Abstract
The interactions between cancer cells and the surrounding host stromal tissue play a critical role in tumor progression and metastasis, but the molecular nature of this relationship remains largely uncharacterized. Furthermore, although genetic changes of neoplastic cells in tumors contribute significantly to tumor progression, it is not known whether similar changes occur in the adjacent host stromal microenvironment and whether they contribute to or inhibit tumorigenesis. To address this question in an unbiased and genome-wide manner, we applied high-resolution DNA copy number analysis to murine stromal DNA isolated from human xenograft tumors that were formed in immunodeficient mice. We show that numerous amplifications and deletions are found within the host stromal microenvironment, suggesting that alterations in host DNA copy number can occur and may play a significant role in modifying tumor–stromal interactions.
Keywords: cancer, stroma, microarray
It has long been understood that tumors are complex tissues. Genetically aberrant “cancer cells,” the ultimate cause of the tumor, are only one component of most epithelial tumors; other “stromal” components include cells of mesenchymal origin that have repeatedly been implicated as potential contributors to the growth of tumors (1, 2). Tumors frequently also contain many kinds of inflammatory cells, suggesting a connection between tumor growth and inflammation (3). Furthermore, the similarity between the cell populations and activities associated with normal wound healing and those found in tumors has often been noted (4, 5). In their influential review, Hanahan and Weinberg emphasize the possibility that stromal contributions may be vital to the growth of tumors, and go so far as to suggest that “… in some tumors, these cooperating cells may eventually depart from normalcy, co-evolving with their malignant neighbors to sustain the growth of the latter” (1).
Genome-scale comprehensive gene expression studies of solid tumors have served to illustrate in striking detail the complexity of tumors and the diversity of nonepithelial cell types they contain. The earliest studies noted that gene expression profiles characteristic of stromal and inflammatory cells are usually detected, as are expression signatures previously associated with wound healing and angiogenesis (4, 6, 7). These and subsequent studies of many different tumor types provided a mass of detailed information, leading to three general observations. First, each anatomical type of cancer is associated with recognizable expression pattern usually closely related to the pattern seen in normal tissues; some types (e.g., breast cancer) are classifiable into a small family of “subtypes” (6, 8, 9). Second, within a cancer “subtype,” the tumors of each individual nevertheless remain distinguishable by differences in gene expression pattern. These patterns are the sum of gene expression in the genetically deranged and the stromal cells comprising the tumor. Third, the expression profiles of distant metastases tend to be even more similar to those of the primary tumor than primary tumors of the same subtype are to each other (breast, refs. 6 and 10; lung, ref. 7; gastric, ref. 11).
Thus, even distant metastases, presumably founded by a single migrant cancer cell, somehow become organized like the primary tumors, acquiring populations of stromal and inflammatory cells very similar to those found in the parent primary. This implies that the genetically deranged cancer cells in both the metastasis and the primary tumor must initiate and benefit from a complex choreography of mutual signaling and support. The detailed expression pattern appears to be characteristic, possibly even unique, for each independently arising cancer cell; the general pattern is characteristic of the particular cancer subtype. Particularly interesting in this context was the finding by Chen et al. (11) that, in the case of hepatocellular carcinoma, where an individual patient might have more than one independently arising tumor as well as metastases of some of them, only the clonally related tumors (on the basis of DNA changes) had similar expression profiles.
Here we describe the results of an experiment designed to address specifically the possibility that the stromal components are involved in some kind of “coevolution” with the tumor, by looking directly for alterations in the genomic DNA of stromal cells during the establishment of human tumors that result from the introduction of clones of malignant human tumor cell lines into nude mice. This cross-species design provided a secure way to distinguish the genetically human cancer cells from the putatively coevolving stromal helper cells. Tumors that arise in this system necessarily have stromal components derived from the mouse, even though the malignant founder cells are human. Any changes in stromal cell genomes would be changes in the mouse genome, and not the human genome.
We used representational oligonucleotide microarray analysis (ROMA), which has proven to be both sensitive and reliable, enabling the detection of copy number changes ranging from a few tens of kilobases to entire chromosome arms (12–14). Briefly, ROMA's sensitivity is derived from its reduction of the complexity of the test and reference genomes analyzed (≈3% of the original genome) by making BglII genomic representations, consisting of small restriction fragments, between 200 and 1,200 base pairs. The BglII restriction fragments are then amplified by adaptor-mediated PCR, fluorescently labeled and the test and reference samples are hybridized to custom fabricated microarrays containing oligonucleotide probes to BglII fragments designed in silico from the mouse genome sequence assembly to be complementary with these fragments. Putative changes detected by ROMA were then confirmed by using quantitative real-time PCR (12–14).
Below, we show evidence that amplifications and deletions of mouse genes of potential biological interest were detected in these xenograft tumors. The detection of amplifications and deletions in the stroma indicates the presence of clones of mutant host cells, and strongly suggests that such cells have been selected for proliferation in the course of the establishment of the tumors, a process that takes between 30 and 150 days from the injection of the human cells.
Results
Detection of Copy Number Alterations in the Genomes of Mouse Stromal Cells in Human Xenograft Tumors.
We reasoned that if genetically aberrant cancer cells coevolve with the stromal cell types that are found in human tumors, then malignant human epithelial cells that establish a complex xenograft tumor that contains stromal participants in a nude mouse might involve the mouse stroma in a similar process. Therefore, we produced a variety of human xenograft tumors in nude mice and examined the genomes of mouse stromal cells from these tumors for DNA copy number changes.
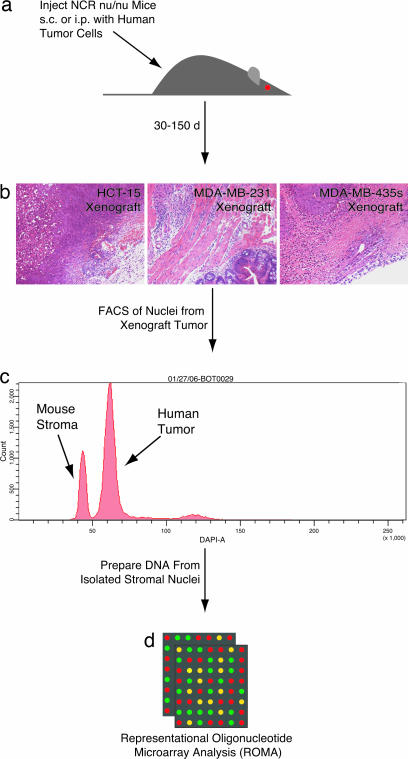
Fig. 1 gives an overview of the experimental system, with additional details available in Materials and Methods and supporting information (SI) Figs. 3 and 4. Human tumor cell lines of various origins (HCT-15, colorectal carcinoma; MDA-MB-231, breast adenocarcinoma; MDA-MB-435s, breast carcinoma) were injected s.c. or i.p. into female NCR nu/nu mice (Fig. 1a). Tumors developed 30–150 days later, and the gross tumors were dissected (Fig. 1b). The xenograft tumors were then subjected to a FACS analysis wherein mouse stromal cell and human cancer cell nuclei were separated based on DNA content (Fig. 1c). We then used ROMA to survey the DNA copy number of the isolated mouse stromal component from the xenograft tumors (Fig. 1d). Signal noise due to sample degradation (SI Fig. 4) was overcome through the comparison of stromal nuclei from one xenograft tumor with the stromal nuclei of a second xenograft tumor caused by the same human tumor cell line. ROMA comparison of tail DNA from the mice used in the stromal analysis was then used to account for any genetic differences between these outbred individual mice (14). Thus, the copy number alterations changes reported here are necessarily stromal differences between two tumors. It is important to note that any consistent changes in the stroma of two tumors would be normalized out, and therefore not observed using our analysis. Identification of a deletion in Ighg (SI Fig. 5b) in the stroma of several tumors (BOT0029, BOT0031) caused by the human MDA-MB-435s breast carcinoma cell line, presumably the result of somatic hypermutation and/or V(D)J recombination of the Ig locus (15, 16) within an inflammatory component of the stroma, provided early validation of our method.
Fig. 1.
Isolation of stromal nuclei from human xenograft tumors in NCR nu/nu mice. (a) Human tumor cell lines of various origins (HCT-15, MDA-MB-231, MDA-MB-435s) were injected s.c. or i.p. into NCR nu/nu mice. (b) Xenograft tumors developed 30–150 days later, and the gross tumors were dissected. (c) The xenograft tumors were then subjected to FACS analysis, and the mouse stroma and human tumor nuclei were separated based on DNA content. (d) ROMA analysis was then performed on isolated stromal material.
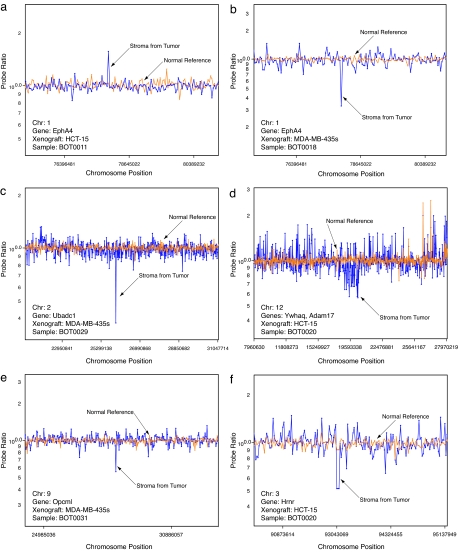
The ROMA survey identified 38 candidate copy number alterations in the stroma associated with seven tumors. Fig. 2 presents array data for six copy number alterations (one amplification and five deletions) contained within the genes EphA4 (Fig. 2 a and b), Ubadc1 (Fig. 2c), Ywhaq and Adam17 (Fig. 2d), Opcml (Fig. 2e), and Hrnr (Fig. 2f). ROMA array data for additional DNA copy number alterations is available in SI Fig. 5. To provide some measure of validation of these and the remaining ROMA predictions, we applied quantitative PCR (qPCR) to measure the DNA copy number of the individual genes encompassed by the array data. Additionally, to delineate the boundaries of candidate amplifications and deletions, we also used qPCR to determine the copy number of genes flanking several of these predictions (SI Table 2).
Fig. 2.
Alterations in stromal DNA copy number identified by ROMA. (a) Amplification in Epha4. (b) Deletion in Epha4. (c) Deletion in Ubadc1. (d) Deletion in Ywhaq and Adam17. (e) Deletion in Opcml. (f) Deletion in Hrnr. Copy number profile of stroma from tumor (blue) and normal reference DNA (orange) are indicated. Probe ratios are plotted on a log10 scale.
Alterations in copy number were widely distributed throughout the genome (Table 1). Significantly, the same genes were not altered in each tumor produced by a given human cancer cell line. If indeed there is coevolution, there must be more than one kind of successful stromal evolutionary path. It is also worth noting that we found changes of such a magnitude that a large number of stromal cells in each tumor must have shared the same amplification and deletion, suggesting strongly a common clonal origin that then was highly selected during the development of the xenograft tumor. Finally, it is important to note that the DNA copy number of the human homologues of the genes altered in the host stroma are not altered in the parental human tumors used in this study (data not shown).
Table 1.
Alterations in stromal DNA copy number validated by qPCR
| Gene symbol(s) | Gene name(s) | Change in copy number | Relative copy number (2ΔCt) | Band | Deletion/amplicon size, kb | Sample name | Tumor type | Name of xenograft tumor |
|---|---|---|---|---|---|---|---|---|
| Adamdec1 | ADAM-like (decysin 1) | Loss | 0.42 | 14D1 | 256.56 | BOT0026 | Breast | MDA-MB-231 |
| EphA4 | Eph receptor A4 | Gain | 2.84 | 1qC4 | 31.22 | BOT0011 | Colon | HCT-15 |
| EphA4 | Eph receptor A4 | Loss | 0.43 | 1qC4 | 31.22 | BOT0018 | Breast | MDA-MB-435s |
| Hrnr | Hornerin | Loss | 0.17 | 3qF2.1 | 6.89 | BOT0020 | Colon | HCT-15 |
| Hrnr | Hornerin | Loss | 0.40 | 3qF2.1 | 6.89 | BOT0028 | Breast | MDA-MB-435s |
| Ighg | Immunoglobulin heavy chain (gamma polypeptide) | Loss | 0.29 | 12qF1 | 553.54 | BOT0029 | Breast | MDA-MB-435s |
| Ighg | Immunoglobulin heavy chain (gamma polypeptide) | Loss | 0.21 | 12qF1 | 553.54 | BOT0031 | Breast | MDA-MB-435s |
| Kcnd2 | Potassium voltage-gated channel, Shal-related family, member 2 (Kv4.2) | Gain | 2.40 | 6qA2 | 69.48 | BOT0028 | Breast | MDA-MB-435s |
| Mup1; Mup2 | Major urinary protein 1; Major urinary protein 2 | Loss | 0.26, 0.26 | 4qB3 | 860.02 | BOT0029 | Breast | MDA-MB-435s |
| Mup1; Mup2 | Major urinary protein 1; Major urinary protein 2 | Loss | 0.10, 0.14 | 4qB3 | 860.02 | BOT0031 | Breast | MDA-MB-435s |
| Opcml | Opioid binding protein/cell adhesion molecule-like | Loss | 0.16 | 9qA4 | 672.87 | BOT0031 | Breast | MDA-MB-435s |
| Rabgap1 | RAB GTPase activating protein 1 | Loss | 0.28 | 2qB | 20.77 | BOT0028 | Breast | MDA-MB-435s |
| Ubadc1 | Ubiquitin associated domain containing 1 (putative glialblastoma cell differentiation-related) | Loss | 0.37 | 2qA3 | 16.13 | BOT0028 | Breast | MDA-MB-435s |
| Ubadc1 | Ubiquitin associated domain containing 1 (putative glialblastoma cell differentiation-related) | Loss | 0.14 | 2qA3 | 16.13 | BOT0029 | Breast | MDA-MB-435s |
| Ubadc1 | Ubiquitin associated domain containing 1 (putative glialblastoma cell differentiation-related) | Loss | 0.18 | 2qA3 | 16.13 | BOT0031 | Breast | MDA-MB-435s |
| V2r14; V2r15; V2r4 | Vomeronasal 2, receptor, 14; Vomeronasal 2, receptor, 15; Vomeronasal 2, receptor, 4 | Loss | 0.43, 0.43, 0.06 | 7qA1 | 1769.45 | BOT0031 | Breast | MDA-MB-435s |
| Ywhaq; Adam17 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide; Disintegrin and metallopeptidase domain 17 | Loss | 0.06, 0.06 | 12qA1.2–1.3 | 2718.49 | BOT0020 | Colon | HCT-15 |
In sum, 17 alterations in DNA copy number, constituting a nonredundant set of 15 unique genes, observed by ROMA analysis were unambiguously confirmed by direct and flanking qPCR analysis (Table 1 and SI Table 2). Two of these alterations in copy number were amplifications, and 15 were deletions (Table 1).
Individual Genes Altered in DNA Copy Number in Tumor Stroma.
A summary is provided of some of the amplification and deletion events whose known biology is of particular interest with respect to carcinogenesis or cancer progression (see SI Text). A complete list of all of the PCR-confirmed events is found in Table 1.
The single most notable event occurred in the stroma of a xenograft tumor (BOT0011) caused by the human colorectal carcinoma cell line HCT-15, which contained a 2.8-fold amplification of the gene encoding EphA4 (Fig. 2a and Table 1). Interestingly, the stroma of a second xenograft tumor (BOT0018) caused by the human breast carcinoma cell line MDA-MB-435s contained a 0.43-fold deletion in EphA4 (Fig. 2a and Table 1). In the ROMA survey, this amplification and deletion were both identified as single-probe events, and based on PCR, the nearest unamplified probes were 31.22 kb apart (Table 1). EphA4 is a receptor tyrosine kinase that belongs to a large family of Ephrin receptors. Although primarily involved in mediating developmental events directed by Ephrin (17), particularly in the nervous system, many studies have indicated a direct role for the ephrins and their receptors in tumor progression and angiogenesis (18). Increased expression of Ephrins and their receptors have been correlated with survival and invasive capacity of colorectal and ovarian cancers (19–21). Additionally, EphA2 and EphB2 have emerged as attractive drug targets for cancer therapy (22, 23). That EphA4 was observed as both amplification and deletion in the stroma caused by two different cell lines may be related to differences in the parental tumor cell lines as well as the anatomical location of the xenograft tumors (s.c. versus i.p.).
Discussion
The data summarized above and in Table 1 show that there are occasional DNA copy number changes in stromal cells of xenograft tumors in nude mice caused by the introduction of cells from cloned human cancer cell lines. The DNA copy number changes are widely dispersed through the mouse genome and include both amplifications and deletions. Genes within the generally quite short regions of DNA that are altered (average length of 497.6 kb) include several whose biology (e.g., EphA4 and Adam17) is immediately interesting with respect to their potential roles in cancer. A few alterations appear in more than one independent tumor (e.g., Hornerin, Ubadc1, and EphA4) and in two cases, the causative cell lines were different.
There are several features to note about our detection of DNA alterations in tumor stromal cells.
Our experimental design allowed us to separate cleanly stromal cells from the causative cancer cells. There was no indication in the FACS-sorting step of possible mouse–human hybrid nuclei (24, 25), nor was there any human genetic material detected in the DNA from the isolated mouse nuclei (SI Fig. 3). The changes we detected are not present in the parental human tumors (data not shown).
Our detection of any changes at all (especially the deletions) implies that there may indeed be a clonal selection process involved in the establishment and progression of the kind of xenograft tumors we studied. Heretofore, there was no direct reason to believe that stromal cells would show a high degree of apparent clonality.
The observation that the same-cell human cell line causes tumors that show different stromal cell DNA alterations suggests that there must be several effective paths for the mouse cells to evolve and be selected to become tumor stroma.
Relatively little is known in detail about the biological or molecular basis for tumor–cell interactions with the stroma, although there are very many suggestive studies implicating each of a legion of growth factors, signaling molecules, and extracellular matrix components. Current knowledge of these interactions is based on studies using coculture systems, reconstitution experiments, histological classification and subsequent microdissection, or subtractive bioinformatics (26–29). Many of these experiments are complicated by difficulties in separating tumor cells from stromal cells, on the one hand, and in defining the cellular source(s) of the many growth factors and signaling molecules that are detected in tumors.
Establishment of the role of angiogenesis in tumor progression stands as a notable exception in which these difficulties were overcome. The stroma has been shown to be a significant source of VEGF, which stimulates endothelial cells to proliferate and migrate, resulting in the formation of the tumor-associated vasculature (30), leading to the emergence of effective therapies based on blocking this activity (31). The paracrine relationship between the tumor and stroma during tumor angiogenesis has also been convincingly demonstrated by the tumor-derived PDGF-mediated recruitment of stromal fibroblasts to the neovasculature of primary tumor (32, 33).
Various studies have demonstrated alterations in gene expression, loss of heterozygosity, and epigenetic changes in the stroma of human cancers (29, 34–41). We did not observe any of these as DNA copy number changes; instead, we found changes in genes, some of whose connection to cancer was previously documented, and some where the connection was just plausible. We believe that the important result here is that this kind of analysis can lead to more direct investigation of genes, because there is evidence that they are, indeed, under selection during the establishment and/or development of complex tumor tissues.
Implications of an Altered Stromal Genome
The causative role of host DNA copy number variation during cancer is an interesting question. It is plausible that the DNA copy number alterations are the result of selective pressures placed on the stroma by the tumor to foster a pro-oncogenic microenvironment. Alternatively, they may represent host selection for mutant stromal cells recruited to suppress tumor progression. Finally, it is possible that our findings are incidental and that during the process of stromal proliferation, the response is often clonal, and we are merely detecting preexisting somatic mutation by our sensitive methods.
This study suggests that a substantial portion of the alterations observed in both DNA copy number and gene expression seen in clinical human tumors may be affected by genomic changes in the tumor-associated stromal tissue. It will be increasingly important in the future to distinguish which features of the genomic profiles (DNA copy number or gene expression) are due to stroma and which are due to primary epithelial cells that caused the cancer. Although our analysis is able to distinguish unambiguously between the tumor and host genomes in a mouse xenograft model, understanding the extent to which genomic alterations occur in the human stroma associated with human tumors may prove to be challenging. However, in favorable cases, where the ploidy of the tumor vastly differs from normal, we can perform similar studies to the ones just described, cleanly separating human tumor and stromal nuclei.
The finding that significant DNA copy number alterations occur in the nontumor stromal tissue at sites of tumor growth has numerous implications. The accumulation of stromal mutations within the primary tumor may provide a mechanism of resistance to antiangiogenic and host-targeted therapies (31). These data underscore the opportunity of attacking cancer-specific activities of stromal tissue as well as those of the primary tumor (42).
Materials and Methods
Cell Culture and Experimental Tumors.
Human HCT-15, MDA-MB-231, and MDA-MB-435s tumor cell lines were cultured in DMEM supplemented with 15% FBS, penicillin/streptomycin, and l-glutamine. For injection into experimental animals, cells were grown to 70% confluence, harvested by trypsinization, washed in PBS, and diluted in PBS at a concentration of 1 × 107 cells per ml. Cell aliquots were kept on ice until injection.
Eight-week-old female NCR nu/nu mice (Taconic, Germantown, NY) were injected with a 26-gauge needle either i.p. or s.c. in the hind flank with 106 cells. Mice were monitored daily for morbidity and killed by cervical dislocation either when the mouse became moribund or when tumors reached a diameter of 1 cm. Tumors were harvested aseptically and immediately frozen in liquid nitrogen for archival storage or fixed in 4% buffered formalin for histological analysis. All animal studies were conducted in accordance with the Institutional Animal Care and Use Committees of Princeton University and Cold Spring Harbor Laboratory.
Isolation of Stromal Nuclei.
Mouse stromal nuclei are isolated from tumor samples by finely mincing grossly dissected tumors in a Petri dish in 0.5–1.0 ml of NST-DAPI buffer [800 ml of NST (146 mM NaCl/10 mM Tris base, pH 7.8/1 mM CaCl2/21 mM MgCl2/0.05% BSA/0.2% Nonidet P-40], 200 ml of 106 mM MgCl2, 10 mg of DAPI, and 0.1% boiled RNase A using two no. 11 scalpels in a cross-hatching motion. Minced tissue is then incubated on ice for 15 min. Before flow cytometric analysis, samples were filtered through 37-μm plastic mesh. Nuclei were sorted with a Becton Dickinson (Franklin Lakes, NJ) FACS DiVa Flow Cytometer and Cell Sorter. Skin samples from non-tumor-implanted mice and normal human subjects were prepared as above and used as reference samples for calibration of FACS collection gates to differentially sort mouse and human nuclei based on DNA content (2.5 vs. 2.85 Gb; see also SI Fig. 3). Sorted stromal nuclei yielded a collection profile identical to that of normal mouse skin nuclei; sorted human tumor nuclei yielded a collection profile identical to that of the parental human tumor cell line.
DNA Digestion, Labeling, and Microarray Hybridization.
Digestion of genomic DNA with BglII, labeling, and hybridization to 85K oligonucleotide microarrays (NimbleGen Systems, Madison, WI) were performed as described (13, 14). Briefly, the complexity of the isolated stromal DNA was reduced by making BglII genomic representations, consisting of small (200–1,200 bp) fragments amplified by adaptor-mediated PCR. Stromal DNA samples and matched reference DNA were then labeled differentially with Cy5-dCTP or Cy3-dCTP using Amersham Pharmacia (Piscataway, NJ) Megaprime labeling kit, and hybridized in comparison with each other. An Axon GenePix 4000B scanner was used to scan processed arrays, and GenePix Pro 4.0 software was used for quantification of intensity for the arrays.
ROMA Data Analysis.
Array data were imported into S-PLUS for further analysis. Data were normalized by using an intensity-based Lowess curve-fitting algorithm (43). Log ratio values were averaged from color reversal experiments. Early iterations of ROMA analysis of mouse stromal nuclei using matched tail DNA as reference were hampered by significant background noise, likely the result of the early stages of apoptosis in the xenograft tumor and stromal DNA degradation (SI Fig. 4a). This obstacle was overcome by performing the ROMA analysis in two stages: (i) Comparison of stromal nuclei from one xenograft tumor with the stromal nuclei of a second xenograft tumor; both xenograft tumors were caused by the same human tumor cell line and thus shared the same pattern of DNA degradation. This step significantly reduced background noise and allowed for the identification of DNA copy number changes in stromal DNA. (ii) Comparison of tail DNA from the mice used in the stromal analysis. This stage accounted for any genetic differences between these outbred individual mice, and facilitated the isolation of bona fide DNA copy number changes in the stroma from other genetic differences such as single-nucleotide or copy number polymorphisms (14). Additional details are available in SI Fig. 4.
Quantitative Real-Time PCR.
qPCR primers and dual-labeled fluorogenic probes used to validate candidate copy number alterations were designed in silico from the mouse genome, NCBI Build 33, and manufactured by Sigma-Proligo (Woodlands, TX) or Integrated DNA Technologies (Coralville, IA). Primer/probe sequences are available in SI Table 3. DNA copy number was quantified by using the Applied Biosystems 7900 sequence detection system (Applied Biosystems, Foster City, CA), and the data were analyzed with SDS 2.1 software (Applied Biosystems) using standard protocols.
Candidate copy number alterations based on 1–2 probe events in ROMA (e.g., EphA4) were validated by using qPCR probes to three distinct regions of the candidate gene. Probes were designed to: (i) the BglII fragments producing the probe event in ROMA, (ii) regions proximal to the BglII fragments, but still within the candidate gene, and (iii) genes directly flanking the candidate gene. Large candidate copy number alterations (>10 probe events, e.g., Adam17) were validated by using qPCR probes to regions within the ROMA prediction, but not specifically to the BglII fragments.
The relative gene copy numbers were derived by using the formula 2ΔCt, where ΔCt is the difference in amplification cycles required to detect amplification product from equal starting concentrations of matched reference tail or skin genomic DNA as compared with genomic DNA from mouse stromal nuclei isolated from gross dissected xenograft tumors (44). β-Actin was used as a reference probe for normalization. PCRs were performed in quadruplicate, and copy number alterations were scored as validated if 2ΔCt (relative copy number) was ≥1.5 (gain) or ≤0.5 (loss) with CV ≤15% of mean 2ΔCt.
Acknowledgments
We are grateful for the assistance of Pam Moody and Jennifer Meth with FACS analysis of stromal and tumor nuclei. Lisa Bianco and the staff of the Cold Spring Harbor Animal Facility provided invaluable support throughout this study. Dr. Peter Siegel (McGill University, Montreal, QC, Canada) provided extensive training in the use of mice as a model system. We thank Drs. Vladimir Grubor, Lakshmi Muthuswamy, members of the Wigler Laboratory and the Cold Spring Harbor Laboratory community-at-large for helpful discussions and kind support. R.J.P. thanks Dr. Becket Feierbach for her love, patience, and support throughout this study. This work was supported by National Institutes of Health Grant 5R01-CA078544-07; Department of the Army Grants W81XWH-06-1-0189, W81XWH04-1-0477, W81XWH-05-1-0068, and W81XWH-04-0905; grants from Miracle Foundation, Breast Cancer Research Foundation, Long Islanders Against Breast Cancer, West Islip Breast Cancer Foundation, Long Island Breast Cancer (1 in 9), and The Karches Foundation; an Elizabeth McFarland Breast Cancer Research Grant; and a grant from Breast Cancer Help, Inc. (to M.W.). M.W. is an American Cancer Society Research Professor.
Abbreviations
- ROMA
representational oligonucleotide microarray analysis
- qPCR
quantitative PCR.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at http://www.pnas.org/cgi/content/full/0609635104/DC1.
References
- 1.Hanahan D, Weinberg RA. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ, Radisky D. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dvorak HF. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 6.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, van't Veer LJ. Cancer Res. 2005;65:9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 11.Chen X, Cheung ST, So S, Fan ST, Barry C, Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 13.Lucito R, Healy J, Alexander J, Reiner A, Esposito D, Chi M, Rodgers L, Brady A, Sebat J, Troge J, et al. Genome Res. 2003;13:2291–2305. doi: 10.1101/gr.1349003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmi B, Hall IM, Egan C, Alexander J, Leotta A, Healy J, Zender L, Spector MS, Xue W, Lowe SW, et al. Proc Natl Acad Sci USA. 2006;103:11234–11239. doi: 10.1073/pnas.0602984103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goossens T, Klein U, Kuppers R. Proc Natl Acad Sci USA. 1998;95:2463–2468. doi: 10.1073/pnas.95.5.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odegard VH, Schatz DG. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson DG. Nat Rev Neurosci. 2001;2:155–164. doi: 10.1038/35058515. [DOI] [PubMed] [Google Scholar]
- 18.Carmeliet P, Tessier-Lavigne M. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 19.Clevers H, Batlle E. Cancer Res. 2006;66:2–5. doi: 10.1158/0008-5472.CAN-05-3849. [DOI] [PubMed] [Google Scholar]
- 20.Guo DL, Zhang J, Yuen ST, Tsui WY, Chan ASY, Ho C, Ji J, Leung SY, Chen X. Carcinogenesis. 2006;27:454–464. doi: 10.1093/carcin/bgi259. [DOI] [PubMed] [Google Scholar]
- 21.Jubb AM, Zhong F, Bheddah S, Grabsch HI, Frantz GD, Mueller W, Kavi V, Quirke P, Polakis P, Koeppen H. Clin Cancer Res. 2005;11:5181–5187. doi: 10.1158/1078-0432.CCR-05-0143. [DOI] [PubMed] [Google Scholar]
- 22.Carles-Kinch K, Kilpatrick KE, Stewart JC, Kinch MS. Cancer Res. 2002;62:2840–2847. [PubMed] [Google Scholar]
- 23.Mao W, Luis E, Ross S, Silva J, Tan C, Crowley C, Chui C, Franz G, Senter P, Koeppen H, Polakis P. Cancer Res. 2004;64:781–788. doi: 10.1158/0008-5472.can-03-1047. [DOI] [PubMed] [Google Scholar]
- 24.Duelli D, Lazebnik Y. Cancer Cell. 2003;3:445–448. doi: 10.1016/s1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 25.Jacobsen BM, Harrell JC, Jedlicka P, Borges VF, Varella-Garcia M, Horwitz KB. Cancer Res. 2006;66:8274–8279. doi: 10.1158/0008-5472.CAN-06-1456. [DOI] [PubMed] [Google Scholar]
- 26.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 27.Schmeichel KL, Bissell MJ. J Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.West RB, Nuyten DSA, Subramanian S, Nielsen TO, Corless CL, Rubin BP, Montgomery K, Zhu S, Patel R, Hernandez-Boussard T, et al. PLoS Biol. 2005;3:e187. doi: 10.1371/journal.pbio.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 30.Ferrara N, Gerber H-P, LeCouter J. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 31.Ferrara N, Kerbel RS. Nature. 2005;438:967–974. doi: 10.1038/nature04483. [DOI] [PubMed] [Google Scholar]
- 32.Dong J, Grunstein J, Tejada M, Peale F, Frantz G, Liang WC, Bai W, Yu L, Kowalski J, Liang X, Fuh G, Gerber HP, Ferrara N. EMBO J. 2004;23:2800–2810. doi: 10.1038/sj.emboj.7600289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tejada ML, Yu L, Dong J, Jung K, Meng G, Peale FV, Frantz GD, Hall L, Liang X, Gerber H-P, Ferrara N. Clin Cancer Res. 2006;12:2676–2688. doi: 10.1158/1078-0432.CCR-05-1770. [DOI] [PubMed] [Google Scholar]
- 34.Tuhkanen H, Anttila M, Kosma VM, Yla-Herttuala S, Heinonen S, Kuronen A, Juhola M, Tammi R, Tammi M, Mannermaa A. Int J Cancer. 2004;109:247–252. doi: 10.1002/ijc.11733. [DOI] [PubMed] [Google Scholar]
- 35.Paterson RF, Ulbright TM, MacLennan GT, Zhang S, Pan CX, Sweeney CJ, Moore CR, Foster RS, Koch MO, Eble JN, Cheng L. Cancer. 2003;98:1830–1836. doi: 10.1002/cncr.11747. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto N, Yoshida T, Yamashita K, Numata Y, Okayasu I. Br J Cancer. 2003;89:707–712. doi: 10.1038/sj.bjc.6601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou X-P, Eng C. Nat Genet. 2002;32:355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- 38.Fukino K, Shen L, Matsumoto S, Morrison CD, Mutter GL, Eng C. Cancer Res. 2004;64:7231–7236. doi: 10.1158/0008-5472.CAN-04-2866. [DOI] [PubMed] [Google Scholar]
- 39.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 40.Hill R, Song Y, Cardiff RD, Van Dyke T. Cell. 2005;123:1001–1011. doi: 10.1016/j.cell.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 41.Hida K, Hida Y, Amin DN, Flint AF, Panigrahy D, Morton CC, Klagsbrun M. Cancer Res. 2004;64:8249–8255. doi: 10.1158/0008-5472.CAN-04-1567. [DOI] [PubMed] [Google Scholar]
- 42.Liotta LA, Kohn EC. J Natl Cancer Inst. 2002;94:1113–1114. doi: 10.1093/jnci/94.15.1113. [DOI] [PubMed] [Google Scholar]
- 43.Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT, et al. Nature. 1998;396:699–703. doi: 10.1038/25387. [DOI] [PubMed] [Google Scholar]