Abstract
Insertion sequences (IS)1397 and ISKpn1, found in Escherichia coli and Klebsiella pneumoniae, respectively, are IS3 family members that insert specifically into short palindromic repeated sequences (palindromic units or PUs). In this paper, we first show that although PUs are naturally absent from extrachromosomal elements, both ISs are able to transpose from the chromosome or from a plasmid into PUs artificially introduced into target plasmids. We also show that ISKpn1 target specificity is restricted to K.pneumoniae Z1 PU type, whereas IS1397 target specificity is less stringent since the IS targets the three E.coli Y, Z1 and Z2 PU types indifferently. Experiments of transposition of both ISs driven by both transposases demonstrate that the inverted repeats flanking the ISs are not responsible for this target specificity, which is entirely due to the transposase itself. Implications on ISs evolution are presented.
INTRODUCTION
Insertion sequences (ISs) are small (800–2500 bp) DNA segments capable of inserting into target DNA molecules with a more or less pronounced target specificity. Over 500 ISs have been identified so far in more than 40 bacterial species and classified into 17 families on the basis of ORF organization, length of the target duplications, similarity of their terminal inverted repeats (IRs) and signature motifs among the transposases (for a review see 1). They play important roles in DNA translocations and other rearrangements in bacteria (2).
Target-site selection differs significantly from element to element. In some systems this process is extremely stringent while in other systems it is not. For example, Tn7 insertion has two alternative pathways for target selection, which relies on transposon encoded proteins (3–5). The TnsABCD pathway leads to transposition into attTn7, a unique site on Escherichia coli chromosome (6), whereas the TnsABCE pathway leads to transposition onto the lagging strand synthesized during DNA replication (7). In the first (TnsABCD) pathway, the target DNA structure plays a critical role in target-site selection (8,9). Similar studies have shown that transposition of Tn10 involves several steps. The interaction between DNA target and the transposase induces DNA conformational changes, which result in an interaction between the transposase and the sequences flanking the target (10). Target-site selection machinery can recognize either specific DNA sequences (11) and/or DNA structures. For example, IS231A recognizes an S-shaped structure (12), and bent and cruciform DNAs are favored retroviral integration sites in vitro (13).
Insertion sequence IS1397, found in several E.coli isolates (14), and ISKpn1, found in Klebsiella pneumoniae (15) belong to the IS3 family. These closely related ISs have 25 bp-long terminal inverted repeats (IRL and IRR) and encode two proteins, OrfA and OrfB, which are in phase 0 and –1, respectively. A –1 translational frameshift leads to a fusion protein, OrfAB, which is the transposase. IS1397 and ISKpn1 have always been found inserted into a PU (palindromic unit, or repetitive extragenic palindromic sequences) (16,17). PUs are imperfect palindromes and constitute the basic motif of BIMEs (Bacterial Interspersed Mosaic Elements), a family of extragenic sequences scattered over the E.coli chromosome (18). BIME consist of an ordered assembly of PUs and extra-PU motifs. A complete description of BIMEs can be found at: http://www.pasteur.fr/recherche/unites/pmtg/repet/index.html. They could play a role in the functional organization of the bacterial nucleoid, associated to proteins. PUs and BIME-like structures have also been described in several enterobacteria (Salmonella enterica serovar Typhimurium and Klebsiella), where slight sequence variations allowed us to define species-specific consensuses (14,15).
Our previous results demonstrated that IS1397 transposes specifically into PUs present on the chromosome of several enterobacteria (E.coli, S.enterica serovar Typhimurium, K.pneumoniae and K.oxytoca), with a preference for E.coli consensus PUs (14,15). In species containing PUs, which differ from E.coli PU consensus, such as Klebsiella, the relative frequency of transposition outside PUs was increased. Analysis of these non-PU insertions revealed the presence of a conserved sequence (5′GCCGG3′) located 11–13 bp upstream from the insertion site. This sequence shares similarities with PUs. However, since both ISs recognize specific PUs more efficiently than the consensus sequence alone, we postulated that the specificity of insertion into PUs could be due to PU DNA secondary structure and/or PU-bound factors (19).
We had also observed in natural insertion events that ISKpn1 was specific for a subclass of K.pneumoniae PUs. In this paper, we show that ISKpn1 is active for transposition into the chromosomes of K.pneumoniae and E.coli. We have also developed a tool allowing us to study IS1397 and ISKpn1 transposition onto a plasmid. We show that IS1397 and ISKpn1 transposases are able to promote not only their own transposition, but surprisingly also transposition of each other. We thus studied IS1397 and ISKpn1 transposition driven by the ISKpn1 transposase into the chromosomes of E.coli and K.pneumoniae, and driven by each transposase onto a plasmid carrying a K.pneumoniae chromosomal DNA fragment containing BIME in which E.coli PUs and K.pneumoniae PUs alternate. Our results allowed us to confirm IS1397 and ISKpn1 specificities for E.coli and K.pneumoniae PU types, respectively, and to demonstrate that target specificities of these two ISs are entirely due to their transposases.
MATERIALS AND METHODS
Media and bacterial strains
Luria–Bertani (LB) medium was used for bacteria growth. Kanamycin (Km) was used at a concentration of 25 µg/ml, ampicillin (Amp) and chloramphenicol (Cm) were used at a concentration of 50 µg/ml. Isopropyl-β-d-thiogalactopyranoside (IPTG) was used at a final concentration of 10–3 M and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) at 40 µg/ml. LBS medium is LB medium without NaCl and containing 10% sucrose. Strains were grown at 37°C.
Strains used were the following: K.pneumoniae MGH78578 (gift from Dr McClelland, GSC, St Louis), K.pneumoniae subspecies pneumoniae (ATCC 13883), E.coli TOP10F’: F’ [lacIq Tn10(TetR)] mrcA, Δ(mrr-hsdRMS-mrcBC), Φ80lacZΔM15 ΔlacX74 recA1 deoR araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG (Invitrogen) and E.coli BW19610: DE3(lac)X74 ΔuidA::pir-116 recA1 ΔphoA532 Δ(phnC?D-P)33-30 (20). R2 and R4 are TOP10F’ carrying chromosomal insertions of IROK into Y type PUs located in pgi-yjbE region (GTT GCCGGATGCGGCG TGAA C[IRR ← IRL]AA CGCCTTATCCGGC CTAC ATA) and yiif-fdhE region (CAT GCCGGATGCGGCG TGAA C[IRL → IRR]AA CGCCTTATCCGGC CTAC AAA), respectively. These strains were obtained and characterized according to Wilde et al. (15).
Oligonucleotides
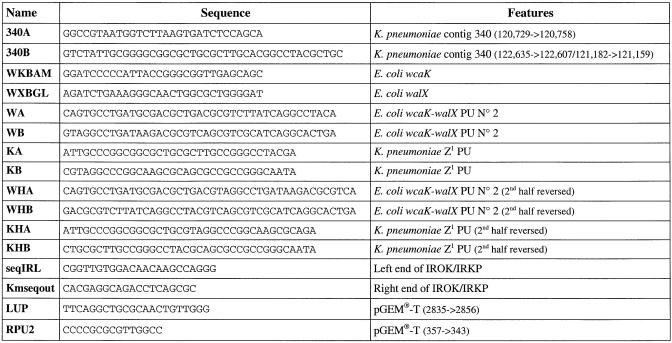
Oligonucleotides were purchased from Genset. Their sequences and main features are presented in Table 1.
Table 1. Oligonucleotides.
Sequence and characteristics of oligonucleotides used for this study. The corresponding coordinates are indicated in the case of contig 340 from K.pneumoniae (oligonucleotides 340A and 340B).
DNA techniques
Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs or Boehringer Mannheim and used as recommended. Plasmid DNA manipulations were carried out by using standard procedures (21). Extraction of total cellular DNA was performed with the DNA Easy Tissue kit (Qiagen). PCRs were performed by using TaKaRa Ex Taq™ PCR kit as recommended with a Mastercycler gradient apparatus (Eppendorf).
DNA sequencing
DNA sequencing was performed either as described previously (14) or by ESGS (Cybergene), or by Génome express S.A.
Plasmids
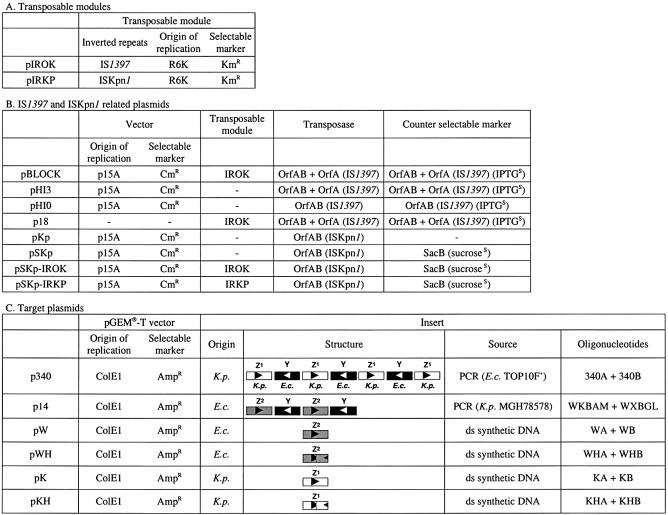
Plasmid constructions involved many steps, which can be obtained upon request (cwilde@pasteur.fr). All PCR fragments were systematically sequenced after cloning. The main characteristics of the plasmids used for this study are indicated in Table 2.
Table 2. Plasmids.
The details for the construction of plasmids are given in Materials and Methods. The main characteristics (origins of replication, antibiotic resistance genes) are indicated. The table has been subdivided in three sections. Transposable modules: pIROK and pIRKP carry the transposable modules derived from IS1397 and ISKpn1, respectively. IS1397 and ISKpn1 transposase plasmids: counter selectable markers and the type (OrfA or OrfAB) and the origin (IS1397 or ISKpn1) of the transposase are indicated. All plasmids express lacI, which allows the repression of the transposases in the absence of IPTG. Target plasmids: they all derive from pGEM®-T (Promega). The origin of the inserts (either a PCR or a double stranded DNA fragment) is indicated as well as their structure. The boxes indicate the PUs and the triangle indicates their orientation. White boxes, K.pneumoniae (K.p.) Z1 PUs; gray boxes, E.coli (E.c.) Z2 PUs; black boxes, E.coli Y PUs.
Transposable modules. IROK (15) and IRKP are two transposable modules where a Km resistance gene and the R6K origin of replication (22,23) are flanked by IRs from IS1397 and ISKpn1, respectively. Plasmids pIROK and pIRKP result from the circularization of the two modules and can replicate only in strains expressing the Pir protein, such as E.coli BW19610.
IS1397 and ISKpn1 transposase plasmids. Plasmid pBLOCK (15) results from the insertion of IROK transposable module into pHI3. Both plasmids express IS1397 OrfAB and an OrfA-LacI fusion under the control of plac promoter. Plasmid pHI0 derives from pHI3 as a result of a XbaI deletion which removed IS1397 orfA gene. Plasmid p18 is a derivative of pBLOCK in which a NheI–SpeI deletion removed the P15A origin of replication and the Cm resistance gene. Plasmid pKp is equivalent to pHI0: the orfAB gene from IS1397 has been replaced by its ISKpn1 homolog. As for IS1397 orfAB fusion, the addition of 1 nt and the disruption of the palindrome destroyed the frameshift window (24). Plasmid pSKp is a derivative of pKp in which Bacillus subtilis sacB gene (19) has been cloned. The two transposable modules IROK and IRKP have been cloned into pSKp, leading to pSKp-IROK and pSKp-IRKP, respectively.
Target plasmids. These plasmids are all derivatives of TA-cloning vector pGEM®-T (Promega). Plasmid p340 carries a composite DNA fragment from K.pneumoniae (contig 340 from strain MGH78578: positions 120 729–121 159 joined to 122 605–122 635; ftp://genome.wustl.edu/pub/seqmgr/bacterial/klebsiella/). This fragment was generated by PCR using K.pneumoniae MGH78578 DNA as a template and oligonucleotides 340A and 340B as primers. It carries a BIME where seven E.coli and K.pneumoniae PUs alternate in opposite orientations. In MGH78578, the last PU is a K.pneumoniae Z1 type interrupted by ISKpn1 (located between positions 121 160 and 122 604 on contig 340). In the cloned fragment, the last PU was reconstituted (deletion of ISKpn1 and of the 3 duplicated bp). Plasmid p14 contains the E.coli K-12 wcaK-walX intergenic region (4-PU BIME) generated by PCR using oligonucleotides WKBAM and WXBGL as primers. Complementary oligonucleotides WA and WB, KA and KB, WHA and WHB, KHA and KHB were annealed and introduced into pGEM®-T, resulting in pW, pK, pWH and pKH, respectively. In pW, the insert corresponds to the third PU (Z2 type) of E.coli K-12 wcaK-walX intergenic region. In pK, the insert corresponds to the K.pneumoniae PU target consensus for ISKpn1 (15, and this study). In pWH, pKH the first halves of the PUs were identical to pWH and pKH, respectively, but the second halves have been inverted.
Selection of chromosomal transposition events
Escherichia coli TOP10F’ and K.pneumoniae were transformed with pSKp-IROK and pSKp-IRKP. Independent transformants (28 E.coli and 12 K.pneumoniae clones of each type) were grown overnight at 37°C, in LB liquid medium containing Km and Cm. One to 150 µl of each culture were plated on LBS containing Km. After a 24 h incubation at 37°C, sucrose resistant-Km resistant colonies were replica plated on LBS plates containing Km or Km and Cm in order to test for plasmid loss. Twenty-nine percent of the pSKp-IROK-containing E.coli colonies, 68% of the pSKp-IRKP-containing E.coli colonies and 99% of K.pneumoniae displayed the correct phenotype, i.e. sucrose resistant, Km resistant and Cm sensitive. Two colonies originating from each independent clone were re-isolated on LBS containing Km and grown overnight at 37°C in LB liquid medium containing Km. Escherichia coli genomic DNAs were digested with MluI (which has no site in the transposable module) and 1–5 µg samples were analyzed by Southern blot with IROK as a probe, as already described (19). Fragments of digested DNAs were circularized with T4 DNA ligase and used to transform BW19610, an E.coli strain, which allows autonomous replication of circles containing R6K origin of replication present in IROK. The same procedure was applied to K.pneumoniae genomic DNAs, except that they were digested by a combination of several enzymes that do not cut into the transposable module, followed by a treatment with Klenow DNA polymerase in order to fill-in protruding 5′-ends before circularization by T4 DNA ligase. BW19610 recombinant clones were selected on LB plates containing Km. After DNA sequencing with oligonucleotides seqIRL (complementary to a region of R6K origin of replication) and Kmseqout (corresponding to a region located between the end of Km resistance gene and IRR of the transposable modules), chromosomal regions flanking the module were identified using FASTA software (25) at Infobiogen (http://www.infobiogen.fr/) and BLAST software at the Washington University of Saint Louis, MO (http://blast.wustl.edu). For transposition events on E.coli chromosome, gene names were identified on the Colibri Web Server (http://genolist.pasteur.fr/Colibri/).
Selection of transposition events on plasmids
The assays rely on the inability of the donor module to replicate in tester strains. R6K origin of replication is present on IROK and IRKP and is active only in Pir+ strains. Km-resistant strains harboring the donor module either on the chromosome (in the case of R2 or R4 strains) or as a free plasmid (p18, pIROK or pIRKP in Pir+ BW19610 strain) were transformed for Cm resistance with a plasmid that expresses the transposase from IS1397 (pHI0 or pHI3) or ISKpn1 (pKp). This second step was not necessary in the case of p18 since in this plasmid IROK is already associated with IS1397 OrfAB expression system. In a last step, strains are transformed with the target plasmids, which confer Amp resistance. In all cases, independent antibiotic-resistant clones were checked when necessary for IPTG sensitivity (due to the toxicity of IS1397 OrfA and OrfAB, a phenotype, which was not observed in the case of ISKpn1 OrfAB) and were grown overnight in liquid at 37°C. Plasmids were extracted and used to electroporate TOP10F’ competent cells. Serial dilutions of electroporated cell suspensions were plated onto LB agar plates containing Amp or Amp and Km. The frequency of transposition was measured as the ratio between the number of Amp- and Km-resistant clones and the number of Amp-resistant clones. Individual colonies were tested for the presence and for the orientation of IROK or IRKP in PGEM-T target plasmids using oligonucleotides Kmseqout (internal to IKOK and IRKP) and either LUP or RPU (flanking the insert) as primers for colony-PCR assay. The size of the fragments was measured on agarose gel electrophoresis, which allowed us to determine approximately the position of the insertion site. A few representative clones were sequenced (using primers SeqIRL and Kmseqout) to define precisely in each case the structure at the junctions between the target and the transposable module.
RESULTS
Transposition of IS1397 and ISKpn1 into K.pneumoniae and E.coli chromosome using ISKpn1 transposase
We first analyzed whether ISKpn1 OrfAB transposase was active in two enterobacteria: K.pneumoniae and E.coli. Our transposition assays relied on two distinct events: the transposition of a transposable module from a donor plasmid into the chromosome of the bacteria, followed by the loss of the donor plasmid. The modules were tracked by Km resistance and the donor plasmids were counter-selected by sucrose resistance. The two different modules used for this study, IROK and IRKP, differ only by the nature of the IRs, which are, respectively, IS1397 and ISKpn1 IRs.
5 × 108 CFU from independent E.coli and K.pneumoniae clones containing pSKp-IRKP or pSKp-IROK were plated on LBS plates containing Km. For K.pneumoniae, each plate contained about 200 sucrose-resistant colonies, whereas for E.coli, plates contained from 7 to about 200 sucrose-resistant colonies. They were replica plated on LBS plates containing Km or Km and Cm, in order to check for plasmid loss. Ninety-nine percent of the K.pneumoniae clones and 68% of the E.coli clones were CmS. Independent KmR, SacR, CmS clones (two per plate) were examined for the presence of the transposable module on the chromosome by Southern blot hybridization. We recovered plasmids encompassing IROK and IRKP after circularization of chromosomal DNA fragments due to the presence of a Km resistance gene and an R6K origin of replication, which is active in E.coli BW19610. The plasmids were used to sequence the new flanking regions of the transposable module.
ISKpn1 and IS1397 targets in K.pneumoniae chromosome
We sequenced nine transposition events of IRKP and 12 transposition events of IROK into the K.pneumoniae chromosome using ISKpn1 transposase (Table 3). All insertions but one (clone IRKP I) occurred into PUs, with a 3 or 4 bp duplication. More precisely, sequence comparison of the consensus insertion site with consensuses of K.pneumoniae PU types (11) showed that the modules inserted exclusively into Z1 type (Table 3). They were distributed between 12 regions. Several independent transposition events occurred in the same region, either in the same PU (Table 3, regions III, IV, VI, VII and X) or in different PUs of the same BIME (Table 3, region VII). Interestingly, IROK and IRKP were targeted to the same hot-spots, showing that the nature of the IRs flanking the transposable module (IS1397 or ISKpn1 IRs) does not influence the target choice. For example, insertions in clones IROK J and IRKP G or clones IROK K and IRKP B occurred in the same PU in the same orientation, and insertions in clones IROK F and IRKP C and D occurred in the same PU, with different orientations. Interestingly, these three clones transposed to the same position as ISKpn1 in the sequenced strain MGH78578 (ISKpn1 is also present in the K.pneumoniae strain that we used for transposition, but not at the same positions) (15). This confirms that this particular PU is a hot-spot for transposition.
Table 3. Insertion sites of IROK and IRKP into K.pneumoniae chromosome.
aInsertion sites, insertion consensus and K.pneumoniae PU consensuses are figured; duplication of the target site (3 or 4 bp) is underlined.
bThe white boxes indicate the K.pneumoniae PUs and the triangles indicate their orientation; the gray boxes symbolize the IROK or IRKP transposable module.
cThe region of K.pneumoniae chromosome (ftp://genome.wustl.edu/pub/seqmgr/bacterial/klebsiella/) is indicated: I (unidentified), II (contig (ctg) 108 coordinates (coord) 66 293), III (ctg 118 coord 48 827), IV (ctg 118 coord 18 576), V (ctg 123 coord 39 555), VI (ctg 125 coord 994), VII (ctg 128 coord 111 632 for clone IROK I and ctg 128 coord 111 385 for clones IROK J and IRKP G), VIII (ctg 129 coord 133 946), IX (ctg 131 coord 149 035), X (ctg 131 coord 192 320), XI (ctg 132 coord 330 297), XII (ctg 101 coord 19 163).
dIn clone IROK L, the sequence of the transposable module is bracketed, the double arrow indicates its orientation, and only the 9 external nt of the IRs are written. ISKpn1 sequence (dark gray box in the drawing) is written in lower case and the nucleotide change in the IROK module is twice underlined.
eKlebsiella pneumoniae PU consensuses have been determined previously (11).
We also observed an unusual transposition event for clone IROK L, in which transposition had occurred next to a resident K.pneumoniae ISKpn1, already inserted into the loop of a PU. The last 3 nt at the end of IROK IRR, 5′-TCA-3′, were changed into 5′-GCA-3′ (change in italic), as found in ISKpn1 IRL. Furthermore, we found an extra 3 nt (GAT) between IS1397 IRL and ISKpn1 IRL. This particular example will be discussed further (see Discussion).
ISKpn1 and IS1397 targets in E.coli chromosome
The observed specificity of ISKpn1 transposase for K.pneumoniae Z1 PU type raised the question of whether other PU types could be targeted. Thus, we used ISKpn1 transposase to study ISKpn1 and IS1397 transposition into E.coli chromosome, which is lacking K.pneumoniae Z1 PU type. In this context, we analyzed eight transposition events of IROK and 19 transposition events of IRKP (Table 4).
Table 4. Insertion sites of IROK and IRKP into E.coli chromosome.
aDuplicated nucleotides are underlined. The consensus sequence 5′-GCCC(GG)-3′ of the 10-bp window of insertion sites is twice underlined.
bGene names have been retrieved from Colibri Web Server (http://genolist.pasteur.fr/Colibri/). Gene orientations are indicated by an arrow, and the module insertion and orientation by a double arrow.
cISKpn1 insertion consensus is taken from Table 3.
All insertions occurred outside PUs, with a 3 or 4 bp duplication (Table 4). The majority of insertions were located inside genes. We observed again hot-spots of insertion, but they were different for each transposable module. Transposition events into the same insertion site always occurred in the same orientation (not shown). repC was the target for IROK (clones IROK 7 and 8) and IRKP (clones IRKP 4, 5 and 6), but not at the same position. Despite the lack of targeting into PUs, insertion sites of IRKP and IROK revealed, in 9 cases out of 13, the presence of a consensus sequence (5′GCCC3′, which can be extended to 5′GCCCGG3′ in two cases) in a 10-bp window upstream of the insertion site. This sequence is also present in the stem of K.pneumoniae Z1 PU type.
Target specificity of the two ISs with the two transposases: transposition events onto a target plasmid
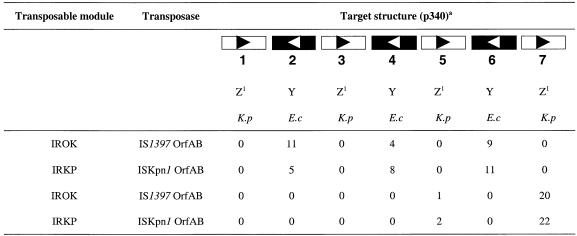
PUs have only been found on chromosomes and not on plasmids or phages. Thus, we wanted to know whether IS1397 and ISKpn1 could transpose into PU targets cloned on a plasmid. For this, we developed an assay as described in Materials and Methods. A favorable situation enabled us to compare in a single experiment E.coli and K.pneumoniae PUs for target selectivity. Indeed, we used a K.pneumoniae intergenic region containing a 7-PU BIME, which alternate K.pneumoniae Z1 PUs with E.coli Y PUs. This BIME was a natural insertion site for ISKpn1 and has been shown to be a target for IS1397 (15). A plasmid containing this BIME (p340) was used a target for IS1397 and ISKpn1 donor modules (present on pIROK and pIRKP, respectively) using either IS1397 or ISKpn1 transposases (respectively, expressed from pHI0 or pKp). Transposition occurred exclusively into PUs in all configurations with comparable frequencies (not shown). We determined insert orientations by PCR and we identified which PU had been targeted. A number of representative examples were checked by DNA sequencing. The results are presented in Table 5. Remarkably, no difference was found between IROK and IRKP, which could transpose indifferently into PUs of both types (i.e. E.coli or K.pneumoniae). On the contrary, we found a clear-cut PU choice according to the type of transposase, namely IS1397 OrfAB being exclusively specific for PUs Nos 2, 4 and 6 (E.coli type) whereas ISKpn1 was found exclusively specific for PU Nos 5 and 7 (K.pneumoniae type). This result confirms that the two ISs are exclusively specific for the PU types corresponding to the species they were detected in, and it demonstrates that this specificity is unambiguously due to the transposases and not to the IRs.
Table 5. Transposition of IROK and IRKP into plasmid p340.
For each configuration (IROK or IRKP transposable module/IS1397 or ISKpn1 OrfAB transposase), the number of transposition events into the different PUs present on plasmid p340 is given.
aAbbreviations and symbols used for the drawing of p340 BIME insert are identical to Table 2.
Impact of BIME and PU arrangement on transposition specificity
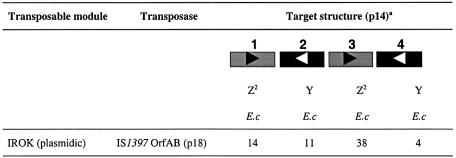
To analyze further the roles of PU sequence and structure on target specificity for IS1397 transposase, we used the wcaK-walX intergenic region from E.coli. This includes a BIME-2 containing four PUs. This region had been shown to be a hot-spot for IS1397 transposition (14). Results presented in Table 6 show that plasmid p14, which carries this region, was an efficient target for transposition. When the donor module was on p18, a high copy number plasmid, which also carries the transposase gene, the transposition frequency was 2 × 10–2. IPTG is an inducer of IS1397 OrfA and OrfAB, which are lethal when over-expressed (14). The addition of sub-lethal concentrations of IPTG did not increase the already high frequency of transposition (not shown). Sixty-seven clones were analyzed by PCR to predict the location and the orientation of IROK inserts. As observed previously, several representative clones from each situation were sequenced. All insertions were found in PUs with a 3 nt duplication and a random module orientation (not shown). As reported in Table 6, a majority of insertions occurred into the third PU (38 out of 67 cases analyzed). It should be noted that this PU is almost identical to the first, with the exception of flanking sequences. This shows that local constraints interfere with the efficiency of targeting.
Table 6. Transposition of IROK into plasmid p14.
The number of IROK transposition events into the different PUs present on plasmid p14 is given.
aAbbreviations and symbols used for the drawing of p14 BIME-2 insert are identical to Table 2.
To determine more precisely whether the environment and the integrity of PUs could play a role in target specificity, we constructed a target plasmid (pW) containing only the third PU from wcaK-walX BIME-2 and a target plasmid (pWH) containing the two halves of the same PU but in opposite orientations. The same approach was followed for a typical solo Z1 K.pneumoniae PU (pK and pKH). We found that only pW was a target for transposition of the IROK donor module carried by plasmid p18, but with a lower efficiency (8.4 × 10–5) than observed before. Transposition into pWH, pK and pKH occurred at an undetectable frequency (<10–6). The same results were observed when IROK donor module was present on the chromosome (strains R2 and R4, data not shown). These results confirm that Z1 K.pneumoniae PUs are not targets for IS1397 transposition. They also show that the overall sequence of the PU (i.e. the presence of the two halves) is not sufficient for targeting IS1397. The correct PU structure (i.e. palindromic) is important, as well as the local environment, since a given PU sequence behaves differently when it is alone and when it is included in a BIME, and even there its position inside the BIME has an influence.
DISCUSSION
We have shown previously that IS1397 was able to transpose from a plasmid onto the E.coli chromosome (14,15,19). In this paper, we show that IS1397 and ISKpn1 are also able to transpose in E.coli either from a plasmid or from the chromosome onto a target plasmid. They can both recognize a BIME as well as a single PU as an exclusive target site for transposition. These results show that although PUs have never been found on natural plasmids or other extra chromosomal elements, they can be recognized as targets by both ISs when they are artificially introduced into a plasmid. This indicates that IS1397 targets similar or identical PU conformations on a plasmid and on the chromosome, suggesting that no specific association between PUs and chromosomal proteins is required for this process.
We show here that ISKpn1 is active for transposition into the chromosomes of K.pneumoniae and E.coli. Natural ISKpn1 insertion sites observed previously on the chromosome of K.pneumoniae (15) were PUs of the K.pneumoniae type, and more precisely Z1 (Table 3). Our results show that IRKP transposes almost exclusively into PUs in the chromosome of K.pneumoniae (eight cases out of nine), and confirm ISKpn1 specificity for K.pneumoniae Z1 PU type. This target choice is more stringent than for IS1397, which is able, as shown previously, to recognize the three E.coli PU types (Y, Z1 and Z2) (14). As a consequence, IS1397 transposition in K.pneumoniae occurs exclusively into PUs that are of the E.coli type, and which are present along the entire chromosome of K.pneumoniae. In parallel, ISKpn1 is specific for K.pneumoniae Z1 PU type, and transposes exclusively outside PUs into the chromosome of E.coli, which does not contain any K.pneumoniae Z1 PU type.
The transposition event in clone IROK L (Table 3) resembles the cases observed in Yersinia pestis atypical transposition events (19) of IROK next to a resident Y.pestis IS whose ends are close to IS1397 IRs (Fig. 1). As suggested, a 3-bp shift of the cutting site on the circular intermediate occurred, leading to the deletion of the last 3 bp of IS1397 IROK (TCA), and 3 extra bp flanking the module have been kept inside. However, since these 3 nt do not correspond to those originally flanking IROK IRs on the donor plasmid, we postulate that this clone results from a second-step transposition event on K.pneumoniae chromosome. In the case of Y.pestis, we hypothesized these transposition events could be due to the co-action of IS1397 and an endogenous transposase. The fact that IS1397 can transpose via ISKpn1 transposase is in favor of this hypothesis since it shows that a given transposase not only recognizes its own IRs, but it is also active on the IR of other similar ISs. This could account for the high instability of the Y.pestis genome.
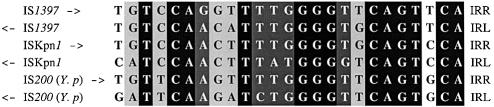
Figure 1.
Comparison of IS1397, ISKpn1 and IS200 IRs. Yersinia pestis IS200 has been described as a target for IS1397 (19). In order to align the six IR sequences, IRLs have been inverted (i.e. insertion sites are located on the right side of the sequence). IS orientations are indicated by an arrow. At a given position, identical nucleotides between all six IRs are shaded in black, in dark gray when identical in five IRs and in light gray when identical in four IRs.
As IS1397 and ISKpn1 are very close, especially in their external IRs sequences (84 and 72% identical for IRR and IRL, repectively; Fig. 1), we wondered whether both transposases would be able to recognize both IRs, and to promote transposition of both ISs. More precisely, we wanted to know if the target specificity was due to the transposases or to the IRs. We used two approaches for this. First, we compared the transposition of two different modules into the chromosomes of K.pneumoniae and E.coli, using ISKpn1 transposase (Tables 3 and 4). The two modules (IROK and IRKP) only differ by the flanking IRs (IS1397 and ISKpn1, respectively). No difference in target choice was observed (i.e. K.pneumoniae Z1 PU type), suggesting, at least for ISKpn1, that transposition specificity resides in the nature of the transposase and not in the origin of the IR. A second approach enabled us to extend this conclusion to the case of IS1397. Indeed, we used the four combinations (transposases versus IRs) in a transposition assay onto a plasmid carrying both PU targets. Unambiguously, we demonstrated that IR types have no effect on target specificity and that this phenomenon is entirely due to the transposase.
We detected a conserved sequence (5′GCCC3′, which can be extended to 5′GCCCGG3′) upstream of IROK and IRKP insertion site in the E.coli chromosome (Table 4). This sequence is also present in the Z1 stem of K.pneumoniae PU type. This suggests that, as hypothesized for IS1397 (19), a small consensus sequence could be recognized by ISKpn1 transposase, and may be sufficient for transposition when the actual PU targets are not found. However, transposition into PUs could involve other factors such as a specific PU conformation or a PU-binding protein.
A strict target specificity can present both advantages and drawbacks for the parasitic equilibrium between ISs and their hosts. PUs are located outside genes and scattered all over the chromosome (18). They are therefore readily available targets and minimize the risk of gene inactivation, which could be deleterious for the host and, as a consequence, for the mobile element. However, the strict specialization of an IS for such a target limits its lateral transfer to other species, which would not share the same type of repeated element. A possible way to bypass this limitation would be to transpose more or less at random into sites that show some similarities with the original one. In order to propagate efficiently within its new host, the IS would have to evolve to finally acquire a transposase endowed with a new specificity. The close similarity between IS1397 and ISKpn1 shows that a limited number of steps have been sufficient for such an adaptation, which has already occurred during evolution.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Chandler for helpful advice about the manuscript. This work was supported in part by the ‘Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires’ from ‘Ministère Français de l’Education Nationale de la Recherche et de la Technologie’. C.W. was supported by a grant from the ‘Fondation pour la Recherche Médicale’.
REFERENCES
- 1.Chandler M. and Mahillon,J. (2001) Insertion sequences revisited. In Craig,N., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, Washington DC, pp. 305–366. [Google Scholar]
- 2.Haren L., Ton-Huang,B. and Chandler,M. (1999) Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- 3.Bainton R.J., Kubo,K.M., Feng,J.N. and Craig,N.L. (1993) Tn7 transposition: target DNA recognition is mediated by multiple Tn7-encoded proteins in a purified in vitro system. Cell, 72, 931–943. [DOI] [PubMed] [Google Scholar]
- 4.Kubo K.M. and Craig,N.L. (1990) Bacterial transposon Tn7. J. Bacteriol., 172, 2774–2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig N.L. (2001) Tn7. In Craig,N., Craigie,R., Gellert,M. and Lambowitz,A. (eds), Mobile DNA II. ASM Press, Washington DC, pp. 423–456. [Google Scholar]
- 6.Skelding Z., Sarnovsky,R. and Craig,N.L. (2002) Formation of a nucleoprotein complex containing Tn7 and its target DNA regulates transposition initiation. EMBO J., 21, 3494–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolkow C.A., DeBoy,R.T. and Craig,N.L. (1996) Conjugating plasmids are preferred targets for Tn7. Genes Dev., 10, 2145–2157. [DOI] [PubMed] [Google Scholar]
- 8.Rao J.E., Miller,P.S. and Craig,N.L. (2000) Recognition of triple-helical DNA structures by transposon Tn7. Proc. Natl Acad. Sci. USA, 97, 3936–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuduvalli P.N., Rao,J.E. and Craig,N.L. (2001) Target DNA structure plays a critical role in Tn7 transposition. EMBO J., 20, 924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pribil P.A. and Haniford,D.B. (2000) Substrate recognition and induced DNA deformation by transposase at the target-capture stage of Tn10 transposition. J. Mol. Biol., 303, 145–159. [DOI] [PubMed] [Google Scholar]
- 11.Wilde C. (2003) PhD thesis. Paris VII-Denis Diderot University.
- 12.Hallet B., Rezsohazy,R., Mahillon,J. and Delcour,J. (1994) IS231A insertion specificity: consensus sequence and DNA bending at the target site. Mol. Microbiol., 14, 131–139. [DOI] [PubMed] [Google Scholar]
- 13.Katz R.A., Gravuer,K. and Skalka,A.M. (1998) A preferred target DNA structure for retroviral integrase in vitro. J. Biol. Chem., 273, 24190–24195. [DOI] [PubMed] [Google Scholar]
- 14.Clément J.-M., Wilde,C., Bachellier,S., Lambert,P. and Hofnung,M. (1999) IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into Palindromic Units of Bacterial Interspersed Mosaic Elements. J. Bacteriol., 181, 6929–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilde C., Bachellier,S., Hofnung,M. and Clément,J.-M. (2001) Transposition of IS1397 in the family Enterobacteriaceae and first characterization of ISKpn1, a new insertion sequence associated with Klebsiella pneumoniae Palindromic Units. J. Bacteriol., 183, 4395–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilson E., Clément,J.-M., Brutlag,D. and Hofnung,M. (1984) A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J., 3, 1417–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern M. J., Prossnitz,E. and Ferro-Luzzi Ames,G. (1988) Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol., 2, 141–152. [DOI] [PubMed] [Google Scholar]
- 18.Bachellier S., Clément,J.-M. and Hofnung,M. (1999) Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol., 150, 627–639. [DOI] [PubMed] [Google Scholar]
- 19.Wilde C., Bachellier,S., Hofnung,M., Carniel,E. and Clément,J.-M. (2002) Palindromic Unit-Independent transposition of IS1397 in Yersinia pestis. J. Bacteriol., 184, 4739–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf W., Jiang,W. and Wanner,B.L. (1994) Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene, 138, 1–7. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J., Frisch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 22.Shafferman A., Kolter,R., Stalker,D. and Helinski,D.R. (1982) Plasmid R6K DNA replication. III. Regulatory properties of the pi initiation protein. J. Mol. Biol., 161, 57–76. [DOI] [PubMed] [Google Scholar]
- 23.Stalker D.M., Kolter,R. and Helinski,D.R. (1982) Plasmid R6K DNA replication. I. Complete nucleotide sequence of an autonomously replicating segment. J. Mol. Biol., 161, 33–43. [DOI] [PubMed] [Google Scholar]
- 24.Polard P., Prere,M.F., Chandler,M. and Fayet,O. (1991) Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol., 222, 465–477. [DOI] [PubMed] [Google Scholar]
- 25.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]