to each other, uracil DNA glycosylase can remove both uracil residues, resulting in a MDS consisting of two closely opposed AP sites. The major AP endonucleases (exonuclease III or endonuclease IV), the AP lyase activities or the NER pathway in the bacterium can then convert the AP sites to SSB-repair intermediates. The repair products shown are those generated by the AP endonucleases and AP lyases of the base excision repair pathway. If the two uracil residues were originally situated ≤7 bp apart, the two SSB-repair intermediates form a DSB. A DSB is not formed if the uracil lesions were originally separated by 13–33 bp. In the absence of uracil DNA glycosylase, repair is not initiated to generate the SSB-repair intermediates and hence a DSB is not formed.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank Dr Susan S. Wallace for critical reading of the manuscript. This work was supported by the NIH grant CA 85693.
REFERENCES
- 1.Goodhead D.T. (1994) Initial events in the cellular effects of ionizing radiations: clustered damage in DNA. Int. J. Radiat. Biol., 65, 7–17. [DOI] [PubMed] [Google Scholar]
- 2.Povirk L.F. and Houlgrave,C.W. (1988) Effect of apurinic/apyrimidinic endonucleases and polyamines on DNA treated with bleomycin and neocarzinostatin: specific formation and cleavage of closely opposed lesions in complementary strands. Biochemistry, 27, 3850–3857. [DOI] [PubMed] [Google Scholar]
- 3.Ward J.F., Evans,J.W., Limoli,C.L. and Calabro-Jones,P.M. (1987) Radiation and hydrogen peroxide induced free radical damage to DNA. Cancer, 55, 105–112. [PMC free article] [PubMed] [Google Scholar]
- 4.Bourdat A.-G., Douki,T., Frelon,S., Gasparutto,D. and Cadet,J. (2000) Tandem base lesions are generated by hydroxyl radical within isolated DNA in aerated aqueous solution. J. Am. Chem. Soc., 122, 4549–4556. [Google Scholar]
- 5.Sutherland B.M., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) Clustered damages and total lesions induced in DNA by ionizing radiation: oxidized bases and strand breaks. Biochemistry, 39, 8026–8031. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland B.M., Bennett,P.V., Sidorkina,O. and Laval,J. (2000) Clustered DNA damages induced in isolated DNA in human cells by low doses of ionizing radiation. Proc. Natl Acad. Sci. USA, 97, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulston M., Fulford,J., Jenner,T., de Lara,C. and O’Neill,P. (2001) Clustered DNA damage induced by γ radiation in human fibroblasts (HF19), hamster (V79-4) cells and plasmid DNA is revealed as Fpg and Nth sensitive sites. Nucleic Acids Res., 30, 3464–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sutherland B.M., Bennett,P.V., Sutherland,J.C. and Laval,J. (2002) Clustered DNA damages induced by X-rays in human cells. Radiat. Res., 157, 611–616. [DOI] [PubMed] [Google Scholar]
- 9.Ward J.F. (1981) Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat. Res., 86, 185–195. [PubMed] [Google Scholar]
- 10.Dianov G.L., Timchenko,T.V., Sinitsina,O.I., Kuzminov,A.V., Medvedev,O.A. and Salganik,R.I. (1991) Repair of uracil residues closely spaced on the opposite strand of plasmid DNA results in double-strand break and deletion formation. Mol. Gen. Genet., 225, 448–452. [DOI] [PubMed] [Google Scholar]
- 11.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 12.Dugle D.L., Gillespie,C.J. and Chapman,J.D. (1976) DNA strand breaks, repair and survival in X-irradiated mammalian cells. Proc. Natl Acad. Sci. USA, 73, 809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahnstrom G. and Bryant,P.E. (1982) DNA double-strand breaks generated by the repair of X-ray damage in Chinese hamster cells. Int. J. Radiat. Biol., 41, 671–676. [DOI] [PubMed] [Google Scholar]
- 14.Bonura T., Smith,K.C. and Kaplan,H.S. (1975) Enzymatic induction of DNA double-strand breaks in gamma-irradiated E.coli K-12. Proc. Natl Acad. Sci. USA, 72, 4265–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaisdell J.O. and Wallace,S.S. (2001) Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc. Natl Acad. Sci. USA, 98, 7426–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaisdell J.O., Harrison,L. and Wallace,S.S. (2001) Base excision repair processing of radiation-induced clustered DNA lesions. Radiat. Protect. Dosimetry, 97, 25–31. [DOI] [PubMed] [Google Scholar]
- 17.Weinfeld M., Rasouli-Nia,A., Chaudhry,M.A. and Britten,R.A. (2001) Response of base excision repair enzymes to complex DNA lesions. Radiat. Res., 156, 584–589. [DOI] [PubMed] [Google Scholar]
- 18.Harrison L. and Malyarchuk,S. (2002) Can DNA repair cause enhanced cell killing following treatment with ionizing radiation? Pathophysiology, 8, 149–159. [DOI] [PubMed] [Google Scholar]
- 19.Harrison L., Hatahet,Z. and Wallace,S.S. (1999) In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J. Mol. Biol., 290, 667–684. [DOI] [PubMed] [Google Scholar]
- 20.Bennett S.E., Sanderson,R.J. and Mosbaugh,D.W. (1995) Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry, 34, 6109–6119. [DOI] [PubMed] [Google Scholar]
- 21.Sung J.S. and Mosbaugh,D.W. (2000) Escherichia coli double-strand uracil-DNA glycosylase: involvement in uracil-mediated DNA base excision repair and stimulation of activity by endonuclease IV. Biochemistry, 39, 10224–10235. [DOI] [PubMed] [Google Scholar]
- 22.Saparbaev M. and Laval,J. (1998) 3,N4-ethenocytosine, a highly mutagenic adduct, is a primary substrate for Escherichia coli double-stranded uracil-DNA glycosylase and human mismatch-specific thymine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 95, 8508–8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis Ltd, London, UK. [Google Scholar]
- 24.Téoule R. (1987) Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol., 51, 573–589. [DOI] [PubMed] [Google Scholar]
- 25.Duncan B.K. (1985) Isolation of insertion, deletion and nonsense mutations of the uracil-DNA glycosylase (ung) gene of Escherichia coli K-12. J. Bacteriol., 164, 689–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seidman C.E., Struhl,K., Sheen,J. and Jessen,T. (1997) Introduction of plasmid DNA into cells. In Current Protocols in Molecular Biology, Vol. 1, (Suppl. 37), John Wiley and Sons, Inc., New York, NY, 1.8.4–1.8.5. [DOI] [PubMed] [Google Scholar]
- 27.Malyarchuk S., Youngblood,R., Landry,A.M., Quillin,E. and Harrison,L. (2003) The mutation frequency of 8-oxo-7,8 dihydroguanine (8-oxodG) situated in a multiply damaged site: comparison of a single and two closely opposed 8-oxodG in Escherichia coli. DNA Rep., 2, 695–705. [DOI] [PubMed] [Google Scholar]
- 28.Gellert M., O’Dea,M.H., Itoh,T. and Tomizawa,J. (1976) Novobiocin and coumermycin inhibit DNA supercoiling catalyzed by DNA gyrase. Proc. Natl Acad. Sci. USA, 73, 4474–4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeMarini D.M. and Lawrence,B.K. (1992) Prophage induction by DNA topoisomerase II poisons and reactive-oxygen species: role of DNA breaks. Mutat. Res., 267, 1–17. [DOI] [PubMed] [Google Scholar]
- 30.Viswanathan A., You,H.J. and Doetsch,P.W. (1999) Phenotypic change caused by transcriptional by-pass of uracil in non-dividing cells. Science, 284, 159–162. [DOI] [PubMed] [Google Scholar]
- 31.Liu J. and Doetsch,P.W. (1996) Template strand gap bypass is a general property of prokaryotic RNA polymerases: Implications for elongation mechanisms. Biochemistry, 35, 14999–15008. [DOI] [PubMed] [Google Scholar]
- 32.Zhou W. and Doetsch,P.W. (1993) Effects of abasic sites and DNA single strand breaks on prokaryotic RNA polymerases. Proc. Natl Acad. Sci. USA, 90, 6601–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viswanathan A. and Doetsch,P.W. (1998) Effects of non-bulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem., 273, 21276–21281. [DOI] [PubMed] [Google Scholar]
- 34.David-Cordonnier M.-H., Laval,J. and O’Neill,P. (2001) Recognition and kinetics for excision of a base lesion within clustered DNA damage by the Escherichia coli proteins Fpg and Nth. Biochemistry, 40, 5738–5746. [DOI] [PubMed] [Google Scholar]
- 35.Chaudhry M.A. and Weinfeld,M. (1997) Reactivity of human apurinic/aprymidinic endonuclease and Escherichia coli exonuclease III with bistranded abasic sites in DNA. J. Biol. Chem., 272, 15650–15655. [DOI] [PubMed] [Google Scholar]
- 36.Lin J.-J. and Sancar,A. (1989) A new mechanism for repairing oxidative DNA damage to DNA: (A)BC exonuclease removes AP sites and thymine glycols from DNA. Biochemistry, 28, 7979–7984. [DOI] [PubMed] [Google Scholar]
- 37.Snowden A., Kow,Y.W. and Van Houten,B. (1990) Damage repertoire of the Escherichia coli UvrABC nuclease complex includes abasic sites, base-damage analogues and lesions containing adjacent 5′ or 3′ nicks. Biochemistry, 29, 7251–7259. [DOI] [PubMed] [Google Scholar]
- 38.Back J.H., Chung,J.H., Park,Y.I., Kim,K.-S. and Han,Y.S. (2003) Endonuclease IV enhances base excision repair of endonuclease III from Methanobacterium thermoautotrophicum. DNA Rep., 2, 455–470. [DOI] [PubMed] [Google Scholar]
- 39.David-Cordonnier M.-H., Laval,J. and O’Neill,P. (2000) Clustered DNA damage, influence on damage excision by XRS 5 nuclear extracts and Escherichia coli Nth and Fpg proteins. J. Biol. Chem., 275, 11865–11873. [DOI] [PubMed] [Google Scholar]
- 40.Sokolsky T.D. and Baker,T.A. (2003) DNA gyrase requirements distinguish the alternate pathways of Mu transposition. Mol. Microbiol., 47, 397–409. [DOI] [PubMed] [Google Scholar]
- 41.Bliska J.B., Benjamin,H.W. and Cozzarelli,N.R. (1991) Mechanism of Tn3 resolvase recombination in vivo. J. Biol. Chem., 266, 2041–2047. [PubMed] [Google Scholar]
- 42.Bliska J.B. and Cozzarelli,N.R. (1987) Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J. Mol. Biol., 194, 205–218. [DOI] [PubMed] [Google Scholar]