Abstract
Molecular mechanisms mediating group A Streptococcus (GAS)-host interactions remain poorly understood but are crucial for diagnostic, therapeutic, and vaccine development. An optimized high-density microarray was used to analyze the transcriptome of GAS during experimental mouse soft tissue infection. The transcriptome of a wild-type serotype M1 GAS strain and an isogenic transcriptional regulator knockout mutant (covR) also were compared. Array datasets were verified by quantitative real-time reverse transcriptase-polymerase chain reaction and in situ immunohistochemistry. The results unambiguously demonstrate that coordinated expression of proven and putative GAS virulence factors is directed toward overwhelming innate host defenses leading to severe cellular damage. We also identified adaptive metabolic responses triggered by nutrient signals and hypoxic/acidic conditions in the host, likely facilitating pathogen persistence and proliferation in soft tissues. Key discoveries included that oxidative stress genes, virulence genes, genes related to amino acid and maltodextrin utilization, and several two-component transcriptional regulators were highly expressed in vivo. This study is the first global analysis of the GAS transcriptome during invasive infection. Coupled with parallel analysis of the covR mutant strain, novel insights have been made into the regulation of GAS virulence in vivo, resulting in new avenues for targeted therapeutic and vaccine research.
Skin and soft tissue infections are most commonly caused by bacteria and account for ∼7 to 10% of hospitalizations in North America.1 Substantial portions are caused by Streptococcus pyogenes (group A streptococci; GAS). A major human pathogen, GAS is associated with millions of skin and throat infections each year.2 Serious complications after infection, namely rheumatic fever, glomerulonephritis, and reactive arthritis, can follow even relatively mild GAS infections and cause significant morbidity worldwide.3,4 Some GAS infections rapidly progress to life-threatening invasive diseases such as septicemia, streptococcal toxic shock syndrome, and necrotizing fasciitis, requiring life-saving interventions in the form of aggressive fluid replacement, general supportive measures, and/or emergent surgical debridement.2–4 Nearly 15,000 cases of invasive GAS disease and an estimated 1300 deaths occur annually in the United States.5 Epidemiological data also suggest increased incidence and severity of GAS infections in recent years.2,6,7 Because progression to systemic toxicity and shock can occur throughout a span of hours, early recognition and initiation of aggressive therapies are essential.
Molecular mechanisms mediating GAS-host interactions remain poorly understood but are crucial for rapid diagnostic, therapeutic, and vaccine development.3,4 As a consequence, concerted efforts have been made to identify regulatory mechanisms involved in coordinating the in vitro expression of GAS virulence determinants. Genes have been identified that regulate production of proven and putative virulence factors, and several of these regulators are known to respond to environmental changes in vitro or to specific phases of the bacterial growth cycle.3,8 However, far less is known about gene expression in vivo during GAS infection of mammalian hosts.9–11 Infection sites or the host circulation environment are complex, and their influence on bacterial gene regulation cannot be simulated in vitro.12 Hence, further understanding of GAS host/pathogen molecular interactions requires analysis of in vivo conditions that better approximate the natural history of infections.
Streptococcal soft tissue infections, such as cellulitis involving the subdermal layers and superficial fascia, often commence with nonspecific symptoms such as erythema, heat, soft tissue edema, generalized pain, and sometimes serosanguinous bullae.6,7 Progression to diffuse necrosis of subcutaneous and deep fascia (necrotizing fasciitis) often occurs without overt skin injury13 and leads to development of vasculitis, intravascular thrombosis, and tissue gangrene. Histopathology of debrided soft tissue reveals acute inflammation of subcutaneous tissue with bacterial aggregates and multifocal necrosis. Mice subcutaneously injected with GAS also develop inflamed bullae that later ulcerate; diffuse inflammatory infiltrates with high bacterial counts, and diffuse tissue necrosis including thrombosed vessels.3 In a previous study,14 we measured a subset (n = 17) of GAS transcripts expressed in vivo 2 days after inoculation with a wild-type (WT) serotype M1 GAS strain and a ΔcovR isogenic deletion mutant of a key GAS two-component regulatory system (TCS) referred to as CovR-CovS (Cov, control of virulence). In this present study we have expanded our investigation by using a custom high-density array for whole GAS transcriptome analysis in the murine soft tissue infection model. The data presented here provide substantial new information about GAS survival strategies in soft tissue. The most abundant GAS transcripts detected were found to encode extracellular proteins that are key virulence determinants. Other expressed GAS genes are potential targets for therapeutic intervention and warrant further study.
Materials and Methods
Bacterial Strains
Serotype M1 GAS strains commonly cause pharyngitis and invasive infections.2 Strain MGAS5005 (no. BAA-947; American Type Culture Collection, Rockville, MD), a WT clinical strain (serotype M1), and its isogenic ΔcovR derivative strain (JRS950) have been described.14–16 Bacteria were cultured statically on Trypticase soy agar containing 5% sheep blood agar (Becton-Dickinson, Cockeysville, MD) or in Todd-Hewitt broth (Becton-Dickinson) containing 0.2% (w/v) yeast extract (THY; Difco Laboratories, Detroit, MI), at 37°C in an atmosphere containing 5% CO2. For generating inoculum, cells were harvested from THY at late-exponential phase (OD600 ∼0.75) to limit infectivity differences across strains associated with up-regulated capsule biosynthesis in strain JRS950, which is maximal in the early-to-mid exponential growth phases.
Mouse Soft Tissue Infection Model
This model of GAS soft tissue infection has been used extensively to study bacterial-host interactions.14,17,18 Our experimental protocol was approved by the Institutional Animal Care and Use Committee, National Institute of Allergy and Infectious Diseases. Outbred, immunocompetent, hairless male Crl:SKH1-hrBR mice (5 weeks old; 20 to 25 g) (Charles River Breeding Laboratories, Bar Harbor, ME), maintained on standard mouse food and water ad libitum, were randomly assigned to one of two treatment groups (WT or ΔcovR mutant GAS strain, n = 27 each; four mice per cage). Immediately before inoculation, the animals were weighed and anesthetized with isoflurane (Aerrane; Ohmeda Caribe, Guayama, Puerto Rico) inhalation. The animals were inoculated subcutaneously in the dorsal side with 0.1 ml of pyrogen-free phosphate-buffered saline (PBS) (diluent) or ∼3 × 107 colony-forming units of MGAS5005 (WT) or JRS950 (ΔcovR). The actual colony-forming units of viable bacteria inoculated were verified by growth on blood agar.
To blind the investigator, cage numbers were reassigned after inoculation, and the blind was broken after data analysis. Methods for clinical measurements and analysis of gross pathologies have been reported previously for this soft tissue model of infection,14,19 and data are found in Supplementary Tables 1 and 2 (see http://ajp.amjpathol.org). Length (L) and width (W) values were used to calculate abscess volume [V = 4/3π(L/2)2 × (W/2)] and area [A = π(L/2) × (W/2)], using equations for a spherical ellipsoid as described.14
At 53 hours after inoculation, mice were euthanized and weighed, the infection site was swabbed to confirm GAS infection, and tissue was obtained from each animal via a biopsy that included dermis and underlying soft tissue lesions. Tissues were wrapped in aluminum foil, snap-frozen in liquid nitrogen, and stored at −80°C until RNA was isolated. Three additional control mice injected with sterile saline failed to show symptoms of clinical infection and did not grow GAS bacterial colonies on plating.
Experimental Design
A one-factor experimental design with two treatment (strain) levels was used. Samples were randomized at the start of procedures, and care was taken to ensure that batches of sample preparation, array hybridizations, and posthybridization washes were not confounded with treatment. Additional experimental design details are outlined in the Supplementary Materials and Methods (see http://ajp.amjpathol.org).
RNA Isolation
Frozen tissue extracts were divided into three aliquots from which RNA was purified. Tissue extracts were pulverized with a series of sharp blows delivered with a 3-pound drill hammer (Razor-Back; Union Tools Inc., Columbus, OH). Cell lysis and RNA isolation were conducted with a FastRNA kit (MP Biomedicals, Irvine, CA) as described.20 The concentrate was fragmented with a Qiashredder (Qiagen, Inc., Valencia, CA) and the isolated total RNA (containing both bacterial and host RNA) was purified in 96-well format using a plate centrifugation system (RNeasy 96; Qiagen), with on-column DNase I treatment and posttreatment with DNAFree (Ambion, Inc., Austin, TX) as described.10,20 RNA integrity was assessed with a 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA)10,20 and by measurement of the A260/A280 ratios. Absence of contaminating bacterial genomic DNA was confirmed by real-time polymerase chain reaction (PCR). Two RNA aliquots were pooled to perform the microarrays; the remaining extract was used for real-time RT-PCR validation.
Bacterial Target Preparation
Fifty-four microarray targets were prepared primarily as described.10,20 Briefly, each RNA sample was divided into two to three aliquots of 10-μg total RNA (each) to which 0.8 μg of bacteriophage MS2 carrier RNA (Roche Applied Biosciences, Indianapolis, IN) was added. Control spike transcripts (130 pmol/L) were added to each RNA aliquot, and 5 μg of random primers (Invitrogen, Carlsbad, CA) were annealed (10 minutes at 70°C, 10 minutes at 25°C). cDNA synthesis reactions and postsynthesis RNA digestion were performed as described.10,20 In brief, the resultant cDNA was purified using Qiaquick 96 kit (Qiagen) according to the manufacturer’s recommendation. For cDNA fragmentation, 3 μg of cDNA and 0.75 U of DNase I (Roche Biosciences) were used (10 minutes at 37°C, 10 minutes at 98°C). The desired cDNA size range of 50 to 200 bases was verified by separating 200 ng of cDNA on a RNA 6000 Nano LabChip (Agilent) using the 2100 BioAnalyzer (Agilent) with no added dye in the loading buffer. The fragmented cDNA was then end-labeled with biotin-ddUTP as per the Enzo BioArray terminal labeling kit (60 minutes at 37°C; Enzo Diagnostics, New York, NY) and pooled with other end-labeled cDNAs of the same sample. Samples were hybridized to 54 Rocky Mountain Laboratory (RML) Affymetrix custom GAS arrays, as described.10,20
Affymetrix GeneChip Custom Array
The Affymetrix Inc. (Santa Clara, CA) custom high-density anti-sense oligonucleotide GeneChip array (designated RMLChip herein) has been described.10,16 It contains redundant probe sets representing more than 90% coverage of predicted coding regions (1869 ORFs) encoded by the MGAS5005 genome.16 Each probe set is used to detect the presence of a single transcript, and several GAS transcripts (genes) are represented by more than one probe set. For in-depth RMLChip details, refer to the Supplementary Materials and Methods (see http://ajp.amjpathol.org).
GeneChip Hybridization
Target hybridizations, washing, staining, and scanning were performed by the National Institute of Allergy and Infectious Diseases Affymetrix core facility (Science Applications International Corp., Frederick, MD) using a GeneChip hybridization oven and the Pseudomonas aeruginosa hybridization protocol (Affymetrix), as described.20 The hybridization solution volume used was 200 μl because the RMLChip is a standard size array. Each array was scanned at 570 nm at 3-μm resolution with a GeneArray scanner. Scanned DAT-image files were analyzed with Affymetrix Microarray Suite (MAS) 5.0 software. The raw CEL files have been submitted to Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). Downstream genome analysis was accomplished using MicrobesOnline (available at http://www.microbesonline.org)21 and in-house bioinformatics analysis.
Statistical Analysis
Model-based expression estimates for each gene were obtained using the PM-MM difference model22 of dCHIP software as described.20 The gene expression estimates were normalized across samples by quadratic scaling to an artificial array with the median expression for each gene.23 Two-dimensional scatterplots of expression estimates were generated for all pairs of samples within the same treatment group to examine uniformity across samples. Hierarchical clustering and principal components analyses also were performed (Supplementary Figure 1, see http://ajp.amjpath.org). Five arrays with poorer quality RNA and low within-strain correlation were removed as outliers. Downstream data analysis was performed with MGAS5005 genome-specific probe sets using Partek Pro (Partek Inc., St. Louis, MO). To evaluate expression rankings, absolute square root-transformed expression estimates were integer-ranked, starting from 1 for the most abundant GAS transcript and increasing correspondingly with decreasing transcript detection. To investigate expression correlations, standard Pearson correlation coefficients were determined for select genes versus all other genes using Partek Pro. To investigate strain effects, expression estimates were analyzed by analysis of variance with treatment (WT versus ΔcovR strain) as a fixed effect. Technical variables included in the analysis included batches for sample preparation, sample hybridization, and posthybridization washes (Supplementary Figure 2, see http://ajp.amjpath.org). Final results were subjected to multiple testing correction using Q ≤ 0.05 false discovery rate cutoff values.24,25 Using rigorous permutation-based statistics, we also performed significance analysis of function and expression (termed SAFE)26 to assess the significance of multiple gene categories in GAS in vivo transcriptional responses across strains. A detailed description of the procedures used is provided in the Supplementary Materials and Methods (see http://ajp.amjpathol.org).
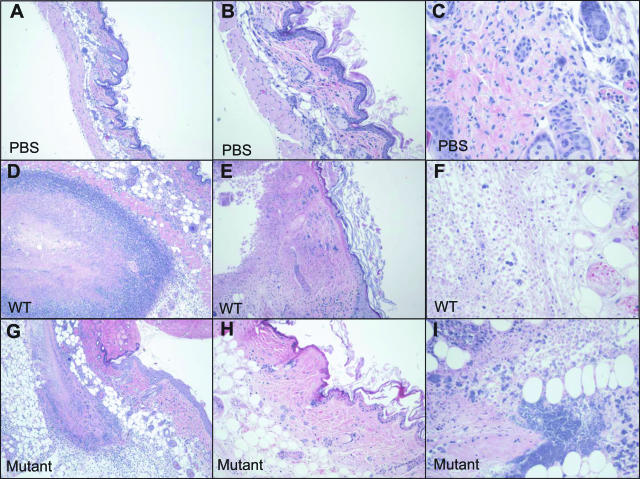
Figure 1.
Histopathological assessment of GAS mouse skin infection. Photomicrographs of H&E-stained, formalin-fixed tissue biopsies taken from PBS-inoculated (control) animals (top row), strain MGAS5005 (WT)-infected animals (middle row), and animals infected with ΔcovR mutant strain JRS950 (bottom row) at 2 days after inoculation. Results are representative of tissues obtained from animals in each of the groups. Additional histopathology and gram stains of biopsies from the same animals (D, G) are depicted in Figure 2, B and E and C and F, respectively. Original magnifications: ×40 (A, D, G); ×100 (B, E, H); ×200 (C, F, I).
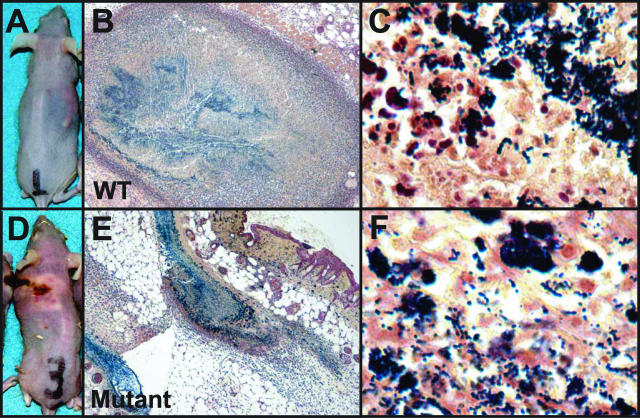
Figure 2.
Gram stains of GAS during invasive soft tissue infection. Depicted are gram-stained skin tissue sections from mice infected with WT GAS strain MGAS5005 (B, C) and ΔcovR mutant strain JRS950 (E, F). Also shown are the external pathologies for the same animals infected with WT (A) and ΔcovR mutant (D). IHC analyses of biopsies from the same animals are depicted in Figure 5, B (WT) and D (Mutant), respectively. Original magnifications: ×40 (B, E); ×1000 (C, F).
Quantitative Real-Time Reverse Transcriptase (RT)-PCR Analysis
Real-time reverse transcriptase (RT)-PCR assays were conducted to validate a subset of the microarray data. Eight oligonucleotide primer pairs and 6FAM-labeled probe sets (specific for cfa, dppA, emm1, sceD, sclA, sic, slo, and speA2) were used to perform target amplification and detection from cDNA templates in 20-μl multiplex two-step RT-PCR reactions as described.20 Targets were selected to encompass the full range of expression signal values identified by array transcriptome analysis. Target abundance was normalized to JOE-labeled internal reference transcript proS, which is transcribed at constant levels throughout the GAS growth cycle in vitro and not affected by covR inactivation.14 Differences in median values were evaluated for statistical significance with the Mann-Whitney rank sum test at the P ≤ 0.001 level.
Sampling and Histological Assessment
Tissue used for histological examination was prepared from mice subcutaneously inoculated with 2.4 × 107 colony-forming units of GAS strains MGAS5005 or ΔcovR JRS950 (n = 16, each strain) as described above, except that 4-week-old (15 to 20 g) female Crl:SKH1-hrBR mice (Charles River Breeding Laboratories) were used. Six animals inoculated with PBS were used as controls. For assessment of bacterial content, histopathology, and bacterial protein expression, mice were euthanized 48 hours after inoculation, and the skin and underlying soft tissue were removed from inoculation sites and fixed in 10% buffered formalin before embedding in paraffin. To assess the presence of bacteria and pathological changes, formalin-fixed tissues were sectioned and stained with gram stain or hematoxylin and eosin (H&E) stain (Sigma, St. Louis, MO), according to standard methodologies. An Olympus model BX51 microscope equipped with a Q-FIRE (Olympus, Tokyo, Japan) camera was used for image capture.
Immunohistochemical Analysis
Rabbit polyclonal anti-GAS antibodies made against purified recombinant GAS proteins27 were used for immunostaining. Paraffin-embedded tissues were cut into 4-μm sections and stained with antibodies specific for 16 bacterial antigens [M5005_Spy ORF numbers designated in square brackets] (AtmB [0271]; PrtS [0342]; MtsA [0368]; IdeS/Mac [0668]; [0942]; PstS [0955]; SpeA2 [0996]; MalE [1058]; PrsA [1133]; [1308]; HtsA/SiaA [1528]; Shp [1529]; DppA [1704]; Lmb [1711]; Fba [1713]; SIC [1718]) using biotinylated secondary antibodies in combination with horseradish peroxidase-coupled streptavidin (DAKO Corp., Carpinteria, CA) and the substrate AEC (BioGenex, San Ramon, CA). To evaluate nonspecific staining, a polyclonal antibody recognizing a control peptide (Bethyl Laboratories, Montgomery, TX) was used as negative control. The M3.1/1-24 control peptide, representing the N-terminal peptide of serotype M3 Emm3.1, was used because this peptide is not encoded within the genome of the serotype M1 WT strain MGAS5005.16 All immunohistochemically (IHC) stained sections were counterstained with Mayer’s hematoxylin and mounted using synthetic aqueous-based mounting medium (DAKO Faramount).
Results
Gross and Microscopic Pathology
PBS-inoculated skin sections from the immunocompetent Crl:SKH1-hrBR mice were histologically normal whereas animals inoculated with either WT or ΔcovR strain demonstrated subcutaneous foci of suppurative inflammation with adjacent infiltrating polymorphonuclear lymphocytes (PMNs), dermatitis, loss of stratum cornea, fibrin thrombi in the small vessels, and varying degrees of dermal and epidermal necrosis with degenerate and necrotic neutrophils (Figure 1). ΔcovR-inoculated mice had more extensive inflammation (panniculitis), ulcerated epidermis and necrosis of subcutaneous tissue fascia with some areas of muscle involvement (myonecrosis), and faster ulcer development (Figure 1), thus confirming previous reports of the hypervirulence of ΔcovR mutant GAS strains. All gram stains of infected tissues revealed aggregates of gram-positive cocci in multiple focal planes (Figure 2). Taken together, these observations provide histopathological evidence of high bacterial loads and diffuse tissue necrosis analogous to progressive human infections.
Abundant in Vivo GAS Transcripts
The vast majority of the top 200 detected GAS transcripts in WT-inoculated tissue extracts from soft tissue infection encode products involved in protein synthesis (n = 22), information processing (n = 15), carbohydrate metabolism (n = 15), membrane transport (n = 15), stress adaptation (n = 15), or cellular processing (n = 14) are shown in Supplementary Table 3 (see http://ajp.amjpathol.org) and discussed in more detail below. In addition, transcripts with virulence (n = 10), cell wall metabolism (n = 8), and unknown functional annotation (n = 9) also were detected at high levels (Supplementary Table 3, see http://ajp.amjpathol.org) implying that replication, protein synthesis, cellular remodeling, and stress adaptation are important functions during soft tissue infection. Proteins encoded by five GAS transcripts found to be abundant in the infected tissue [penicillin-binding protein (PBP) 1A (pbp1a), a protein translation initiation factor (tdcF), a putative lipoprotein (atmB), a hypothetical protein of unknown function (M5005_Spy1104), and DNA polymerase III (polC) involved in DNA replication] have been previously detected by in vivo-induced antigen technology (IVIAT) using convalescent human and mouse28 sera.
The virulence-associated sic and emm transcripts, encoding the anti-phagocytic extracellular streptococcal inhibitor of complement (SIC) and hypervariable surface antigen designated M protein, ranked within the top seven most abundant transcripts in WT-inoculated animals and within the top 11 in mutant-inoculated mice (Supplementary Table 3, see http://ajp.amjpathol.org). Six cotranscribed ORFs (M5005_Spy0971-6), encoding hypothetical and general stress proteins related to Gls24 of Enterococcus faecalis, were among the 20 most highly expressed GAS transcripts detected with both inoculating strains, an important finding given that Gls24 may be important for virulence in the murine invasive model of S. pneumoniae infection.29 Other high-level GAS transcripts detected in WT-infected tissues include the chromosomal spd locus (ranked 14th) encoding a secreted DNase17; sagA (ranked 28th) encoding a potent host cytolysin called streptolysin S (SLS); dppA (ranked 47th) encoding dipeptide binding protein; gap/plr (ranked 71st) and eno (ranked 117th) encoding glycolytic enzymes glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and α-enolase, respectively; scpA (ranked 84th) encoding C5a peptidase; spyCEP (ranked 97th) encoding an IL-8-cleaving surface proteinase30; lmb (ranked 192nd) encoding a laminin-binding surface adhesin,31 and spyA (ranked 211th) encoding an ADP-ribosyl transferring exotoxin that disrupts host cell cytoskeletal structures.32 Several GAS regulatory genes also had highly expressed transcripts, including the repressor of class I heat shock genes hrcA33 (ranked 15th), transcriptional regulator RofA (ranked 30th) involved in modulating adhesin expression,8 ferric iron transport regulator PerR34 (ranked 130th), and the TCS designated Ihk/Irr (ranked 162nd/288th), which is essential to GAS survival responses during PMN interactions.35 Among the top 200 detected GAS transcripts, 20 encode for products predicted (by their amino acid sequences) as cell surface-associated or secreted, whereas transcripts encoding experimentally confirmed extracellular gene products such as SIC, streptodornase (ranked 14th), Mn2+-binding surface lipoprotein designated MtsA (ranked 88th), and an immunogenic secreted protein designated Isp2 (ranked 188th) also were observed.
GAS Protective Response Augmented in the Soft Tissue Environment
As described above, we detected elevated transcript levels in WT-infected mouse tissues for genes involved in oxidative stress protection and stress adaptation. These transcripts also included genes encoding the reactive oxygen-reducing enzymes superoxide dismutase SodA (ranked 114th),36 peroxiredoxin reductase AhpC and AhpF (ranked 115th and 147th),37 and NADH oxidase NOX (ranked 129th),38 molecular scavengers such as the antioxidant Dpr protein (ranked 76th),39 clpC (ranked 158th) encoding heat shock protease ClpC involved in tolerance to environmental stress,40 recA (ranked 169th) encoding the RecA regulator of bacterial SOS responses, and sortase SrtA (ranked 131st) involved in the maturation of extracellular GAS proteins. Molecular chaperones as well as DNA and protein repair functions (chaperone proteases) also were elevated in soft tissues (Supplementary Table 3, see http://ajp.amjpathol.org). Importantly, we also detected abundant transcripts encoding products involved in GAS cell wall formation and modification, including the murein transpeptidase-transglycosylases PBP 1B and PBP 2A (ranked 29th and 149th, respectively) and UDP-N-acetylmuramate-alanine ligase (murC; ranked 151st) among others suggesting an important role in cell wall modifications or repair during tissue infection.
Co-Expressed Transcripts
Numerous in vitro studies have demonstrated that emm, sic, and mga are coregulated by the multigene transcriptional activator called Mga. High correlation (r > 0.9) between co-expressed transcripts suggested that in addition to emm, sic, and mga, chromosomally adjacent genes M5005_Spy1721-8 also are likely Mga regulon members (Supplementary Table 4, see http://ajp.amjpathol.org). These adjacent coding regions contain an immunogenic secreted protein designated Isp; the Ihk/Irr TCS; and members of an ATP-binding cassette transport system of unknown function. High correlation (r = 0.91) between transcripts also was observed between eno, encoding the surface-localized glycolytic enzyme α-enolase, and emm encoding the anti-phagocytic M1 protein (data not shown). The latter represents an interesting and new observation because these genes are distant on the GAS chromosome, yet both encode proteins that interact directly with host plasminogen.41
Analysis of Strain Effect in the Soft Tissue Environment
GAS transcript correlations with transcription of covR/S are shown in Supplementary Table 5 (see http://ajp.amjpathol.org). In total, 76 GAS transcripts were differentially expressed across WT versus covR mutant in the lesions (Q < 0.05; P < 0.002; Figure 3; Supplementary Table 3; Supplementary Figures 3 and 4, see http://ajp.amjpathol.org). CovR acts as a transcriptional repressor14,20,42–44, and therefore it was not unexpected that the vast majority (n = 50) of differentially expressed transcripts were more abundant in tissues infected by the ΔcovR strain as compared with the WT-type strain. Genes or transcripts up-regulated as compared with WT conditions (1.4-fold to 2.8 × 103-fold higher expression) encoded the secreted DNase streptodornase (spd); plasminogen activator streptokinase (ska); cysteine protease (speB), and the entire streptolysin S (SLS) biosynthesis locus (sagA-I). Up-regulated transcripts in the ΔcovR mutant are generally associated with unknown functional annotations (n = 14), but only six were common with up-regulated transcripts after ex vivo blood culture.20 However, carbohydrate metabolism (n = 9); virulence (n = 8), and membrane transport (n = 7) functions also were statistically more significantly up-regulated than in the WT. Noteworthy up-regulated transcripts include genes coding for the 67-kd myosin-crossreactive streptococcal antigen (M5005_Spy0385); hyaluronan (capsule) synthesis enzyme HasA, and a cell wall-modifying enzyme peptidoglycan amidohydrolase (M5005_Spy0500). Conversely, one third of differentially expressed transcripts (n = 26) were significantly less abundant in ΔcovR versus the WT-inoculated tissues (1.5- to 12.6-fold reduced expression; Supplementary Table 3, see http://ajp.amjpathol.org). To further investigate transcriptome differences between WT and covR-minus, we analyzed functional categories that were overrepresented by gene-specific analysis based on 10,000 permutations of array assignments, a process previously described.26 Sixty-five GAS transcripts achieved the minimum cutoff value (false discovery rate ≤0.003). Numerous GAS transcripts encoding virulence-associated extracellular products exhibited overexpression in ΔcovR-inoculated tissues versus WT (Supplementary Figure 5; Supplementary Table 6, see http://ajp.amjpathol.org), a finding seen previously in in vitro experimental conditions.14,20 In contrast, most transcripts involved in stress adaptation were underexpressed in ΔcovR- versus WT-inoculated tissues (Supplementary Figure 6; Supplementary Table 6, see http://ajp.amjpathol.org), suggesting that although CovR acts primarily as a transcriptional repressor for tissue damaging virulence factors, lack of this repression may mean less stress adaptation is required within the localized environment. Virtually no transcripts were detected for the activator of CovR expression (designated RocA; Supplementary Table 3, see http://ajp.amjpathol.org) suggesting that CovR is modulated in vivo. Although we detected mid-range levels of transcripts encoding CovR (ranked 269th) in WT-infected tissues, CovR phosphorylation status determines its DNA-binding (repressing) activity.45
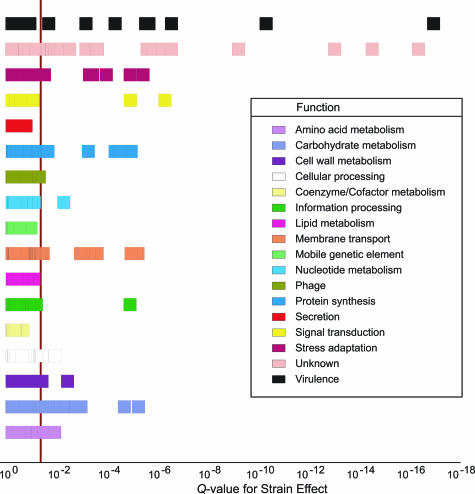
Figure 3.
Analysis of variance revealed a significant strain effect in the in vivo microarray expression data. The depicted box plot shows the confidence of calls for strain effect in Q values (P values adjusted for multiple testing using false discovery rate) on the horizontal axis versus GAS functional category. A vertical line delineates the statistical significance threshold (Q < 0.05) for differentially expressed GAS transcripts in WT versus ΔcovR mutant strain. The 76 identified differential transcripts in the in vivo transcriptome data are primarily associated with carbohydrate metabolism, membrane transport, signal transduction, stress adaptation, unknown functions, and virulence.
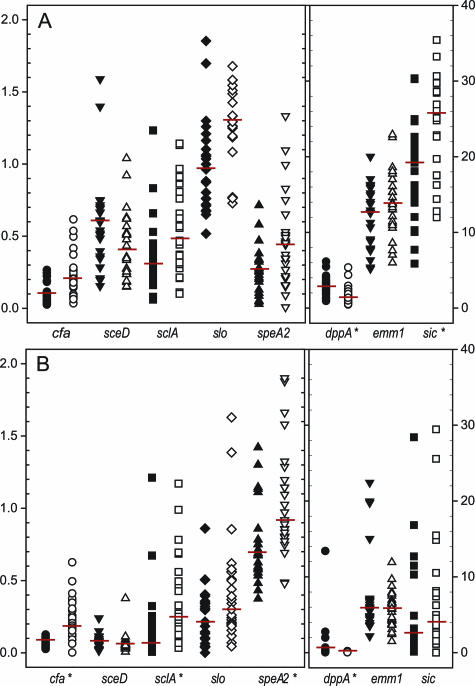
Figure 4.
Verification of representative oligonucleotide microarray transcriptome data. Depicted are GAS expression estimates for eight selected GAS transcripts, as obtained by RMLChip microarray experiments (A) and real-time RT-PCR (TaqMan) assays (B). Results depicted summarize the analysis of 49 mice infected with either WT strain MGAS5005 (n = 23; closed symbols) or the isogenic covR mutant (n = 26; open symbols) and are expressed as median expression relative to the reference transcript proS (M5005_spy1673). Asterisks depict transcripts detected as differentially expressed at Q ≤ 0.001 in arrays (A) and in TaqMan assays at P ≤ 0.001 according to the Mann-Whitney rank sum test (B).
Validation of Oligonucleotide-Array Gene Expression (Transcript) Results
To validate the microarray data, we performed quantitative reverse transcription RT-PCR (TaqMan) assays with eight selected GAS transcripts (Figure 4). The transcript detection rank orders were nearly identical with only dppA and speA reciprocated across the two methods (in rank order of abundance: sic, emm, dppA, slo, sceD, sclA, speA, cfa in microarray dataset (Figure 4A) versus emm, sic, speA, slo, sceD, sclA, dppA, cfa by TaqMan analysis (Figure 4B). TaqMan assays detected much higher levels of speA transcripts encoding the pyrogenic toxin superantigen PTSAg SpeA2, possibly resulting from use of the single endogenous transcript (proS) in TaqMan assays as opposed to use of all transcripts for normalizing microarrays. Strong positive correlation (r = 0.87) was observed overall for expression measurements obtained by the two methods, and for all transcripts, concordance was observed between methods for the direction (increase versus decrease) of expression differences across inoculating GAS strains.
Immunohistochemical Analysis of Mouse Tissue
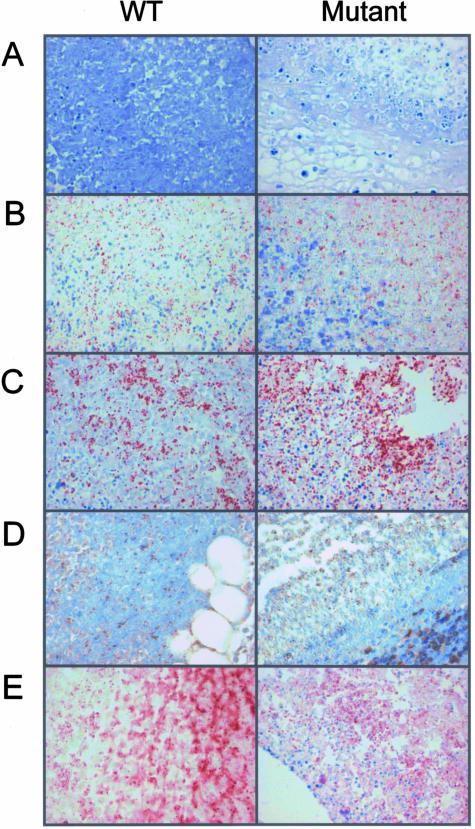
In situ IHC with specific antibodies directed against 16 GAS proteins was used to detect proteins made from identified transcripts. Absence of specific staining in negative control tissues confirmed lack of antibody cross-reactivity (Figure 5). Seven of the 16 GAS-specific antibodies were reactive in situ with several GAS antigens demonstrating high levels of immunoreactivity or abundance (Figure 5; Supplementary Table 7, see http://ajp.amjpathol.org). Our detection of immunoreactivity of PrsA protein (M5005_SPy1133; ranked 121st transcript) supports previous observations of in vivo expression of a PrtM homolog involved in protease maturation,27 a significant note, given that PrtM-immunized mice are protected against S. pneumoniae infection.46 Despite limited detection of malE RNA transcripts (M5005_Spy1058; ranked 561st); a surprisingly high degree of immunoreactivity was observed for the solute-binding surface lipoprotein MalE, known to be involved in maltodextrin/maltosaccharide uptake (see below). Immunoreactivity also was noted in the IHCs for the extracellular virulence-associated proteins PTSAg SpeA2 (M5005_Spy0996) and SIC (Supplementary Table 7, see http://ajp.amjpathol.org), confirming at the protein level the RNA expression of these two important virulence genes.
Figure 5.
Immunohistochemical confirmation of GAS protein expression during soft tissue infection. Representative IHC results for tissue obtained from mice infected with GAS WT MGAS5005 (left) and ΔcovR mutant strain JRS950 (right) analyzed with anti-M3.1/1-24 antibodies (negative control) (A); anti-SPE A2; M5005_spy0996 (B); anti-MalE; M5005_spy 1058 (C); anti-Obp; M5005_spy1308 (D); and anti-SIC; M5005_spy1718 rabbit polyclonal antisera (E) at 2 days after inoculation.
Carbohydrate and Energy Metabolism
Contiguous coding regions (M5005_spy1062-7) involved in the transport and catabolism of complex carbohydrates known as maltodextrins were among the highest-expressed GAS transcripts detected in our in vivo data (Supplementary Table 3, see http://ajp.amjpathol.org). The corresponding genes encode a maltose/maltodextrin utilization protein (MalA); a maltodextrin ATP-binding cassette transport system (MalC and MalD); two enzymes involved in maltodextrin rearrangement (cyclomaltodextrin glucanotransferase AmyA and cyclomaltodextrinase AmyB), and a maltose/maltodextrin solute-binding protein (MalX), respectively. Synchronous with these results, low transcript levels were detected for two nearby genes encoding putative transcriptional repressors, the maltose operon regulator MalR (M5005_spy1057) and an inferred LacI-family regulator (designated herein as MalT; M5005_spy1061), suggesting a cause and effect relationship. Functionally related transcripts (M5005_spy1055-60) also were detected adjacent to chromosome region (M5005_spy1062-7), including genes encoding enzymes maltodextrin phosphorylase (MalP/GlgP) and amylomaltase (MalQ/MalM), and a maltose/maltosaccharide/maltodextrin-binding ATP-binding cassette transport system (MalEFG). Our analysis of coordinated expression of transcripts discovered another functionally related locus nearby (M5005_spy1680-2), that encoded the Mga-activated polysaccharidase designated pullulanase47; a dextran glucosidase, and a maltose/maltodextrin solute-binding protein MsmK, suggesting that maltodextrin-related transcript co-expression is likely important in GAS soft tissue infection.
Recent published findings suggested that transcripts involved in maltodextrin metabolism (M5005_spy1061-7) may be positively regulated by the TCS designated SptR/S loci (M5005_Spy0680/1).48 However, we found no correlation of malT-malX expression with either sptR or sptS transcripts (r < 0.5), whereas positive correlations were revealed with transcripts encoding the TCS designated LytR/S, whose homologs affect cell wall metabolism in Staphylococcus aureus49 (r > 0.7; Supplementary Table 4, see http://ajp.amjpathol.org). Strong correlation with lytRS (r > 0.85) was similarly observed for the chromosomal region flanking lytRS (M5005_Spy1303-10), which coincidentally encodes yet another putative complex sugar transport system. Together, these findings provide supporting evidence for involvement of the CovR-regulated LytR/S TCS in mediating GAS complex carbohydrate metabolism during soft tissue infection.
GAS ferments carbohydrates via the Embden-Meyerhof-Parnas (EMP) pathway to form pyruvate for energy metabolism. As with many bacteria, glucose is the preferred catabolic substrate and high glucose concentrations repress expression of alternative catabolic pathways via the catabolite repressor CcpA.50 We discovered that ccpA was expressed at low range (ranked 342nd) in the mouse tissues suggesting that glucose supplies were limiting at that time. In agreement with this finding, high levels of lactate oxidase (lctO; ranked 63rd) transcripts, of which the protein product is known to convert lactate to acetate thereby generating ATP via acetyl phosphate,51 were detected simultaneously (Supplementary Table 3, see http://ajp.amjpathol.org). Together, these results suggest a glucose-poor environment with adaptation by GAS for using lactate as a carbon energy source during soft tissue infection. Elevated transcripts also were detected for pfl and pta encoding pyruvate formate-lyase and phosphotransacetylase (ranked 81st and 102nd, respectively), which are involved in pyruvate oxidation to coenzyme A and formate51 suggesting that metabolic pathways downstream of pyruvate were activated. However, GAS also is capable of fermenting nitrogen-containing compounds for energy metabolism generating 1 mol of ATP per mol of arginine or histidine metabolized and high levels of transcripts were detected for the histidine catabolic pathway and histidine deaminase (hutH; ranked 90th), which generates urocanate and ammonia. Coordinate with histidine catabolic pathway expression, a polycistronic transcript (M5005_Spy1270-5) encoding arginine deiminase (ADI; ranked 177th) along with transcripts encoding other related functions within the arginine catabolic pathway (Supplementary Table 3, see http://ajp.amjpathol.org) were detected.
GAS Transcripts Correlated with Gross Cutaneous Injury
Analysis of WT-inoculated tissue array data revealed 18 GAS transcripts whose expression correlated directly with lesion volume (Q < 0.05; Supplementary Table 8, see http://ajp.amjpathol.org). Of these, six encode genes of unknown function, whereas seven are localized in the bacterial cytoplasm and involved in information processing, including transcriptional repressor ArgR1 (M5005_Spy1229; ranked 313th), the multigene transcriptional activator Mga (ranked 418th), and the DNA mismatch repair protein MutL (ranked 592nd). No GAS transcripts exhibited significant correlation with erythema volume (Q > 0.05; data not shown).
Discussion
Bacterial pathogens use coordinated gene expression to respond to environmental change and to elude host defenses, thereby facilitating persistence, proliferation, invasion, and dissemination. As GAS infects humans using a variety of different routes (skin, mucosal surfaces, and blood), it likely expresses distinct sets of genes that enable it to adapt to each diverse and changing host microenvironment.11 Current understanding of bacterial gene regulation is based primarily on studies conducted under controlled laboratory conditions, in which typically only one variable is altered at a time. Although this approach is powerful in pathogenesis studies, it does not adequately mimic the extensive variety of signals experienced simultaneously when bacteria are growing in the host.12 Global expression microarray (transcriptome) analysis of bacteria grown in vivo provides a unique opportunity to identify how pathogens adjust gene expression genome-wide in response to complex host environments.10
Abundant GAS Transcripts in Soft Tissue Infection
WT GAS is a superbly adapted pathogen, able to protect itself against damaging host effectors during murine soft tissue infection by expressing numerous anti-oxidant, molecular chaperone, cytotoxin, and cell wall/lipoteichoic acid modifying proteins. For example, we observed emm transcript-encoding M protein, the hypervariable major surface antigen used for serological typing of GAS strains, predominate in lesions. This well-studied bacterial virulence determinant acts as an adhesin; promotes inflammation; impedes phagocytosis by binding complement control factors, fibrinogen, kininogen, and also plasminogen;3,4 and is abundantly transcribed during GAS culture ex vivo in blood.20 Host fibrinogen levels rise sharply during tissue inflammation and injury, and M protein/fibrinogen complexes activate PMNs through β2-crosslinking triggering release of heparin-binding protein (HBP), a proinflammatory mediator which in turn promotes excessive vascular leakage.52 Another important molecule whose RNA was present in high amounts in our tissue model was GAS cysteine protease SpeB, which has been shown to cleave many host proteins including cytokine precursors, cell receptors, fibrin, vitronectin, matrix proteoglycans, cationic antimicrobial peptides, and immunoglobulins, thereby contributing to endothelial and epidermal damage, tissue destruction, and bacterial dissemination.53–55 SpeB enzymatic activity also releases fragments of fibrinogen-complexed M1 protein [fibrinogen-M1], thereby contributing to vascular leakage and host tissue damage.52 However, at the same time, we detected virtually no grab transcripts, which encode a host protease inhibitor α2-macroglobulin-binding bacterial surface protein designated GRAB. α2-Macroglobulin-GRAB complexes concentrate SpeB activity at the bacterial surface and against smaller substrates, thereby protecting GAS against killing by the antibacterial peptide LL-37.56 Because SpeB also degrades many GAS extracellular proteins in the absence of GRAB, not surprisingly we detected modulated, mid-ranging speB transcripts (ranked 304th in WT) at 7- to 25-fold lower levels than the most abundantly detected GAS transcripts in tissues. Therefore, in the soft tissue model we propose that low levels of grab, moderate levels of speB, and high emm1.0 transcript production occurs, providing sufficient SpeB proteolytic activity and hence M1-fibrinogen complex formation all of which leads to substantial tissue damage. Increasing transcription of speB throughout time during soft tissue infection has recently been described.11 These observations, coupled with postinfectious induction of tissue edema, which leads to depletion of vascular volume, may account for the rapid onset of life-threatening hypotension and shock in severe GAS infections.7 Interruption of this pathophysiological pathway could be critical for therapeutic intervention of aggressive, invasive GAS infections.
During soft tissue infection, WT GAS expresses high levels of transcripts for products that inhibit host innate defenses and protect GAS against neutrophil-derived reactive oxygen species and antimicrobial peptides. For example, sic (ranked first) and spd (ranked 14th) were among the most abundantly expressed GAS transcripts measured. We confirmed the high expression levels of SIC by IHC analysis of infected tissues (Figure 4). SIC is known to inhibit complement-mediated bacterial lysis via C5b67 binding, innate defenses such as lysozyme and PMNs, and can eliminate chemotactic recruitment of leukocytes, T cells, and mast cells by inactivating antimicrobial peptides such as LL-37 and β-defensins.57,58 Similar findings were observed in vivo in nonhuman primates and after GAS culture ex vivo in whole blood where Mga regulon transcripts were the highest expressed of all detected regulons.10,20 On the other hand, streptodornase (spd) is a secreted DNase that may interfere with PMN function, by degradation of innate immune structures known as neutrophil extracellular traps.17,59 In addition, high levels of expression of sagA (ranked 28th), which encodes the potent cytolysin SLS, was notable because SLS is thought to accelerate necrotic fascial injury through cytotoxic or apoptotic effects on many cell types including, but not limited to, neutrophils.60 The untranslated mRNA of the pleiotropic effect locus (pel), which incidentally contains sagA, also acts as an anti-sense RNA transcriptional regulator, augmenting GAS virulence factor expression [SIC, M-protein, extracellular NAD-glycohydrolase (NADase encoded by nga/spn), plasminogen activator streptokinase encoded by ska, and SpeB] when pel RNA expression is induced.61,62 We detected abundant sagA transcripts; thus pel-induced virulence gene expression may be predicted, as was observed in this study for all but nga (ranked 780th in WT extracts). Therefore, these results, when coupled with the elevated expression of the neutrophil chemoattractant (interleukin-8)-cleaving SpyCEP protease (ranked 97th),30 support the idea that coordinated, increased expression of sic, spd, sagA, speB, and spyCEP likely accounts for lack of inflammatory cells present at GAS sites in deep tissues.63 Expression of the aforementioned GAS products, along with M1 protein, streptokinase, and the host cytolysins streptolysin O (SLO; ranked 268th) and NADase likely contributes to much of the cutaneous and subcutaneous pathologies observed in our animal model tissue infections (Supplementary Table 3; reviewed in Supplementary Table 9,64–84 see http://ajp.amjpathol.org).
Serotype M1 strains are often associated with postinfectious sequelae, possibly resulting from immunological cross-reactivity of GAS antigens with host tissues.3,4 Virtaneva and colleagues,10 observed during nonhuman primate experimental pharyngitis, high in vivo levels of GAS transcripts, such as emm, cardiolipin synthetase, and myosin-cross reactive streptococcal antigen, results we have now confirmed in our in vivo study; M protein (emm, ranked seventh), hydroxylated phospholipid cardiolipin synthetase (CL synthase, M5005_Spy0926; ranked 111th), and 67-kd myosin-crossreactive streptococcal antigen85 (M5005_Spy0385, ranked 125th). Increased production of these transcripts in both the oro-pharynx model and now the soft-tissue model suggests an increased risk of nonsuppurative sequelae, a prospect that warrants further study.
Three proteins encoded by up-regulated WT transcripts; gap/plr, eno, and ska, ranked 71st, 117th, and 453rd, respectively, are known to interact or bind with host plasminogen.3 Fibrinogen binding to M-protein (emm) leads to host plasminogen binding. Bound plasminogen interacts with GAS streptokinase to generate a bacterial-bound, serine protease-designated plasmin.86 GAS surface-localized plasmin has been implicated in GAS dissemination.3,41,74–86 Statistically higher expression of ska transcripts in the ΔcovR mutant likely contributed to lesion size differences between mutant and WT. Our observation of abundant plr transcripts is also noteworthy given that the encoded protein also has been identified as an acute poststreptococcal glomerulonephritis-associated antigen (designated NAPlr).87,88
GAS is sensitive to penicillin in vitro, yet high-dose penicillin demonstrates reduced bactericidal activity in vivo, attributable in part to inadequate drug perfusion in poorly vascularized or necrotic fascia.89 In addition, large GAS inocula refractory to penicillin have been suggested as metabolically inactive or demonstrating decreased presence of penicillin-binding proteins (PBPs).7,90,91 At the time point of our collected tissue samples, we detected abundant levels of PBP1A- and PBP2A-encoding transcripts and found no evidence suggesting inactive or reduced metabolic activity in vivo. Previously published studies have demonstrated that our custom high-density array approach is capable of sensitive detection of low copy transcripts.10 Therefore, the mechanisms responsible for reduced efficacy of penicillin treatment in vivo may be more complex than previously thought, perhaps warranting focused study. Although we observed abundant GAS transcripts encoding proinflammatory M protein, SLS cytotoxin, and plasmin receptor Plr (ranked seventh, 28th, and 71st, respectively), decreased protein expression encoded by these genes may explain why co-administration of clindamycin (a protein synthesis inhibitor) seems to enhance bacterial clearance in progressive cases, relative to penicillin treatment alone.4,89 Future investigations are warranted to elucidate this clindamycin phenomenon.
Transcripts encoding GAS PTSAg exotoxins were virtually undetectable at the time of our tissue sampling. PTSAgs are potent T-cell mitogens that stimulate a massive release of inflammatory cytokines and that correlate with severity of systemic clinical manifestations in GAS infections.92 However, data observed in nonhuman primates suggest that PTSAgs SpeA2, SpeJ, and SmeZ demonstrate differential expression before, during, and after pharyngeal colonization.10 Our findings therefore support clinical and experimental data revealing reciprocal, or temporal expression of SpeA and SpeB in vivo.93 Recent observations suggest that direct contact with human PBMCs, which occurs early in infection, induces SpeA expression94 whereas cysteine protease SpeB expression increases temporally during later stages of soft tissue infections.11 Our in situ immunohistochemical data showed immunoreactivity for SpeA at 48 hours after inoculation. Numerous GAS proteins are known to contribute to the induction of neutrophil apoptosis,69 leading to an absence of proinflammatory cells at GAS sites in deep tissues.63 Together, these findings do not preclude the expression of PTSAg exotoxins at early stages of soft tissue infection, but rather suggest that the time we harvested the tissue (2.5 days after infection) was more likely a mid point during the GAS soft tissue infection time course.
Adaptive Metabolism
Central and adaptive metabolism profoundly affects pathogen persistence in vivo and likely accounts for why GAS regulatory- and metabolism-associated transcripts (primarily cytoplasmic) predominate in soft tissues. We found that in vivo GAS abundantly transcribes genes in two chromosomal regions (M5005_spy1062-7 and 1680-2), which are involved in metabolizing complex carbohydrates such as maltodextrins, which may include but not be limited to glycogen, maltotetraose, maltotriose, and maltose. These expression results are consistent with a report in which 81% of convalescent-phase sera from patients with invasive GAS infections had antibodies reactive with M5005_SPy1680, also known as pullulanase (PulA).95 PulA is a polysaccharidase that is positively regulated by the multigene transcriptional activator called Mga and which uses host glycoproteins as its enzymatic substrate.47 GAS metabolism of complex host-derived carbohydrates may be particularly important during soft tissue infections because of abundant host glycoproteins and host cell contents released during cell lysis. Degradation of host proteins in necrotic fascia may contribute to fermentation of arginine and histidine during soft tissue infection. In turn, fermentation of amino acids produces ammonia, which could beneficially neutralize surrounding pH thereby protecting GAS from acid-induced damage.96 GAS arginine deaminase activity has been linked to inhibition of human peripheral blood mononuclear cell proliferation.97 The arginine deiminase system of Streptococcus suis is induced by arginine, elevated temperature, and reduced oxygen tension, and is subject to carbon catabolite repression.98 Therefore, low glucose and hypoxic microenvironments, after vascular injury, may serve as signals that trigger the GAS adaptive response, which in turn enables bacterial persistence and proliferation in soft tissues.
GAS Transcript Expression Is Not Correlated with Cutaneous Injury
Correlation analysis of GAS transcripts with cutaneous lesion volume produced only a few candidates (n = 18), most of which appear to encode proteins predicted to be cytoplasmic or membrane-bound in function. Although these discoveries are attractive, lack of biochemical functional annotation for these genes limits our ability to hypothesize their role in lesion development. These findings are consistent with the clinical observation that substantial necrosis occurs subcutaneously in some human infections without overt injury of the overlying skin.13 Further studies are needed to identify bacterial genes associated with soft tissue disease severity and the functions they may impart in this important aspect of disease.
GAS Strain Comparison in Vivo
The CovR response regulator has been shown to play a central role in GAS regulatory networks by directly or indirectly influencing expression of 15% of all GAS chromosomal genes during in vitro growth.14 CovR negatively regulates biosynthesis of the anti-phagocytic hyaluronic acid capsule42 and the expression of numerous cell-surface-associated and secreted proteins known to promote survival or impart virulence in humans.14,42–44 Hyperencapsulated GAS variants with spontaneous covR/S mutations have been isolated after WT bacterial passage in human blood, passage in mice, or after natural-acquired human infections.99–101 Importantly, high GAS bacterial colony-forming unit numbers has been found to negatively correlate with covR expression in vivo during acute phase GAS pharyngitis in primates.10 Despite absence of CovR function in the mutant strain, we found fewer differentially expressed transcripts (n = 76) between mutant and WT during soft tissue infection than we had extrapolated from our previous in vitro studies.14,20 This led us to hypothesize that transcription of covR in WT GAS occurs at lower levels in vivo during soft tissue infection at the time point studied than under in vitro growth conditions. Indeed, covR and covS transcripts ranked 269th and 755th, respectively, in GAS-infected lesion extracts, whereas covR/covS transcripts were more abundant (transcripts ranked 30th to 280th) during a 90-minute time course of culturing in human blood ex vivo.20 This finding supports in vivo down-regulation of covR expression,10,99–101 leading to increased virulence factor production14 and resultant tissue pathology and suggests that soft tissue effectors not present in whole blood are likely responsible for this down-regulation. Based on in vitro studies, high [Mg2+] concentration has been proposed as an environmental stimulus responsible for up-regulation of CovR/S expression and therefore repression of the expression of CovR-regulated genes.102 In vivo microenvironment analysis of magnesium concentrations throughout time, during the course of soft tissue infections may provide insight into the effectors regulating CovR/S expression.
In contrast to in vitro findings,14,20 more than half of the total number of transcripts functionally related to stress adaptation were transcribed at lower levels in vivo in the ΔcovR mutant as compared with the WT. Given that we detected more transcripts encoding the transcriptional repressor of class I stress genes (designated HrcA)33 in WT (ranked 15th) than in mutant (ranked 42nd), we would predict the converse; that transcript levels encoding the negatively regulated class I molecular chaperones (DnaJ, DnaK, and GroES) and chaperone proteases (GroEL, ClpP, and ClpL) would be higher in ΔcovR-infected tissues as compared with WT extracts. These results imply other peripheral regulatory mechanisms may be in place, and suggest that less stress adaptation may be required in the localized environment when host cell damaging virulence factors are derepressed. This finding serves to highlight the importance of conducting in vivo analyses to improve understanding of pathogen-host interactions.12
Emerging Model of GAS Gene Regulation in Vivo
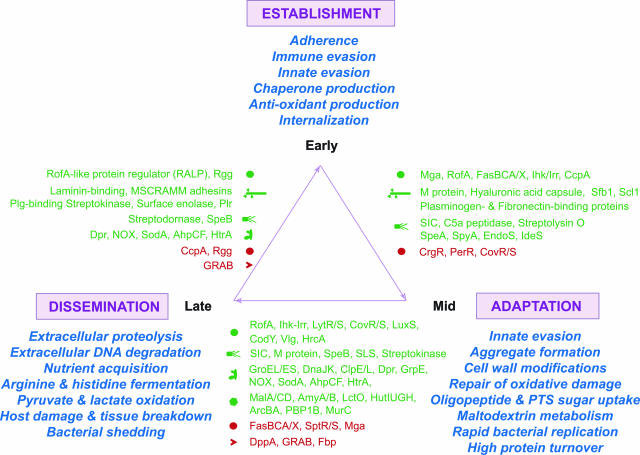
Our study has provided the first in vivo analysis of GAS gene expression during soft tissue infection. By differential gene expression, GAS successfully transitions through three stages of host infection: namely, 1) establishment of infection, 2) adaptation, and 3) dissemination (Figure 6). Information about stage 1 is based on an expanding body of work,8,10,11,34,103 and our results support this information by combined early expression of Mga, Ihk/Irr, and FasBCA/X regulatory systems that promote evasion of immune and innate host defenses and enhance host cell contact through surface adhesin production. Down-regulation of the negative regulatory systems of PerR, CovR/S, and CrgR promote production of reactive oxygen species detoxifying enzymes, hyaluronic acid capsule, and antimicrobial peptide resistance mechanisms, respectively,8,35,103–107 and our soft tissue model demonstrates similar results. Interestingly, we have discovered that transcription of transport and central metabolism genes appears tailored to unique GAS environments where host glycopeptides and complex carbohydrates are available, including maltodextrins, but which must be used under acidic and hypoxic conditions. Alteration of these conditions ultimately enables GAS to persist and proliferate. GAS cell wall modifications can reduce cell permeability and thereby increases antimicrobial peptide resistance.108 The reduced efficacy of penicillin-based antibiotics in treating circulation-poor deep-seated GAS infections also may be related to this GAS adaptation. Throughout time, production and accumulation of the SLS cytolysin, SpeB cysteine protease, proinflammatory PTSAgs, and fibrinogen-M1 protein complexes have the ability to cause cumulative damage to host cells, resulting in disruption of host barriers leading to bacterial dissemination. Evidence suggests that dissemination of GAS from localized tissues, occurs as GAS undergoes additional remodeling, which can include late-stage cysteine protease expression,11 (Figure 6). A carbon catabolite response element (cre) operator site precedes the Mga promoter, and thus expression of Mga (and virulence factors it transcriptionally activates) may be subject to carbon catabolite control.109 Linkage of the Mga regulon to GAS nutritional requirements may help explain why in our study we observed correlated expression between eno encoding glycolytic and surface α-enolase and emm encoding M1 protein. The linkage of GAS metabolism regulation and virulence factor expression to nutritional signals provides an intuitive model that helps explain GAS movement from chronic or acute sites to naïve sites.
Figure 6.
Emerging model of GAS gene regulation in soft tissue infection. Depicted schematically is the emerging model of GAS gene expression in vivo during suppurative infections. By finely modulating gene expression, GAS transitions through three general stages during host infection: namely, 1) establishment (top), 2) adaptation (bottom right), and 3) dissemination (bottom left). Establishment is achieved through bacterial adhesin production and evasion of immune and innate host defenses. Proliferation in large bacterial aggregates provides enhanced opportunity for bacterial resistance and concentration of extracellular products aimed at disrupting various host functions. Adaptive expression of nutrient transport and central metabolism systems enables GAS to acquire and use host peptides and complex carbohydrate substrates, such as maltodextrins. Cumulative damage to host cells, tissue injury, and disruption of host barriers results in enhanced potential for bacterial dissemination to naïve host sites. GAS undergoes additional transcription changes that promote persistence elsewhere, initiating new foci of infection. Green text (up-regulation); red text (down-regulation).
Few studies to date have succeeded in measuring global transcription in the in vivo host environment.10,110–113 This study has overcome difficult technical hurdles to provide a unique dataset on the GAS transcriptome during soft tissue infection. Twenty to thirty percent of many pathogen genome ORFs lack biochemical functional annotation. Our transcriptome data suggest that many of these hypothetical ORFs of unknown function are expressed in the soft tissue model, often with good correlation to other virulence factors and pathology. Our data provides new avenues of study for determining the role of these hypotheticals and whether diagnostic or vaccine candidates exist within this group of genes. Owing to the fact that bacterial sampling required animal sacrifice, our end-point study probed mid and late stages of infection (stages 2 and 3, Figure 6). The proportion of adapting versus disseminating bacteria in the tissue extracts cannot be known and thus, the transcriptomes obtained represent averages for the bacterial population. Future studies will involve optimization and protocol development for sensitive array analysis of microscopic tissue biopsies collected throughout time after low-dose intradermal inoculation. These experiments will provide a GAS transcriptome profile of the temporal natural history of soft tissue infection. In addition, we anticipate future tissue model studies will also focus on the comparison of in vivo GAS transcriptomes of epidemiologically distinct strains. Last, although our infection model mimics conditions present in infected humans, we acknowledge that mice cannot adequately replicate either the highly complex environment that GAS encounters in the infected human host or the human host response. Future bacterial expression studies should also be performed during natural human infections, provided appropriate precautions are taken to minimize technical variation and to adequately account for patient-to-patient variability associated with the nature and logistics of clinical specimen sampling, which often occurs under life-saving and time-constrained circumstances involving patients recently treated with antimicrobial agents. Correlation analysis will help pinpoint those GAS genes or ORFs expressed in common during different stages of disease, which may lead to novel, broad-spectrum therapeutic, vaccine, or diagnostic candidates against this important human pathogen.
Concluding Statement
A full understanding of host/pathogen interactions requires knowledge of gene expression in vivo.10,12,114–116 Our data provide new information about GAS gene expression during soft tissue infection, and highlight the benefit of using sensitive high-density microarray analysis. The in vivo, soft tissue GAS transcriptome differs significantly from that obtained in blood ex vivo,20 in isolated cell populations,35 and during colonization and acute pharyngitis,10 stressing the ability of GAS to alter its transcriptome to permit growth in the different host environments. Our results unambiguously demonstrate that many proven and putative GAS virulence factors are transcribed in vivo and contribute to extensive cellular pathology observed in GAS soft tissue infections. Our study provides the first genome-wide view of the GAS transcriptome, in vivo, during skin infection and suggests numerous avenues for investigation into targeted therapeutics against this important human pathogen. This analysis framework is cross-applicable for examining other pathogens causing invasive infections.
Supplementary Material
Acknowledgments
We thank J. Yang and R. Lempicki of the Science Application International Corporation-Frederick Affymetrix core facility for hybridizations; R. Larson and J. Kupko (Rocky Mountain Laboratories) for providing technical expertise with animal handling and database submission, respectively; T.J. Downey and S. Jing (Partek, Inc.) and E. Spitznagel (University of Washington) for statistical assistance; J.R. Scott (Emory School of Medicine) for providing the ΔcovR mutant derivative of MGAS5005 (strain JRS950); P. Schlievert (University of Minnesota) for anti-SpeA antibody; and G. McClarty and T.G. Schwan for critical review of the manuscript.
Footnotes
Address reprint requests to James M. Musser, M.D., Ph.D., Center for Molecular and Translational Human Infectious Diseases Research, The Methodist Hospital Research Institute, and Department of Pathology, Methodist Hospital, 6565 Fannin St., B490, Houston, TX 77030. E-mail: jmmusser@tmh.tmc.edu.
Supported in part by National Institute of Allergy and Infectious Diseases intramural funding.
Supplementary material for this article can be found on http://ajp.amjpathol.org.
References
- DiNubile MJ, Lipsky BA. Complicated infections of skin and skin structures: when the infection is more than skin deep. J Antimicrob Chemother. 2004;53:37–50. doi: 10.1093/jac/dkh202. [DOI] [PubMed] [Google Scholar]
- Musser JM, Krause RM. Krause RM, editor. New York: Academic Press,; The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome. Emerging Infections. 1998:pp 185–218. [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- O’Brien KL, Beall B, Barrett NL, Cieslak PR, Reingold A, Farley MM, Danila R, Zell ER, Facklam R, Schwartz B, Schuchat A. Epidemiology of invasive group A Streptococcus disease in the United States, 1995–1999. Clin Infect Dis. 2002;35:268–276. doi: 10.1086/341409. [DOI] [PubMed] [Google Scholar]
- Stevens DL. The flesh-eating bacterium: what’s next? J Infect Dis. 1999;179:S366–S374. doi: 10.1086/513851. [DOI] [PubMed] [Google Scholar]
- Stevens D. Streptococcal toxic shock syndrome associated with necrotizing fasciitis. Annu Rev Med. 2000;51:271–288. doi: 10.1146/annurev.med.51.1.271. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Hynes W. Virulence factors of the group A streptococci and genes that regulate their expression. Front Biosci. 2004;9:3399–3433. doi: 10.2741/1491. [DOI] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Gardner DJ, Bailey JR, Parnell MJ, Musser JM. Longitudinal analysis of group Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J Bacteriol. 2006;188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handfield M, Progulske-Fox A, Hillman J. In vivo induced genes in human diseases. Periodontol 2000. 2005;38:123–134. doi: 10.1111/j.1600-0757.2005.00110.x. [DOI] [PubMed] [Google Scholar]
- Headley AJ. Necrotizing soft tissue infections: a primary care review. Am Fam Physician. 2003;68:323–328. [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Migliaccio CAL, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe NP, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou SJ, Pan X, Vuopio-Varkila J, Salmelinna S, McGeer A, Low DE, Schwartz B, Schuchat A, Naidich S, De Lorenzo D, Fu YX, Musser JM. Rapid selection of complement-inhibiting protein variants in group A Streptococcus epidemic waves. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, DeLeo FR, Musser JM. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci USA. 2005;102:1679–1684. doi: 10.1073/pnas.0406641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Montgomery CA, Rurangirwa J, Geske RS, Barrish JP, Adams GJ, Musser JM. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Liyanarachchi S, Wright FA, Davuluri R, Lockman JC, de la Chapelle A, Pellegata NS. Gene expression profiling of isogenic cells with different TP53 gene dosage reveals numerous genes that are affected by TP53 dosage and identifies CSPG2 as a direct target of p53. Proc Natl Acad Sci USA. 2002;99:15632–15637. doi: 10.1073/pnas.242597299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekutieli D, Benjamini Y. Resampling based FDR controlling multiple hypotheses testing. J Stat Plan Infer. 1999;82:171–196. [Google Scholar]
- Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- Lei B, Liu M, Chesney GL, Musser JM. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis. 2004;189:79–89. doi: 10.1086/380491. [DOI] [PubMed] [Google Scholar]
- Salim KY, Cvitkovitch DG, Chang P, Bast DJ, Handfield M, Hillman JD, de Azavedo JCS. Identification of group A Streptococcus antigenic determinants upregulated in vivo. Infect Immun. 2005;73:6026–6038. doi: 10.1128/IAI.73.9.6026-6038.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng F, Nannini EC, Murray BE. Importance of gls24 in virulence and stress response of Enterococcus faecalis and use of the Gls24 protein as a possible immunotherapy target. J Infect Dis. 2005;191:472–480. doi: 10.1086/427191. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Taylor GW, Ferguson M, Murray S, Rendell N, Wrigley A, Bai Z, Boyle J, Finney SJ, Jones A, Russell HH, Turner C, Cohen J, Faulkner L, Sriskandan S. Specific C-terminal cleavage and inactivation of interleukin-8 by invasive disease isolates of Streptococcus pyogenes. J Infect Dis. 2005;192:783–790. doi: 10.1086/432485. [DOI] [PubMed] [Google Scholar]
- Terao Y, Kawabata S, Kunitomo E, Nakagawa I, Hamada S. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect Immun. 2002;70:993–997. doi: 10.1128/iai.70.2.993-997.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coye LH, Collins CM. Identification of SpyA, a novel ADP-ribosyltransferase of Streptococcus pyogenes. Mol Microbiol. 2004;54:89–98. doi: 10.1111/j.1365-2958.2004.04262.x. [DOI] [PubMed] [Google Scholar]
- Woodbury R, Haldenwang WG. HrcA is a negative regulator of the dnaK and groESL operons of Streptococcus pyogenes. Biochem Biophys Res Commun. 2003;302:722–727. doi: 10.1016/s0006-291x(03)00254-7. [DOI] [PubMed] [Google Scholar]
- Ricci S, Janulczyk R, Bjorck L. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect Immun. 2002;70:4968–4976. doi: 10.1128/IAI.70.9.4968-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Vuong C, Kobayashi SD, Porcella SF, Otto M, Musser JM, DeLeo FR. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J Immunol. 2004;173:1194–1201. doi: 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- Janulczyk R, Ricci S, Bjorck L. MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect Immun. 2003;71:2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King KY, Horenstein JA, Caparon MG. Aerotolerance and peroxide resistance in peroxidase and perR mutants of Streptococcus pyogenes. J Bacteriol. 2000;182:5290–5299. doi: 10.1128/jb.182.19.5290-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CM, Mallett TC, Claiborne A, Caparon MG. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J Bacteriol. 2000;182:448–455. doi: 10.1128/jb.182.2.448-455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Fukui K, Koujin N, Ohya H, Kimura K, Kamio Y. Regulation of the intracellular free iron pool by Dpr provides oxygen tolerance to Streptococcus mutans. J Bacteriol. 2004;186:5997–6002. doi: 10.1128/JB.186.18.5997-6002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim YM, Kerr AR, Silva NA, Mitchell TJ. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect Immun. 2005;73:730–740. doi: 10.1128/IAI.73.2.730-740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, McArthur J, McKay F, Ranson M. Is plasminogen deployed as a Streptococcus pyogenes virulence factor? Trends Microbiol. 2005;13:308–313. doi: 10.1016/j.tim.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Heath A, DiRita VJ, Barg NL, Engleberg NC. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J Bacteriol. 2004;186:3928–3937. doi: 10.1128/JB.186.12.3928-3937.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overweg K, Kerr A, Sluijter M, Jackson MH, Mitchell TJ, de Jong APJM, de Groot R, Hermans PWM. The putative proteinase maturation protein A of Streptococcus pneumoniae is a conserved surface protein with potential to elicit protective immune responses. Infect Immun. 2000;68:4180–4188. doi: 10.1128/iai.68.7.4180-4188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytonen J, Haataja S, Finne J. Streptococcus pyogenes glycoprotein-binding strepadhesin activity is mediated by a surface-associated carbohydrate-degrading enzyme, pullulanase. Infect Immun. 2003;71:784–793. doi: 10.1128/IAI.71.2.784-793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, III, Sumby P, Sitkiewicz I, Granville C, DeLeo FR, Musser JM. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611–618. doi: 10.1128/jb.178.3.611-618.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titgemeyer F, Hillen W. Global control of sugar metabolism: a gram-positive solution. Antonie Van Leeuwenhoek. 2002;82:59–71. [PubMed] [Google Scholar]
- Seki M, Iida K, Saito M, Nakayama H, Yoshida S. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J Bacteriol. 2004;186:2046–2051. doi: 10.1128/JB.186.7.2046-2051.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herwald H, Cramer H, Morgelin M, Russell W, Sollenberg U, Norrby-Teglund A, Flodgaard H, Lindbom L, Bjorck L. M protein, a classical bacterial virulence determinant, forms complexes with fibrinogen that induce vascular leakage. Cell. 2004;116:367–379. doi: 10.1016/s0092-8674(04)00057-1. [DOI] [PubMed] [Google Scholar]
- Kapur V, Majesky M, Li L, Black R, Musser J. Cleavage of interleukin 1{beta} (IL-1{beta}) precursor to produce active IL-1{beta} by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur V, Topouzis S, Majesky MW, Li LL, Hamrick MR, Hamill RJ, Patti JM, Musser JM. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- Svensson M, Scaramuzzino D, Sjobring U, Olsen A, Frank C, Bessen D. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol. 2000;38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- Nyberg P, Rasmussen M, Bjorck L. {alpha}2-Macroglobulin-proteinase complexes protect Streptococcus pyogenes from killing by the antimicrobial peptide LL-37. J Biol Chem. 2004;279:52820–52823. doi: 10.1074/jbc.C400485200. [DOI] [PubMed] [Google Scholar]
- Fernie-King BA, Seilly DJ, Willers C, Wurzner R, Davies A, Lachmann PJ. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunology. 2001;103:390–398. doi: 10.1046/j.1365-2567.2001.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T, Takamatsu D, Koyanagi M, Zhao J, Imanishi K, Uchiyama T. Cytocidal effect of Streptococcus pyogenes on mouse neutrophils in vivo and the critical role of streptolysin S. J Infect Dis. 2005;192:107–116. doi: 10.1086/430617. [DOI] [PubMed] [Google Scholar]
- Li Z, Sledjeski DD, Kreikemeyer B, Podbielski A, Boyle MD. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J Bacteriol. 1999;181:6019–6027. doi: 10.1128/jb.181.19.6019-6027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold M, Siller M, Roppenser B, Vlaminckx B, Penfound T, Klein R, Novak R, Novick R, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol. 2004;53:1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- Cockerill FR, Thompson R, Musser J, Schlievert P, Talbot J, Holley K, Harmsen W, Ilstrup D, Kohner P, Kim M, Frankfort B, Manahan J, Steckelberg J, Roberson F, Wilson W. Molecular, serological, and clinical features of 16 consecutive cases of invasive streptococcal disease. Southeastern Minnesota Streptococcal Working Group. Clin Infect Dis. 1998;26:1448–1458. doi: 10.1086/516376. [DOI] [PubMed] [Google Scholar]
- Bhakdi S, Tranum-Jensen J, Sziegoleit A. Mechanism of membrane damage by streptolysin-O. Infect Immun. 1985;47:52–60. doi: 10.1128/iai.47.1.52-60.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Wang B, Pentland A, Caparon M. Streptolysin O and adherence synergistically modulate proinflammatory responses of keratinocytes to group A streptococci. Mol Microbiol. 1998;27:337–346. doi: 10.1046/j.1365-2958.1998.00681.x. [DOI] [PubMed] [Google Scholar]
- Limbago B, Penumalli V, Weinrick B, Scott JR. Role of streptolysin O in a mouse model of invasive group A streptococcal disease. Infect Immun. 2000;68:6384–6390. doi: 10.1128/iai.68.11.6384-6390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschel SD, Borgia SM, Barg NL, Low DE, De Azavedo JCS. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Im6mun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsburg I. Is streptolysin S of group A streptococci a virulence factor? APMIS. 1999;107:1051–1059. doi: 10.1111/j.1699-0463.1999.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci USA. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ EA, Meals E, English BK. Streptococcal pyrogenic exotoxins A (SpeA) and C (SpeC) stimulate the production of inducible nitric oxide synthase (iNOS) protein in RAW 264.7 macrophages. Shock. 1997;8:450–453. [PubMed] [Google Scholar]
- Cusumano V, Fera MT, Carbone M, Anzani Ciliberti F, Bellantoni A. Synergic activities of streptococcal pyrogenic exotoxin A and lipoteichoic acid in cytokine induction. New Microbiologica. 2000;23:37–45. [PubMed] [Google Scholar]
- Rasmussen M, Müller HP, Björck L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at the bacterial surface by binding alpha2-macroglobulin. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- Lottenberg R, Desjardins L, Wang H, Boyle M. Streptokinase-producing streptococci grown in human plasma acquire unregulated cell-associated plasmin activity. J Infect Dis. 1992;166:436–440. doi: 10.1093/infdis/166.2.436. [DOI] [PubMed] [Google Scholar]
- Sun H, Ringdahl U, Homeister JW, Fay WP, Engleberg NC, Yang AY, Rozek LS, Wang X, Sjobring U, DG. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–1286. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- Elsner A, Kreikemeyer B, Braun-Kiewnick A, Spellerberg B, Buttaro BA, Podbielski A. Involvement of Lsp, a member of the LraI lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect Immun. 2002;70:4859–4869. doi: 10.1128/IAI.70.9.4859-4869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, McLandsborough L, Kondagunta A, Cleary PP. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe NP, Ireland RM, DeLeo FR, Gowen BB, Dorward DW, Voyich JM, Liu M, Burns EH, Culnan DM, Bretscher A, Musser JM. Insight into the molecular basis of pathogen abundance: group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. Proc Natl Acad Sci USA. 2002;99:7646–7651. doi: 10.1073/pnas.112039899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson F, Berggard K, Stalhammar-Carlemalm M, Lindahl G. Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp Med. 2003;198:1057–1068. doi: 10.1084/jem.20030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick IM, Schmidtchen A, Sjobring U. Interactions between M proteins of Streptococcus pyogenes and glycosaminoglycans promote bacterial adhesion to host cells. Eur J Biochem. 2003;270:2303–2311. doi: 10.1046/j.1432-1033.2003.03600.x. [DOI] [PubMed] [Google Scholar]
- Carlsson F, Sandin C, Lindahl G. Human fibrinogen bound to Streptococcus pyogenes M protein inhibits complement deposition via the classical pathway. Mol Microbiol. 2005;56:28–39. doi: 10.1111/j.1365-2958.2005.04527.x. [DOI] [PubMed] [Google Scholar]
- Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser JM. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Burns EHJ, Wyde PR, Podbielski A, Rurangirwa J, Moore-Poveda DK, Musser JM. Genetic inactivation of an extracellular cysteine protease (SpeB) expressed by Streptococcus pyogenes decreases resistance to phagocytosis and dissemination to organs. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary PP, Larkin A. Hyaluronic acid capsule: strategy for oxygen resistance in group A streptococci. J Bacteriol. 1979;140:1090–1097. doi: 10.1128/jb.140.3.1090-1097.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywes C, Wessels M. Group A Streptococcus tissue invasion by CD44-mediated cell signalling. Nature. 2001;414:648–652. doi: 10.1038/414648a. [DOI] [PubMed] [Google Scholar]
- Kil KS, Cunningham MW, Barnett LA. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect Immun. 1994;62:2440–2449. doi: 10.1128/iai.62.6.2440-2449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khil J, Im M, Heath A, Ringdahl U, Mundada L, Cary Engleberg N, Fay W. Plasminogen enhances virulence of group A streptococci by streptokinase-dependent and streptokinase-independent mechanisms. J Infect Dis. 2003;188:497–505. doi: 10.1086/377100. [DOI] [PubMed] [Google Scholar]
- Yoshizawa N, Yamakami K, Fujino M, Oda T, Tamura K, Matsumoto K, Sugisaki T, Boyle MDP. Nephritis-associated plasmin receptor and acute poststreptococcal glomerulonephritis: characterization of the antigen and associated immune response. J Am Soc Nephrol. 2004;15:1785–1793. doi: 10.1097/01.asn.0000130624.94920.6b. [DOI] [PubMed] [Google Scholar]
- Oda T, Yamakami K, Omasu F, Suzuki S, Miura S, Sugisaki T, Yoshizawa N. Glomerular plasmin-like activity in relation to nephritis-associated plasmin receptor in acute poststreptococcal glomerulonephritis. J Am Soc Nephrol. 2005;16:247–254. doi: 10.1681/ASN.2004040341. [DOI] [PubMed] [Google Scholar]
- Zimbelman J, Palmer A, Todd J. Improved outcome of clindamycin compared with beta-lactam antibiotic treatment for invasive Streptococcus pyogenes infection. Pediatr Infect Dis J. 1999;18:1096–1100. doi: 10.1097/00006454-199912000-00014. [DOI] [PubMed] [Google Scholar]
- Gemmell C, Peterson P, Schmeling D, Kim Y, Mathews J, Wannamaker L, Quie P. Potentiation of opsonization and phagocytosis of Streptococcus pyogenes following growth in the presence of clindamycin. J Clin Invest. 1981;67:1249–1256. doi: 10.1172/JCI110152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskandan S, McKee A, Hall L, Cohen J. Comparative effects of clindamycin and ampicillin on superantigenic activity of Streptococcus pyogenes. J Antimicrob Chemother. 1997;40:275–277. doi: 10.1093/jac/40.2.275. [DOI] [PubMed] [Google Scholar]
- Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak M, Khurshid A, Green K, Peeples J, Wade J, Thomson G, Schwartz B, Low DE. An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–1404. doi: 10.1038/nm1202-800. [DOI] [PubMed] [Google Scholar]
- Kazmi SU, Kansal R, Aziz RK, Hooshdaran M, Norrby-Teglund A, Low DE, Halim AB, Kotb M. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group A streptococcal isolates in vivo. Infect Immun. 2001;69:4988–4995. doi: 10.1128/IAI.69.8.4988-4995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansal RG, Aziz RK, Kotb M. Modulation of expression of superantigens by human transferrin and lactoferrin: a novel mechanism in host-Streptococcus interactions. J Infect Dis. 2005;191:2121–2129. doi: 10.1086/430386. [DOI] [PubMed] [Google Scholar]
- Reid SD, Green NM, Sylva GL, Voyich JM, Stenseth ET, DeLeo FR, Palzkill T, Low DE, Hill HR, Musser JM. Postgenomic analysis of four novel antigens of group A Streptococcus: growth phase-dependent gene transcription and human serologic response. J Bacteriol. 2002;184:6316–6324. doi: 10.1128/JB.184.22.6316-6324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun. 2000;68:2441–2448. doi: 10.1128/iai.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BA, Palmer JM, Robson T, Jones CED, Fischer M, Glanville M, Mellor GD, Diamond AG, Kehoe MA, Goodacre JA. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extract is associated with arginine deiminase activity. Infect Immun. 1998;66:3050–3058. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruening P, Fulde M, Valentin-Weigand P, Goethe R. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J Bacteriol. 2006;188:361–369. doi: 10.1128/JB.188.2.361-369.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravins M, Jaffe J, Hanski E, Shetzigovski I, Natanson-Yaron S, Moses AE. Characterization of a mouse-passaged, highly encapsulated variant of group A streptococcus in in vitro and in vivo studies. J Infect Dis. 2000;182:1702–1711. doi: 10.1086/317635. [DOI] [PubMed] [Google Scholar]
- Raeder R, Harokopakis E, Hollingshead S, Boyle MD. Absence of SpeB production in virulent large capsular forms of group A streptococcal strain 64. Infect Immun. 2000;68:744–751. doi: 10.1128/iai.68.2.744-751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg CN, Heath A, Miller A, Rivera C, DiRita VJ. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J Infect Dis. 2001;183:1043–1054. doi: 10.1086/319291. [DOI] [PubMed] [Google Scholar]
- Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+. Proc Natl Acad Sci USA. 2003;100:4227–4232. doi: 10.1073/pnas.0636231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DW, Musser JM, DeLeo FR. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 2003;100:1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Geist RT, Perez-Casal J, Scott JR. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHeyningen T, Fogg G, Yates D, Hanski E, Caparon M. Adherence and fibronectin binding are environmentally regulated in the group A streptococci. Mol Microbiol. 1993;9:1213–1222. doi: 10.1111/j.1365-2958.1993.tb01250.x. [DOI] [PubMed] [Google Scholar]
- Smoot LM, Smoot JC, Graham MR, Somerville GA, Sturdevant DE, Migliaccio CA, Sylva GL, Musser JM. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, Boyle MD, Buttaro BA, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- Kristian SA, Datta V, Weidenmaier C, Kansal R, Fedtke I, Peschel A, Gallo RL, Nizet V. D-Alanylation of teichoic acids promotes group A Streptococcus antimicrobial peptide resistance, neutrophil survival, and epithelial cell invasion. J Bacteriol. 2005;187:6719–6725. doi: 10.1128/JB.187.19.6719-6725.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr–a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Boyce JD, Wilkie I, Harper M, Paustian ML, Kapur V, Adler B. Genomic scale analysis of Pasteurella multocida gene expression during growth within the natural chicken host. Infect Immun. 2002;70:6871–6879. doi: 10.1128/IAI.70.12.6871-6879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A. Host-induced epidemic spread of the cholera bacterium. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Dziejman M, Mekalanos JJ. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci USA. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Marlow D, Palyada K, Naikare H, Panciera R, Whitworth L, Clarke C. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect Immun. 2005;73:1797–1810. doi: 10.1128/IAI.73.3.1797-1810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T, Schoolnik GK. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol Microbiol. 2003;47:879–889. doi: 10.1046/j.1365-2958.2003.03338.x. [DOI] [PubMed] [Google Scholar]
- Shelburne S, Musser J. Virulence gene expression in vivo. Curr Opin Microbiol. 2004;7:283–289. doi: 10.1016/j.mib.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Boyce J, Cullen P, Adler B. Genomic-scale analysis of bacterial gene and protein expression in the host. Emerg Infect Dis. 2004;10:1357–1362. doi: 10.3201/eid1008.031036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.