Abstract
Long terminal repeat (LTR) retrotransposons and DNA transposons are transposable elements (TEs) that perform cleavage and transfer at precise DNA positions. Here, we present statistical analyses of sequences found at the termini of precise TEs in the human genome. The results show that the terminal di- and trinucleotides of these TEs are highly conserved. 5′TG…CA3′ occurs most frequently at the termini of LTR retrotransposons, while 5′CAG…CTG3′ occurs most frequently in DNA transposons. Interestingly, these sequences are the most flexible base pair steps in DNA. Both the sequence preference and the degree of conservation of each position within the human LTR dinucleotide termini are remarkably similar to those experimentally demonstrated in transposable phage Mu. We discuss the significance of these observations and their implication for the function of terminal residues in the transposition of precise TEs.
INTRODUCTION
In the post-genomic era, transposable elements (TEs) are demanding attention by their abundant presence in all three biological kingdoms (1). The proportion of TEs in different genomes varies; from only several insertion sequences in bacteria to more than 70% TEs in some plant genomes. For instance, the genomic portion of TEs is 2% in worm, 3% in yeast, 15% in fruit fly, 14–25% in rice (2), 45% in humans (3), 50–80% in maize and more than 70% in barley (2). TEs have been labeled ‘selfish (or parasitic) DNA’ (4,5). Having acquired the capacity to multiply, they continue to increase their copy number in host genomes as long as the increased genetic load is tolerated by the host. Co-evolution and co-adaptation of TEs with their host genomes is essential for their long-term survival, and many beneficial effects of this co-existence are evident (6). Many if not most TEs in large genomes are no longer active. This is also true for the human genome (3), where there is little evidence of their continuing mobility today (7). The importance of TEs in host genome evolution is comprehensively discussed in recent reviews by Kidwell and Lisch (1,2,6).
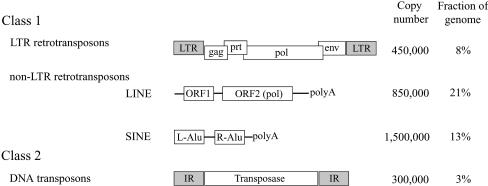
TEs are divided into two major classes based on whether their transposition intermediate is RNA (class 1 or retrotransposons) or DNA (class 2 or DNA transposons) (8) (Fig. 1). Class 1 TEs can be divided into two sub-classes, long terminal repeat (LTR) retrotransposons and non-LTR retrotransposons, which are further divided into long interspersed nuclear elements (LINEs) and short interspersed nuclear elements (SINEs). While both LTR retrotransposons and DNA transposons transpose via cleavage and transfer at precise nucleotide positions (9), non-LTR retrotransposons move by imprecise mechanisms such as ‘target-primed reverse transcription’ (10). The latter elements have a random length of poly(A) sequence at their 3′ end, and usually harbor truncations at their 5′ end due to the failure of reverse transcription (11).
Figure 1.
Classes of TE-derived interspersed repeats, their copy number and fraction in the human genome (3). LTRs and IRs are indicated by shaded boxes.
Transposable phage Mu has long served as a paradigm for the study of TEs (12). Mu is a DNA transposon but, interestingly, its transposition biochemistry is very similar to that of LTR retrotransposons, such as HIV (13). Virtually all retroviruses, many LTR retrotransposons, and some bacterial DNA transposons including Mu, encode (harbor) the dinucleotide 5′TG…CA3′ at their termini (this representation denotes terminal nucleotides on the same DNA strand) (14–20). Genetic and biochemical studies using mutants of the terminal dinucleotide in both Mu and retroviruses have shown that these two nucleotides play essential roles in transposition (21–29). In the case of Mu, examination of the activity of all 16 combinations of the dinucleotide sequence showed a clear hierarchy of reactivity in both in vivo and in vitro transposition (30). The +1 position (5′T…A3′) was seen to be far more important than the +2 position (5′G…C3′). The substrate preference of the Mu transposase for dinucleotides that retained the wild-type residue at the +1 position was 5′TG > TA > TT > TC (30). This decreasing order of reactivity can be clearly superimposed on the decreasing flexibility of their dinucleotide steps (30,31). The terminal nucleotides play an early role in transposition, since all deviations from the wild-type sequence affect assembly of the active transposase complex or transpososome (30,32–34). Given that transpososome assembly is associated with melting of DNA around the Mu ends (35,36), we have proposed that the terminal 5′TG…CA3′ sequence has been selected at Mu termini primarily for its conformational flexibility, which assists in DNA opening at the termini prior to initiation of transposition chemistry (30). Strong support for this notion has come from the finding that mismatched or unpaired termini are indifferent to the sequence on the cleaved strand and are extremely proficient in assembly and transposition (34). ‘Open termini’ formation is likely to be the rate-limiting event during assembly, and the T residue on the bottom strand is the primary determinant of transpososome stability (34). A role for DNA deformation around the termini has also been suggested for transposition in HIV (37) and in V(D)J recombination systems (38,39).
To define the sequence conservation of terminal nucleotides in precise TEs, we have availed ourselves of the large human genome sequence database deposited in the public domain (3), and analyzed the data using some statistical approaches. The abundance of LTR retrotransposons and DNA transposons in the human genome (∼8 and ∼3%, respectively; see Fig. 1) is well suited to our studies. Our analysis reveals that a common characteristic of precise TEs is the high conservation of their terminal two to three nucleotides. These conserved nucleotides are ends of inverted repeats (IRs) and LTRs in DNA transposons and LTR retrotransposons, respectively (Fig. 1). We have found that, similar to the observed hierarchy of reactivity of dinucleotide sequences at Mu termini (30), the most abundant species at the ends of LTR retrotransposons is 5′TG…CA3′ followed by 5′TA…TA3′. For DNA transposons, the most abundant species is 5′CAG…CTG3′. The hallmark of these base pair steps is their greater conformational flexibility. The +1 position shows the highest degree of sequence conservation, with the +2 and +3 positions showing a decreasing gradient of conservation. In light of experimental conclusions based on Mu as well as other transposons, these data allow us to propose that the function of the +1 position is a critical feature of the transposition machinery of precise TEs, molecular interactions which influence a rate-limiting event other than precise cleavage.
MATERIALS AND METHODS
Graphical representations of consensus sequence conservation patterns of human TEs were generated by WebLogo (http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi) using 243 human LTR elements (excluding internal sequences) and 113 human DNA transposon elements from Repbase Update, which maintains a current listing of all reported repeat sequences (version 8.17 released on December 2001; see http://www.girinst.org/Repbase_Update.html) (40). The sequence of each element represents a consensus derived from multiple repeats. For the human genome sequence, database hg5 (frozen data set of October 2000) at UCSC (http://genome.ucsc.edu) was used as the data source because of its high degree of completion at the time we launched this project. All human repeats detectable by the RepeatMasker program (http://repeatmasker.genome.washington.edu) are collected in MySQL tables of hg5. Our program for sequence extraction, written in C++ language, was designed to search for repeat sequences matching given parametric values of the table, read information of its chromosomal position, and then extract a designated number of nucleotides from the border region between the repeat and its flanking genomic DNA at either 5′ or 3′ ends. Terminal sequences for each element were collected and analyzed separately because a large portion of identified TEs have truncations at either termini. According to our analysis, ∼2.5% of extracted repeat sequences based on the hg5 data set and clustered into several genomic regions, were incorrect. These incorrectly masked genomic regions, corresponding to ∼3% of the entire human genome, were ignored from input MySQL tables. This modification reduced the fraction of incorrect sequence output down to <0.1%. The UCSC bioinformatics group also estimates that ∼5% of the genome had coordinates of the repeats offset incorrectly, but that this problem has been fixed after hg10, the frozen data set of December 2001 (communication with Dr Jim Kent). Various analyses of the collected sequences were performed using scripts written in Perl.
The MySQL tables of hg5 contain 227 different LTR elements (excluding internal sequences) and 115 different DNA transposons elements. For sequence analysis, only those with more than 20 (arbitrarily decided significant sample size) extracted sequences for both ends (210 LTRs and 95 DNA transposons) have been considered. Vectors with n dimensions based on genomic sequence frequency of the terminal two to three base pairs were generated for all TEs of interest, and then subjected to the programs Cluster and TreeView (available from http://rana.lbl.gov/EisenSoftware.htm), for hierarchical clustering analysis (under average linkage clustering setting) and its graphical display.
RESULTS
Analysis of terminal dinucleotides of human LTR retrotransposons
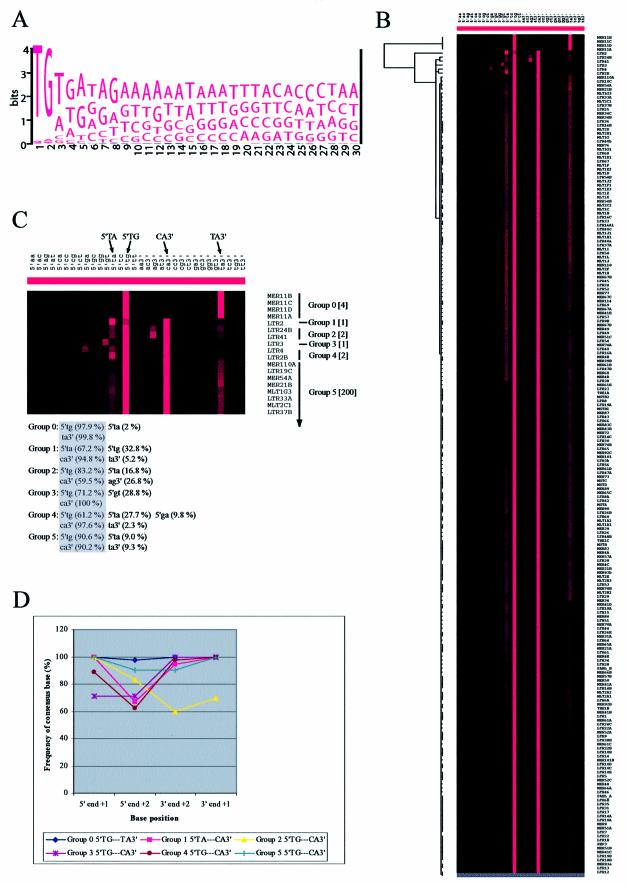
243 LTR element consensus sequences covering all copies of these elements in the human genome are compiled in the Repbase Update (see Materials and Methods). A WebLogo alignment of these sequences shows that the terminal two bases are highly conserved among the entire class of human LTRs (Fig. 2A, only the 5′ end is shown). From an evolutionary point of view, the degree of sequence conservation positively correlates with its functional importance, implying that these two terminal base pairs play a critical common role in transposition. Profiling TEs by genomic frequencies of all possible terminal dinucleotide sequences for both termini is a persuasive approach for developing new biological notions in the study of TEs. The WebLogo alignment shown in Figure 2A provides information on relative frequency for each base position only, and does not provide the relative order of dinucleotide frequencies. Towards this end, therefore, we performed an aggregative hierarchical clustering analysis designed for large sets of objects (i.e. TEs) (41). This statistical analysis procedure consists of several steps. First, measuring similarities in a particular quantified property between individual objects (e.g. frequencies of all possible terminal dinucleotide sequences). Secondly, grouping of the most similar objects into clusters. Thirdly, merging these clusters according to their similarities until all are fused into one single cluster. With a suitable graphical representation method, cluster analysis provides a powerful tool for data mining. Similar approaches have been successfully employed in the analysis of microarray gene expression data (42).
Figure 2.
(Opposite) Analysis of terminal dinucleotide steps of human LTRs. (A) Display of pattern in aligned consensus sequences at 5′ ends of human LTRs. The illustration was generated by WebLogo using 243 human LTR consensus sequences from Repbase Update. (B) Display of hierarchical clustering of human LTRs taken from MySQL tables (see Materials and Methods). The dendrogram and color image were generated as described in the text. Each human LTR is represented by a single row, and each different terminal dinucleotide sequence is represented by a single column; of the 32 columns, the left 16 are for the 5′ end and the right 16 for the 3′ end. The brightness of the red color indicates the frequency of the given dinucleotide sequence in the multiple copies of each element. Two major red columns are evident for each terminus. (C) Magnified image of the top part of (B) showing six different groups of human LTRs based on patterns of terminal dinucleotide conservation. The number of elements belonging to each group is indicated in brackets. The two most frequent patterns are indicated at the top. The average sequence frequencies of the conservation pattern in each group is indicated at the bottom, where 5′ and 3′ ends of the highest frequency pattern are included in a shaded box. (D) Graph representing the degree of consensus sequence conservation for terminal base positions in each group of human LTRs.
All human repeats reported to the Repbase Update have been identified and mapped by the RepeatMasker program and maintained in MySQL tables (see Materials and Methods). For calculation of genomic frequencies of all 16 possible terminal dinucleotide sequences within a specific human LTR retrotransposon, the necessary data set was collected by simply using these informative tables and the corresponding genome sequences. A program for data collection was designed to identify human repeats satisfying given parametric values of the tables, e.g. name of the element and the range of sequence extraction. Eighty nucleotides were extracted at both the 5′ and 3′ ends from the border region between the element and its flanking genomic DNA (i.e. 40 nt within the element and 40 nt of the flanking DNA). During data collection, the internal sequences of these retrotransposon elements were ignored (Repbase Update maintains separate files of internal sequences and LTR ends). Using the collected raw sequence data sets, we characterized individual human LTRs by a 32-dimensional vector, in which each dimension corresponds to the genomic frequency of one of 16 different kinds of terminal dinucleotides at both 5′ and 3′ termini. These quantitative data were organized and graphically represented using Cluster and TreeView programs (42). In illustrations generated by these programs, TEs representing the most similar patterns in terminal dinucleotide frequencies are located next to each other.
Figure 2B shows the TreeView illustration of hierarchical clustering of 210 different human LTR elements by their genomic terminal dinucleotide frequency patterns. Six groups (Groups 0–5) emerged from this analysis (Fig. 2C). The main feature of this illustration is that most LTR elements (205 out of 210 total analyzed; Groups 2–5) have 5′TG…CA3′ as the most frequent terminal dinucleotide; a majority of these show 5′TA…TA3′ as the second most frequent terminal dinucleotide (elements in Groups 4 and 5; note that each element represents a consensus of hundreds of repeats, see Materials and Methods). All groups, with the exception of Groups 0 and 1, show a canonical inversion of the terminal dinucleotide sequence consensus (Fig. 2C, shaded box). Members of Group 5, which includes the majority of human LTRs, show 5′TG…CA3′ (with a range of 80–100%) and 5′TA…TA3′ (with a range of 0–20%) as the consensus and the most tolerated non-consensus terminal dinucleotide, respectively.
Finally, we looked at the degree of base conservation at each of the two dinucleotide positions. As shown in Figure 2D, all human LTRs show the highest degree of conservation of the +1 position, irrespective of their terminal dinucleotide sequence, with the exception of Group 3, which shows equal conservation of the +1 and +2 positions.
Analysis of terminal trinucleotides of human DNA transposons
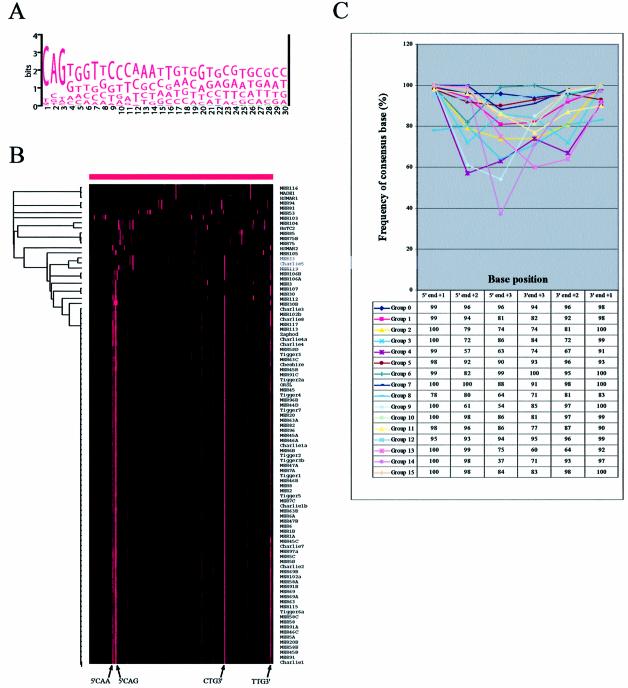
The WebLogo alignment of all the human DNA transposon element repeat sequences shows that three nucleotides at their termini are highly conserved (Fig. 3A; only the 5′ end is shown). This suggests that, unlike human LTRs that have conserved only two terminal base residues among the entire class, three terminal base residues are functionally important in human DNA transposons. Therefore, the same approaches of data collection and statistical analysis as described for dinucleotides, were used for the analysis of terminal trinucleotides of human DNA transposons. For the analysis, we generated a 128-dimensional vector, with 64 dimensions for each terminus. Hierarchical clustering analysis resulted in a far more complex illustration reflecting more divergent sequence patterns for DNA transposon termini (Fig. 3B). This is not just due to the larger number of dimensions in the vector inasmuch as an analysis considering only the terminal dinucleotide in these transposons also generated a much more complicated tree view (data not shown).
Figure 3.
Analysis of terminal trinucleotide steps of human DNA transposons. (A) WebLogo-generated pattern of aligned consensus sequences at 5′ ends of 113 human DNA transposons. (B) Display of hierarchical clustering of human DNA transposons taken from MySQL tables (see Materials and Methods). There are a total 128 columns, the left 64 columns for the 5′ end and the right 64 for the 3′ end. The two most frequent trinucleotide sequences for both termini of the majority group (Group 15) are marked on the bottom. (C) Graph representing the degree of consensus sequence conservation for terminal base positions in each group of human DNA transposons.
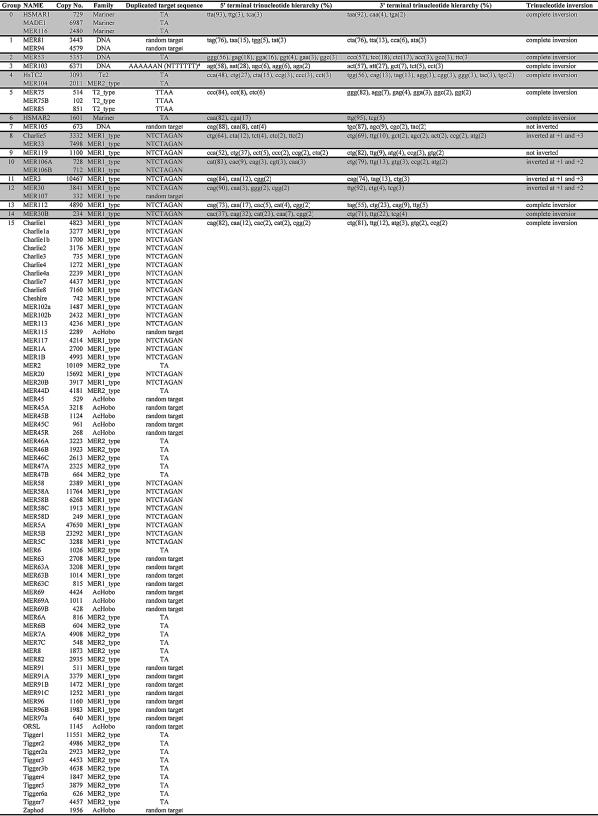
Hierarchical clustering based on the genomic terminal trinucleotide sequence frequency of human DNA transposons produced 16 groups (Table 1). The majority group (Group 15; 71 out of 95 total elements belong to this group) has a consensus sequence of 5′CAG…CTG3′ (with a range of 75–90%; Table 1 and Fig. 3B). The second most frequent trinucleotide for the same group is 5′CAA…TTG3′ (with a range of 5–20%). We note that our classification of human DNA transposon groups based on terminal trinucleotide sequence patterns parallels their independent classification into families by sequence homologies based on the full-length element (see Table 1). This parallelism may reflect a specific feature of the transposition machinery of each group (see Discussion).
Table 1. Conservation patterns of terminal trinucleotides of human DNA transposons.
Ninety-five human DNA transposons are divided into 16 different groups (Groups 0–15) based on the conservation pattern of the terminal trinucleotide. Copy no. indicates the total number of detectable copies of each DNA transposon in the entire human genome. Families are adopted from Repbase Update. *Refers to a target site duplication which is not identical at the 5′ and 3′ ends, in contrast to the rest of the DNA transposons. The trinucleotide sequences for each conserved pattern are presented in decreasing order of average frequency (in parentheses).
An interesting finding, similar to that observed with LTR retrotransposons, is that, within each group, the highest degree of base conservation is for the +1 position, irrespective of both sequence and orientation of the terminal trinucleotide (Fig. 3C). A significant sequence conservation is observed for all three positions, although the degree of conservation generally decreases at inner positions, with +1 > +2 > +3 for both termini (exceptions include Groups 3, 4 and 6 which show a reverse gradient of conservation between +2 and +3 at both termini, and Group 8 which shows a slightly higher conservation of +2 compared with + 1 at the 5′ terminus).
Although the +1 position is the most highly conserved within each group of human DNA transposons, the data presented in Figure 3 show that these transposons can have any of the four different bases at the +1 position, and that their terminal trinucleotide sequences are not necessarily inverted. There are three different patterns of terminal trinucleotide sequence orientation (Table 1)—complete inversion (Groups 0, 1, 2, 3, 4, 5, 6, 13, 14, 15), partial inversion (Groups 8, 10, 11, 12) and non-inversion (Groups 7, 9). This observation indicates that the different degrees of base conservation between terminal nucleotide positions are not associated with base specificity, but rather may be due to their differential positional importance, i.e. molecular interactions at the +1 position are more important than those at the internal positions.
DISCUSSION
Conservation of terminal dinucleotides in human LTR retrotransposons: lessons from Mu
We have shown in this study that the terminal two base pairs are highly conserved in human LTRs (Fig. 2). The remarkable aspect of the conservation pattern is that out of 16 possible dinucleotide combinations, 5′TG…CA3′ is the most frequent dinucleotide and 5′TA…TA3′ is the most tolerated non-consensus terminal dinucleotide in most of the LTRs. We will first discuss these results in light of similar findings from studies on the transposable phage Mu, and then extract general principles that can be extended to the termini of DNA transposons.
Experiments with phage Mu showed that only four dinucleotide combinations were active in the presence of the transposase MuA only (30). The most active of these was the wild-type 5′TG…CA3′, followed by 5′TA…TA3′, and then 5′TT…AA3′, with 5′TC…GA3′ being the least active. The in vivo activity of 5′TA…TA3′ was 0.4% of that of the wild-type 5′TG…CA3′. The dinucleotide steps of these two most active sequences are extremely flexible as revealed by computational modeling studies or examination of crystal structures in public databases (31,43–45). All of these studies have classified all pyrimidine/purine steps, CA/TG, TA/TA, CG/CG, as well as GG/CC, as flexible dinucleotide steps with minor variations in the order of flexibility. However, the conformational properties of a dinucleotide step are sensitive to its immediate sequence context, and this context effect has been examined in computer modeling approaches using tetranucleotide steps, i.e. there is an additional base pair at both ends of the dinucleotide step (46). According to this examination, the flexibility of GG/CC and CG/CG steps has a strong context dependency, i.e. their conformational properties vary depending on neighboring base pair steps. On the other hand, CA/TG and TA/TA are either weakly dependent or quite independent of neighboring base steps, respectively. This context independence is probably crucial in maintaining the deformability of the dinucleotide step during the nomadic life of TEs in the host genome because nucleotide sequences immediately flanking the outside region of TE termini keep changing every time they transpose into new genomic locations. Excluding GG/CC and CG/CG, the context-dependent dinucleotide steps, CA/TG is more flexible than TA/TA, and this order of flexibility follows the order of preference of these dinucleotides in Mu transposition (30). The strong preference in all human LTR retrotransposons for the same two flexible base pair steps that are the most active at the ends of Mu makes a compelling argument that, like Mu, the human LTRs have evolved their terminal dinucleotide sequence preferences through an intrinsic structural property of the DNA itself. In this context, it is interesting to note that Mu and LTR retrotransposons have a similar transposition mechanism (47,48). In addition, X-ray crystallization studies have shown that there is great structural similarity among the catalytic domains of Mu transposase, HIV integrase and ASV integrase (49–52).
In the study of Mu, while four dinucleotide combinations were active when provided the MuA transposase alone, transposition of the remaining 12 dinucleotide variants was detected only upon inclusion of MuB protein, which stimulates the activity of MuA (30). In contrast, the analysis of the genome fossil record shows only two or three significantly detectable dinucleotides at the termini of human LTRs (Fig. 2C). This might be due to insufficient transposition activity of less favorable dinucleotides in the case of LTRs, or disappearance of low copy LTRs (because of the low reactivity of their terminal dinucleotides) through sequence erosion during evolutionary time.
The mechanisms responsible for maintenance, dispersion, fixation and genomic clearance of TEs remain largely unknown. While it is known that retrotransposons amplify their genomes by converting RNA transcripts into DNA using an error-prone polymerase, reverse transcriptase, and may be expected to generate favorable mutations at a rapid rate to improve their fertility, it is not possible to determine exactly what percentage of the time they spend at RNA (high mutation rate) versus passive host DNA replication (low mutation rate). Many of the LTR elements have been around long before the mammalian radiation, some have argued since early vertebrate evolution; repeated horizontal acquisition of some families also seems to have occurred during human evolution (53,54). Whether the present LTRs originated from a single element in the distant past, or whether they were acquired at different times, their internal sequences are varied enough to classify them into distinct elements and families. It is therefore striking that the termini of half a million copies of these elements have a bias for only two particular dinucleotide sequences. We argue that the demand for flexibility may have acted as a major selective pressure in the evolution of LTR retrotransposon termini.
Conservation of terminal trinucleotides in human DNA transposons
DNA transposons in the human genome have a conserved trinucleotide at their termini (Fig. 3), the most frequent of which is 5′CAG…CTG3′ found in the majority group (Group 15; Table 1 and Fig. 3). Because the currently available crystal structure data set in the public domain used for the study of dinucleotide step properties is not large enough for deriving a trinucleotide step model, a much larger experimentally generated data set has been employed to measure flexibility of the trinucleotide step. One approach uses DNase I, considered to be a good molecular probe of bendable (flexible) sequence regions (55), because flexible sequences should be more accessible to DNase I cleavage. Thus, DNase I cleavage frequency on naked DNA can be interpreted as a quantitative measure of flexibility. The other approach uses differential preferences in positioning of DNA sequences in nucleosomes. Because flexible sequences may occupy any position in nucleosomes, they will show little positional preference (56). Interestingly, both approaches determined CAG/CTG to be the most highly flexible trinucleotide step. Thus, flexibility, the same criterion as determined for dinucleotide steps at the ends of LTRs, appears to be applicable to the trinucleotide step at the ends of DNA transposons as well. This is a strong indication that DNA transposons have also evolved terminal sequence preferences through a structural property of the DNA itself.
DNA transposons move via a mechanism that uses DNA as an intermediate. They insert into new genomic locations by a cut-and-paste mechanism. Although their transposition is not replicative at the molecular level, they are still capable of propagation in the genome by collaboration with the host replication and recombination machinery (57). Interestingly, our classification of human DNA transposon groups based on their conserved terminal trinucleotide property was seen to parallel an independent family classification based on full-length sequence homology (Table 1; families are adopted from Repbase Update). Given that consensus sequences of DNA transposons represent their ancient forms, and that class 2 elements (DNA transposons) are much more diverse than class 1 elements (retrotransposons) in their transposition mechanism (9), it is possible that the observed parallelism between these two types of classification implies that each terminal trinucleotide consensus signifies a specific feature of the transposition machinery of each group. The prevalence of the most flexible trinucleotide step 5′CAG…CTG3′ in a large majority of human DNA transposon termini is another indication of the beneficial effect of such steps for transposition.
Gradient of base conservation in the terminal base pair step of precise TEs
Another interesting observation from this work is the consistently maximum degree of base conservation at the +1 position in nearly all TEs analyzed (except Group 8 of DNA transposons) (see Figs 2D and 3C). This coincides with the previous observation that mutation of the +1 position is far more severe than that of the +2 position in Mu transposition (30) and in HIV integration (29). Putting together the observations from biological and statistical analyses, we infer that the base at the +1 position has evolved as a common determinant of transposition in both LTR retrotransposons and their cousin, transposable phage Mu, possibly by sharing similar types of DNA–protein interactions.
A differential base conservation between +2 and +3 positions was also observed in the case of conserved terminal trinucleotides, with a generally more conserved +2 position (only Groups 3, 4 and 6 of DNA transposons show a reverse gradient between +2 and +3 positions). The gradient of base importance in terminal trinucleotide steps has been experimentally demonstrated as well in Tn10 (58) and in the TE-derived V(D)J recombination system (39). In both systems, the first strand-nicking step is not inhibited by a mutation at any of the three base pairs in the outside end of Tn10 and in the recombination signal sequence end of the V(D)J system. In contrast, the subsequent hairpin formation step is severely defective with either a +1 or +2 base mutation, but much less with a +3 base mutation. Interestingly, the steeper gradient of functional importance between the +2 and +3 positions observed for both Tn10 and V(D)J systems, coincides with the base conservation gradient pattern of the majority group of human DNA transposons, Group 15 (Fig. 3). Taken together, the above observations indicate a common feature among precise TEs—a gradient of base conservation in the terminal base pair step that has a peak at the +1 position and slopes inside.
The +1 position and the importance of DNA flexibility at termini of precise TEs
Precise TEs employ transposases or integrases encoding a signature DDE motif to bring about movement (59). The DDE motif comprises the catalytic pocket of both transposases and integrases, and thus far, was the only widely shared feature among these elements whose similar transposition chemistry is in accordance with this shared DDE motif. Results from the present statistical study of human TEs have identified a flexible terminal base pair step as another hallmark of precise TEs. Intuition would suggest that conservation of terminal sequences in these elements is related to the chemistry of cleavage and transfer at precise nucleotide positions. However, experimental data with Mu and other precise elements show otherwise.
DNA melting/distortion has been demonstrated around the termini of both assembled and cleaved Mu transpososomes (35,36,60), and around the terminal nucleotides in the Tn5 and Tn10 transpososomes (58,61). The crystal structure of the cleaved Tn5 synaptic complex shows that the terminal two base pairs are open, the phosphodiester backbone of the non-cleaved strand is distorted, and the thymine residue at the +2 position of the non-cleaved strand is ‘flipped out’ (62). The advantage of DNA deformation around TE-host junction regions for transposition has also been experimentally derived in other transposition model systems, such as HIV (37) and V(D)J recombination (38,39). Assuming that this DNA distortion is crucial for transposition, we may hypothesize that the flexible terminal base pair step contributes to transposition by facilitating DNA distortion at the junction region. Indeed, pre-melted Mu termini are indifferent to the sequence of the strand that undergoes chemistry (34), and in both Tn10 (58) and V(D)J recombination systems (39), the first strand-nicking step is not inhibited by a mutation at any of the terminal three base pairs.
Molecular interactions between the terminal nucleotides of a TE and amino acid residues of a transposase is seen in the crystal structure of the cleaved Tn5 synaptic complex, where the ‘flipped’ thymine residue at +2 on the non-cleaved strand is held by multiple amino acid residues of the transposase (62). In the case of Mu also, the +1 and +2 residues on the non-cleaved strand likely contact the transposase, with the thymine residue at +1 being important for stable assembly of the transpososome (34). Considering the immediate proximity of these conserved base pairs, one may imagine that they collaborate through establishing an extended area to allow multiple interactions with the transposase, achieving a DNA distortion large enough to precisely deposit the cleavage site in the catalytic pocket. One may ask why the +1 position is more important than the next conserved position. DNA distortion at the flexible base pair step is ‘permissive’, that is, distortion of the flexible DNA region requires some kind of force, or the local DNA structure remains unchanged (43). A reasonable model is that the DNA deformation takes place gradually and not simultaneously, with the rate-limiting step being application of the initial force. Therefore, the checkpoint (base specificity) for the first contact is most strict and this strictness decreases as the energy barrier lowers, allowing some sequence tolerance. Indeed, it has been experimentally demonstrated with Mu that mismatches at the +1 position are more proficient at transpososome assembly than those at +2, and that mismatches at both positions are far more superior than those at either one (34). Assuming that the first contact occurs at the +1 position and subsequent contact points gradually move inside, we can explain the observed gradient pattern of base conservation (+1 > +2 > +3) at the termini of both Mu and precise TEs.
In summary, conservation of the most flexible two to three base pair steps at the termini of precise TEs implicates a functional role for these sequences in structural transitions in DNA. We propose that base pairs at the +1 position have the highest degree of conservation because molecular interactions occurring here are the rate-limiting step in this process.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Edward Marcotte for comments on the manuscript and Jef Boeke for helpful discussions. This work was supported by a grant from the National Institutes of Health (GM33247). Partial support was provided by the Robert Welch Foundation (F-1531).
REFERENCES
- 1.Kidwell M.G. and Lisch,D.R. (2000) Transposable elements and host genome evolution. Trends Ecol. Evol., 15, 95–99. [DOI] [PubMed] [Google Scholar]
- 2.Kidwell M.G. and Lisch,D. (2002) Transposable elements as sources of genomic variation. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA II. American Society for Microbiology, Washington, DC, pp. 59–90. [Google Scholar]
- 3.Lander E.S., Linton,L.M., Birren,B., Nusbaum,C., Zody,M.C., Baldwin,J., Devon,K., Dewar,K., Doyle,M., FitzHugh,W. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 4.Doolittle W.F. and Sapienza,C. (1980) Selfish genes, the phenotype paradigm and genome evolution. Nature, 284, 601–603. [DOI] [PubMed] [Google Scholar]
- 5.Orgel L.E. and Crick,F.H. (1980) Selfish DNA: the ultimate parasite. Nature, 284, 604–607. [DOI] [PubMed] [Google Scholar]
- 6.Kidwell M.G. and Lisch,D. (1997) Transposable elements as sources of variation in animals and plants. Proc. Natl Acad. Sci. USA, 94, 7704–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltimore D. (2001) Our genome unveiled. Nature, 409, 814–816. [DOI] [PubMed] [Google Scholar]
- 8.Finnegan D.J. (1989) Eukaryotic transposable elements and genome evolution. Trends Genet., 5, 103–107. [DOI] [PubMed] [Google Scholar]
- 9.Craig N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (2002) Mobile DNA II. American Society for Microbiology, Washington, DC. [Google Scholar]
- 10.Luan D.D., Korman,M.H., Jakubczak,J.L. and Eickbush,T.H. (1993) Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 11.Eickbush T. (1999) Exon shuffling in retrospect. Science, 283, 1465–1467. [DOI] [PubMed] [Google Scholar]
- 12.Chaconas G. and Harshey,R.M. (2002) Transposition of phage Mu DNA. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA II. American Society for Microbiology, Washington, DC, pp. 384–402. [Google Scholar]
- 13.Engelman A., Mizuuchi,K. and Craigie,R. (1991) HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell, 67, 1211–1221. [DOI] [PubMed] [Google Scholar]
- 14.Bingham P.M. and Zachar,Z. (1989) Retrotransposons and the FD transposon from Drosophila melanogaster. In Berg,E.D. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 485–502. [Google Scholar]
- 15.Boeke J.D. (1989) Transposable elements in Saccharomyces cerevisiae. In Berg,E.D. and Howe,M.M. (eds), Mobile DNA. American Society for Microbiology, Washington, DC, pp. 335–374. [Google Scholar]
- 16.Coffin J.M. (1991) Retroviridae and their replication. In Fields,B.N. and Knipe,D.M. (eds), Fundamental Virology, 2nd Edn. Raven Press, New York, pp. 645–708. [Google Scholar]
- 17.Craig N.L. (1996) Transposon Tn7. Curr. Top. Microbiol. Immunol., 204, 27–48. [DOI] [PubMed] [Google Scholar]
- 18.Asante-Appiah E. and Skalka,A.M. (1997) Molecular mechanisms in retrovirus DNA integration. Antiviral Res., 36, 139–156. [DOI] [PubMed] [Google Scholar]
- 19.Brown P.O. (1997) Integration. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, pp. 161–203. [PubMed] [Google Scholar]
- 20.Mahillon J. and Chandler,M. (1998) Insertion sequences. Microbiol. Mol. Biol. Rev., 62, 725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roth M.J., Schwartzberg,P.L. and Goff,S.P. (1989) Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell, 58, 47–54. [DOI] [PubMed] [Google Scholar]
- 22.Craigie R., Fujiwara,T. and Bushman,F. (1990) The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell, 62, 829–837. [DOI] [PubMed] [Google Scholar]
- 23.LaFemina R.L., Callahan,P.L. and Cordingley,M.G. (1991) Substrate specificity of recombinant human immunodeficiency virus integrase protein. J. Virol., 65, 5624–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Surette M.G. and Chaconas,G. (1991) Stimulation of the Mu DNA strand cleavage and intramolecular strand transfer reactions by the Mu B protein is independent of stable binding of the Mu B protein to DNA. J. Biol. Chem., 266, 17306–17313. [PubMed] [Google Scholar]
- 25.Vink C., van Gent,D.C., Elgersma,Y. and Plasterk,R.H. (1991) Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol., 65, 4636–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leavitt A.D., Rose,R.B. and Varmus,H.E. (1992) Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J. Virol., 66, 2359–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman P.A., Dickson,M.L. and Fyfe,J.A. (1992) Human immunodeficiency virus type 1 integration protein: DNA sequence requirements for cleaving and joining reactions. J. Virol., 66, 3593–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushman F.D., Engelman,A., Palmer,I., Wingfield,P. and Craigie,R. (1993) Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl Acad. Sci. USA, 90, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esposito D. and Craigie,R. (1998) Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein–DNA interaction. EMBO J., 17, 5832–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee I. and Harshey,R.M. (2001) Importance of the conserved CA dinucleotide at Mu termini. J. Mol. Biol., 314, 433–444. [DOI] [PubMed] [Google Scholar]
- 31.El Hassan M.A. and Calladine,C.R. (1997) Conformational characteristics of DNA: Empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Phil. Trans. R. Soc. Lond., 355, 43–100. [Google Scholar]
- 32.Watson M.A. and Chaconas,G. (1996) Three-site synapsis during Mu DNA transposition: a critical intermediate preceding engagement of the active site. Cell, 85, 435–445. [DOI] [PubMed] [Google Scholar]
- 33.Coros C.J. and Chaconas,G. (2001) Effect of mutations in the Mu-host junction region on transpososome assembly. J. Mol. Biol., 310, 299–309. [DOI] [PubMed] [Google Scholar]
- 34.Lee I. and Harshey,R.M. (2003) The conserved CA/TG motif at Mu termini: T specifies stable transpososome assembly. J. Mol. Biol., 330, 261–275. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z., Namgoong,S.Y., Zhang,X. and Harshey,R.M. (1996) Kinetic and structural probing of the precleavage synaptic complex (type 0) formed during phage Mu transposition. Action of metal ions and reagents specific to single-stranded DNA. J. Biol. Chem., 271, 9619–9626. [DOI] [PubMed] [Google Scholar]
- 36.Kobryn K., Watson,M.A., Allison,R.G. and Chaconas,G. (2002) The Mu three site synaptic complex: a strained assembly platform in which delivery of the L1 transposase binding site triggers catalytic commitment. Mol. Cell, 10, 659–669. [DOI] [PubMed] [Google Scholar]
- 37.Scottoline B.P., Chow,S., Ellison,V. and Brown,P.O. (1997) Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev., 11, 371–382. [DOI] [PubMed] [Google Scholar]
- 38.Cuomo C.A., Mundy,C.L. and Oettinger,M.A. (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsden D.A., McBlane,J.F., van Gent,D.C. and Gellert,M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 40.Jurka J. (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet., 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 41.Sneath P.H.A. and Sokal,R.R. (1973) Numerical Taxonomy: The Principles and Practice of Numerical Classification. W.H. Freeman, New York. [Google Scholar]
- 42.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickerson R.E. (1998) DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res., 26, 1906–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson W.K., Gorin,A.A., Lu,X.J., Hock,L.M. and Zhurkin,V.B. (1998) DNA sequence-dependent deformability deduced from protein–DNA crystal complexes. Proc. Natl Acad. Sci. USA, 95, 11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Packer M.J., Dauncey,M.P. and Hunter,C.A. (2000) Sequence-dependent DNA structure: dinucleotide conformational maps. J. Mol. Biol., 295, 71–83. [DOI] [PubMed] [Google Scholar]
- 46.Packer M.J., Dauncey,M.P. and Hunter,C.A. (2000) Sequence-dependent DNA structure: tetranucleotide conformational maps. J. Mol. Biol., 295, 85–103. [DOI] [PubMed] [Google Scholar]
- 47.Grindley N.D.F. and Leschziner,A.E. (1995) DNA transposition: from a black box to a color monitor. Cell, 83, 1063–1066. [DOI] [PubMed] [Google Scholar]
- 48.Mizuuchi K. and Baker,T.A. (2002) Chemical mechanisms for mobilizing DNA. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA II. American Society for Microbiology, Washington, DC, pp. 12–23. [Google Scholar]
- 49.Dyda F., Hickman,A.B., Jenkins,T.M., Engelman,A., Craigie,R. and Davies,D.R. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science, 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- 50.Bujacz G., Jaskolski,M., Alexandratos,J., Wlodawer,A., Merkel,G., Katz,R.A. and Skalka,A.M. (1995) High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol., 253, 333–346. [DOI] [PubMed] [Google Scholar]
- 51.Rice P. and Mizuuchi,K. (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- 52.Bujacz G., Jaskolski,M., Alexandratos,J., Wlodawer,A., Merkel,G., Katz,R.A. and Skalka,A.M. (1996) The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure, 4, 89–96. [DOI] [PubMed] [Google Scholar]
- 53.Smit A.F. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 54.Medstrand P., van de Lagemaat,L.N. and Mager,D.L. (2002) Retroelement distributions in the human genome: variations associated with age and proximity to genes. Genome Res., 12, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brukner I., Sanchez,R., Suck,D. and Pongor,S. (1995) Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J., 14, 1812–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Satchwell S.C., Drew,H.R. and Travers,A.A. (1986) Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol., 191, 659–675. [DOI] [PubMed] [Google Scholar]
- 57.Brookfield J.F.Y. (1995) Transposable elements as selfish DNA. In Sherratt,D.J. (ed.), Mobile Genetic Elements. Oxford University Press, Oxford, pp. 130–153. [Google Scholar]
- 58.Allingham J.S., Wardle,S.J. and Haniford,D.B. (2001) Determinants for hairpin formation in Tn10 transposition. EMBO J., 20, 2931–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haren L., Ton-Hoang,B. and Chandler,M. (1999) Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- 60.Lavoie B.D., Chan,B.S., Allison,R.G. and Chaconas,G. (1991) Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu-host junction of the Mu type 1 transpososome. EMBO J., 10, 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhasin A., Goryshin,I.Y., Steiniger-White,M., York,D. and Reznikoff,W.S. (2000) Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol., 302, 49–63. [DOI] [PubMed] [Google Scholar]
- 62.Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]