Abstract
Oxidation of membrane-bound quinol molecules is a central step in the respiratory electron transport chains used by biological cells to generate ATP by oxidative phosphorylation. A novel family of cytochrome c quinol dehydrogenases that play an important role in bacterial respiratory chains was recognised in recent years. Here, we describe the first structure of a cytochrome from this family, NrfH from Desulfovibrio vulgaris, which forms a stable complex with its electron partner, the cytochrome c nitrite reductase NrfA. One NrfH molecule interacts with one NrfA dimer in an asymmetrical manner, forming a large membrane-bound complex with an overall α4β2 quaternary arrangement. The menaquinol-interacting NrfH haem is pentacoordinated, bound by a methionine from the CXXCHXM sequence, with an aspartate residue occupying the distal position. The NrfH haem that transfers electrons to NrfA has a lysine residue from the closest NrfA molecule as distal ligand. A likely menaquinol binding site, containing several conserved and essential residues, is identified.
Keywords: cytochrome c nitrite reductase, NapC-NirT family, NrfH/NrfA membrane protein complex, quinol dehydrogenase
Introduction
Prokaryotes are key players in the terrestrial recycling of many elements, such as carbon, nitrogen, sulphur and iron. They are capable of transforming inorganic molecules containing these elements, and have an astounding metabolic diversity. These microorganisms explore almost every possible energy source and oxidant available on earth, and are able to sustain life in the most extreme environments. In contrast to higher organisms, prokaryotes also display a great versatility in their respiratory chains (Richardson, 2000). This is due to the coordinated expression of a diversity of dehydrogenases and reductases that feed electrons to, or receive electrons from, membrane-bound quinone electron carriers. Reduction and oxidation of quinone/quinol molecules are central steps in respiratory electron transport chains, used by biological cells to generate a proton-motive force across the membrane that drives synthesis of ATP. The study of bacterial membrane complexes involved in these energy-conserving steps has permitted a molecular understanding of such fundamental energy-generating biological processes.
A new group of proteins that oxidise menaquinol, and transfer electrons to periplasmic reductases of various inorganic compounds, were identified in Proteobacteria (Roldan et al, 1998; Simon, 2002), one of the main lineages of the Bacteria domain. They are membrane-associated cytochromes comprising an N-terminal transmembrane helix and a hydrophilic globular domain that binds four haems c. This contrasts with the better known quinone-interacting cytochromes b found in several respiratory complexes, which are integral membrane proteins (Berks et al, 1995). Phylogenetic analyses show that these cytochromes c are more widely distributed among the Proteobacteria than the bc1 complex, reflecting the important role they play in their flexible respiratory chains. Examples include NrfH, the electron donor to the pentahaem cytochrome c nitrite reductase NrfA (Simon et al, 2000); NapC, the electron donor to the nitrate reductase NapA (Roldan et al, 1998; Cartron et al, 2002); NirT, thought to donate electrons to the cd1 nitrite reductase (Jungst et al, 1991); and CymA, which is apparently required in electron transfer for the reduction of various substrates like nitrate, fumarate, FeIII and MnIV (Schwalb et al, 2003). A subgroup of this family includes the cytochromes TorC, DmsC and DorC that are involved in electron transfer to the trimethylamine N-oxide and dimethylsulphoxide reductases (Ujiiye et al, 1996; Shaw et al, 1999; Gon et al, 2001), and which include an additional monohaem domain that is responsible for electron transfer from the tetrahaem domain to the reductases. The NrfH cytochrome is more widespread among bacteria than other members of the family (Gross et al, 2005). In Wolinella succinogenes, NrfH has been shown to mediate electron transport from menaquinol to NrfA (Simon et al, 2000). A mutant lacking NrfH was unable to grow by nitrite respiration and had NrfA exclusively in the soluble cell fraction, showing that NrfH anchors the NrfHA complex to the membrane and is essential for electron transport (Simon et al, 2001).
Many quinone-reacting cytochromes b participate in the generation of a proton-motive force through a redox-loop mechanism in enzymes that have the active sites for the substrate and the quinone on opposite sides of the membrane (Jones et al, 1980; Richardson and Sawers, 2002; Jormakka et al, 2003). The fundamental question of whether energy conservation can be associated with electron transport in cytochrome c quinol dehydrogenases has so far not been answered, as the site of menaquinol oxidation was unknown. Results with W. succinogenes NrfHA complex reconstituted in proteoliposomes indicated that reduction of nitrite by menaquinol is an electroneutral process, and therefore protons resulting from menaquinol oxidation are thought to be liberated to the periplasm where they balance the protons consumed by nitrite reduction (Simon, 2002).
Here, we describe the first X-ray structure of the cytochrome c quinol dehydrogenase, NrfH, from the sulphate-reducing δ-proteobacterium Desulfovibrio vulgaris Hildenborough (Pereira et al, 2000). NrfH was crystallised as a membrane complex with its electron acceptor NrfA (Rodrigues et al, 2006). In most organisms, the NrfA nitrite reductase catalyses the last step in the anaerobic respiratory process of nitrate (or nitrite) ammonification (Simon, 2002). However, some sulphate-reducing bacteria, such as D. vulgaris, express an NrfHA nitrite reductase even though they cannot grow by nitrite ammonification (Pereira et al, 2000). In these organisms, the nitrite reductase enables them to overcome inhibition by nitrite, produced by sulphide-oxidising nitrate-reducing bacteria in their common habitat (Greene et al, 2003).
The structure reveals that D. vulgaris NrfH has a highly unusual haem coordination, and forms a strong complex with the NrfA dimer displaying an asymmetrical haem arrangement. NrfH haem 1 is a methionine-coordinated high-spin haem that is unique in biological systems. We propose a binding site for the menaquinol molecule close to haem 1 that includes several conserved and essential residues. This binding site is at the periplasmic interface of the membrane, indicating that protons from menaquinol oxidation are liberated to the periplasm so that reduction of nitrite by NrfHA is not associated with energy conservation.
Results and discussion
Complex architecture
The structure of the NrfHA complex was determined by a combination of molecular replacement and multiwavelength anomalous dispersion (MAD) methods (Rodrigues et al, 2006) and was refined to an R-factor of 20.1% (R-free 24.0%) at 2.3 Å resolution (Table I). The asymmetric unit contains six NrfH and 12 NrfA molecules forming three dimers of NrfHA2 units, where each membrane-anchored NrfH is tightly bound to two NrfA molecules (Figure 1A). This provides a rationale for the physiological relevance of the NrfA dimer, which has been highlighted in all NrfA crystal structures determined so far (Einsle et al, 1999, 2000; Bamford et al, 2002; Cunha et al, 2003). The large α4β2 assembly, corresponding to a dimer of NrfHA2 units, has overall dimensions of approximately 150 × 120 × 95 Å (Figure 2) and a total surface area of about 76 000 Å2. This dimer is predicted by PISA server (Krissinel and Henrick, 2004) as the most probable multimeric form. The interface area between two NrfHA2 units is 3175 Å2 (about 7.7% of the total surface area of this unit), and includes mainly electrostatic interactions between molecules of different NrfHA2 units. These observations indicate that the striking α4β2 arrangement corresponds to the biologically active form of the complex, which is further supported by the determined molecular mass of the complex in solution (ca. 300 kDa).
Table 1.
Data collection, phasing and refinement statistics
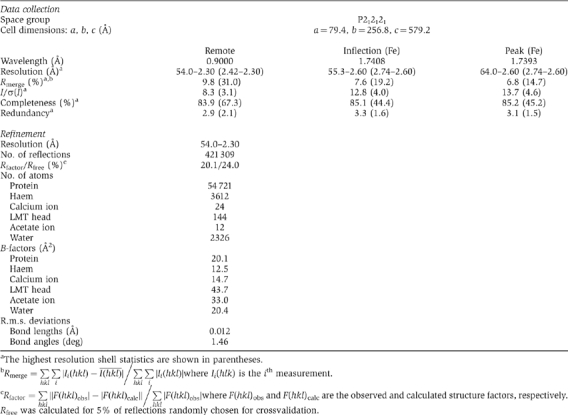 |
Figure 1.
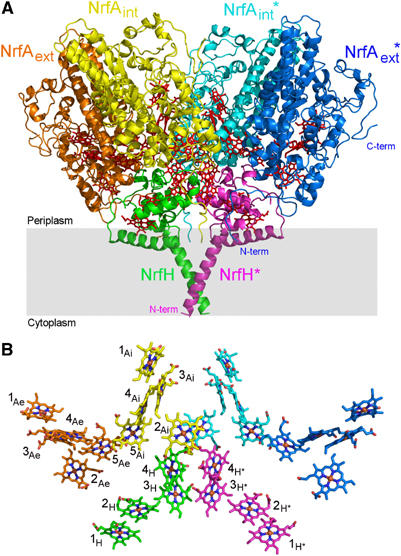
(A) Secondary structure of NrfHA viewed parallel to the membrane (grey rectangle) with haems drawn as red sticks. Each NrfH subunit, shown in green and magenta, is tightly bound to one NrfA dimer shown in orange/yellow and light blue/dark blue. The NrfA monomers located in the core of the complex are referred as internal (NrfAint), whereas the others are named external (NrfAext). Molecules from one NrfHA2 unit are labelled with *. (B) Ensemble of 28 haem groups in the same orientation as in (A). Iron, oxygen and nitrogen atoms are shown in brown, red and dark blue, respectively. Carbon atoms are coloured as each monomer in (A). Haems are numbered according to the order of their binding motifs in the protein chain. NrfAint, NrfAext and NrfH haems are labelled with Ai, Ae and H, respectively. All figures, except Figure 4, were prepared with Pymol (DeLano, 2002).
Figure 2.
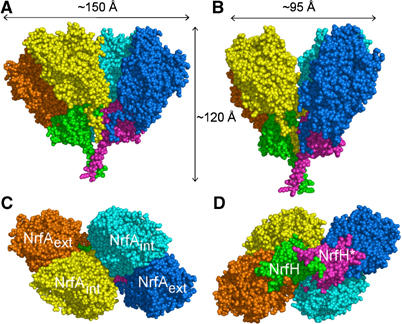
Representation of NrfHA complex in CPK rendering. The monomers are coloured according to Figure 1. (A) A view of the complex in a parallel orientation with respect to the membrane. (B) The complex viewed parallel to the membrane and rotated ∼60° from (A). (C) The complex perpendicular to the membrane normal and viewed from the periplasmic space. (D) The complex perpendicular to the membrane normal and rotated ∼180° from (C).
NrfH structure
NrfH is an α-helical protein (Figure 3A) with a novel protein fold, as no significantly similar structure is found by fold recognition analysis using DALI (Holm and Sander, 1999). The transmembrane helix, comprising residues Lys14 to Asp38, is included in our crystallographic model, whereas the cytoplasmic N-terminal region (residues 1–13) is not observed in the electron density maps. The helix from Leu75 to Leu95, which is approximately perpendicular to the transmembrane one (Figure 3A), should also be embedded in the membrane as it is mainly formed by hydrophobic residues. This helix is likely to contribute to the membrane attachment of NrfH as previous studies with NapC (Cartron et al, 2002) and CymA (Schwalb et al, 2003) showed that truncated forms without the transmembrane helix are still capable of oxidising menaquinol and transferring electrons to the corresponding periplasmic reductases. The two NrfH molecules in the complex are oriented so that the transmembrane helices are tilted in the membrane, crossing each other around Gly19. This arrangement brings NrfH haem 1 in close contact with the membrane, in a position that should be optimal for its interaction with menaquinol.
Figure 3.
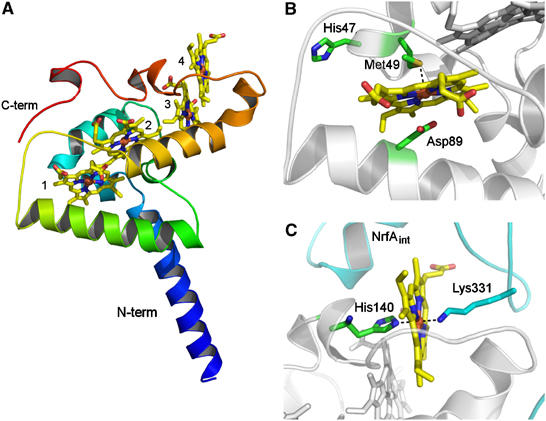
(A) Overall fold of NrfH. The polypeptide chain is ramp-coloured from blue (N-terminal) to red (C-terminal). The four c-type haems are shown in stick rendering and coloured by atom type (iron—brown, oxygen—red, nitrogen—blue, carbon—yellow). The two longer α-helices displayed in blue and green are inserted in the membrane. The haems, numbered according to their attachment to the protein, are arranged in a di-haem parallel motif, where the two pairs of haems are perpendicularly packed. (B) Zoomed view of haem 1 showing the proximal ligand, Met49 (∼2.8 Å distance from Fe), Asp89 (∼3 Å distance from Fe) and His47 from the CXXCHXM motif. These residues show C atoms in green, N in blue, O in red and S in gold. (C) Zoomed view of haem 4 showing its coordination (proximal ligand is His140 and distal one is Lys331 from an internal NrfA subunit, whose carbon atoms are coloured in cyan).
The globular domain of NrfH binds four haems c, which are arranged in two pairs (haems 1/2 and 3/4, numbered according to their order in the sequence) displaying a typical di-haem parallel stacking motif, where haems 2 and 3 are almost perpendicular to each other (Figure 3A). This reveals that the haem arrangement of NrfH is characteristic of a diverse family of multi-haem cytochromes c, whose haem groups are arranged in alternated stacking and perpendicular di-haem motifs (Iverson et al, 1998; Pereira and Xavier, 2005). This family includes NrfA and other proteins such as hydroxylamine oxidoreductase or the recently reported tetrathionate reductase (Mowat et al, 2004). Members of this family show no sequence identity or overall structural similarity, but the haem arrangement is conserved and some limited local similarity can be observed for the polypeptide fold surrounding the haems. These features are present in NrfH, with the closest relative in the family being cytochrome c554 of Nitrosomonas europea (Iverson et al, 1998) (DaliLite; Holm and Park, 2000; Z score around 3.5), whose haems 1, 3 and 4 can be superimposed with haems 1, 2 and 3 of NrfH, respectively.
NrfH haem coordination
The most notable feature of the NrfH structure is its surprising haem coordination. In particular, haem 1 displays unprecedented ligation, with a methionine residue, Met49 from the CXXCHXM motif, as proximal axial ligand (Sδ–Fe distance of about 2.8 Å) rather than the histidine of the CXXCH haem c-binding motif (Figure 3B). There is only one precedent in which this histidine is not an iron ligand: in Shewanella oneidensis tetrathionate reductase, where a distant lysine replaces the histidine as the proximal ligand to the catalytic haem (Mowat et al, 2004). In the case of NrfH, Met49 is located two residues downstream of the presumed histidine ligand. Met49 is conserved in NrfH proteins (Figure 4) and in most sequences of similar cytochromes like NapC (Roldan et al, 1998). Mutagenesis studies on W. succinogenes NrfH showed that this residue is essential for menaquinol oxidation (Gross et al, 2005). The second unexpected feature of NrfH haem 1 is the presence of an aspartate residue (Asp89) occupying the distal ligand position, with its carboxylate group approximately parallel to the haem plane and with its Oδ2 atom at ∼3 Å from the iron (Figure 3B). No continuous electron density is observed between the iron and the oxygen atoms, showing that Asp89 is not coordinated to the iron (Figure 5). Interestingly, this aspartate residue is replaced by a histidine in many NrfH proteins (a residue that is also essential for menaquinol oxidation in W. succinogenes NrfH; Gross et al, 2005) or a glutamate among NapC proteins (Figure 4). Thus, NrfH haem 1 is a methionine-coordinated high-spin haem, which, to the best of our knowledge, has never been described in biological systems.
Figure 4.
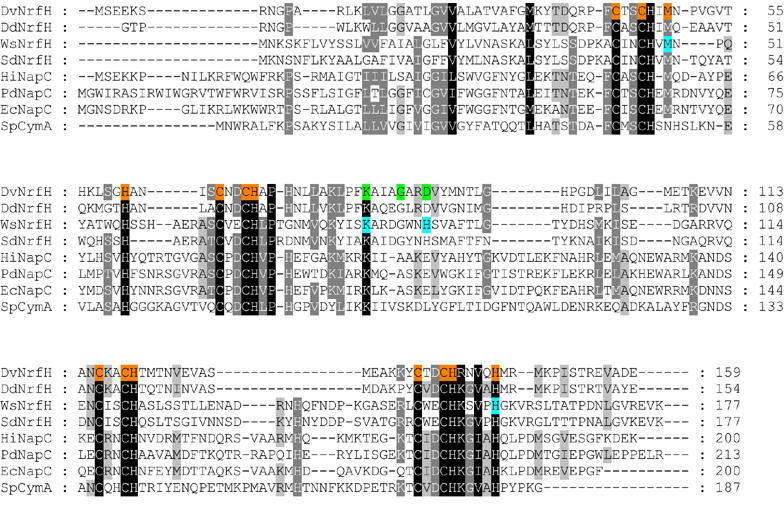
Multiple sequence alignment of NrfH homologous proteins. The primary sequence of D. vulgaris Hildenborough NrfH (DvNrfH), D. desulfuricans ATCC 27774 NrfH (DdNrfH), W. succinogenes NrfH (WsNrfH), Sulfurospirillum deleyianum NrfH (SdNrfH), Haemophilus influenzae NapC (HiNapC), Paracoccus denitrificans NapC (PdNapC), E. coli NapC (EcNapC) and Shewanella putrefaciens CymA (SpCymA) were aligned using ClustalW (Chenna et al, 2003). The PdNapC sequence was truncated at residue 213. D. vulgaris NrfH haem binding sites and ligands are highlighted in orange, key residues in the putative menaquinol binding site are shown in green and W. succinogenes NrfH residues that were shown to be essential for menaquinol oxidation (Gross et al, 2005) are indicated in cyan.
Figure 5.
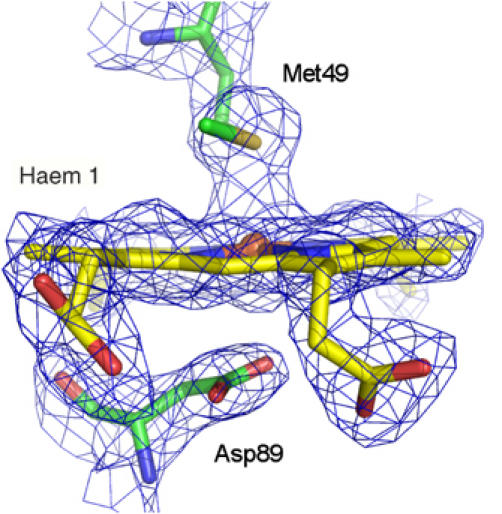
Electron density map around NrfH haem 1 (2Fo−Fc map contoured at 1.2σ). Haem 1 and its axial ligands are shown in sticks model. Same colour code as Figure 3B).
Previous EPR studies on the D. vulgaris NrfHA complex provided evidence for the presence of two high-spin haems (Pereira et al, 2000). One of these is the NrfA catalytic haem 1 and the other can now be assigned to NrfH haem 1. The signals for the high-spin haems are only observed during a redox titration, as in the oxidised native state both haems are EPR silent because they are spin-coupled to their neighbouring low-spin haems (Schumacher et al, 1994; Pereira et al, 2000; Bamford et al, 2002). High-spin haems are generally catalytic haems of enzymes where a substrate may bind, or ligand-binding haems in sensor and transport proteins. Neither of these cases seems to be applicable to NrfH haem 1, and its unique characteristics are probably related to its function as menaquinol-reacting haem. The fifth ligand of high-spin haems is most commonly a histidine, but cysteine thiolate ligation is present in cytochromes P-450 and tyrosine coordination in catalases. The presence of a methionine as fifth ligand has not been previously reported and may be associated with a higher redox potential for NrfH haem 1, which can favour its reduction by menaquinol.
Another remarkable feature of NrfH is the coordination of haem 4 by a lysine residue (Lys331) from an NrfA protein (Figure 3C). This lysine residue, which belongs to the NrfA molecule that is closer to NrfH haems (named internal NrfA, NrfAint), is the distal haem ligand, whereas NrfH His140 is the proximal haem ligand. Haem 4, which is the gateway for electrons in transit from NrfH to NrfAint, is notably protruding from the NrfH structure towards a groove composed by NrfAint residues (Figure 3C). This finding reinforces the structural complementarity between NrfH and NrfA subunits and the stability of the complex, which cannot be dissociated under non-denaturing conditions (Pereira et al, 2000; Almeida et al, 2003). Lys331 is conserved in NrfH-interacting NrfA proteins, suggesting that this lysine residue should also be the distal ligand in other NrfH proteins. In NrfA proteins that interact with NrfB, there is also a conserved lysine. However, this residue should not be involved in NrfB haem ligation, as all the haems of this cytochrome are known to be bis-histidinyl coordinated (Clarke et al, 2004). Moreover, NrfA and NrfB proteins form a transient complex. Haem coordination by a lysine is also unusual for a haem, and was first reported for the catalytic haem of NrfA (Einsle et al, 1999), and more recently for tetrathionate reductase (Mowat et al, 2004). However, in both cases, the haems are five-coordinated catalytic haems, in contrast with NrfH haem 4, which shows a novel His–Lys coordination.
The NrfH haems 2 and 3 are bis-histidinyl coordinated, with His70 and His120 as proximal ligands, and His145 and His61 as distal ligands, respectively. These residues are apparently conserved in similar cytochromes like NapC, suggesting analogous coordination for haems 2 and 3 in all members of the family. There is evidence that all haems in NapC (Cartron et al, 2002) and CymA (Field et al, 2000) are bis-His ligated. Four conserved His in NapC and CymA proteins have been identified as the probable haem distal ligands (Cartron et al, 2002). Two of these His, the distal ligands to haems 1 and 4, are not conserved in NrfH proteins. It could be expected that the coordination of NapC or CymA haem 4 is different from NrfH, as these proteins do not form stable complexes with their redox partners. The His that probably coordinates this haem in NapC and CymA is found in a sequence segment that is absent in NrfH. Interestingly, the methionine residue, which coordinates haem 1, is also conserved in most NapC proteins. This leaves open the question of whether this Met could also coordinate haem 1 in these proteins upon ligand switching. Future studies are required to reveal if the unusual coordination of NrfH haem 1 is a general feature of the family or not. The sequence homology between NapC and NrfH is not high enough to permit construction of a model for NapC based on the NrfH structure. The overall fold of NapC and NrfH proteins is likely to be similar, although some variations are expected, as they couple with different electron acceptor proteins (Simon, 2002; Gross et al, 2005).
Possible menaquinol binding site
So far, D. vulgaris NrfHA complex had not been shown to use menaquinol as electron donor for nitrite reduction. We have confirmed that this complex shows a high level of activity for this reaction (12.2 U mg−1), using the reduced form of the menaquinone analogue 2,3-dimethyl-1,4-naphtoquinone (DMNH2) as electron donor, indicating that NrfH can indeed oxidise menaquinol. We have identified a pronounced cavity in NrfH located close to haem 1 that we propose to be the menaquinol-binding site (Figure 6A). This cavity, located between haem 1 and the second membrane NrfH helix, is wide enough to accommodate the menaquinol head group (Figure 6), and has an entrance that is notably directed towards the membrane region. Several of the residues that form the cavity are highly conserved, such as Gly86 and Lys82, and to a lesser extent Asp89, the residue that occupies the haem 1 distal coordination site (a His in W. succinogenes) (Figure 4). Previous studies with W. succinogenes NrfH showed that point mutations on the equivalent residues of Lys82 and Asp89 led to a drastic decrease in specific electron transport activity from menaquinol (1–4% of that of wild type) and no growth by nitrite respiration (Gross et al, 2005). In Escherichia coli formate dehydrogenase (Jormakka et al, 2002) and nitrate reductase (Bertero et al, 2005), the histidine haem ligands were reported to be involved in quinone binding. In the particular case of the menaquinol-oxidising nitrate reductase, His66 that coordinates the haem bD forms a hydrogen bond with the hydroxyl group of pentachlorophenol, a quinol-binding inhibitor (Bertero et al, 2005). The structure of NrfH suggests that this role may be performed by Asp89, which in some other NrfH proteins is replaced by a histidine.
Figure 6.
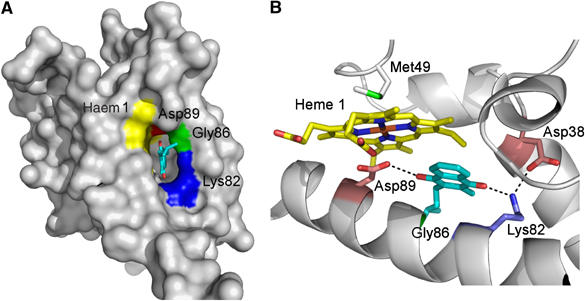
(A) NrfH molecular surface displaying the entrance for the proposed menaquinol binding pocket (390 Å2 of surface area and 430 Å3 of volume, calculations with CASTp server; Binkowski et al, 2003). The view is approximately perpendicular to the membrane from the cytoplasmic side. This pocket lies in the vicinity of haem 1 (yellow) with conserved residues Lys82 and Gly86 depicted in blue and green, respectively. Menaquinol is drawn with C atoms in cyan and O atoms in red. The non-coordinating Asp89, which occupies the distal axial site, is coloured in red. (B) A menaquinol head (aliphatic chain to C15) was manually docked into the cavity near haem 1, followed by structural idealisation with Refmac5, showing that it may establish H-bonds with Asp89 (Oδ2 at 3.0 Å to the O4 oxygen atom) and Lys82 (NZ at 2.8 Å to the O1 oxygen atom). Haem 1, Asp38, Met49, Lys82, Gly86, Asp89 and menaquinol head are represented in sticks rendering. Haem 1 is coloured by atom type and the C atoms of the other protein residues are coloured according to A.
An acidic residue, proposed to work as a proton shuttle, is also a recurrent theme in quinol-binding sites of respiratory proteins such as bc1 complex (Crofts et al, 1999), E. coli bo3 quinol oxidase (Abramson et al, 2000), W. succinogenes and E. coli fumarate reductase (Lancaster et al, 2000; Iverson et al, 2002) or E. coli DMSO reductase (Geijer and Weiner, 2004). A second acidic residue of NrfH, Asp38, lies close to the cavity in hydrogen-bonding distance to Lys82, and may also be involved in the proton transfer to the periplasm. The conserved Lys82 may establish a hydrogen bond with the hydroxyl group of menaquinol, as proposed for the essential Lys86 of E. coli nitrate reductase that lies close to the menaquinol binding site (Bertero et al, 2005). A role of proton shuttling has also been proposed for a similar lysine residue in E. coli fumarate reductase (Iverson et al, 2002).
The remarkable conservation of Gly86 in the centre of the helix strongly suggests that this residue is required to avoid steric clashes upon menaquinol binding. Several conserved glycine residues are also observed in the menaquinol-binding site of the nitrate reductase (Bertero et al, 2005). The polar head of the menaquinol molecule was docked into this cavity, so that its oxygen atoms are within H-bonding distances of Lys82 and Asp89 (Figure 6B). We propose a possible mechanism for menaquinol oxidation: after binding to the cavity with its hydroxyl groups H-bonded to Lys82 and Asp89, menaquinol transfers one electron to NrfH haem 1. One proton is transferred to the periplasm possibly through Lys82 and Asp38, forming a semiquinone intermediate. The second electron is transferred to haem 1 and a second proton moves to the periplasm, possibly through Asp89, forming the menaquinone product. However, the order of events concerning the proton and electron transfers cannot be precisely defined with the available data.
The finding that the NrfH menaquinol-binding site is at the membrane–periplasm interface confirms predictions that electron transport through the NrfH cytochromes is electroneutral and not associated with energy conservation (Simon, 2002), as the protons generated upon oxidation of menaquinol are most likely released directly to the periplasm. This finding is of no relevance to the metabolism of D. vulgaris, which only uses NrfHA to detoxify nitrite, but is important for other Desulfovibrio spp like D. gigas and D. desulfuricans, which grow by respiratory nitrite ammonification with H2 or formate (Steenkamp and Peck, 1981; Barton et al, 1983). In this growth mode, a proton-motive force has to be generated upon reduction of menaquinone by either H2 or formate. In Desulfovibrio spp, the periplasmic hydrogenases and formate dehydrogenases are unusual, as they lack the quinone-reducing cytochrome b subunit found in other similar proteins, and instead transfer electrons to a network of soluble cytochromes c (Heidelberg et al, 2004; Pereira et al, 2006). The soluble cytochromes then are proposed to transfer electrons to one of several possible transmembrane electron transfer complexes that include quinone-interacting proteins (Rossi et al, 1993; Pereira et al, 1998; Saraiva et al, 2001; Pereira et al, 2006). Thus, the requirement of energy conservation on the oxidation of H2 or formate suggests that these transmembrane complexes can reduce menaquinone at the negative side of the membrane with uptake of protons from the cytoplasm.
Overall haem arrangement of the complex
The formation of a stable complex between the NrfH protein with its physiological partner, NrfA, enables important insights into the electron transfer pathway from menaquinol oxidation to nitrite reduction. The overall haem arrangement of the NrfHA complex is quite surprising, as the NrfH haem groups are positioned non-symmetrically with respect to the two NrfA molecules (Figure 1). Only one of these, the NrfAint, has the haems in close proximity with NrfH haems (Figure 1B and Table II). On the contrary, the haems from the external NrfA (NrfAext) are further apart from the NrfH haems, with minimum edge-to-edge distances of ∼17 Å. The proximity of NrfH haem 4 to both haems 2 and 5 of NrfAint, with edge-to-edge distances of 12.1 and 8.5 Å, respectively (Table II), suggests that both haems can accept electrons through direct electron tunnelling (Page et al, 2003). The electron transfer from NrfAint to the NrfAext molecule should then occur through both haems 5 of the NrfA dimer, which are in very close contact (Figure 1B). Another notable characteristic regarding haem packing is the close contact and parallel stacking of haems 3 and 4 of the two NrfH molecules in the complex (edge-to-edge distances vary between 4.6 and 10.4 Å; Table II), which should allow for fast electron transfer between them. This close proximity and haem arrangement strongly suggests a physiological role for this packing (Figure 1B).
Table 2.
Distances between relevant haem groups
| NrfH–NrfH haems | Distance (Å) | NrfH–NrfAint haems | Distance (Å) | NrfH–NrfH* haems | Distance (Å) |
|---|---|---|---|---|---|
| 1–2 | 9.1 (3.4) | 4–2 | 17.7 (12.1) | 3–3 | 10.7 (4.6) |
| 2–3 | 11.9 (5.8) | 4–5 | 15.3 (8.5) | 4–4 | 18.7 (10.4) |
| 3–4 | 9.6 (4.2) | 3–4 and 4–3 | 16.0 (8.2) | ||
| Distances are referred to as centre-to-centre and edge-to-edge (in parentheses). | |||||
| The values were determined for NrfAint, NrfH and NrfH* proteins corresponding to chains A, C and F, respectively. | |||||
Overall, the picture that emerges from analysis of the 28 haem cluster of the α4β2 complex is that the two NrfH haems 1 are the electron entry points from menaquinol oxidation, from where the electrons can travel very fast through the haem network to the four catalytic haems in the complex (NrfA haem 1). The 28 haems may be required for electron storage during the 24-electron reduction of four nitrite molecules occurring at the four catalytic sites.
NrfA structure and intermolecular interactions
The three-dimensional structure of the NrfA dimer from D. vulgaris Hildenborough is similar to other NrfA structures determined so far (Einsle et al, 1999, 2000; Bamford et al, 2002; Cunha et al, 2003). The most homologous one is NrfA from D. desulfuricans (Cunha et al, 2003), showing an r.m.s. deviation of 0.9 Å for 474 superimposed Cα atoms. The specific characteristics of the other NrfA catalytic subunits are also observed in D. vulgaris, namely the three-helix bundle at the dimer interface, the catalytic haem with a lysine as the proximal ligand from a CXXCK motif and a water molecule at the distal side, four bis-histidinyl-coordinated haems, a calcium-binding site near haem 1 and the putative channels for substrate access and product efflux. A second Ca2+ is present in the vicinity of haems 3 and 4, as also observed in E. coli (Bamford et al, 2002) and D. desulfuricans (Cunha et al, 2003) NrfA proteins.
A relevant feature shown by the D. vulgaris NrfA structure is the elucidation of the conformation adopted by the first amino acids (25–40) of the mature protein, which were not visible in any of the available NrfA crystal structures, but are ordered in the D. vulgaris NrfHA complex. These amino-acid residues form an extended tail that embraces NrfH subunits, allowing for several intersubunit interactions and thus contributing to the complex stability (Figures 1A and 2). The N-terminal tail of the NrfAint monomer interacts with both NrfH subunits of the α4β2 complex, whereas the tail of the NrfAext interacts with only one NrfH molecule (Figure 2A).
The NrfA dimer binds tightly to the NrfH protein through both of the monomers. The NrfA2/NrfH contact surface has an area of ∼3000 Å2, which covers about 30% of the solvent-accessible area of NrfH, where approximately one-third of the NrfH residues are at van der Waals contacts with NrfA residues. This interface exhibits a remarkable complementarily and there is a strong electrostatic contribution due to many salt-bridge and hydrogen-bond interactions.
The two NrfAint molecules are not strongly interacting with each other (Figure 2B), as the interfacial area (860 Å2) corresponds to only ∼4% of the solvent-accessible area of NrfA. In contrast, the two NrfH molecules establish significant interactions over their N-terminal transmembrane helices and through residues surrounding haems 3 and 4. According to PISA server predictions (Krissinel and Henrick, 2004), the two NrfH molecules are able to form a stable dimer, in which the contact area (∼870 Å2) is about 8% of the monomer surface. It would be interesting to verify if dimerisation has any general relevance within this family of membrane-bound cytochromes c.
Concluding remarks
The 2.3 Å X-ray structure of the NrfHA complex, isolated from D. vulgaris, represents an important contribution to the limited number of available structures of quinone-interacting membrane complexes. It reveals, for the first time, the structure of a cytochrome c quinol dehydrogenase, NrfH, which belongs to a family of cytochromes widespread in bacterial respiratory chains. The NrfH cytochrome displays a very unique haem coordination. Contrary to what would be expected for a classical haem c binding motif (CXXCH), the proximal ligand of NrfH haem 1 is not the histidine, but instead a methionine, which is found two positions downstream (CXXCHXM). Moreover, the distal axial position in haem 1 is occupied by a non-coordinating aspartate residue. Thus, this haem is a methionine-bound high-spin haem, which is unprecedented in biological systems. These special features of NrfH haem 1 are likely related to its function as electron acceptor for menaquinol oxidation. The coordination of NrfH haem 4 is also exceptional, with a histidine as proximal ligand (His140) and a lysine (Lys331) from the NrfAint subunit as distal ligand. A cavity found near NrfH haem 1, which includes several highly conserved residues, is proposed to be the menaquinol-binding site. The NrfHA complex shows a striking quaternary structure, consisting of four NrfA subunits and two NrfH subunits, which should constitute the biological unit of the enzyme. The disposition of the NrfA molecules with respect to NrfH is not symmetrical, as only one of the monomers from the NrfA dimer receives electrons directly from the NrfH protein. The biological action of the NrfHA complex is essential for D. vulgaris to survive in the presence of other anaerobic bacteria that reduce nitrate to nitrite (Greene et al, 2003). This may be a crucial property in the complex ecosystems found in sediments where D. vulgaris and other sulphate reducers are important targets for bioremediation of toxic metals and radionuclides (Lovley, 2003).
Materials and methods
Crystallisation, data collection and phasing
Protein production, crystallisation, X-ray data collection and preliminary structure determination have already been described in detail (Rodrigues et al, 2006). In summary, 5 mg ml−1 of native protein complex, solubilised in 0.015% (w/v) dodecyl-β-D-maltoside (DDM) detergent, was crystallised at 277 K using 10% PEG 4K (4 kDa polyethylene glycol) at pH 7.5. Diffraction data were measured at the Swiss-Light-Source synchrotron at three wavelengths suitable for a MAD experiment based on iron atoms. Data were collected at 2.3 Å resolution, with overall completeness of 83.9% (67.3% in outer shell).
NrfHA crystals belong to P212121 space group, with cell parameters a=79.4, b=256.8 and c=579.2 Å. The crystal asymmetric unit contains 12 NrfA and six NrfH molecules. The NrfHA complex structure was solved by a combination of molecular replacement and MAD methods. The structure of the larger subunit NrfA (∼60 kDa) was solved using the coordinates of the NrfA dimer (∼120 kDa) from D. desulfuricans (PDB code: 1OAH; Cunha et al, 2003) as search model. An anomalous Fourier map was then calculated using the phases from the molecular replacement solution of the NrfA dimer substructure and the anomalous difference coefficients obtained from the peak data set, which allowed the location of the iron atoms from the smaller and unknown NrfH subunit (∼20 kDa) (Rodrigues et al, 2006). Relevant data collection statistics are given in Table I.
Structure refinement
The electron density map calculated with SHARP (de la Fortelle and Bricogne, 1997) at 2.3 Å, using only the experimental phases, was of very good quality and enabled the manual model building of the unknown NrfH protein. Residues 14–158 (out of 159) were built using COOT (Emsley and Cowtan, 2004). GUISIDE from the CCP4 suite of programs (Collaborative Computational Project, 1994) was used for the correction of NrfA side chains. Model rebuilding of two NrfA molecules, which form the physiological dimer, was performed with COOT. The other molecules of the crystal asymmetric unit were generated from this NrfHA2 unit with LSQKAB within CCP4, using the NrfA substructure obtained by molecular replacement. Restrained refinement at 2.4 Å resolution was carried out with Refmac5 (Murshudov et al, 1999), using the experimental phases. At this stage, tight NCS restraints were applied for NrfAint, NrfAext and NrfH molecules (three NCS groups of six monomers each). This procedure decreased the initial R and Rfree of 35 to 24.8 and 27.1%, respectively. Further cycles of manual fitting and restrained refinement were followed before the addition of water molecules, which was performed using both COOT and ARP/wARP (Perrakis et al, 1999) programs. The strength of NCS restraints slightly decreased at the end of the refinement, which converged to R and Rfree values of 20.1 and 24.0%, respectively, at 2.3 Å resolution.
The final model comprises 6813 amino-acid residues, 84 haem groups, 24 calcium ions, belonging to 18 protein chains (chains A, D, G, J, M, P were assigned to NrfAint; chains B, E, H, K, N, Q to NrfAext and chains C, F, I, L, O, R to NrfH). A calcium-binding site is found near each NrfA haem 1 and a second Ca2+ ion is octahedrally coordinated in the vicinity of haems 3 and 4. A blob of electron density, located in a cavity surrounded by the N-terminal tail of NrfAint (residues 26–29) and an inter-helical loop of NrfH (residues 73–74), was found in each NrfHA2 unit. This electron density was assigned as the polar head of dodecylmaltoside detergent (designated as LMT—(laurylmaltoside) monomer). In addition, 2326 water molecules and three acetate ions were also included in the crystallographic model.
The different NrfA molecules start at residues 25 or 26 and end at residues 519–522, depending on the protein chain. A few residues that belong to the β-sheet loop formed by residues 325–331 from NrfAext monomers were poorly defined in the electron density maps. Their average temperature factors are around 45 Å2, which are considerably above the average temperature factor of all NrfA protein residues (20.1 Å2).
The model for the electron-donor subunit NrfH comprises residues from 14 to 158 and has an averaged overall temperature factor of 27.4 Å2. NrfH transmembrane helix is not well ordered, as most of its residues (from 14 to 31) have high-temperature factors (between ∼100 and 50 Å2). Ramachandran plot analysis using the program PROCHECK (Laskowski et al, 1993) showed that 89.8% of the total protein residues are within most favoured regions, 9.6% are in additionally allowed regions, 0.4% in the generously allowed regions and only 0.2% in disallowed ones. The refinement statistics are summarised in Table I.
Molecular mass determination
Molecular mass determination by native size-exclusion chromatography was performed on a GE Healthcare Superose 6 PC3.2/30 column, calibrated with the Amersham Bioscience high molecular mass calibration kit (molecular weight range from 158 to 669 kDa). Elution was performed with buffer 10 mM K phosphate pH 6.5, 0.15 M NaCl and 0.025% DDM. The determined molecular mass of the complex is 300 kDa, which corresponds to two NrfHA2 units (α4β2 arrangement).
Nitrite reductase activity
A 2 mM stock solution of DMN was reduced with NaBH4 and the reduction was followed spectrophotometrically (Snyder and Trumpower, 1999). The anoxic reaction mixture included 50 mM Tris–HCl buffer, pH 7.6, 0.2 mM DMNH2 and enzyme (same sample used for crystallisation), and the reaction was started by addition of 10 mM NaNO2. Oxidation of DMNH2 was followed at 270 nm (Weiner et al, 1986). One unit of activity (U) is equivalent to oxidation of 1 μmol min−1 of DMNH2. All reactions were performed inside an anaerobic chamber. The determined activity is 12.2 U mg−1.
Accession code
Coordinates have been deposited in the Protein Data Bank under the accession code 2J7A.
Acknowledgments
Filipa Valente and Inês Martins are acknowledged for purification of the NrfHA complex. We thank Clemens Schulze-Briese for assistance with data collection at the Swiss Light Source, PSI, Villigen. Travel to SLS was supported by the European Commission under the 6th Framework Programme through the Key Action: Strengthening the European Research Area, Research Infrastructures under contract no. RII3-CT-2004-506008. We acknowledge Fundação para a Ciência e Tecnologia for FCT-POCTI fellowship to MLR (SFRH/BPD/24372/2005), FCT grant POCTI/2002/ESP/44782 (to ICP) and POCI/BIA-PRO/55621/2004 (to MA and BI fellowship to TFO).
References
- Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikstrom M (2000) The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol 7: 910–917 [DOI] [PubMed] [Google Scholar]
- Almeida MG, Macieira S, Goncalves LL, Huber R, Cunha CA, Romao MJ, Costa C, Lampreia J, Moura JJ, Moura I (2003) The isolation and characterization of cytochrome c nitrite reductase subunits (NrfA and NrfH) from Desulfovibrio desulfuricans ATCC 27774. Re-evaluation of the spectroscopic data and redox properties. Eur J Biochem 270: 3904–3915 [DOI] [PubMed] [Google Scholar]
- Bamford VA, Angove HC, Seward HE, Thomson AJ, Cole JA, Butt JN, Hemmings AM, Richardson DJ (2002) Structure and spectroscopy of the periplasmic cytochrome c nitrite reductase from Escherichia coli. Biochemistry 41: 2921–2931 [DOI] [PubMed] [Google Scholar]
- Barton LL, LeGall J, Odom JM, Peck HD Jr (1983) Energy coupling to nitrite respiration in the sulfate-reducing bacterium Desulfovibrio gigas. J Bacteriol 153: 867–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks BC, Page MD, Richardson DJ, Reilly A, Cavill A, Outen F, Ferguson SJ (1995) Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol Microbiol 15: 319–331 [DOI] [PubMed] [Google Scholar]
- Bertero MG, Rothery RA, Boroumand N, Palak M, Blasco F, Ginet N, Weiner JH, Strynadka NC (2005) Structural and biochemical characterization of a quinol binding site of Escherichia coli nitrate reductase A. J Biol Chem 280: 14836–14843 [DOI] [PubMed] [Google Scholar]
- Binkowski TA, Naghibzadeh S, Liang J (2003) CASTp: computed atlas of surface topography of proteins. Nucleic Acids Res 31: 3352–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron ML, Roldan MD, Ferguson SJ, Berks BC, Richardson DJ (2002) Identification of two domains and distal histidine ligands to the four haems in the bacterial c-type cytochrome NapC; the prototype connector between quinol/quinone and periplasmic oxido-reductases. Biochem J 368: 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TA, Dennison V, Seward HE, Burlat B, Cole JA, Hemmings AM, Richardson DJ (2004) Purification and spectropotentiometric characterization of Escherichia coli NrfB, a decaheme homodimer that transfers electrons to the decaheme periplasmic nitrite reductase complex. J Biol Chem 279: 41333–41339 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Crofts AR, Barquera B, Gennis RB, Kuras R, Guergova-Kuras M, Berry EA (1999) Mechanism of ubiquinol oxidation by the bc(1) complex: different domains of the quinol binding pocket and their role in the mechanism and binding of inhibitors. Biochemistry 38: 15807–15826 [DOI] [PubMed] [Google Scholar]
- Cunha CA, Macieira S, Dias JM, Almeida G, Goncalves LL, Costa C, Lampreia J, Huber R, Moura JJ, Moura I, Romao MJ (2003) Cytochrome c nitrite reductase from Desulfovibrio desulfuricans ATCC 27774. The relevance of the two calcium sites in the structure of the catalytic subunit (NrfA). J Biol Chem 278: 17455–17465 [DOI] [PubMed] [Google Scholar]
- de la Fortelle E, Bricogne G (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Macromol Crystallogr A 276: 472–494 [DOI] [PubMed] [Google Scholar]
- DeLano WLT (2002) The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific [Google Scholar]
- Einsle O, Messerschmidt A, Stach P, Bourenkov GP, Bartunik HD, Huber R, Kroneck PM (1999) Structure of cytochrome c nitrite reductase. Nature 400: 476–480 [DOI] [PubMed] [Google Scholar]
- Einsle O, Stach P, Messerschmidt A, Simon J, Kroger A, Huber R, Kroneck PM (2000) Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 ˚ resolution, inhibitor binding, and heme-packing motifs. J Biol Chem 275: 39608–39616 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Field SJ, Dobbin PS, Cheesman MR, Watmough NJ, Thomson AJ, Richardson DJ (2000) Purification and magneto-optical spectroscopic characterization of cytoplasmic membrane and outer membrane multiheme c-type cytochromes from Shewanella frigidimarina NCIMB400. J Biol Chem 275: 8515–8522 [DOI] [PubMed] [Google Scholar]
- Geijer P, Weiner JH (2004) Glutamate 87 is important for menaquinol binding in DmsC of the DMSO reductase (DmsABC) from Escherichia coli. Biochim Biophys Acta 1660: 66–74 [DOI] [PubMed] [Google Scholar]
- Gon S, Giudici-Orticoni MT, Mejean V, Iobbi-Nivol C (2001) Electron transfer and binding of the c-type cytochrome TorC to the trimethylamine N-oxide reductase in Escherichia coli. J Biol Chem 276: 11545–11551 [DOI] [PubMed] [Google Scholar]
- Greene EA, Hubert C, Nemati M, Jenneman GE, Voordouw G (2003) Nitrite reductase activity of sulphate-reducing bacteria prevents their inhibition by nitrate-reducing, sulphide-oxidizing bacteria. Environ Microbiol 5: 607–617 [DOI] [PubMed] [Google Scholar]
- Gross R, Eichler R, Simon J (2005) Site-directed modifications indicate differences in axial haem c iron ligation between the related NrfH and NapC families of multihaem c-type cytochromes. Biochem J 390: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Seshadri R, Haveman SA, Hemme CL, Paulsen IT, Kolonay JF, Eisen JA, Ward N, Methe B, Brinkac LM, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Fouts D, Haft DH, Selengut J, Peterson JD, Davidsen TM, Zafar N, Zhou L, Radune D, Dimitrov G, Hance M, Tran K, Khouri H, Gill J, Utterback TR, Feldblyum TV, Wall JD, Voordouw G, Fraser CM (2004) The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat Biotechnol 22: 554–559 [DOI] [PubMed] [Google Scholar]
- Holm L, Park J (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16: 566–567 [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C (1999) Protein folds and families: sequence and structure alignments. Nucleic Acids Res 27: 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson TM, Arciero DM, Hsu BT, Logan MSP, Hooper AB, Rees DC (1998) Heme packing motifs revealed by the crystal structure of the tetra-heme cytochrome c554 from Nitrosomonas europaea. Nat Struct Biol 5: 1005–1012 [DOI] [PubMed] [Google Scholar]
- Iverson TM, Luna-Chavez C, Croal LR, Cecchini G, Rees DC (2002) Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J Biol Chem 277: 16124–16130 [DOI] [PubMed] [Google Scholar]
- Jones RW, Lamont A, Garland PB (1980) The mechanism of proton translocation driven by the respiratory nitrate reductase complex of Escherichia coli. Biochem J 190: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M, Byrne B, Iwata S (2003) Protonmotive force generation by a redox loop mechanism. FEBS Lett 545: 25–30 [DOI] [PubMed] [Google Scholar]
- Jormakka M, Tornroth S, Byrne B, Iwata S (2002) Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295: 1863–1868 [DOI] [PubMed] [Google Scholar]
- Jungst A, Wakabayashi S, Matsubara H, Zumft WG (1991) The nirSTBM region coding for cytochrome cd1-dependent nitrite respiration of Pseudomonas stutzeri consists of a cluster of mono-, di-, and tetraheme proteins. FEBS Lett 279: 205–209 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Lancaster CR, Gorss R, Haas A, Ritter M, Mantele W, Simon J, Kroger A (2000) Essential role of Glu-C66 for menaquinol oxidation indicates transmembrane electrochemical potential generation by Wolinella succinogenes fumarate reductase. Proc Natl Acad Sci USA 97: 13051–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lovley DR (2003) Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol 1: 35–44 [DOI] [PubMed] [Google Scholar]
- Mowat CG, Rothery E, Miles CS, McIver L, Doherty MK, Drewette K, Taylor P, Walkinshaw MD, Chapman SK, Reid GA (2004) Octaheme tetrathionate reductase is a respiratory enzyme with novel heme ligation. Nat Struct Mol Biol 11: 1023–1024 [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ (1999) Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Page CC, Moser CC, Dutton PL (2003) Mechanism for electron transfer within and between proteins. Curr Opin Chem Biol 7: 551–556 [DOI] [PubMed] [Google Scholar]
- Pereira IA, LeGall J, Xavier AV, Teixeira M (2000) Characterization of a heme c nitrite reductase from a non-ammonifying microorganism, Desulfovibrio vulgaris Hildenborough. Biochim Biophys Acta 1481: 119–130 [DOI] [PubMed] [Google Scholar]
- Pereira IA, Xavier AV (2005) Multi-heme c cytochromes and enzymes. In Encyclopedia of Inorganic Chemistry, King RB (ed) John Wiley & Sons [Google Scholar]
- Pereira IAC, Romão CV, Xavier AV, LeGall J, Teixeira M (1998) Electron transfer between hydrogenases and mono and multiheme cytochromes in Desulfovibrio spp. J Biol Inorg Chem 3: 494–498 [Google Scholar]
- Pereira PM, Teixeira M, Xavier AV, Louro RO, Pereira IA (2006) The Tmc complex from Desulfovibrio vulgaris Hildenborough is involved in transmembrane electron transfer from periplasmic hydrogen oxidation. Biochemistry 45: 10359–10367 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Richardson D, Sawers G (2002) Structural biology. PMF through the redox loop. Science 295: 1842–1843 [DOI] [PubMed] [Google Scholar]
- Richardson DJ (2000) Bacterial respiration: a flexible process for a changing environment. Microbiology 146 (Part 3): 551–571 [DOI] [PubMed] [Google Scholar]
- Rodrigues ML, Oliveira T, Matias PM, Martins IC, Valente FM, Pereira IA, Archer M (2006) Crystallization and preliminary structure determination of the membrane-bound complex cytochrome c nitrite reductase from Desulfovibrio vulgaris Hildenborough. Acta Crystallogr F Struct Biol Cryst Commun 62: 565–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan MD, Sears HJ, Cheesman MR, Ferguson SJ, Thomson AJ, Berks BC, Richardson DJ (1998) Spectroscopic characterization of a novel multiheme c-type cytochrome widely implicated in bacterial electron transport. J Biol Chem 273: 28785–28790 [DOI] [PubMed] [Google Scholar]
- Rossi M, Pollock WB, Reij MW, Keon RG, Fu R, Voordouw G (1993) The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J Bacteriol 175: 4699–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva LM, da Costa PN, Conte C, Xavier AV, LeGall J (2001) In the facultative sulphate/nitrate reducer Desulfovibrio desulfuricans ATCC 27774, the nine-haem cytochrome c is part of a membrane-bound redox complex mainly expressed in sulphate-grown cells. Biochim Biophys Acta 1520: 63–70 [DOI] [PubMed] [Google Scholar]
- Schumacher W, Hole U, Kroneck MH (1994) Ammonia-forming cytochrome-c nitrite reductase from Sulfurospirillum deleyianum is a tetraheme protein—new aspects of the molecular composition and spectroscopic properties. Biochem Biophys Res Commun 205: 911–916 [DOI] [PubMed] [Google Scholar]
- Schwalb C, Chapman SK, Reid GA (2003) The tetraheme cytochrome CymA is required for anaerobic respiration with dimethyl sulfoxide and nitrite in Shewanella oneidensis. Biochemistry 42: 9491–9497 [DOI] [PubMed] [Google Scholar]
- Shaw AL, Hochkoeppler A, Bonora P, Zannoni D, Hanson GR, McEwan AG (1999) Characterization of DorC from Rhodobacter capsulatus, a c-type cytochrome involved in electron transfer to dimethyl sulfoxide reductase. J Biol Chem 274: 9911–9914 [DOI] [PubMed] [Google Scholar]
- Simon J (2002) Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol Rev 26: 285–309 [DOI] [PubMed] [Google Scholar]
- Simon J, Gross R, Einsle O, Kroneck PM, Kroger A, Klimmek O (2000) A NapC/NirT-type cytochrome c (NrfH) is the mediator between the quinone pool and the cytochrome c nitrite reductase of Wolinella succinogenes. Mol Microbiol 35: 686–696 [DOI] [PubMed] [Google Scholar]
- Simon J, Pisa R, Stein T, Eichler R, Klimmek O, Gross R (2001) The tetraheme cytochrome c NrfH is required to anchor the cytochrome c nitrite reductase (NrfA) in the membrane of Wolinella succinogenes. Eur J Biochem 268: 5776–5782 [DOI] [PubMed] [Google Scholar]
- Snyder CH, Trumpower BL (1999) Ubiquinone at center N is responsible for triphasic reduction of cytochrome b in the cytochrome bc(1) complex. J Biol Chem 274: 31209–31216 [DOI] [PubMed] [Google Scholar]
- Steenkamp DJ, Peck HD Jr (1981) Proton translocation associated with nitrite respiration in Desulfovibrio desulfuricans. J Biol Chem 256: 5450–5458 [PubMed] [Google Scholar]
- Ujiiye T, Yamamoto I, Nakama H, Okubo A, Yamazaki S, Satoh T (1996) Nucleotide sequence of the genes, encoding the pentaheme cytochrome (dmsC) and the transmembrane protein (dmsB), involved in dimethyl sulfoxide respiration from Rhodobacter sphaeroides f. sp. denitrificans. Biochim Biophys Acta 1277: 1–5 [DOI] [PubMed] [Google Scholar]
- Weiner JH, Cammack R, Cole ST, Condon C, Honore N, Lemire BD, Shaw G (1986) A mutant of Escherichia coli fumarate reductase decoupled from electron transport. Proc Natl Acad Sci USA 83: 2056–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]


