Abstract
The histological detection of laminin-rich vasculogenic mimicry patterns in human primary uveal melanomas is associated with death from metastases. We therefore hypothesized that highly invasive uveal melanoma cells forming vasculogenic mimicry patterns after exposure to a laminin-rich three-dimensional microenvironment would differentially express genes associated with invasive and metastatic behavior. However, we discovered that genes associated with differentiation (GDF15 and ATF3) and suppression of proliferation (CDKNa1/p21) were up-regulated in highly invasive uveal melanoma cells forming vasculogenic mimicry patterns, and genes associated with promotion of invasive and metastatic behavior such as CD44, CCNE2 (cyclin E2), THBS1 (thrombospondin 1), and CSPG2 (chondroitin sulfate proteoglycan; versican) were down-regulated. After forming vasculogenic mimicry patterns, uveal melanoma cells invaded only short distances, failed to replicate, and changed morphologically from the invasive epithelioid to the indolent spindle A phenotype. In human tissue samples, uveal melanoma cells within vasculogenic mimicry patterns assumed the spindle A morphology, and the expression of Ki67 was significantly reduced in adjacent melanoma cells. Thus, the generation of vasculogenic mimicry patterns is accompanied by dampening of the invasive and metastatic uveal melanoma genotype and phenotype and underscores the plasticity of these cells in response to cues from the microenvironment.
The term vasculogenic mimicry describes the formation of perfusion pathways in tumors by highly invasive, genetically deregulated tumor cells1,2: vasculogenic because, although these pathways do not form from pre-existing vessels, they distribute plasma and may contain red blood cells and mimicry because the pathways are not blood vessels and merely mimic vascular function. In vasculogenic mimicry of the patterned matrix type,3 highly invasive tumor cells form looping patterns rich in extracellular matrix (ECM) in three-dimensional (3D) culture conditions.1 Highly invasive tumor cells do not generate these patterns when grown under monolayer conditions or on thin matrix, and poorly invasive tumor cells do not generate these patterns under any culture condition.1,4
These looping patterns, termed the “fluid-conducting meshwork,”5 connect to endothelial cell-lined blood vessels4,6 and conduct fluid in vitro1,4 and in animal models of melanoma.5,7,8 Vasculogenic mimicry patterns are composed of laminin,5,9,10 collagen types IV11 and VI,12 fibronectin,13 and glycosaminoglycans, especially heparan sulfate proteoglycan.7,14 These patterns stain only focally and weakly for collagen I and are structurally different from stromal host-derived fibrovascular septa. There is a strong association between the histological detection of vasculogenic mimicry patterns in primary uveal15–21 and cutaneous22,23 melanomas and subsequent death from metastasis, consistent with the in vitro observations that these patterns are generated exclusively by highly invasive tumor cells.1,4
Although differences in gene expression between poorly invasive and highly invasive melanoma cells capable of generating vasculogenic mimicry patterns have been identified,1,24,25 there are no reports of gene expression in tumor cells that have generated patterns. Maniotis and colleagues26 recently described the sequestration of digestion by AluI and MspI binding sites in highly invasive epithelioid uveal melanoma cells compared with poorly invasive spindled tumor cells and normal uveal melanocytes. Changes in the sequestration and exposure of AluI binding sites were also detected when highly invasive melanoma cells were exposed to a microenvironment permissive of vasculogenic mimicry pattern formation, suggesting that significant shifts in gene expression may accompany the generation of these patterns.
We therefore studied gene expression profiles in uveal melanoma cells generating vasculogenic mimicry patterns in two stages. Because pattern formation requires highly invasive tumor cells in a permissive ECM microenvironment,4 we first compared gene expression between poorly invasive and highly invasive uveal melanoma cells in monolayer conditions on plastic.1,24,25 We then studied gene expression in highly invasive uveal melanoma cells under paired culture conditions—monolayer conditions on plastic (vasculogenic mimicry patterns are not formed even by highly invasive uveal melanoma cells in this condition), and 3D Matrigel cultures that permit highly invasive cells to generate vasculogenic mimicry patterns. We expected to discover under these conditions, highly invasive uveal melanoma cells forming vasculogenic mimicry patterns differentially expressed genes associated with invasive and metastatic behavior because the histological detection of these patterns in tissue sections of human primary uveal melanomas is associated with death from metastatic melanoma.16
Contrary to our expectations, highly invasive uveal melanoma cells engaged in the formation of vasculogenic mimicry patterns in 3D Matrigel cultures overexpressed genes associated with suppression of transcription and proliferation. Because highly invasive uveal melanoma cells featuring large nucleoli formed vasculogenic mimicry patterns in vitro, these cells elongated, nucleoli diminished in size, the cells invaded only short distances, and they failed to replicate. In human uveal melanoma tissues, tumor cells within vasculogenic mimicry patterns assumed the morphological characteristics of spindle A melanoma cells that are associated with indolent behavior27 whereas epithelioid cells with prominent nucleoli were identified within the population of cells that were circumscribed by the patterns. Additionally, the percentage of tumor cells expressing Ki67 in the vicinity of vasculogenic mimicry patterns in tissue sections was less than half of that detected in tumor cells not associated with these patterns. Therefore, the interactions between the ECM microenvironment and highly invasive tumor cells that engage in the formation of vasculogenic mimicry patterns are accompanied by shifts in gene expression, morphology, and behavior that reflect dampening rather than the enhancement of the invasive uveal melanoma phenotype.
Materials and Methods
Overall Experimental Design
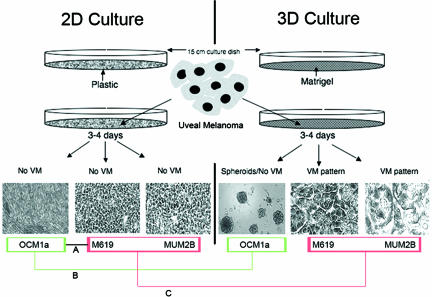
Experiments were designed to study changes in gene expression signatures, invasive behavior, and morphology in highly invasive uveal melanoma cells that generate vasculogenic mimicry patterns. Figure 1 illustrates schematically the experimental strategies used in gene expression studies. In brief, differential gene expression was first determined in two-dimensional (2D) culture conditions in plastic to allow for comparisons between the low invasive OCM1a and the highly invasive M619 and MUM2B uveal melanoma cell lines (Figure 1, comparison A). Next, differential gene expression was determined in all three cell lines between 2D culture conditions in plastic and in 3D culture conditions in Matrigel (Figure 1, comparisons B and C). The directionality of differential expression was confirmed for selected genes by real-time reverse transcriptase-polymerase chain reaction (RT-PCR).
FIGURE 1.
Schematic of the experimental design for comparing gene expression in uveal cells in 2D culture conditions on plastic versus 3D cultures on thick Matrigel, permissive of vasculogenic mimicry pattern formation. After 3 to 4 days, neither the poorly invasive OCM1a primary uveal melanoma cells, the highly invasive M619 primary uveal melanoma cells, nor the highly invasive MUM2B metastatic uveal melanoma cells generated vasculogenic mimicry (VM) patterns in 2D cultures on plastic. However, in the same time period, M619 and MUM2B cells generated looping patterns surrounding clusters of tumor cells characteristic of VM of the patterned matrix type in 3D Matrigel cultures. Cells were harvested at 3 to 4 days and prepared for gene expression profiling as described in the Materials and Methods. First, gene expression in OCM1a (bordered in green) was compared with gene expression in M619 and MUM2B cells (bordered in red, comparison A; also see Supplementary Data Table 1, http://ajp.amjpathol.org). Next, gene expression in OCM1a cells in 2D was compared with the same cells in 3D Matrigel cultures that failed to generate VM patterns in these conditions (comparison B). Finally, gene expression of M619 and MUM2B cells that did not generate VM patterns in 2D culture conditions was compared with the same cells after they generated VM patterns in 3D culture conditions (comparison C). Genes from comparison B were subtracted from comparison C to generate Supplementary Data Table 2 (see http://ajp.amjpathol.org).
To study functional changes that accompanied the generation of vasculogenic mimicry patterns, we compared the ability of highly invasive M619 and MUM2B cells to invade in 2D culture conditions on plastic and after the formation of vasculogenic mimicry in 3D Matrigel cultures using high-density and low-density culture techniques as described below. Because cell morphology (the Callender classification) is a well-established and reliable independent attribute of uveal melanoma associated with death from metastatic uveal melanoma,27–29 morphological features of M619 and MUM2B cells were documented in 2D and 3D culture conditions before and after generation of vasculogenic mimicry patterns, respectively.
Finally, to test if changes in gene expression and in the functional and morphological characteristics of cells generating vasculogenic mimicry patterns are present in vivo, we developed a uveal melanoma progression tissue microarray. The tissue microarray was designed to test statistically for associations or lack of associations between the histological presence in human tissue samples of vasculogenic mimicry patterns, cell morphology, and Ki67 as detected by immunohistochemistry. Additionally, the morphology of invasive uveal melanoma cells generating vasculogenic mimicry patterns in 3D Matrigel cultures was compared with melanoma cells from human tissue samples containing vasculogenic mimicry patterns. Specific methods for each of the experiments are presented next.
2D Cell Cultures
The OCM1a primary uveal melanoma cell line30 was a generous gift from Dr. June Kan-Mitchell (Karmanos Cancer Institute, Wayne State University, Detroit, MI). The highly invasive primary (M619) and liver metastatic uveal melanoma (MUM2B) cell lines have been characterized previously.1 Briefly, the primary uveal OCM1a melanoma cells are poorly invasive in membrane invasion culture system assays,31 do not deform floating collagen gels, and do not form vasculogenic mimicry patterns in 3D cultures. By contrast, the M619 primary and MUM2B metastatic uveal melanoma cells are highly invasive in membrane invasion culture system assays, deform floating collagen gels, and form vasculogenic mimicry patterns in 3D cultures (Table 1).1 Melanoma cells were grown in 15-cm plastic dishes in Eagle’s minimum essential medium (EMEM) (BioWhittaker, Walkersville, MD) supplemented with 15% heat-inactivated fetal bovine serum (Fisher, Ontario, ON, Canada), 1% nonessential-amino acids (NEAA-Mixture, 100×; BioWhittaker) 2 mmol/L l-glutamine, and penicillin/streptomycin without the addition of exogenous ECM molecules.
TABLE 1.
In Vitro Characteristics of Melanocytic Cell Lines
| Culture designation | Callender cell type | Invasive potential | Deformation of floating collagen gels | Formation of vasculogenic mimicry patterns in thick three-dimensional Matrigel culture conditions |
|---|---|---|---|---|
| OCM1a (primary) | Spindle B | Poor | − | − |
| M619 (primary) | Spindle B | High | + | + |
| MUM2B (metastatic) | Spindle B and epithelioid | High | + | + |
3D Cultures for Transcriptional Profiling
3D cultures were established by growing cells on Matrigel (BD Biosciences, Bedford, MA) poured onto plastic tissue culture dishes to a depth of ∼0.2 mm followed by polymerization for 1 hour at 37°C. Cells were seeded at saturating densities (50 million cells/150-mm dish) on the polymerized 3D gel coatings, and the cultures were checked daily for the presence of looping patterns that are characteristic for highly invasive melanoma cells in these conditions.
Transcriptional Profiling
For each cell line and each culture condition, RNA isolation, labeling, and hybridization were performed independently for each technical replication. Each cell line was replicated six times under each of the two culture conditions. Thus, for each cell line, a total of 12 samples were analyzed. An Affymetrix Human Genome U133A Gene Chip array (Affymetrix, Santa Clara, CA) was used for measuring the transcription profiles. The GeneChip U133 set (A+B) contains 45,000 probe sets probes covering 39,000 transcripts. Cells were harvested and collected by centrifugation followed by solubilization using TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA was isolated from the Trizol extracts using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Labeling reactions and hybridizations were performed according to the standard Affymetrix GeneChip eukaryotic target labeling protocol. In brief, 5 to 15 μg of total cellular RNA per sample was used to synthesize the double-stranded cDNA, which was then transcribed in vitro in the presence of biotinylated dNTPs (Enzo Diognostics, Farmingdale, NY). Biotinylated target cRNA was fragmented and brought up in hybridization mix. Successful labeling of all of the samples (a minimum of 15 μg of IVT product per sample) was followed by the test array hybridizations. Test hybridizations were performed with the use of Affymetrix Test3 arrays to ensure quality of the biotinylated target. Test3 arrays contain probe sets corresponding to commonly expressed genes from the human, mouse, rat, and yeast genomes, along with prokaryotic control genes. Successful test hybridizations indicating efficient cRNA amplification and strong target hybridization activity were followed by actual experimental hybridizations. Hybridizations were followed by binding of the streptavidin-conjugated fluorescent marker. Detection of the bound probe was achieved by laser excitation of the fluorescent marker and scanning of the resultant emission spectra using a confocal laser microscope (ProbeArray Scan; Agilent Technologies, Palo Alto, CA). Data acquisition was performed using Affymetrix Microarray Suite 5.0. Labeling and hybridization quality was controlled through addition of exogenous labeling and hybridization spike controls. Collected hybridization images were subjected to quality control to remove arrays from analysis that failed to meet criteria both suggested by Affymetrix and developed internally in the laboratory. These quality requirements included low Q-noise (1 to 10), low background (less than a 100), sample-dependent percentage of probes detected as present (20 to 50% for mammals), 3′/5′ ratio of hybridization intensities for control probe sets of no more than 3, and hybridization efficiency defined by intensities detected for the CRE spike control probe set preferably higher than 2000 fluorescent units and minimal deviation of scaling factors for the whole set of arrays to be analyzed.
Statistical Analyses of Microarray Data
Background correction, data normalization, and statistical significance analysis were conducted with the use of commercially available software packages for microarray data analysis (ArrayAssist; Iobion, Inc., La Jolla, CA) and GeneCluster 2.0 (MIT, Boston, MA). Input CEL files were grouped for normalization and further analysis based on the particular pair-wise comparisons to be performed. No significant changes in expression profiles of replicated samples were observed (data not shown).
To examine differential gene expression between melanoma cells of low invasive behavior (OCM1a) with melanoma cells of high invasive behavior (M619, MUM2B), all grown in 2D culture conditions (Figure 1, comparison A), input Affymetrix CEL files were grouped as follows: OCM1a and M619, OCM1a and MUM2B, and M619 and MUM2B. Normalization and background correction of hybridization intensities was performed within each of the three comparison groups using RMA algorithm32 followed by a significance analysis test. Differentially expressed genes were identified based on P values calculated using a permutation test (GeneCluster 2.0, MIT) and a two-class t-test (ArrayAssist; Iobion, Inc.). No significant differences between the results of these two statistical methods were observed (data not shown). Supplementary Data (see http://ajp.amjpathol.org) files that accompany this report include only P values calculated using a two-class Student’s t-test.
We next investigated differential gene expression in each of the three uveal melanoma cell lines under paired culture conditions: 2D versus 3D culture conditions (Figure 1, comparisons B and C). CEL files were divided into groups based on the cell line designation (OCM1a, M619, or MUM2B) and culture condition for each cell line, using identical normalization and background correction strategies and statistical methods. Differentially expressed genes were annotated with the use of Affymetrix NetAffx analysis center (http://www.affymetrix.com/analysis/index.affx). Additional functional gene annota-tion defined by Locus Link was obtained with the use of DAVID—Gene Ontology supported annotation tool (http://david.niaid.nih.gov/david/version2/index.htm). Text search with the use of terms of interest was performed to identify and group genes associated with corresponding functions.
Real-Time RT-PCR
Real-time RT-PCR analysis was used to confirm the differences in expression levels obtained from microarray analysis on five representative transcripts. The same RNA samples that were subjected to microarray analysis were used for real-time PCR analysis. The following transcripts were analyzed from both 2D and 3D conditions: GDF15, CDKN1A (p21WAF1/CIP1), CD44, and ATF3. One-step RT-PCR was performed using the Quantitect SYBR Green RT-PCR kit (Qiagen) according to the manufacturer’s instructions on a Bio-Rad iCycler iQ instrument (Bio-Rad, Hercules, CA). Each 50-μl reaction contained 1× buffer with dNTPs, reverse transcriptase, HotStarTaq DNA polymerase, 2.5 mmol/L MgCl2, 20 pmol of each primer, and 10 nmol/L fluorescein dye. A melt-curve analysis after PCR and agarose gel electrophoresis was used to ensure amplification specificity. Primers were designed and optimized using Oligo v6.63 software (Molecular Biology Insights Inc., Cascade, CO). Primer sequences and conditions for real-time PCR are shown in Table 2. Each sample was run in duplicate. Higher Ct values indicated lower levels of expression and vice versa. An unpaired Student’s t-test was performed to analyze differences in mean Ct values (Prism software version 4; GraphPad, San Diego, CA).
TABLE 2.
Primer Sequences and Reaction Conditions for Quantitative Real-Time RT-PCR
| Transcript | GenBank Accession No. | Primer sequences | Reaction conditions |
|---|---|---|---|
| GDF15 | NM004864 | Forward:5′-GTGTTGCTGGTGCTCTCGTG-3′ | RT: 52°C, 30 minutes; 95°C 13:30 minutes PCR: 95°C 35 seconds, 60°C 35 seconds; 40 cycles of PCR, melt curve |
| ATF3 | NM001674 | Reverse: 5′-GAATCTGGAGTCTTCGGAGTG-3′ | RT: 50°C, 30 minutes; 95°C 13:30 minutes; PCR: 95°C 35 seconds, 60°C 40 seconds; 40 cycles of PCR, melt curve |
| Forward: 5′-CTGCCACCGGATGTCCTCT-3′ | |||
| CDKN1A p21WAF1/CIP1 | NM078467 | Reverse: 5′-CTCGTCGCCTCTTTTTCCTTTC-3′ | RT: 52°C, 30 minutes; 95°C 13:30 minutes; PCR: 95°C 35 seconds, 62°C 35 seconds; 40 cycles of PCR, melt curve |
| Forward: 5′-ACTGTGATGCGCTAATGGC-3′ | |||
| CD44 | AJ251595 | Reverse: 5′-CCCGTGGGAAGGTAGAGCTTG-3′ | RT: 52°C, 35 minutes; 95°C 13:30 minutes; PCR: 95°C 35 seconds, 65°C 35 seconds; annealing temperature <1°C after cycle 10, every 10 cycles; 42 cycles of PCR, melt curve |
| Forward: 5′-AGCGGACCCCAGCCTCTG-3′ | |||
| Reverse: 5′-GGAGGGCGCGGGCGAAAG-3′ |
High-Density 3D Halo Cultures and Very Low-Density 3D Cultures
After comparing gene expression in MUM2B cells under paired culture conditions (2D cultures on plastic and 3D cultures on Matrigel), we compared the morphology and migration of MUM2B metastatic malignant uveal melanoma cells placed in a constrained locations in plastic wells and Matrigel. To develop compact, well demarcated, high-density cultures on plastic, we placed a sterilized 3-mm cloning ring (Corning, Corning, NY) in the center of each of six 35-mm-diameter well plates. Cloning rings were filled with MUM2B cells at a concentration of 0.5 million cells. Cultures were incubated for 2 to 3 hours, allowing for the cells to securely adhere to the bottom of the wells, followed by aspiration of extra medium and addition of fresh complete medium in the cloning rings. The wells were washed with the media once to remove cells that failed to attach. This process left a dense yet compact and well-demarcated plug of cells in the center of the wells. Supplemented EMEM was added to each well and the six-well plate was incubated at 37°C with 5% CO2.
To develop compact, well-constrained, high-density cultures on Matrigel for comparison with MUM2B and M619 cells on plastic, we allowed these cells to form vasculogenic mimicry patterns on 3-mm-diameter Matrigel disks and grafted the disks onto a large recipient Matrigel bed. Grafts were fashioned by pouring Matrigel at a thickness of 0.2 mm onto a 60-mm culture dish, allowing the Matrigel to polymerize for 1 hour at 37°C. Sterile coverslips were used to transfer each of the six seeded Matrigel grafts to the center of a 35-mm well that was coated by a 0.2-mm-thick layer of polymerized Matrigel. The well plate housing each of the six grafts was placed in an incubator for 5 minutes at 37°C to anneal the grafts to the underlying Matrigel so that the grafts would not slip when adding or changing media. The demarcated edge between the cells in the graft and the surrounding virgin Matrigel was clearly visualized, allowing for the serial documentation of invasion from the graft into the peripheral matrix. Each well was filled with 3 ml of filtered supplemented EMEM, and the plate was returned to the incubator (37°C, with 5% CO2), and the cultures were observed by phase microscopy.
Because it is difficult to observe the behavior of individual cells in mass culture conditions, M619 and MUM2B cells that had formed patterns in Matrigel were harvested and seeded at low density (5 cells per 5× field) and observed by phase microscopy throughout a period of 10 days. Cultures were then seeded with MUM2B or M619 cells in a very low-cell density suspension in EMEM supplemented as described above. The low-density cultures were then placed back in incubation for 30 minutes to allow for cell adhesion followed by the addition of 12 ml of supplemented EMEM to each dish, and the culture dishes were returned to the incubator.
Matrigel Dot Cultures
Dots of Matrigel, 1 mm in diameter, were placed on glass coverslips in a 20 × 20 grid pattern spaced 2.5 mm apart using a standard mechanical micropipetter. Coverslips were placed in an incubator at 37°C for 24 hours to allow the Matrigel dots to polymerize. Dot cultures were also used to observe changes in morphology of MUM2B cells as they began to form vasculogenic mimicry patterns. By seeding 5000 MUM2B cells over the grid, it was possible to compare cell shape, nuclear morphology, and nucleolar morphology—hallmarks of the Callender classification of uveal melanoma cells33—between melanoma cells forming vasculogenic mimicry patterns on Matrigel and wild-type MUM2B cells not in contact with Matrigel. MUM2B cells were observed for 5 days, at which time vasculogenic mimicry patterns began to form at the periphery of Matrigel dots.
Changes in MUM2B morphology were documented by phase contrast microscopy and by laser-scanning confocal microscopy after preparations were stained with a polyclonal antibody to fibrillarin, a nucleolar protein. Cultures were fixed in 4% paraformaldehyde for 5 minutes, permeabilized with 0.5% Triton X-100 for 5 minutes, and blocked for 1 hour with 5% goat serum/0.2% Tween, followed by staining with polyclonal anti-fibrillarin (ab5821; Abcam, Cambridge, MA) at a dilution of 1:50 for 1 hour in 37°C. Alexa Fluor 546 goat anti-rabbit IgG (A11010; Molecular Probes, Eugene, OR) was used as secondary antibody at a dilution of 1:200 for 30 minutes in 37°C, followed by mounting on coverslide using Vectashield mounting medium containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA). Cell preparations were examined by laser-scanning confocal microscopy (Zeiss LSM; Carl Zeiss MicroImaging, Thornwood, NY). Cell shape was assessed by differential interference contrast (DIC) with ×25 and ×63 water immersion objectives. Nuclear and nucleolar size and shape were examined on the same preparations by applying the Argon laser for red fluorescence (Alexa 546, 546 nm wavelength) and UV laser (for blue fluorescent DAPI, 405 nm) using ×25 and ×63 objectives at 2-μm-thick intervals. Images were processed using LSM Software Release 2.5 (Carl Zeiss).
Human Tissue Studies
To study the morphology of uveal melanoma cells adjacent to looping vasculogenic mimicry patterns in human tissue samples, we studied tumors that had been shown by us to contain laminin-positive vasculogenic mimicry patterns but not fibrovascular septa.13 Unstained sections from amelanotic melanomas in this set were stained for S100 protein (polyclonal rabbit antibody at a dilution titer of 1:200; DAKO, Carpinteria, CA; conjugated goat anti-rabbit IgG at a dilution titer of 1:200), labeled with diaminobenzidine (DAKO), and counterstained with Harris hematoxylin (Richard-Allan, Kalamazoo, MI). We have shown previously that S100 protein stains most tumor cells in primary uveal melanomas in contrast to other melanoma markers such as HMB45 and MELAN-A that tend to stain subpopulations of tumor cells.10
A progression uveal melanoma tissue microarray containing 190 cores from 91 blocks of primary uveal melanomas was stained with Ki67 to study the spatial relationships between tumor cell proliferation and vasculogenic mimicry patterns. It is known that both increased proliferation and the detection of vasculogenic mimicry patterns detected from histological human material are independently associated with death from metastatic uveal melanoma.34 In the construction of this microarray, one to three pairs of cores were obtained from different areas of the tumor to sample for different tumor locations (ciliary body versus choroid), cell type by the modified Callender classification (epithelioid versus spindle),33 and the presence or absence of looping vasculogenic mimicry patterns. Thus, for each core, the location, cell type, and presence of vasculogenic mimicry patterns were known. Tumor location, cell type, and vasculogenic mimicry patterns are independently associated with death from metastatic uveal melanoma.34 Donor cores measuring 0.6 mm in diameter were placed into the recipient block using a tissue microarrayer (Beecher Instruments, Inc., Sun Prairie, WI) coupled to a semiautomated instrument.35
Paraffin sections of the tissue microarray slides were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol. All sections underwent antigen retrieval for 30 minutes using citrate target retrieval (DAKO). Ki-67 clone (MIB-1, diluted 1:100; DAKO) was used as primary antibody for 60 minutes. LSAB+System-AP (DAKO) was used as the detection system with Permanent Red (DAKO) as chromogen. Slides were air-dried, counterstained with hematoxylin, and mounted in Permount (Biomeda, Foster City, CA).
Tissue microarray analysis was performed using the BioGenex quantitation system (BioGenex, San Ramon, CA). Test slides of pigmented uveal melanoma tissue stained with Ki67 indicated that the BioGenex system discriminated the permanent red chromogen from melanin at high intensity, obviating the need to bleach tissue sections with possible loss of cores. Each core was scored for the percentage of positive population (the tissue area divided by the population area).
Results
Comparing Gene Expression in Poorly Invasive and Highly Invasive Melanoma Cells in 2D Culture Conditions
In the comparison of differential gene expression between poorly invasive OCM1a primary uveal melanoma cells and highly invasive M619 primary MUM2B metastatic uveal melanoma cells (Figure 1, comparison A), three separate lists of differential genes with a P value cutoff of 0.001 were generated: 7680 genes for the OCM1a versus M619 comparison, 7632 genes for the OCM1a versus MUM2B comparison, and 2204 genes for the M619 versus MUM2B comparison. The latter comparison represents the population of differentially expressed genes with inconsistent directionality between the two highly invasive cell lines. A comparison between OCM1a and M619 and between OCM1a and MUM2B yielded 6302 genes with similar differential expression changes (ie, up-regulated or down-regulated in both comparisons). From this list, the 2204 genes that were not up-regulated or down-regulated in both M619 and MUM2B were subtracted to achieve a final list of 5331 genes that appears in Supplementary Data Table 1 (see http://ajp.amjpathol.org).
Many of the genes up-regulated in the two invasive cell lines compared with OCM1a were identical to genes up-regulated in the highly invasive MUM2B line compared with the poorly invasive MUM2C line as reported by us and our colleagues previously1,36: keratin 7 (KRT7, 94-fold increase), plasminogen activator kinase (PLAU, 49-fold increase), Rho GDP dissociation inhibitor (ARHGDIB, 41-fold increase), tissue factor pathway inhibitor 1 (TFPI, 39-fold increase) and 2 (TFPI2, 36-fold increase), VE-cadherin (CDH5, 38-fold increase), TIE-1 (TIE, 34-fold increase), membrane metalloendopeptidase (MME/CALLA, 28-fold increase), connective tissue growth factor (CTGF, 27-fold increase), and paired box gene 8 (PAX8, 20-fold increase), with P values less than 10−4 for all genes. Genes up-regulated in the poorly invasive uveal melanoma cell line compared with the two highly invasive uveal melanoma cell lines also mirrored our previous results1,36: melan-A (MLANA, 102-fold increase), glycoprotein M6B (GPM6B, 39-fold increase), and S100B (17-fold increase), with P values less than 10−4 for all genes.
Gene Expression in Highly Invasive Melanoma Cells in 3D Culture Conditions after Forming Vasculogenic Mimicry Patterns
Tables of genes differentially expressed in the 3D culture condition compared with 2D were generated separately for each of the cell lines: OCM1a, M619, and MUM2B (data not shown). Next, differential transcripts with the same direction of the change and a P value <0.001 in both of the highly invasive cell lines (MUM2 and M619) were identified. From this list, we subtracted genes differentially expressed in poorly invasive OCM1a cells in 3D versus 2D conditions with P < 0.001 (OCM1a cells did not form vasculogenic mimicry patterns in 3D culture conditions (Figure 1). The resulting list of 990 genes is provided in Supplementary Data Table 2 (see http://ajp.amjpathol.org).
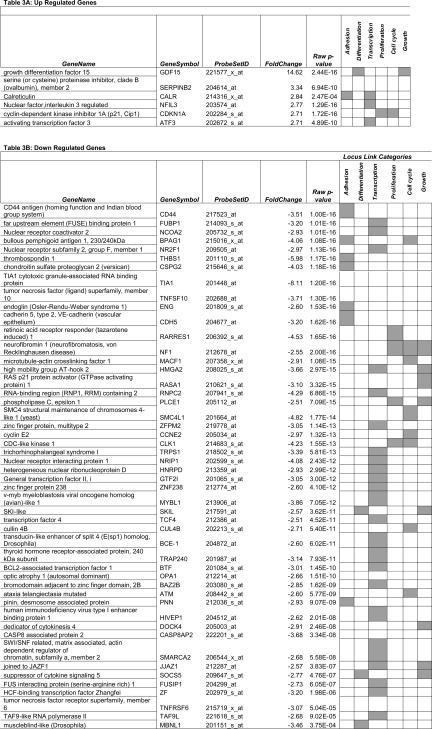
We conjectured that the formation of vasculogenic mimicry patterns by highly invasive cells in 3D cultures would facilitate metastasis and therefore focused on genes within the following Locus Link-defined categories: adhesion, differentiation, transcription, proliferation, cell cycle, and growth. Genes represented in any one of these categories with a fold increase of 2.5 up-regulation or down-regulation in 3D culture conditions are listed in Table 3. Unexpectedly, genes associated with cell cycle arrest (CDKN1A/p21) and differentiation (GDF15, ATF3) were up-regulated (Table 3), whereas genes associated with the promotion of invasive and metastatic behavior such as CD44, cyclin E2 (CCNE2), thrombospondin 1 (THBS1), and chondroitin sulfate proteoglycan (versican; CSPG2) were down-regulated in 3D culture conditions (Table 3).
TABLE 3.
Selected Transcripts Differentially Expressed in Vasculogenic Mimicry Pattern-Forming Cells
Lists genes up-regulated and down-regulated in three-dimensional culture conditions compared to flat culture conditions with a fold change greater than 2.5 times. Genes were annotated based on the locus link defined functional categories: adhesion, differentiation, transcription, proliferation, cell cycle, and growth. Genes annotated with corresponding locus link terms are indicated by gray shading. Complete listing of genes is provided in Supplementary Data Tables 1 and 2 (see http://ajp.amjpathol.org).
Real-Time RT-PCR
Relative expression levels for three genes up-regulated in 3D (GDF15, ATF3, and CDKN1A) and one gene down-regulated in 3D (CD44) by gene expression array studies were quantified based on the Ct values for each transcript. For the four transcripts analyzed, real-time PCR confirmed the direction of the difference in expression levels observed by microarray analysis (Table 4).
TABLE 4.
Validation of Selected Differential Genes by Real-Time RT-PCR
| Transcript | Cell line | Flat | Three-dimensional | t-test |
|---|---|---|---|---|
| GDF15 | MUM2b | 24.2 (SD ± 0.23, n = 6) | 20.1 (SD ± 0.62, n = 5) | P < 0.0001* |
| M619 | 25.98 (SD ± 1.018, n = 4) | 20.2 (n = 1) | ||
| OCM-1 | 19.4 (SD ± 0.23, n = 4) | 20.7 (SD ± 1.089, n = 4) | P = 0.06 | |
| ATF3 | MUM2b | 23.42 (SD ± 0.53, n = 6) | 21.72 (SD ± 0.32, n = 5) | P = 0.0014* |
| M619 | 25.0 (SD ± 0.21, n = 4) | 20.8 (n = 1) | ||
| OCM-1 | 21.15 (SD ± 0.28, n = 4) | 23.15 (SD ± 1.1, n = 4) | P = 0.0184* | |
| CDKN1A | MUM2b | 21.52 (SD ± 0.19, n = 6) | 20.72 (SD ± 0.57, n = 5) | P < 0.01* |
| M619 | 26.53 (SD ± 0.12, n = 4) | 23.7 (n = 1) | ||
| OCM-1 | 23.88 (SD ± 0.20, n = 4) | 25.83 (SD ± 1.49, n = 4) | P = 0.04* | |
| CD44 | MUM2b | 17.4 (SD ± 0.49, n = 6) | 18.28 (SD ± 0.31, n = 5) | P < 0.0079* |
| M619 | 18.33 (SD ± 0.22, n = 4) | 19.40 (n = 1) | ||
| OCM-1 | 18.53 (SD ± 0.22, n = 4) | 19.38 (SD ± 0.47, n = 4) | P = 0.0172* |
Higher Ctvalues indicate lower expression levels and vice versa.
Student’s t-test, P value significant (P < 0.05).
The in Vitro Behavior of Melanoma Cells Forming Vasculogenic Mimicry patterns
To test if the biological behavior of highly invasive uveal melanoma cells in 3D culture conditions was consistent with the expression of cell cycle arrest genes and differentiation genes and the down-regulation of genes associated with proliferation and invasion, we developed a high-density 3D halo culture assay. This assay permitted us to observe the behavior of highly invasive uveal melanoma cells constrained to a defined circular area throughout defined periods of time.
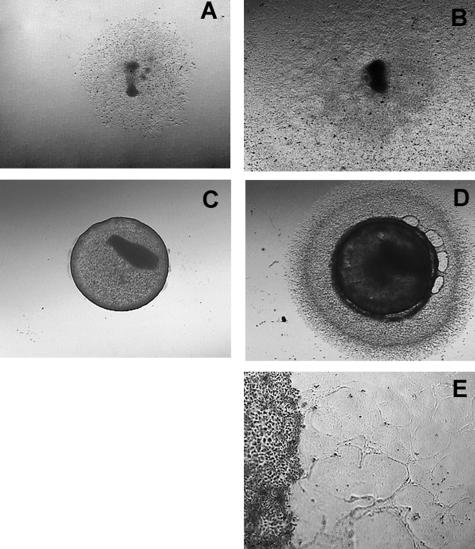
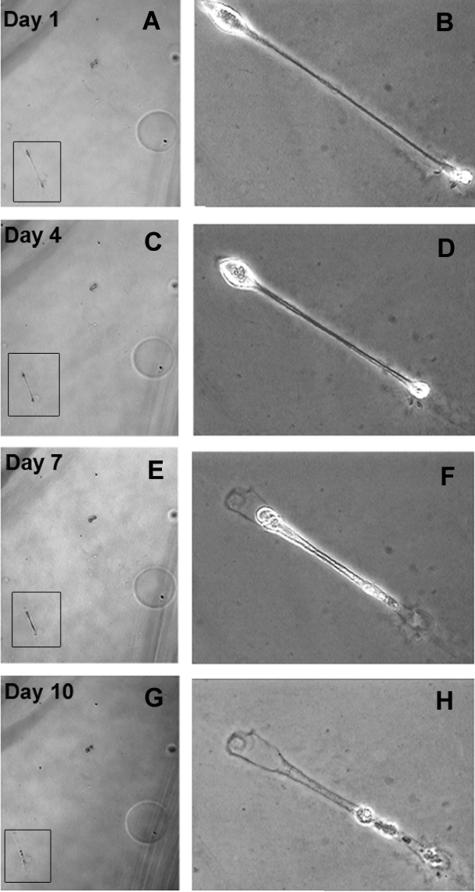
Within 10 days, highly invasive M619 and MUM2B cells that were initially constrained in 2D culture conditions became completely confluent to the edge of the well (Figure 2, A and B). By contrast, M619 or MUM2B cells that had formed vasculogenic mimicry patterns on donor Matrigel grafts invaded only short distances into the recipient Matrigel beds, forming fine reticular patterns. Throughout the 10 days, these cells retracted back toward the graft, leaving track-like impressions on the Matrigel (Figure 2, C–E). To observe more closely the changing morphology of pattern-forming cells, we seeded five MUM2B cells on Matrigel and observed them for 10 days. As in the high-density 3D halo cultures (Figure 2) the cells at low density elongated, did not divide, and retracted (Figure 3, A–H).
FIGURE 2.
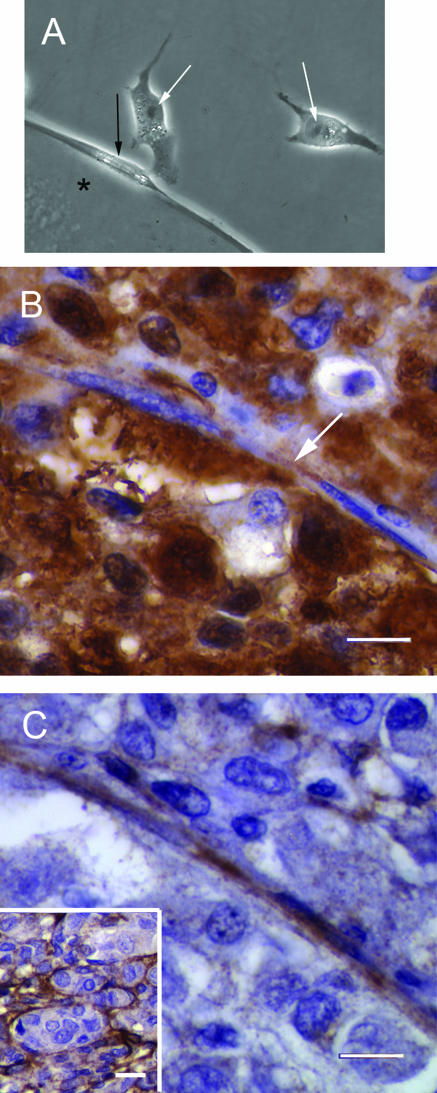
Halo culture system. A: MUM2B metastatic melanoma cells were constrained on plastic culture dishes by a 3-mm-diameter cloning ring. B: Ten days after initial plating, the MUM2B cells did not form vasculogenic mimicry patterns and expanded from the initial constrained area to fill the well. C: MUM2B cells were seeded on a raft of Matrigel until vasculogenic mimicry patterns formed. A 3-mm-diameter punch was taken from the seeded raft and grafted onto a virgin recipient bed of Matrigel. D: Ten days after the graft in C was placed, MUM2B melanoma cells migrated only a short distance onto the recipient Matrigel bed. E: Higher magnification of the edge of the graft illustrated in D. The melanoma cells in the recipient bed are elongated in contrast to the normal epithelioid shape of MUM2B cells. The MUM2B cells formed reticulated looping patterns adjacent to the graft. Note that the cells formed patterns and retracted back toward the graft: impressions of the vacant patterns are visible in the top right area of the picture, just below and to the left of the label. Original magnifications: ×50 (A–D); ×200 (E).
FIGURE 3.
A–H: Low-density cultures of MUM2B cells in Matrigel followed throughout 10 days. The cell of interest appears in boxes at low magnification in A, C, E, and G and at higher magnification in B, D, F, and H. Throughout the 10 days, the elongated MUM2B cell retracted, leaving behind a footprint in the Matrigel. Original magnifications: ×50 (A, C, E, G); ×100 (B, D, F, H).
MUM2B cells were placed in plastic dishes containing discrete dots of Matrigel to compare the morphology of melanoma cells on and off Matrigel. At 5 days after seeding, MUM2B cells that did not contact Matrigel assumed a spindle B to epithelioid cell morphology, and a prominent nucleolus was detected in each cell. At the same time, MUM2B cells at the edge of the Matrigel dots began to form vasculogenic mimicry patterns at the same time point. By DIC (Figure 4) and phase contrast microscopy (Figure 5), pattern-forming MUM2B cells were exceptionally thin and elongated. Minute nucleoli were detected in spindled MUM2B melanoma cells forming patterns only after staining with an antibody to fibrillarin: these small nucleoli oriented along the long axis of the nucleus (Figure 4). Thus, as vasculogenic mimicry patterns began to form, MUM2B cells reverted to a morphology identical to indolent Callender spindle A melanoma cells27 (Figure 5A). By 10 days, almost all cells on the Matrigel dot that had formed vasculogenic mimicry patterns had assumed a Callender spindle A morphology.
FIGURE 4.
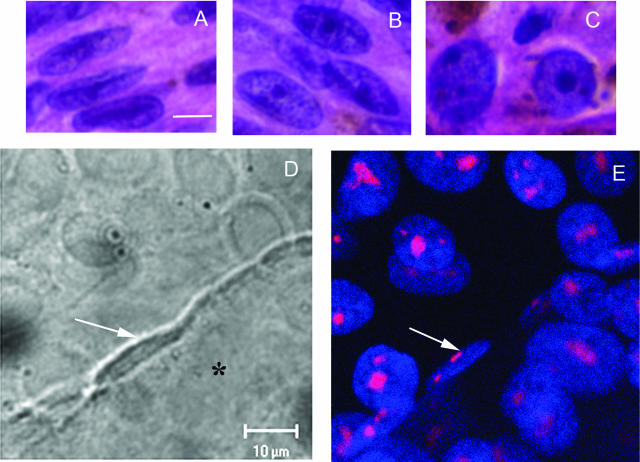
A–C: Melanoma cells from primary uveal melanoma tissue. A: Spindle A cells, featuring a longitudinal fold through the long axis of the nuclei and inconspicuous nucleoli. B: Spindle B cells, featuring a more open nucleus than seen in spindle A cells and prominent nucleoli. C: Epithelioid cells, featuring round cytoplasm, large nuclei, and exceptionally large pleomorphic nucleoli. D: DIC micrograph of a dot culture. Epithelioid MUM2B melanoma cells were seeded dots of Matrigel arranged in a grid on a glass coverslip. At day 5, MUM2B cells in contact with the edge of the dot of Matrigel (asterisk) began to form vasculogenic mimicry patterns. Note the elongated shape and the longitudinal fold through the long axis of the nucleus (white arrow). In contrast to MUM2B cells on plastic that feature a round nucleus with prominent nucleoli, the nucleolus of the spindled MUM2B melanoma cells is barely detectable by DIC microscopy. E: Same field as in D after staining for fibrillarin and DAPI counterstaining. The nucleoli of the spindled MUM2B cells on the edge of the Matrigel dot are extremely small (white arrow) compared with epithelioid cells on the coverslip. Epithelioid cells on the Matrigel dot (to the right of the spindled cell denoted by the arrow) formed vasculogenic mimicry patterns between days 5 and 10 and also assumed a spindled morphology. A–C: H&E. Scale bars = 10 μm. D: DIC microscopy captured through laser-scanning confocal microscope. E: Laser scanning confocal microscopy, section plane thickness = 2 μm.
FIGURE 5.
Comparing morphology of MUM2B cells in vitro with human tissue samples. A: Three MUM2B melanoma cells are illustrated. Two epithelioid MUM2B cells (white arrows) containing prominent nucleoli are positioned on plastic. The spindled MUM2B cell (black arrow), adherent to the dot of Matrigel (asterisk), lacks a prominent nucleolus and features a longitudinal nuclear fold found in many spindle A melanoma cells in vivo. B: Primary amelanotic uveal melanoma stained with antibody to S100 protein. A segment of a looping vasculogenic mimicry pattern bisects the figure. A spindled cell with condensed chromatin lacking a nucleolus is adherent to the pattern. The cell is identifiable as a melanoma cell by the long, slender S100 protein-positive process that extends from the nucleus (arrow). C: Section stained for laminin adjacent to the section illustrated in B. The thin spindled melanoma cells illustrated in J are adherent to the edge of the laminin-positive looping pattern. Inset illustrates laminin-positive vasculogenic mimicry patterns in the same primary uveal melanoma illustrated in B and C. B: S100 protein-hematoxylin; C: laminin-hematoxylin; inset to C stained for laminin. Scale bars = 100 (B, C); 25 μm (C, inset). Original magnification, ×630 (A).
Tissue Immunohistochemistry and Tissue Microarray Study of Ki67 Expression
In tissue sections of primary human uveal melanoma containing vasculogenic mimicry patterns but not fibrovascular septa, S100-positive tumor cells in contact with the looping matrix patterns had a spindled morphology, and the nucleus did not contain prominent nucleoli (Figure 5, B and C), typical of Callender spindle A cells. Cells spatially removed from the patterns tended to feature prominent nucleoli, characteristic of spindle B and epithelioid cells. Epithelioid cell morphology27,28,33 and the increasing size of the nucleolus37,38 in cells from histological sections of primary uveal melanomas are features associated with death from metastatic melanoma.
In the tissue microarray analysis, there was no association between the percentage of cells in the core staining for Ki67 and cell type (mean percent Ki67-positive cells in cores without epithelioid cells = 11.08; mean percent Ki67-positive with epithelioid cells present = 8.583, t = 1.64, P = 0.1029). There was also no association between the percentage of Ki67-positive cells and the location within the eye from which the cores were extracted (mean percent Ki67-positive cells in tumor tissue within the choroid = 9.893; mean percent Ki67 cells in tumor tissue involving the ciliary body = 9.267, t = 0.32, P = 0.7481). However, when vasculogenic mimicry patterns were not present, the mean percentage of cells in the core positive for Ki67 was 11.568, and when vasculogenic mimicry patterns were present, the mean percentage of Ki67-positive cells in the core was only 5.231 (t = 4.71, P < 0.0001). The Cochran-Mantel-Haenszel statistic was applied to the analysis of the percentage of Ki67-positive cells within cores stratified by the first quartile (Ki67 ≤ 3, Ki67 > 3): there is a strong association between the percentage of Ki67-positive cells below 3% and the presence of vasculogenic mimicry patterns (P = 0.0003), even after controlling for cell type and location.
Discussion
Based on the strong association between the presence of vasculogenic mimicry of the patterned matrix type in histological sections of uveal melanoma and death from metastatic melanoma,16–21 it was reasonable to predict that gene expression in pattern-forming melanoma cells would reflect a molecular signature consistent with an aggressive metastatic phenotype. However, we discovered that genes associated with a metastatic phenotype such as THBS1 (thrombospondin 1), CSPG2 (versican), CD44, and CCNE2 (cyclin E2) are down-regulated in pattern-forming cells in 3D culture conditions. By contrast genes associated with suppression of proliferation are up-regulated in pattern-forming cells: there is a nearly 15-fold increase in the expression of GDF15 and a 2.7-fold increase in CDKN1A (p21) in highly invasive pattern-forming melanoma cells compared with the same cells in 2D culture conditions. Although both GDF15 and p21 are downstream targets of p53,39 we detected no differential expression of p53 between poorly and highly invasive melanoma cells, and no difference in expression between highly invasive cells in flat or 3D culture conditions. In melanoma cells, there appears to be no predictable relationship between p21 expression and p53 expression, suggesting that p53-independent pathways may be important for the regulation of p21 in these cells.40 It is remarkable that up-regulation of GDF15 and p21 was induced only by changing the ECM environment.
Additional observations in the gene expression profiles suggest that pattern-forming melanoma cells in 3D culture conditions may have diminished rather than an enhanced metastatic molecular signature. For example, two genes up-regulated in 3D pattern-forming culture conditions down-regulate transcription: NFIL3 (nuclear factor interleukin 3 regulated)41 and ATF3 (activating transcription factor 3).42 Additionally, the up-regulation of SERPINB2 (also known as plasminogen activator inhibitor 2, PAI-2) in 3D pattern-forming conditions has been associated with decreased metastatic potential. A metastatic human melanoma cell line that produces urokinase-type plasminogen activator was stably transfected with cDNA encoding human plasminogen activator inhibitor 2 (PAI-2), and PAI-2-transfected cell lines produced significantly fewer or no metastases.43 The down-regulation of chondroitin sulfate proteoglycan (versican; CSPG1) has been associated previously with melanocytic differentiation.44
Highly invasive melanoma cells that have generated vasculogenic mimicry patterns have only a limited capacity for invasion and proliferation (Figures 2 and 3). The changes in gene expression and functional behavior that accompany pattern formation are also associated with profound alternations in cell shape. In 2D nonpattern-forming conditions, highly invasive melanoma cells featured either a spindle B or epithelioid morphology with prominent nucleoli. By contrast, pattern-forming MUM2B cells on Matrigel assumed a morphology identical to Callender spindle A cells (Figures 4 and 5).
Studies of human tissue samples also confirmed a reduction in proliferative activity in the vicinity of vasculogenic mimicry patterns: the percentage of Ki67-positive cells in vasculogenic mimicry patterns was more than 50% less than tissue not containing these patterns. Furthermore, melanoma cells within vasculogenic mimicry patterns in histological samples of human tumor specimens assumed a spindle A morphology identical to pattern-forming cells identified in vitro (Figures 4 and 5). These findings are consistent with ultrastructural localization of thin spindled melanoma cells within the electron-dense matrix of vasculogenic mimicry patterns.4
It has been known that cellular morphology is associated with outcome in uveal melanomas for more than 70 years.28,29,33 Uveal melanomas containing spindle B or epithelioid cells tend to follow an aggressive clinical course whereas tumors composed exclusively of spindle A cells tend to behave as nevi.27 The pathologist’s classification of cell type depends on not only the shape of the cell but also the size of the nucleolus: epithelioid cells have larger and more pleomorphic nucleoli than spindle B cells, and nucleoli are barely perceptible in spindle A cells. Cytomorphometric studies indicate that the larger the nucleolus in uveal melanoma cells, the more aggressive the clinical course.38,45 The decrease in the size of nucleoli in pattern-forming MUM2B and M619 melanoma cells and in cells in human tissue within vasculogenic mimicry patterns is consistent with gene expression data (up-regulation of NFIL3 and ATF3), suggesting a decrease in transcription in pattern-forming cells.
The in vitro generation of vasculogenic mimicry patterns requires the immersion of highly invasive melanoma cells in an ECM environment rich in laminin. The reversion of epithelioid and spindle B melanoma cells to the nevus-like spindle A morphology that accompanies the generation of vasculogenic mimicry patterns invites a comparison to the in vitro reversion of breast cancer cells to a benign phenotype including the formation of polarized acini. The breast reversion model described by Bissell and associates46–50 requires not only exposure to a laminin-rich ECM environment but also exposure to signaling inhibitors. That highly invasive uveal melanoma cells revert to a nevus-like phenotype on exposure to a 3D microenvironment rich in laminin without the addition of chemically mediated signaling inhibition is remarkable and consistent with the effects of a mechanical (not chemical) signaling pathway mediated through the cytoskeleton that was recently demonstrated by Maniotis and colleagues26 to effect global changes in chromatin organization in a variety of cancer cells, including melanoma and breast cancer, on contact with laminin. Moreover, this reversion may well be yet another manifestation of tumor cell plasticity in response to microenvironmental cues. Recently, Kulesa and colleagues51 reported that adult human metastatic melanoma cells responded to the microenvironmental milieu of the chick embryo by reverting to neural crest-like morphologies and populating host structures derived from neural crest.
Although highly invasive melanoma cells may co-opt ECM elements in the tumor in vivo, these cells are capable of producing the ECM microenvironment required for the generation of vasculogenic mimicry patterns: highly invasive uveal melanoma cells generate laminin,9 collagen IV,11 collagen VI,12 and fibronectin.13 Therefore, after highly invasive melanoma cells elaborate the ECM microenvironment, the tumor cell-generated microenvironment is capable of inducing the development of vasculogenic mimicry patterns. As the highly invasive tumor cells form these patterns, their shapes change to the morphology (spindle A) associated with indolent behavior, proliferation diminishes, and the invasive capacity of these cells weakens significantly.
We and others1,4,5,52,53 have demonstrated previously that vasculogenic mimicry patterns provide tumor perfusion pathways (the “fluid-conducting meshwork”5) outside of the endothelial cell-lined microcirculation. We now propose that vasculogenic mimicry of the patterned matrix type represents the development of a tumor biofilm. Just as multicellular colonies of microorganisms generate a polysaccharide-rich matrix (biofilm) that renders them relatively resistant to antibiotics, metals, radiation, and extreme changes in oxygen tension, pH, and heat compared with planktonic organisms,54 highly invasive tumor cells produce an ECM rich in laminin and other glycosylated ECM proteins. As these patterns form and tumor cells are embedded within them, highly invasive uveal melanoma cells revert to an indolent spindle A morphology with significantly diminished invasive potential. Moreover, as the tissue microarray component of this study indicates, the growth fraction of tumor cells in the vicinity of vasculogenic mimicry patterns decreases, thus providing at least one explanation for the relative resistance of uveal melanomas to some conventional chemotherapeutic strategies targeting the cell cycle. Moreover, the presence of vasculogenic mimicry patterns in uveal melanomas and the accompanying reversion of cell type from the highly aggressive spindle B and epithelioid cells to more indolent spindle A cells may also provide a partial explanation for the variable response of these tumors to radiation. It is well known from clinical studies that patients whose tumors respond immediately and rapidly to brachytherapy are significantly more likely to die of metastatic melanoma than patients whose tumors respond much more slowly.55 Therefore, relative successes in treating planktonic malignancies such as the leukemias compared with solid malignancies may be related in part to the generation of vasculogenic mimicry patterns now documented3 in a wide variety of solid cancers.
It is possible that in some tumors, highly invasive melanoma cells do not generate vasculogenic mimicry patterns or the patterns form only focally (vasculogenic mimicry patterns tend to form in discrete zones, typically at the tumor periphery at the interface between the tumor and the surrounding stroma56 and only seldom encompass the entire tumor57). Therefore, highly invasive tumor cells spatially remote from vasculogenic mimicry patterning may contribute to metastatic behavior. In some patients, generation of vasculogenic mimicry patterns may contribute to complete dampening of metastatic behavior in the primary tumor. Finally, it is noteworthy that ∼10% of patients with uveal melanoma die of metastatic melanoma more than 15 years after treatment of the primary lesion.58 Uveal melanoma tends to disseminate first almost exclusively to the liver,59 and the formation of vasculogenic mimicry patterns in hepatic metastases60 may contribute to subclinical dormancy.
Dampening of the invasive properties of aggressive melanoma cells as they generate vasculogenic mimicry patterns demonstrated in this study seems to contradict associations between the histological detection of these patterns and death from metastatic melanoma. However, although patients seldom die of metastatic uveal melanoma unless vasculogenic mimicry patterns are present (consistent with the observation that poorly invasive melanoma cells do not generate vasculogenic mimicry patterns1,4), only ∼50% of uveal melanoma patients whose tumors contain vasculogenic mimicry patterns die of metastatic melanoma.16 Therefore, the histological detection of vasculogenic mimicry patterns may be a surrogate marker for the presence of invasive tumor cell populations somewhere in the tumor because the generation of these patterns requires the presence of an invasive tumor cell population.1,4
Supplementary Material
Acknowledgments
We thank Richard A. Domanick, Ph.D., and Andre Kajdacsy-Balla, M.D., Ph.D., for many helpful suggestions.
Footnotes
Address reprint requests to Robert Folberg, Department of Pathology, University of Illinois at Chicago, 840 S. Wood St., 110 CSN (MC 847), Chicago, IL 60612. E-mail: rfolberg@uic.edu.
Supported by the National Institutes of Health (grant EY10457).
Supplementary information for this article can be found on http://ajp.amjpathol.org.
References
- Maniotis AJ, Folberg R, Hess A, Seftor EA, Gardner LMG, Pe’er J, Trent JM, Meltzer PS, Hendrix MJC. Vascular channel formation by human melanoma cells in vivo and in vitro: vasculogenic mimicry. Am J Pathol. 1999;155:739–752. doi: 10.1016/S0002-9440(10)65173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folberg R, Hendrix MJ, Maniotis AJ. Vasculogenic mimicry and tumor angiogenesis. Am J Pathol. 2000;156:361–381. doi: 10.1016/S0002-9440(10)64739-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folberg R, Maniotis AJ. Vasculogenic mimicry. APMIS. 2004;112:508–525. doi: 10.1111/j.1600-0463.2004.apm11207-0810.x. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Chen X, Garcia C, DeChristopher PJ, Wu D, Pe’er J, Folberg R. Control of melanoma morphogenesis, endothelial survival, and perfusion by extracellular matrix. Lab Invest. 2002;82:1031–1043. doi: 10.1097/01.lab.0000024362.12721.67. [DOI] [PubMed] [Google Scholar]
- Clarijs R, Otte-Holler I, Ruiter DJ, de Waal RM. Presence of a fluid-conducting meshwork in xenografted cutaneous and primary human uveal melanoma. Invest Ophthalmol Vis Sci. 2002;43:912–918. [PubMed] [Google Scholar]
- Chen X, Ai Z, Rasmussen M, Bajcsy P, Auvil L, Welge M, Leach L, Vangveeravong S, Maniotis AJ, Folberg R. Three-dimensional reconstruction of extravascular matrix patterns and blood vessels in human uveal melanoma tissue: techniques and preliminary findings. Invest Ophthalmol Vis Sci. 2003;44:2834–2840. doi: 10.1167/iovs.02-1333. [DOI] [PubMed] [Google Scholar]
- Potgens AJG, van Altena MC, Lubsen NH, Ruiter DJ, de Waal RMW. Analysis of the tumor vasculature and metastatic behavior of xenografts of human melanoma cell lines transfected with vascular permeability factor. Am J Pathol. 1996;148:1203–1217. [PMC free article] [PubMed] [Google Scholar]
- Ruf W, Seftor EA, Petrovan RJ, Weiss RM, Gruman LM, Margaryan NV, Seftor REB, Miyagi Y, Hendrix MJC. Differential role of tissue factor pathway inhibitors 1 and 2 in melanoma vasculogenic mimicry. Cancer Res. 2003;63:5381–5389. [PubMed] [Google Scholar]
- Chen HB, Chen L, Zhung JK, Chow VW, Wu BQ, Wang ZH, Cheng SB, Chew EC. Expression of laminin in metastatic melanoma cell lines with different metastatic potential. Anticancer Res. 2001;21:505–508. [PubMed] [Google Scholar]
- Chen X, Maniotis AJ, Majumdar D, Pe’er J, Folberg R. Uveal melanoma cell staining for CD34 and assessment of tumor vascularity. Invest Ophthalmol Visual Sci. 2002;43:2533–2539. [PubMed] [Google Scholar]
- Hao XS, Sun BC, Zhang SW, Zhao XL. Correlation between the expression of collagen IV, VEGF and vasculogenic mimicry. Zhonghua Zhong Liu Za Zhi. 2003;25:524–526. [PubMed] [Google Scholar]
- Daniels KJ, Boldt HC, Martin JA, Gardner LM, Meyer M, Folberg R. Expression of type VI collagen in uveal melanoma: role in pattern formation and tumor progression. Lab Invest. 1996;75:55–66. [PubMed] [Google Scholar]
- Lin AY, Maniotis AJ, Valyi-Nagy K, Majumdar D, Setty S, Kadkol S, Leach L, Pe’er J, Folberg R. Distinguishing fibrovascular septa from vasculogenic mimicry patterns. Arch Pathol Lab Med. 2005;129:884–892. doi: 10.5858/2005-129-884-DFSFVM. [DOI] [PubMed] [Google Scholar]
- Smetsers TFCM, van de Westerlo EMA, ten Dam GB, Clarijs R, Versteeg E, van Geloof WL, Veerkamp JH, van Muijen GNP, van Kuppevelt TH. Localization and characterization of melanoma-associated glycosaminoglycans: differential expression of chondroitin and heparan sulfate epitopes in melanoma. Cancer Res. 2003;63:2965–2970. [PubMed] [Google Scholar]
- Folberg R, Pe’er J, Gruman LM, Woolson RF, Jeng G, Montague PR, Moninger TO, Yi H, Moore KC. The morphologic charateristics of tumor blood vessels as a marker of tumor progression in primary human uveal melanoma: a matched case-control study. Hum Pathol. 1992;23:1298–1305. doi: 10.1016/0046-8177(92)90299-i. [DOI] [PubMed] [Google Scholar]
- Folberg R, Rummelt V, Parys-Van Ginderdeuren R, Hwang T, Woolson RF, Pe’er J, Gruman LM. The prognostic value of tumor blood vessel morphology in primary uveal melanoma. Ophthalmology. 1993;100:1389–1398. doi: 10.1016/s0161-6420(93)31470-3. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Sakamoto M, Yoshikawa H, Hata Y, Ishibashi T, Ohnishi Y, Inomata H. Histologic findings and prognosis of uveal malignant melanoma in Japanese patients. Am J Ophthalmol. 1996;121:276–283. doi: 10.1016/s0002-9394(14)70275-2. [DOI] [PubMed] [Google Scholar]
- Schaling DF, van der Pol JP, Schlingemann RO, Parys-Van Ginderdeuren R, Jager MJ, Schaling DF. Rijksuniversiteit te Leiden: Leiden,; Vascular Density and Vascular Patterns in the Prognosis of Choroidal Melanoma. 1996:pp 43–54. [Google Scholar]
- McLean IW, Keefe KS, Burnier MN. Uveal melanoma: comparison of the prognostic value of fibrovascular loops, mean of the ten largest nucleoli, cell type and tumor size. Ophthalmology. 1997;104:777–780. doi: 10.1016/s0161-6420(97)30234-6. [DOI] [PubMed] [Google Scholar]
- Seregard S, Spangberg B, Juul C, Oskarsson M. Prognostic accuracy of the mean of the largest nucleoli, vascular patterns, and PC-10 in posterior uveal melanoma. Ophthalmology. 1998;105:485–491. doi: 10.1016/S0161-6420(98)93032-9. [DOI] [PubMed] [Google Scholar]
- Makitie T, Summanen P, Tarkkanen A, Kivela T. Microvascular density in predicting survival of patients with choroidal and ciliary body melanoma. Invest Ophthalmol Vis Sci. 1999;40:2471–2480. [PubMed] [Google Scholar]
- Warso MA, Maniotis AJ, Chen X, Majumdar D, Patel MK, Shilkaitis A, Gupta TK, Folberg R. Prognostic significance of periodic acid-Schiff-positive patterns in primary cutaneous melanoma. Clin Cancer Res. 2001;7:473–477. [PubMed] [Google Scholar]
- Thies A, Mangold U, Moll I, Schumacher U. PAS-positive loops and networks as a prognostic indicator in cutaneous malignant melanoma. J Pathol. 2001;195:537–542. doi: 10.1002/path.988. [DOI] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, Dougherty E, Wang E, Marincola F, Gooden C, Leuders J, Glatfelter A, Pollock P, Carpten J, Gillanders E, Leja D, Dietrich K, Beaudry C, Berens M, Alberts D, Sondak V, Hayward N, Trent J. Molecular classification of cutaneous melanoma by gene expression profiling. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Seftor EA, Meltzer PS, Kirschmann DA, Pe’er J, Maniotis AJ, Trent JM, Folberg R, Hendrix MJC. Molecular determinants of human uveal melanoma invasion and metastasis. Clin Exp Metastasis. 2002;19:233–246. doi: 10.1023/a:1015591624171. [DOI] [PubMed] [Google Scholar]
- Maniotis AJ, Valyi-Nagy K, Karavitis J, Moses J, Boddipali V, Nunez R, Bissell MJ, Folberg R. Chromatin sensitivity to Alu I endonuclease is regulated by extracellular matrix and the cytoskeleton. Am J Pathol. 2005;166:1187–1203. doi: 10.1016/S0002-9440(10)62338-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean IW, Zimmerman LE, Evans RM. Reappraisal of Callender’s spindle A type of malignant melanoma of choroid and ciliary body. Am J Ophthalmol. 1978;86:557–564. doi: 10.1016/0002-9394(78)90307-0. [DOI] [PubMed] [Google Scholar]
- Callender GR. Malignant melanotic tumors of the eye: a study of histologic types in 111 cases. Trans Am Acad Ophthalmol Otolaryngol. 1931;36:131–142. [Google Scholar]
- Callender GR, Wilder HC, Ash JE. Five hundred malignant melanomas of the choroid and ciliary body followed five years or longer. Trans Am Acad Ophthalmol Otolaryngol. 1942;46:223–230. [Google Scholar]
- Kan-Mitchell J, Mitchell MS, Rao N, Liggett PE. Characterization of uveal melanoma cell lines that grow as xenografts in rabbit eyes. Invest Ophthalmol Vis Sci. 1989;30:829–834. [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Fidler IJ. A simple quantitative assay for studying the invasive potential of high and low human metastatic variants. Cancer Lett. 1987;38:137–147. doi: 10.1016/0304-3835(87)90209-6. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- McLean IW, Foster WD, Zimmerman LE, Gamel JW. Modifications of Callender’s classification of uveal melanoma at the Armed Forces Institute of Pathology. Am J Ophthalmol. 1983;96:502–509. doi: 10.1016/s0002-9394(14)77914-0. [DOI] [PubMed] [Google Scholar]
- Rummelt V, Folberg R, Woolson RF, Hwang T, Pe’er J. Relation between the microcirculation architecture and the aggressive behavior of ciliary body melanomas. Ophthalmology. 1995;102:844–851. doi: 10.1016/s0161-6420(95)30947-5. [DOI] [PubMed] [Google Scholar]
- Matysiak BE, Brodzeller T, Buck S, French A, Counts C, Boorsma B, Datta MW, Kajdacsy-Balla AA. Simple, inexpensive method for automating tissue microarray production provides enhanced micro-array reproducibility. Appl Immunohistochem Mol Morphol. 2003;11:269–273. doi: 10.1097/00129039-200309000-00011. [DOI] [PubMed] [Google Scholar]
- Seftor EA, Meltzer PS, Kirschmann DA, Pe’er J, Maniotis AJ, Trent JM, Folberg R, Hendrix MJC. Molecular determinants of uveal melanoma metastasis. Clin Exp Metastasis. 2002;19:233–246. doi: 10.1023/a:1015591624171. [DOI] [PubMed] [Google Scholar]
- McLean IW, Gamel JW. Prediction of metastasis of uveal melanoma: comparison of morphometric determination of nucleolar size and spectrophotometric determination of DNA. Invest Ophthalmol Vis Sci. 1988;29:507–511. [PubMed] [Google Scholar]
- Moshari A, McLean IW. Uveal melanoma: mean of the longest nucleoli measured on silver-stained sections. Invest Ophthalmol Vis Sci. 2001;42:1160–1163. [PubMed] [Google Scholar]
- Albertoni M, Shaw PH, Nozaki M, Godard S, Tenan M, Hamou MF, Fairlie DW, Breit SN, Paralkar VM, de Tribolet N, Van Meir EG, Hegi ME. Anoxia induces macrophage inhibitory cytokine-1 (MIC-1) in glioblastoma cells independently of p53 and HIF-1. Oncogene. 2002;21:4212–4219. doi: 10.1038/sj.onc.1205610. [DOI] [PubMed] [Google Scholar]
- Vidal MJ, Loganzo F, Jr, de Oliveira AR, Hayward NK, Albino AP. Mutations and defective expression of the WAF1 p21 tumour-suppressor gene in malignant melanomas. Melanoma Res. 1995;5:243–250. doi: 10.1097/00008390-199508000-00006. [DOI] [PubMed] [Google Scholar]
- Ozkurt IC, Tetradis S. Parathyroid hormone-induced E4BP4/NFIL3 down-regulates transcription in osteoblasts. J Biol Chem. 2003;278:26803–26809. doi: 10.1074/jbc.M212652200. [DOI] [PubMed] [Google Scholar]
- Wolfgang CD, Liang G, Okamoto Y, Allen AE, Hai T. Transcriptional autorepression of the stress-inducible gene ATF3. J Biol Chem. 2000;275:16865–16870. doi: 10.1074/jbc.M909637199. [DOI] [PubMed] [Google Scholar]
- Mueller BM, Yu YB, Laug WE. Overexpression of plasminogen activator inhibitor 2 in human melanoma cells inhibits spontaneous metastasis in scid/scid mice. Proc Natl Acad Sci USA. 1995;92:205–209. doi: 10.1073/pnas.92.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domenzain C, Docampo MJ, Serra M, Miquel L, Bassols A. Differential expression of versican isoforms is a component of the human melanoma cell differentiation process. Biochim Biophys Acta. 2003;1642:107–114. doi: 10.1016/s0167-4889(03)00104-6. [DOI] [PubMed] [Google Scholar]
- McCurdy J, Gamel J, McLean I. A simple, efficient, and reproducible method for estimating the malignant potential of uveal melanoma from routine H, E slides. Pathol Res Pract. 1991;187:1025–1027. doi: 10.1016/S0344-0338(11)81076-9. [DOI] [PubMed] [Google Scholar]
- Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PA, Bissell MJ. Tumor reversion: correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Hansen RK, Radisky D, Yoneda T, Barcellos-Hoff MH, Petersen OW, Turley EA, Bissell MJ. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Kasemeier-Kulesa JC, Teddy JM, Margaryan NV, Seftor EA, Seftor RE, Hendrix MJ. Reprogramming metastatic melanoma cells to assume a neural crest cell-like phenotype in an embryonic microenvironment. Proc Natl Acad Sci USA. 2006;103:3752–3757. doi: 10.1073/pnas.0506977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AJ, Freeman WR, Folberg R, Bartsch DU, Scheider A, Schaller, Kampik A. Evaluation of microvascularization pattern visibility in human choroidal melanomas: comparison of confocal fluorescein with indocyanine green angiography. Graefes Arch Clin Exp Ophthalmol. 1999;237:448–456. doi: 10.1007/s004170050260. [DOI] [PubMed] [Google Scholar]
- Clarijs R, van Dijk M, Ruiter DJ, de Waal RMW. Functional and morphologic analysis of the fluid-conducting meshwork in xenografted cutaneous and primary uveal melanoma. Invest Ophthalmol Vis Sci. 2005;46:3013–3020. doi: 10.1167/iovs.04-0876. [DOI] [PubMed] [Google Scholar]
- Harrison JJ, Turner RJ, Ceri H. Persister cells, the biofilm matrix and tolerance to metal cations in biofilm and planktonic Pseudomonas aeruginosa. Environ Microbiol. 2005;7:981–994. doi: 10.1111/j.1462-2920.2005.00777.x. [DOI] [PubMed] [Google Scholar]
- Kaiserman I, Anteby I, Chowers I, Blumenthal EZ, Kliers I, Pe’er J. Post-brachytherapy initial tumour regression rate correlates with metastatic spread in posterior uveal melanoma. Br J Ophthalmol. 2004;88:892–895. doi: 10.1136/bjo.2003.036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folberg R, Fleck M, Mehaffey MG, Meyer M, Bentler SE, Woolson RF, Pe’er J. Mapping prognostically significant vascular patterns in ciliary body and choroidal melanomas. Pathol Oncol Res. 1996;2:229–236. doi: 10.1007/BF02904815. [DOI] [PubMed] [Google Scholar]
- Mehaffey MG, Folberg R, Meyer M, Bentler SE, Hwang T, Woolson RF, Moore KC. Relative importance of quantifying area and vascular patterns in uveal melanoma. Am J Ophthalmol. 1997;123:798–809. doi: 10.1016/s0002-9394(14)71129-8. [DOI] [PubMed] [Google Scholar]
- Kujala E, Makitie T, Kivela T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003;44:4651–4659. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- McLean IW. The biology of haematogenous metastasis in human uveal malignant melanoma. Virchows Arch A Pathol Anat. 1993;422:433–437. doi: 10.1007/BF01606450. [DOI] [PubMed] [Google Scholar]
- Rummelt V, Mehaffey MG, Campbell RJ, Pe’er J, Bentler SE, Woolson RF, Naumann GOH, Folberg R. Microcirculation architecture of metastases from primary ciliary body and choroidal melanomas. Am J Ophthalmol. 1998;126:303–305. doi: 10.1016/s0002-9394(98)00164-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.