Abstract
Single-stranded DNA binding (SSB) protein binds to single-stranded DNA (ssDNA) at the lagging strand of the replication fork in Escherichia coli cells. This protein is essential for the survival of the E.coli cell, presumably because it shields the ssDNA and holds it in a suitable conformation for replication by DNA polymerase III. In this study we undertook a biophysical analysis of the interaction between the SSB protein of E.coli and the χ subunit of DNA polymerase III. Using analytical ultracentrifugation we show that at low salt concentrations there is an increase in the stability in the physical interaction between χ and an EcoSSB/ssDNA complex when compared to that of χ to EcoSSB alone. This increase in stability disappeared in high salt conditions. The sedimentation of an EcoSSB protein lacking its C-terminal 26 amino acids remains unchanged in the presence of χ, showing that χ interacts specifically with the C-terminus of EcoSSB. In DNA melting experiments we demonstrate that χ specifically enhances the ssDNA stabilization by EcoSSB. Thus, the binding of EcoSSB to χ at the replication fork prevents premature dissociation of EcoSSB from the lagging strand and thereby enhances the processivity of DNA polymerase III.
INTRODUCTION
Single-stranded DNA binding (SSB) proteins have been defined as proteins which bind selectively to single- and not double-stranded DNA and have no direct catalytic function (1–3). With these properties in mind, the role of these proteins is clearly one of stabilizing and protecting the single-stranded state of DNA and thus providing a template for DNA polymerization, recombination or repair, thus earning them the title of single-stranded DNA (ssDNA) chaperones.
However, it has become clear that these proteins may have other roles, besides that of binding to ssDNA, through their interactions with proteins involved in DNA metabolism (see e.g. 4). Details of these functions depend upon the class and organism these SSB proteins reside in.
Homotetrameric SSB proteins have been found in prokaryotes and eukaryotic mitochondria. These homotetrameric SSB proteins are essential for the survival of prokaryote cells (5), and the fact that genes coding for such homotetrameric SSB proteins can even be found in higher plants and animals (e.g. Arabidopsis thaliana and Homo sapiens) (6) suggests that they are evolutionarily conserved and play a vital role in the sustainment of life.
The primary structure of prokaryotic homotetrameric SSB proteins can be broken down into three distinct regions. The N-terminal two-thirds of the sequence make up the ssDNA binding region (7), the crystal structure of which has been determined for both prokaryotic and human mitochondrial SSB. These crystal structures have demonstrated no decisive differences between the SSB proteins from these organisms (8–10). In prokaryotes this ssDNA binding domain is followed by a region containing many G, Q and P residues. This region most probably forms no definite secondary structure and hence could not be identified in the crystal structure analysis. At the ultimate C-terminal end prokaryotic homotetrameric SSB proteins possess a negatively charged consensus sequence (-DDDIPF) (11). The last two regions are missing from the mitochondrial proteins (12). A temperature-sensitive Escherichia coli strain (ssb-113) carrying a mutant SSB protein (EcoSSB P176S) has been described (13). This mutation alters the C-terminal consensus sequence to -DDDISF. While the corresponding SSB protein has normal ssDNA binding functions, the phenotype was attributed to an impairment of some protein–protein interaction mediated by the C-terminal region (13). We have previously demonstrated that this mutation drastically reduces the stability of the interaction between EcoSSB and exonuclease I and showed that the C-terminal region is responsible for this interaction (11).
To date, several proteins have been shown to physically interact with E.coli SSB (4,11,14,15). In E.coli genomic replication the cooperation of SSB and the DNA polymerase III holoenzyme is important for DNA replication of the lagging strand (16), as EcoSSB protein protects the vulnerable ssDNA strand and prevents hairpin formation (17). The precise target of the EcoSSB interactions with DNA polymerase III holoenzyme has been identified as the χ subunit. A direct interaction between these two proteins has been shown using gel filtration and surface plasmon resonance (15,18). The C-terminal consensus sequence of EcoSSB has been identified as taking part in this interaction (15). This study analyses the physicochemical details of this protein–protein interaction by analytical ultracentrifugation, surface plasmon resonance and DNA melting experiments. We present quantitative details of this interaction and provide a model of the role of this interaction in bacterial replication.
MATERIALS AND METHODS
Buffers and reagents
Poly(dT) and poly(dA-dT) were purchased from Amersham Pharmacia Biotech. Nucleotide concentrations were determined spectrophotometrically using an absorption coefficient of 8600 M–1cm–1 for poly(dT) at maximum (17) and of 6700 M–1cm–1 for poly(dA-dT) at 260 nm (7). Protein concentrations were determined using the following absorption coefficients at 280 nm: 29 280 M–1cm–1 for DNA polymerase III χ subunit protein (calculated from the amino acid composition) (19), 113 000 M–1cm–1 for EcoSSB wild-type (20) and EcoSSB Q152* (an EcoSSB mutant missing the last 26 amino acids) (21), 99 000 M–1cm–1 for HsmtSSB (22). SSB concentrations are given in tetramers throughout the text. Measurements were carried out in a standard buffer containing 20 mM potassium phosphate pH 7.4 and the indicated amount of NaCl. For low salt measurements the buffer contained 5 mM NaCl, 5 mM potassium phosphate at pH 7.4 and 0.67 M (5% v/v) glycerol (to prevent non-specific aggregation of the proteins).
Protein preparation
Plasmids used for overproduction of SSB proteins were used to transform the E.coli strain BT317 (23) containing the plasmid pRK248 coding for the thermosensitive λcI857 repressor. As they carry the ssb gene under control of the λ pL promotor, protein expression was induced by a temperature shift from 30 to 42°C and cells were harvested 3 h after induction. EcoSSB wild type was purified as described by Lohman et al. (24), whereas HsmtSSB and EcoSSB Q152* were purified as previously described for HsmtSSB (22).
The χ gene was amplified from genomic DNA of the E.coli strain JM109 (Stratagene, La Jolla, CA) by PCR using Pfu DNA polymerase (forward primer, 5′-CGG AAG CCC ATG GCA AAC GCG ACG TTC TAC CTT CTG GAC AAT-3′; reverse primer, 5′-GGA AGC TCG AGT CAT TTC CAG GTT GCC GTA TTC AGG TTG AAA-3′) and cloned into the NcoI and XhoI sites of pET-15b (Novagen, Madison, WI). Due to the usage of the NcoI site the second amino acid of χ was changed from Lys to Ala. After cloning the gene was completely sequenced and no further mutations were detected. For protein expression the plasmid was used to transform BL21(DE3) pLysS (Novagen). Protein expression and purification was carried out according to Xiao et al. (25) with the following modifications. The ATP–agarose column was omitted; after purification the protein was precipitated with 450 g/l (NH4)2SO4, dissolved in 20 mM HEPES pH 6.9, 0.5 mM EDTA, 2 mM DTT, 20% (v/v) glycerol and dialysed against 1 M NaCl, 1 mM EDTA, 2 mM DTT, 60% (v/v) glycerol, 20 mM potassium phosphate pH 7.4 and stored at –20°C. Analytical equilibrium ultracentrifugation gave a molar mass of 16.5 kg/mol, proving the χ protein to be monomeric.
Analytical ultracentrifugation
Analytical ultracentrifugation analysis of protein–protein interactions was carried out with a Beckman/Coulter XL-A analytical ultracentrifuge using the AN 50 Ti rotor at 20°C. Concentration profiles were measured with a photoelectric scanner. Sedimentation rate constants and concentrations of the interacting species were determined by fitting an integrated multi-Gaussian distribution to the observed concentration profiles, as described previously (26). n and K for the interaction of χ and EcoSSB were derived from non-linear least squares fitting of the observed χbound/EcoSSBtotal data to a simple interaction model of n χ molecules with one tetrameric EcoSSB using
(χbound/EcoSSBtotal) = [(nKAssχfree)/(1 + KAssχfree)].
To analyze the distribution of species in a sedimentation velocity experiment the radial positions in all sedimentation profiles of an experiment were transformed to a sedimentation constant coordinate s with
s(x,t) = ln(x/xm)/(∫t0ω2dt′) (27)
and
c(s) = c(x,t)·(x/xm)2.
where x(xm) is the radial position (of the meniscus), s is the sedimentation rate constant (as the new independent variable), t is time and ω is the angular velocity. This procedure causes all profiles to coincide on the abscissa and to be corrected for radial dilution. For ease of comparison the first derivative of c(s) with respect to s is plotted and the result of this parameterization is a distribution of sedimentation constants.
Surface plasmon resonance
Experiments to detect macromolecular interactions by surface plasmon resonance (SPR) were performed with a BIAcore™ biosensor system 3000 (BIAcore Inc., Uppsala, Sweden). EcoSSB protein corresponding to 1028 response units (RU) was attached to a CM5 research grade sensor chip (BIAcore Inc.) using 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide and N-hydroxysuccinimide activation chemistry as described in the manufacturer’s instructions. After protein coupling remaining carbodiimide groups were blocked with ethanolamine. One of the four flow cells (FC) of each four flow cell chip was reacted with ethanolamine alone to serve as a control for non-specific binding. Data collected from this reference cell, FC1, were subtracted as a blank from the data of the EcoSSB-coated FC. Thus, all data shown are difference signals (e.g. FC2 – FC1). Binding experiments were carried out in standard buffer containing 0.3 M NaCl or 0.15 M NaCl (SPR buffer) [to make data comparable to those reported by Kelman et al. (15)] at 20°C at a flow rate of 10 µl/min. Measurements at 5 mM NaCl were done with the low salt buffer mentioned in Buffers and reagents. The interaction between χ and EcoSSB was investigated by injecting different concentrations of χ (e.g. from 40 nM up to 29 µM in random order) over the EcoSSB surface followed by a 60 s washing step. The amount of bound χ protein was determined from the steady-state plateau height 20 s after injection start. After data collection 0.1 M glycine pH 9.5 was injected as a regeneration step to remove the bound χ protein.
The interaction of EcoSSB and χ in the presence of ssDNA was investigated by immobilization of 686 RU of biotinylated-(dT)65 (MWG Biotech, Germany) on a research grade SA (streptavidin) sensor chip. The EcoSSB/ssDNA complex was prepared by injection of EcoSSB, yielding 2893 RU of bound EcoSSB. After EcoSSB injection a 360 s washing step was included. Interaction between χ and the EcoSSB/(dT)65 complex was investigated by injection of different concentrations of χ (0.5–30 µM in random order) followed by a 90 s washing step. The amount of bound χ protein was determined as described above. After regeneration using 0.1 M glycine pH 9.5 the EcoSSB/(dT)65 complex was reloaded (v.s.) with EcoSSB to ensure complete saturation of the ssDNA for the next injection of χ. As a reference signal for non-specific binding we substracted data from an empty FC. Buffers, temperature and flow rate were as described above.
Data collection and programming of the BIAcore™ 3000 were carried out using BIAcore 3000 Control Software ver. 3.2, data analysis with BIAevaluation ver. 3.2. RUmax and KA were determined by fitting a binding isotherm to the data:
RU([χ]) = RUmax·KA·[χ]/(KA·[χ] + 1)
The stoichiometry n was derived from the ratio of RUmax to RU(EcoSSBbound) assuming measured RU values to be proportional to the mass increase on the surface independent of the sort of protein bound:
n = [RUmax/RU(EcoSSBbound)] · [MEcoSSB/Mχ]
with M denoting the molar masses of the respective proteins.
DNA melting curves
DNA melting curves were measured in a DMR 10 (Zeiss, Oberkochen, Germany) spectrophotometer as described previously (28) using a heating rate of 20 K/h. The experiments were carried out with 38 µM nucleotides of poly(dA-dT) in standard buffer containing 0.075 M NaCl. Depression of the melting point of poly(dA-dT) was induced by addition of 1.27 µM of the respective SSB protein and varying concentrations of χ protein. No significant differences between heating and cooling curves could be observed, confirming the reversibility of melting. The melting curves in the presence of χ could only be measured up to 42°C since heat denaturation of χ occurs at higher temperatures.
RESULTS
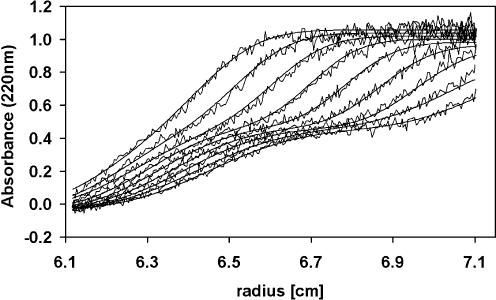
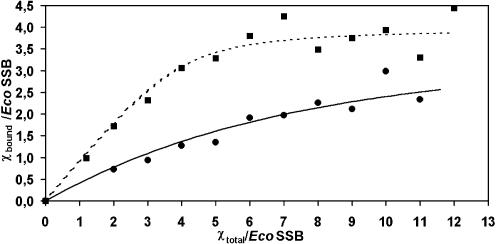
Whenever two macromolecules interact with each other the resulting complex will have a larger mass than any of its constituents and thus will in almost every case sediment faster than any of its components. Figure 1 shows the sedimentation of a complex between EcoSSB and the χ subunit of DNA polymerase III under low salt conditions. The sedimentation coefficients of EcoSSB and χ were determined to be s20,w = 4.4 S and s20,w = 1.8 S, respectively, whereas the saturated complex of EcoSSB and χ sediments at ∼5.6 S. Whenever free χ protein is present in an EcoSSB/χ mixture, two sedimenting boundaries will be observed. The slow sedimenting boundary represents free χ protein while the fast sedimenting one represents the EcoSSB/χ complex as well as free EcoSSB. A general and quantitative description of such a sedimentation of interacting macromolecules deduced from Lamm’s differential equation (29) has been presented before (30). Analyzing the sedimenting boundaries in terms of integral Gaussian distributions using previously described methods (26) yields the relative amounts of free and bound χ protein. These were used to construct a binding isotherm as depicted in Figure 2. From this binding isotherm an equilibrium constant of 4.0 × 105 M–1 for independent binding of four χ proteins to a single tetrameric EcoSSB molecule was deduced.
Figure 1.
Analytical ultracentrifugation of EcoSSB and χ in low salt. Sedimentation velocity experiment with 0.5 µM EcoSSB and 3 µM χ in 5 mM NaCl, 5 mM potassium phosphate pH 7.4, 0.67 M glycerol at 45 000 r.p.m. The absorbance was detected at 220 nm at intervals of 23 min. The smooth lines represent theoretical values calculated as the sum of two integrated Gaussian distributions (26).
Figure 2.
Interaction of EcoSSB or EcoSSB/poly(dT) complex with χ in low salt. 0.5 µM EcoSSB (circles) or a complex of 0.5 µM EcoSSB and 17.5 µM nucleotides of poly(dT) (squares) in 5 mM NaCl, 5 mM potassium phosphate pH 7.4, 0.67 M glycerol were mixed with different concentrations of χ and analyzed in an analytical ultracentrifuge. Free and bound χ concentrations were determined from the sedimentation velocity profiles fitted to a sum of two integrated Gaussian distributions (26). The lines represent the best non-linear least-squares fit with a simple interaction model of n χ molecules with one EcoSSB tetramer using the following parameters: n = 4, K = (4.0 ± 1) × 105 M–1 (solid line); n = 4, K = (7.4 ± 1) × 106 M–1 (dotted line).
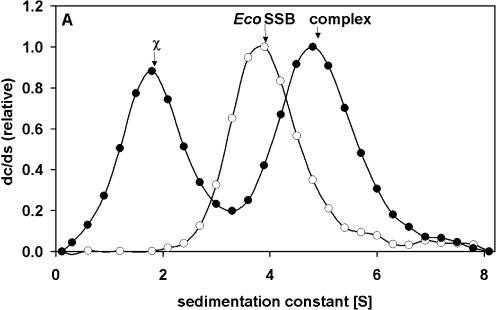
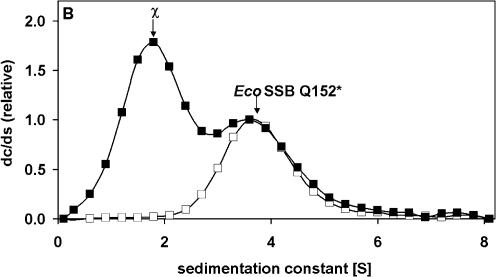
In low salt buffer a stoichiometric complex of EcoSSB and poly(dT) (length ∼900 nt) sediments with s20,w = 22.3 S. In the presence of χ a ternary complex is formed, which at saturation sediments at ∼35 S. From the concentration of free ligand observed in a slow sedimenting boundary and the known total amount of proteins present, a binding isotherm (as shown in Fig. 2) can be evaluated. Assuming four χ proteins bind to a single EcoSSB [in turn engaged in a complex with poly(dT)], an affinity constant of 7.4 × 106 M–1 can be estimated from the best fit shown in Figure 2. At low salt concentrations (5 mM NaCl) the affinity of χ for an EcoSSB/ssDNA complex is enhanced compared to the affinity for EcoSSB alone. Using high salt conditions (0.3 M NaCl) no differences in the binding isotherms in the absence or presence of poly(dT) could be detected, the affinity of (2 ± 1) × 105 M–1(data not shown) is similar to the affinity found in low salt experiments without ssDNA. A sedimentation coefficient distribution of such a complex is depicted in Figure 3. EcoSSB alone sediments with an s value of 3.8 S; in the presence of an excess of χ (1.8 S) a complex is formed which sediments at 4.8 S (Fig. 3A). The sedimentation coefficient of EcoSSB Q152*, a truncated version of EcoSSB missing the last 26 amino acids (21), remains unchanged in the presence of χ (Fig. 3B). Thus, the C-terminus of EcoSSB is essential for the interaction with χ.
Figure 3.
Specific interaction of EcoSSB and χ in high salt detected by analytical ultracentrifugation. (A) 2.5 µM EcoSSB (open circles), mixture of 2.5 µM EcoSSB and 20 µM χ (filled circles). (B) 2.5 µM EcoSSB Q152* (open squares), mixture of 2.5 µM EcoSSB Q152* and 20 µM χ (filled squares). Data were normalized relative to the height of the free SSB peak. Lines were drawn just to guide the eye. The differential sedimentation coefficient distributions clearly show that a complex between EcoSSB and χ sediments faster than EcoSSB alone, whereas the sedimentation of EcoSSB Q152* is not influenced by the presence of χ.
In sedimentation analysis we tested whether χ can directly interact with single-stranded nucleic acid. Using 0.83 µM (dT)65 and a 10-fold excess of χ no complex formation could be detected in standard buffer containing 0.05 M NaCl (data not shown). This confirms the findings of previous studies which failed to detect an interaction between χ and single-stranded nucleic acid using SPR (15,18).
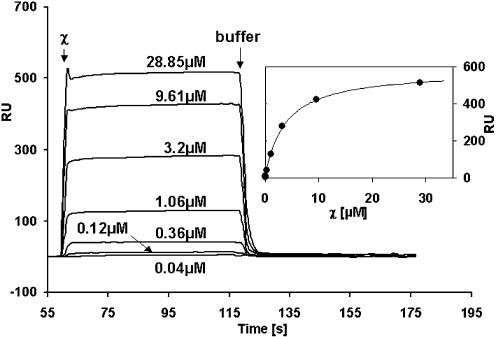
A further possibility to test the interaction of two proteins is SPR. We used this method to test the binding of χ to EcoSSB which had been immobilized on a SPR chip. Figure 4 shows measured SPR profiles using varying χ concentrations in standard buffer containing 0.15 M NaCl (SPR buffer). At concentrations at which the magnitude of the SPR signal created by binding of χ to the EcoSSB-coated surface was large enough to be detected, the rate of binding to the surface was too fast to be measured (cf. Figs 4 and 5). Thus, equilibrium (plateau) values of the individual profiles as a function of χ concentration were used to determine binding affinities (Fig. 4, inset). Assuming a model of independent identical binding sites for χ on the EcoSSB tetramer, a least- squares fit yielded a binding constant of 2.7 × 105 M–1. A similar result was obtained at 0.3 M NaCl (data not shown). From the RUmax value obtained in the fit a stoichiometry of approximately 2.5 χ per EcoSSB tetramer can be deduced. In a similar SPR experiment Kelman et al. (15) observed a stoichiometry of 2.2. Both values are significantly lower than the stoichiometry deduced in this study, using analytical ultracentrifugation. Since our analytical ultracentrifugation data are incompatible with the apparent stoichiometry observed in SPR experiments, it is most likely that the chemical coupling of EcoSSB to the chip surface results in an occlusion of possible binding sites.
Figure 4.
χ interacts with immobilized EcoSSB. In SPR experiments varying concentrations of χ in standard buffer containing 0.15 M NaCl were passed over 1028 RU of EcoSSB immobilized on a CM5 sensor chip. Data collected from control injections of χ over a reference cell reacted with ethanolamine were subtracted as a blank. After 60 s buffer was injected to wash off the χ protein. (Inset) From plateau height the amount of bound χ protein was determined and plotted versus the injected χ concentration. The solid line represents the best fit to a simple interaction model using the following parameters: K = (2.7 ± 0.5) × 105 M–1, RUmax = 580 RU.
Figure 5.
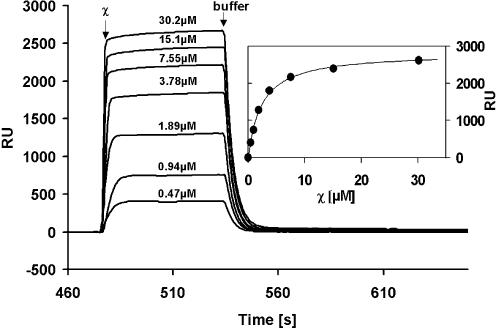
χ interacts with EcoSSB bound to immobilized (dT)65. In SPR experiments varying concentrations of χ in standard buffer containing 0.15 M NaCl were passed over 2893 RU of EcoSSB bound to 686 RU of biotinylated (dT)65 immobilized on a streptavidin sensor chip. Data collected from control injections of χ over an empty reference cell were substracted as a blank. After 60 s buffer was injected to wash off the χ protein. (Inset) From plateau height the amount of bound χ protein was determined and plotted versus the injected χ concentration. The solid line represents the best fit to a simple interaction model using the following parameters: K = (4.2 ± 0.5) × 105 M–1, RUmax =2823 RU.
EcoSSB binds to the oligonucleotide (dT)65 with affinites considerably larger than 109 M–1 (31,32). However, when trying to bind the oligonucleotide to EcoSSB, which had been covalently attached to an SPR chip, we could not detect an appreciable signal, probably due to the electrostatic repulsion between the negatively charged surface of the chip and (dT)65. Thus, to investigate the influence of ssDNA on the EcoSSB/χ interaction, biotinylated (dT)65 was bound to a streptavidin chip and saturated with EcoSSB, before varying concentrations of χ were passed over the resultant immobilized complex (Fig. 5). The evaluation of the binding isotherm (v.s.) yielded a binding constant of 4.2 × 105 M–1 and a stoichiometry of approximately 4.4 χ per EcoSSB tetramer (Fig. 5, inset). Thus, the measured binding constants with and without nucleic acid were seen to be in the same range. The measured stoichiometry of the EcoSSB/χ complex in the presence of DNA is approximately twice that observed in its absence, which is consistent with previous studies (15). This apparent increase in stoichiometry is most likely due to the fact that EcoSSB was attached to the SPR chip via its natural oligonucleotide ligand in this experiment and hence all binding sites are accessible for χ. This interpretation is supported by the results obtained in solution using analytical ultracentrifugation (v.s.).
At 5 mM NaCl, binding of χ to a surface on which EcoSSB has been immobilized could be detected qualitatively as an increase in RU signal, whether or not the EcoSSB had been immobilized to the SPR chip directly or via (dT)65 oligonucleotide. However, these experiments failed to produce reliable quantitative results. A possible reason for this failure is the inherent negative charge of the dextran matrix on the chip, whose effects become overwhelmingly large at low ionic strengths.
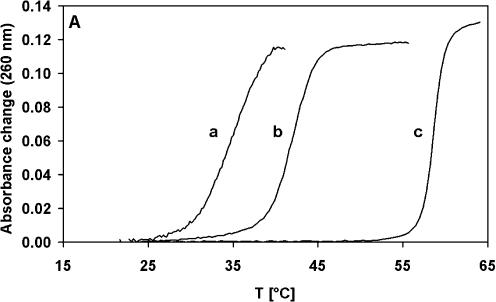
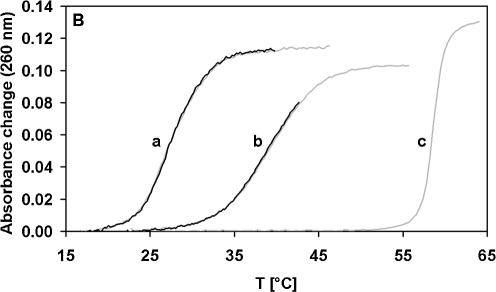
EcoSSB specifically binds single- but not double-stranded DNA and, as a consequence, destabilizes the double strand. Thus, the depression of double-stranded DNA melting temperature by EcoSSB is a measure of the affinity of the protein for ssDNA (7). In the case of poly(dA-dT), saturating amounts of EcoSSB reduce the melting temperature from 59 to 42°C. Addition of an excess of χ over EcoSSB results in a further depression of the melting temperature to 35°C (Fig. 6A). In the presence of EcoSSB Q152*, a truncated version of EcoSSB missing the last 26 amino acids (21), or the human mitochondrial SSB protein (22) the melting point of poly(dA-dT) is also depressed, but addition of χ (Fig. 6B) has no further influence on the melting behavior. Thus, we conclude that binding of χ to EcoSSB enhances the ssDNA binding affinity of EcoSSB and that a specific interaction between the C-terminus of EcoSSB and χ is responsible for this effect.
Figure 6.
χ enhances the stability of the EcoSSB/ssDNA complex. 38 µM nucleotides of poly(dA-dT) were melted in standard buffer containing 0.075 M NaCl in the presence of 1.26 µM SSB protein with and without 6.33 µM χ. (A) EcoSSB and χ (a), EcoSSB (b), no protein (c). (B) EcoSSB Q152* (a), HsmtSSB (b), no protein (c). The gray curves are measured in the absence, the black ones in the presence of χ.
DISCUSSION
In this study we have demonstrated a direct physical interaction between EcoSSB and the DNA polymerase III χ subunit independent of the conditions used. We went on to demonstrate that this interaction stabilizes the complex formed between EcoSSB and ssDNA.
Under low salt (5 mM NaCl) conditions in an analytical ultracentrifuge the interaction of χ with an EcoSSB/ssDNA complex is ∼20-fold stronger than the interaction between χ and EcoSSB alone. Since we and others (15,18) have not been able to observe a direct interaction between χ and ssDNA, this increase in affinity may be an enhancement of the protein–protein interaction. When binding to ssDNA a conformational change of EcoSSB was observed which makes the C-terminus more prone to proteolytic digestion (7). We show that χ interacts with this C-terminus of EcoSSB and thus the conformational change is a likely mechanism for the enhancement of EcoSSB/χ interaction by ssDNA. However, a very weak binding of χ to the ssDNA in this complex cannot be excluded. Addition of salt (0.3 M NaCl) renders the interaction of EcoSSB and χ independent of the presence of ssDNA. We observed a similar result at 0.15 M NaCl using SPR. At 5 mM NaCl our SPR experiments showed binding, but could only be interpreted qualitatively.
In a similar study Kelman and co-workers demonstrated an interaction of χ and EcoSSB at 0.15 M NaCl by SPR experiments (15). However, under low salt conditions using gel filtration experiments the same authors failed to show an interaction between χ and EcoSSB unless ssDNA was present. One possible reason for the discrepancy between these experiments and our demonstration of an EcoSSB/χ interaction at low salt conditions using analytical ultracentrifugation may be due to the fact that the weakly interacting proteins cannot migrate as a complex in a single band in the gel filtration column if the dissociation is too fast. However, in our sedimentation velocity experiments the complex between EcoSSB and χ sediments in the presence of a constant concentration of free χ protein. This allows the detection of a weak and rapidly equilibrating complex.
At low salt concentrations EcoSSB and χ do not influence the activity of DNA polymerase III in a replication assay. At high salt concentrations the presence of EcoSSB and χ increases the processivity of the polymerase (15,18). Thus, the interaction between EcoSSB and χ is responsible for an increase in activity of DNA polymerase III at elevated salt concentrations. In this study we could demonstrate a physical interaction between EcoSSB and χ even at high salt concentrations (0.3 M NaCl).
We could also show that the interaction between EcoSSB and χ at high salt concentrations is highly specific. A truncated version of EcoSSB missing the last 26 amino acids (EcoSSB Q152*) does not interact with the χ protein. Thus, the C-terminus of EcoSSB is essential for the interaction of EcoSSB and χ. This adds to the finding that the interaction of χ and EcoSSB is weakened by the exchange of the penultimate amino acid of EcoSSB (EcoSSB P176S) (15).
The interaction of EcoSSB and a complex of DNA polymerase III subunits χ and ψ has been determined by Glover and McHenry to have a binding constant of 3.7 × 105 M–1 (18). Our present study shows that this interaction is a result of the direct formation of a SSB/χ complex that has a similar affinity (2.7 × 105 M–1). We also show by analytical ultracentrifugation that including ssDNA in the χ/EcoSSB binding assay leads to an ∼20-fold enhancement of the affinity at low salt and leaves the affinity almost unchanged at high salt. Thus, the 1000-fold increase reported (18) for the interaction of EcoSSB and χ in the presence of ssDNA and the ψ and τδδ′ subunits of DNA polymerase III is most probably an effect of the additional protein subunits.
Since the interaction of EcoSSB and χ is influenced by the presence of ssDNA, and χ does not interact measurably with ssDNA, we questioned how the interaction between EcoSSB and χ influences the DNA binding properties of EcoSSB. Our DNA melting curves show that the stability of the ternary complex of EcoSSB, ssDNA and χ is greater than the stability of the binary complex without χ. The DNA binding properties of other SSB proteins (HsmtSSB and EcoSSB Q152*) which do not physically interact with χ remain unchanged. Interestingly, the double-stranded DNA destabilization effect of χ and EcoSSB together is similar to the effect of the C-terminal deletion mutant EcoSSB Q152* alone. The electrostatic repulsion between this negatively charged C-terminus and the ssDNA weakens the SSB–DNA interaction (7,21). This weakening is possibly overcome by shielding of the C-terminus through χ binding.
Through the enhancement of ssDNA binding affinity, χ ties EcoSSB to the replication fork. During DNA synthesis at the lagging strand, DNA polymerase III removes nucleotide by nucleotide from the adjacent EcoSSB protein. EcoSSB can bind up to 65 nt and the binding affinity decreases drastically with decreasing length of the oligonucleotide (33). This lowered affinity would result in the dissociation of EcoSSB from the lagging strand in the absence of interactions between χ and EcoSSB protein. The released DNA could build a hairpin structure and the DNA polymerase would be inhibited. Stabilization of the EcoSSB/DNA complex through interaction with χ could prevent the premature dissociation of EcoSSB and therefore the inhibition of DNA polymerase III. This would result in an increase in the processivity of the polymerase.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Lidia Litz for expert technical assistance, Dr Joachim Greipel for valuable discussions and Dr Dan Mulvihill for critically reading the manuscript. The BIAcore™ machine was kindly put at the disposal of G.W. as a lottery win.
REFERENCES
- 1.Greipel J., Urbanke,C. and Maass,G. (1989) The single-stranded DNA binding protein of Escherichia coli. Physicochemical properties and biological functions. In Saenger,W. and Heinemann,U. (eds), Protein-Nucleic Acid Interaction. Macmillan, London, UK, pp. 61–86. [Google Scholar]
- 2.Meyer R.R. and Laine,P.S. (1990) The single stranded DNA binding protein of E. coli. Microbiol. Rev., 54, 342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sigal N., Delius,J., Kornberg,T., Gefter,M.L. and Alberts,B.M. (1972) A DNA-unwinding protein isolated from Escherichia coli: its interaction with DNA and with DNA polymerases. Proc. Natl Acad. Sci. USA, 69, 3537–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molineux I.J. and Gefter,M.L. (1975) Properties of the Escherichia coli DNA binding (unwinding) protein: interaction with nucleolytic enzymes and DNA. J. Mol. Biol., 98, 811–825. [DOI] [PubMed] [Google Scholar]
- 5.Porter R.D., Black,S., Pannuri,S. and Carlson,A. (1990) Use of the Escherichia coli ssb gene to prevent bioreactor takeover by plasmidless cells. Biotechnology, 8, 47–51. [DOI] [PubMed] [Google Scholar]
- 6.Tomaska L., Nosek,J. and Kucejova,B. (2001) Mitochondrial single-stranded DNA-binding proteins: in search for new functions. Biol. Chem., 382, 179–186. [DOI] [PubMed] [Google Scholar]
- 7.Williams K.R., Spicer,E.K., Lopresti,M.B., Gugenheimer,R.A. and Chase,J.W. (1983) Limited proteolysis studies on the E. coli single-stranded DNA binding protein. J. Biol. Chem., 258, 3346–3355. [PubMed] [Google Scholar]
- 8.Webster G., Genschel,J., Curth,U., Urbanke,C., Kang,C.H. and Hilgenfeld,R. (1997) A common core for binding single-stranded DNA: structural comparison of the single-stranded DNA-binding proteins (SSB) from E.coli and human mitochondria. FEBS Lett., 411, 313–316. [DOI] [PubMed] [Google Scholar]
- 9.Yang C., Curth,U., Urbanke,C. and Kang,C.H. (1997) Crystal structure of human mitochondrial single stranded DNA binding protein at 2.4 Å resolution. Nature Struct. Biol., 4, 153–157. [DOI] [PubMed] [Google Scholar]
- 10.Raghunathan S., Ricard,C.S., Lohman,T.M. and Waksman,G. (1997) Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-Å resolution. Proc. Natl Acad. Sci. USA, 94, 6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genschel J., Curth,U. and Urbanke,C. (2000) Interaction of E.coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol. Chem., 381, 183–192. [DOI] [PubMed] [Google Scholar]
- 12.Tiranti V., Rocchi,M., DiDonato,S. and Zeviani,M. (1993) Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB). Gene, 126, 219–225. [DOI] [PubMed] [Google Scholar]
- 13.Chase J.W., l’Italien,J.J., Murphy,J.B., Spicer,E.K. and Williams,K.R. (1984) Characterization of the E. coli ssb113 mutant single-stranded DNA binding protein. Cloning of the gene, DNA and protein sequence analysis, HPLC, peptide mapping and DNA binding studies. J. Biol. Chem., 259, 805–814. [PubMed] [Google Scholar]
- 14.Umezu K. and Kolodner,R.D. (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem., 269, 30005–30013. [PubMed] [Google Scholar]
- 15.Kelman Z., Yuzhakov,A., Andjelkovic,J. and O’Donnell,M. (1998) Devoted to the lagging strand—the χ subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J., 17, 2436–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelman Z. and O’Donnell,M. (1995) DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu. Rev. Biochem., 64, 171–200. [DOI] [PubMed] [Google Scholar]
- 17.Urbanke C. and Schaper,A. (1990) Kinetics of binding of single-stranded DNA binding protein from Escherichia coli to single-stranded nucleic acids. Biochemistry, 29, 1744–1749. [DOI] [PubMed] [Google Scholar]
- 18.Glover B.P. and McHenry,C.S. (1998) The chi psi subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J. Biol. Chem., 273, 23476–23484. [DOI] [PubMed] [Google Scholar]
- 19.Gill S.C. and von Hippel,P.H. (1989) Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem., 182, 319–326. [DOI] [PubMed] [Google Scholar]
- 20.Lohman T.M. and Overman,L.B. (1985) Two binding modes in E. coli single-strand binding protein (SSB) single-stranded DNA complexes: modulation by NaCl concentration. J. Biol. Chem., 260, 3594–3603. [PubMed] [Google Scholar]
- 21.Curth U., Genschel,J., Urbanke,C. and Greipel,J. (1996) In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res., 24, 2706–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curth U., Urbanke,C., Greipel,J., Gerberding,H., Tiranti,V. and Zeviani,M. (1994) Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur. J. Biochem., 221, 435–443. [DOI] [PubMed] [Google Scholar]
- 23.Landwehr M., Curth,U. and Urbanke,C. (2002) A dimeric mutant of the homotetrameric single-stranded DNA binding protein from Escherichia coli. Biol. Chem., 383, 1325–1333. [DOI] [PubMed] [Google Scholar]
- 24.Lohman T.M., Green,J.M. and Beyer,R.S. (1986) Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry, 25, 21–25. [DOI] [PubMed] [Google Scholar]
- 25.Xiao H., Crombie,R., Dong,Z., Onrust,R. and O’Donnell,M. (1993) DNA polymerase III accessory proteins. III. holC and holD encoding chi and psi. J. Biol. Chem., 268, 11773–11778. [PubMed] [Google Scholar]
- 26.Machner M.P., Urbanke,C., Barzik,M., Otten,S., Sechi,A.S., Wehland,J. and Heinz,D.W. (2001) ActA from Listeria monocytogenes can interact with up to four Ena/VASP homology 1 domains simultaneously. J. Biol. Chem., 276, 40096–40103. [DOI] [PubMed] [Google Scholar]
- 27.Gralén N. and Lagermalm,G. (1952) A contribution to the knowledge of some physico-chemical properties of polystyrene. J. Phys. Chem., 56, 514–523. [Google Scholar]
- 28.Augustyns K., Van Aerschot,A., Van Schepdael,A., Urbanke,C. and Herdewijn,P. (1991) Influence of the incorporation of (S)-9-(3,4-dihydroxybutyl)adenine on the enzymatic stability and base-pairing properties of oligodeoxynucleotides. Nucleic Acids Res., 19, 2587–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamm O. (1929) Die Differentialgleichung der Ultrazentrifugierung. Arkiv Matematik Astronomi Fysik, 21B, 1–4. [Google Scholar]
- 30.Krauss G., Pingoud,A., Boehme,D., Riesner,D., Peters,F. and Maass,G. (1975) Equivalent and non-equivalent binding sites for tRNA on aminoacyl-tRNA synthetases. Eur. J. Biochem., 55, 517–529. [DOI] [PubMed] [Google Scholar]
- 31.Bujalowski W. and Lohman,T.M. (1989) Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I. Evidence and a quantitative model. J. Mol. Biol., 207, 249–268. [DOI] [PubMed] [Google Scholar]
- 32.Bujalowski W. and Lohman,T.M. (1989) Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II. Salt, temperature and oligonucleotide length effects. J. Mol. Biol., 207, 269–288. [DOI] [PubMed] [Google Scholar]
- 33.Lohman T.M. and Bujalowski,W. (1990) E.coli single strand binding protein: multiple single stranded DNA binding modes and cooperativities. In Revzin,A. (ed.), The Biology of Non-specific Protein DNA Interactions. CRC Press, Boca Raton, FL, pp. 131–168. [Google Scholar]