Abstract
During neuroblast (NB) divisions, cell fate determinants Prospero (Pros) and Numb, together with their adaptor proteins Miranda (Mira) and Partner of Numb, localize to the basal cell cortex at metaphase and segregate exclusively to the future ganglion mother cells (GMCs) at telophase. In inscuteable mutant NBs, these basal proteins are mislocalized during metaphase. However, during anaphase/telophase, these mutant NBs can partially correct these earlier localization defects and redistribute cell fate determinants as crescents to the region where the future GMC ‘buds' off. This compensatory mechanism has been referred to as ‘telophase rescue'. We demonstrate that the Drosophila homolog of the mammalian tumor-necrosis factor (TNF) receptor-associated factor (DTRAF1) and Eiger (Egr), the homolog of the mammalian TNF, are required for telophase rescue of Mira/Pros. DTRAF1 localizes as an apical crescent in metaphase NBs and this apical localization requires Bazooka (Baz) and Egr. The Mira/Pros telophase rescue seen in inscuteable mutant NBs requires DTRAF1. Our data suggest that DTRAF1 binds to Baz and acts downstream of Egr in the Mira/Pros telophase rescue pathway.
Keywords: Baz function, DTRAF1, Egr, neuroblast asymmetric division, telophase rescue
Introduction
During Drosophila embryonic neuroblast (NB) division, two groups of proteins are asymmetrically localized to opposite cortical regions along the apical–basal axis of NBs. Bazooka (Baz) (Schober et al, 1999; Wodarz et al, 1999), the Drosophila homolog of Par6 (Par6) (Petronczki and Knoblich, 2001), atypical protein kinase C (DaPKC) (Wodarz et al, 2000), Inscuteable (Insc) (Kraut et al, 1996), Partner of Inscuteable (Pins) (Parmentier et al, 2000; Schaefer et al, 2000; Yu et al, 2000) and the α subunit of heterotrimeric G protein (Gαi) (Schaefer et al, 2000, 2001; Yu et al, 2003), as well as Locomotion defects (Loco) (Yu et al, 2005), localize to the apical cortex of dividing NBs where they are thought to form a multiprotein complex (apical complex). The cell fate determinants Prospero (Pros) (Doe et al, 1991; Vaessin et al, 1991, Matsuzaki et al, 1992) and Numb (Uemura et al, 1989; Rhyu et al, 1994), together with their respective adapter proteins Miranda (Mira) (Ikeshima-Kataoka et al, 1997; Shen et al, 1997) and Partner of Numb (Pon) (Lu et al, 1998), localize to the basal cortex of NBs in a cell-cycle-dependent manner. In late G2 or early prophase, Mira/Pros are transiently localized to the apical cortex but by late prophase Mira/Pros are seen as basal cortical crescents. Similarly, Pon and Numb are cortical in early prophase and colocalize with Mira/Pros basally by late prophase. At telophase, all apical proteins remain in the large daughter cell and Mira/Pros and Pon/Numb are exclusively segregated into the future ganglion mother cell (GMC) (reviewed in Lu et al, 2000).
The cell-cycle-dependent localization of Mira/Pros and Pon/Numb requires the functions of apical complex proteins. Mutation analyses have demonstrated that the apical complex plays a central role in the NB asymmetric division. It controls and coordinates the basal localization of cell fate determinants with the apicobasal orientation of the mitotic spindle at metaphase and the generation of asymmetric spindle geometry late in mitosis (review in Betschinger and Knoblich, 2004). One prominent phenotype seen in mutations of apical proteins is the mislocalization of the cell fate determinants. For example, in insc mutant NBs, cell fate determinants are often delocalized (the crescent occupies more than 50% of the NB cortex, sometimes even becomes uniformly cortical) or mislocalized (the crescent localizes to the lateral or basal-lateral side of the NB cortex) during prophase and metaphase. However, starting from anaphase, the great majority of the mutant NBs redistribute the cell fate determinants Mira/Pros and Pon/Numb as cortical crescents overlying one of the spindle poles, in the region where the future GMC ‘buds off'. This apical protein-independent self-correcting phenomenon was observed in insc and baz mutant NBs (Schober et al, 1999; Wodarz et al, 1999; Peng et al, 2000) and has been referred to as ‘telophase rescue' (Peng et al, 2000). Similar observations were also seen in pins and Gαi mutants (our unpublished data).
Our earlier study on the functions of snail family genes in asymmetric NB divisions proposed that in wild-type (WT) NBs two parallel and independent mechanisms were responsible for the Mira/Pros and Pon/Numb basal localization during NB divisions (Cai et al, 2001). The dominant Insc-dependent pathway, whose members include Insc, Baz and Pins, functions throughout mitosis, whereas the cryptic Insc-independent pathway only acts late in mitosis (anaphase and telophase) and is required for telophase rescue. The process of telophase rescue is ill-understood, although Discs Large (Dlg) has been implicated recently to be involved (Siegrist and Doe, 2005).
We conducted a search for potential new NB markers based on the published RNA in situ patterns. Several potential candidates including the Drosophila homolog of tumor-necrosis factor (TNF) receptor-associated factor (DTRAF1) exhibiting interesting NB expression patterns were selected. DTRAF1 was first identified as a Misshapen (Msn) interacting protein in a yeast two-hybrid screen (Liu et al, 1999). Msn functions genetically upstream of the c-Jun amino-terminal kinase (JNK) mitogen-activated protein kinase module. Failure to activate the JNK pathway results in embryonic lethality owing to defective dorsal closure (Su et al, 1998). DTRAF1 has been suggested to activate the JNK pathway by interacting with Msn in Drosophila embryos. DTRAF1 is also involved in JNK-mediated cell death induced by ectopic Reaper in the Drosophila compound eye (Kuranaga et al, 2002).
Other Drosophila homologs of TNF signaling pathway were also reported. Unlike the mammalian TNF/TNFR family, which contains more than 20 ligands and receptors, the Drosophila counterparts appear to have only one ligand, which is Eiger (Egr) (Igaki et al, 2002; Moreno et al, 2002; Kauppila et al, 2003), and one receptor, which is Wengen (Wgn) (Kanda et al, 2002; Kuranaga et al, 2002; Kauppila et al, 2003). Egr is a type II membrane glycosylated protein, which can be cleaved and released as a soluble ligand. Ectopic expression of both Egr and Wgn induced apoptosis in S2 cells and the Drosophila compound eye (Kauppila et al, 2003). RNA in situ hybridization showed that egr (Kauppila et al, 2003) and DTRAF1 were expressed in NBs (Preiss et al, 2001). On the other hand wgn was expressed in the mesoderm in early embryos and in the ventral cord only in late embryos (Igaki et al, 2002; Kauppila et al, 2003). The possible functions of DTRAF1 and Egr in Drosophila embryonic CNS development are yet to be established.
Here, we analyze the telophase rescue phenomenon in insc mutant NBs and report that DTRAF1 is apically localized and is required for telophase rescue specifically for Mira/Pros but not for Pon/Numb. We demonstrate that the apical localization of DTRAF1 requires Baz and Erg, and DTRAF1 interacts with Baz in vitro. We also show that Egr, the Drosophila homolog of TNF, is a member of the Mira/Pros telophase rescue pathway. Our data suggest that DTRAF1 binds to Baz and acts downstream of Egr in the Mira/Pros telophase rescue pathway.
Results
DTRAF1 is apically localized in mitotic NBs
In our search for genes that are expressed in NBs, DTRAF1 (Preiss et al, 2001) and other candidates were selected for further analyses. Antibody raised against the C-terminal region of DTRAF1 showed that the protein was expressed in NBs, epithelial cells and axon tracks during late embryonic stages (Figure 1). In the early developing CNS, DTRAF1 was enriched on the apical cortex of the dividing NBs. This asymmetric localization was cell-cycle-dependent. During interphase, DTRAF1 was cytoplasmic (Figure 1A). After NBs entered mitosis, DTRAF1 was apically enriched during prophase (Figure 1B) and formed a tight cortical crescent overlying the apical spindle pole at metaphase (Figure 1C). At anaphase, DTRAF1 apical crescent became less prominent and an additional apically enriched, punctated cytoplasmic staining was visible (Figure 1D). At telophase, DTRAF1, both cortical and cytoplasmic, remained visible only in the large daughter cell (Figure 1E).
Figure 1.
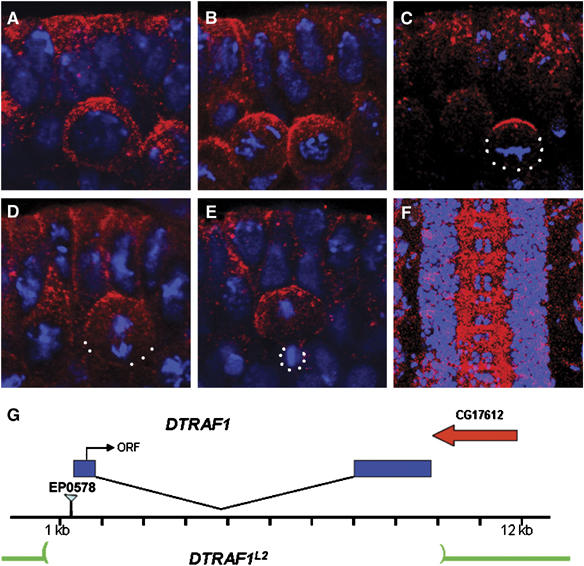
DTRAF1 expression in the embryonic CNS. Stage 10 (A–E) and stage 15 (F) WT embryos are double stained with anti-DTRAF1 (red) and DNA (blue). In interphase, DTRAF1 is cytoplasmic in NBs (A). After NB enters mitosis, DTRAF1 is apically enriched during prophase (B) and forms a tight apical crescent at metaphase (C). At anaphase, the DTRAF1 apical crescent becomes less obvious and additional punctated cytoplasmic staining is visible (D). By telophase DTRAF1, both cortical and cytoplasmic, remains only in the large daughter cell (E). High levels of DTRAF1 are present in the axonal tracts of the ventral nerve cord (F). (G) A schematic diagram showing the DTRAF1 genomic region. The P-element EP0578 is inserted at 59 bp 5′ to the DTRAF1 transcribed region. The entire DTRAF1 coding region is removed in DTRAF1L2. White dots outline the cell body. Apical is up.
We generated DTRAF1 mutants by imprecise excision of the P-element in EP0578, which was inserted 59 bp upstream of the 5′ end of the gene. One of the revertants, DTRAF1L2, is pupal lethal and carries a deletion of about 9 kb (Figure 1G), uncovering the entire DTRAF1 gene. Southern blot analysis indicated that the 3′ deletion did not extend to its neighboring gene CG17612 (data not shown), and only DTRAF1 appeared to be removed. In germline clone embryos lacking both maternal and zygotic DTRAF1 made from the DTRAF1L2 allele (henceforth termed DTRAF1), anti-DTRAF1 antibody did not detect any obvious signals (data not shown), confirming the specificity of our antibody.
Apical localization of DTRAF1 is dependent on Baz
The apical localization of DTRAF1 suggests that it could be a new member of the apical complex that controls NB asymmetric divisions. To investigate this possibility, we examined the localization of apical complex proteins in DTRAF1 mutant NBs. Baz, Insc and Pins localized normally in DTRAF1 mutant NBs (Figure 2A–C), indicating that DTRAF1 was not required for the localization of apical complex members. We further looked at the DTRAF1 expression in baz, insc and pins mutants. In insc or pins mutant NBs, DTRAF1 was normally localized to the apical cortex (Figure 2E and F). However, DTRAF1 was no longer apical but cytoplasmic in baz mutant metaphase NBs (Figure 2D). These observations indicate that DTRAF1 requires Baz, but not Insc and Pins, for its apical localization.
Figure 2.
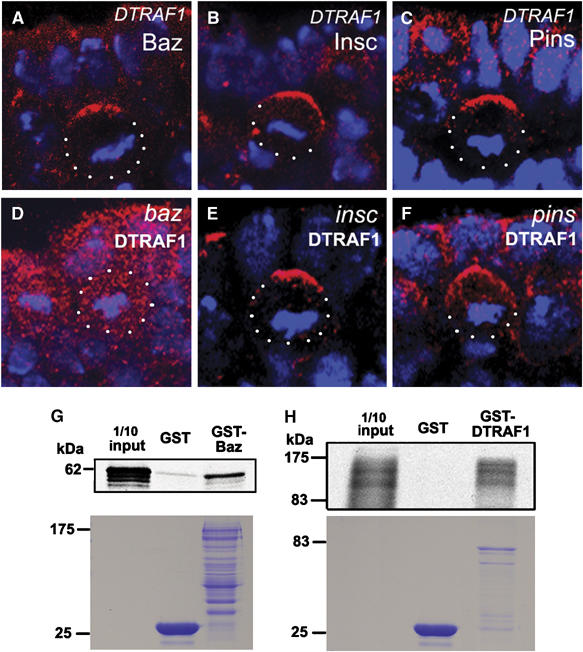
DTRAF1 interacts with Baz in vitro and its apical localization is dependent on Baz but not Pins and Insc. In DTRAF1 mutant embryos, Baz (red, A), Insc(red, B) and Pins (red, C) form crescents in the dividing NBs. The asymmetric localization of DTRAF1 (red) is dependent on Baz (D), but not Insc (E) and Pins (F). Cell body is outlined with dots. DNA is labeled in blue. Apical is up. GST-fusion protein pull-down assays suggest the possible physical interaction between Baz and DTRAF1 (G, H). The full-length GST-Baz fusion protein can specifically pull down the in vitro translated and 35S labeled full-length DTRAF1 (G). Similarly, the full-length GST-DTRAF1 specifically pulls down the in vitro translated and 35S labeled full-length Baz (H). The lower panels are the duplicated SDS gels stained with Coomassie brilliant blue, showing the molecular weights and quantities of recombinant GST, GST–Baz and GST-DTRAF1 applied on each lane. The diffused appearance of 35S labeled Baz is most likely due to the degradation of the in vitro translated Baz.
The dependence of DTRAF1 apical localization on Baz suggests a possible functional relationship between these two proteins. We wanted to know whether DTRAF1 can directly interact with Baz. To explore this issue, we conducted GST fusion protein pull-down assays. GST fusion constructs of full-length DTRAF1 and Baz were made and fusion proteins were expressed and purified. In our assays, 35S-labeled DTRAF1 was pulled down by the GST–Baz fusion protein, but not by the GST protein control (Figure 2G). The reciprocal pull-down experiment further confirmed the direct physical interaction between DTRAF1 and Baz in vitro (Figure 2H). Therefore, it is possible that Baz recruits DTRAF1 to the apical cortex through direct physical interaction.
Cell fate determinants Mira/Pros and Pon/Numb localize normally in DTRAF1 mutant NBs
The apical localization of DTRAF1 in dividing NBs prompted us to investigate the basal localization of the cell fate determinants such as Mira/Pros and Pon/Numb in mutant embryos. As Mira and Pros are always colocalized in embryonic NBs and so are Pon and Numb, we only present anti-Mira and anti-Pon data to represent Pros and Numb localization, respectively. Anti-Mira and anti-Pon staining indicated that removal of both maternal and zygotic DTRAF1 did not affect the basal localization of cell fate determinants (Figure 3B and D). In telophase, the cell fate determinants were exclusively segregated into GMCs (Figure 3F and H) as in the WT embryos. Failure to detect any obvious defects in cell fate determinant localization and segregation in DTRAF1 mutant NBs raised the possibility that DTRAF1 might not be involved in the basal localization of cell fate determinants in the NBs, or the function of DTRAF1 was cryptic or redundant owing to other players.
Figure 3.
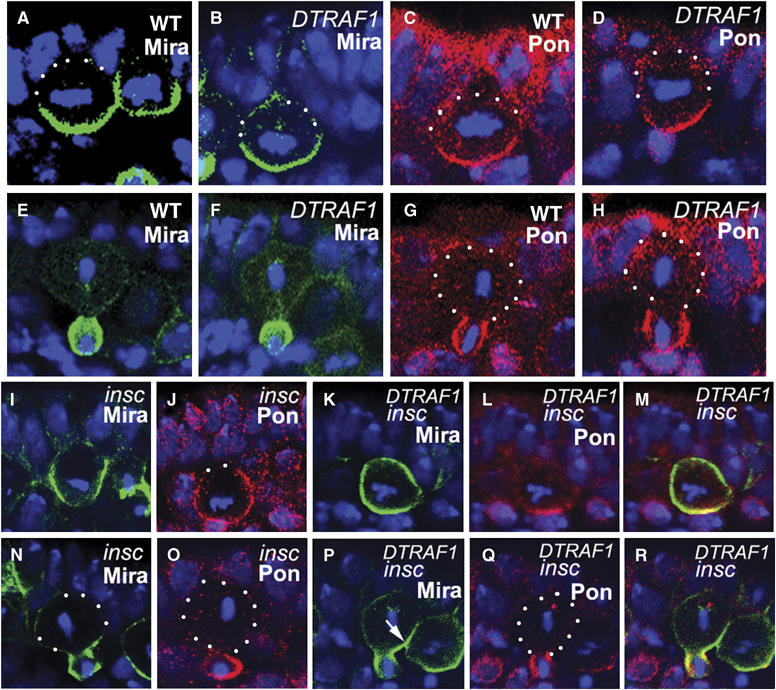
DTRAF1 is required specifically for Mira, but not for Pon, telophase rescue in insc NBs (A–R). Lateral view of stage 10 NBs labeled with anti-Mira (green) or anti-Pon (red). In DTRAF1 mutant NBs, Mira and Pon form basal crescents at metaphase (B, D) and are segregated into future GMCs during telophase (F, H) as in WT NBs (A, C, E, G). In insc mutant NBs, Mira and Pon form extended crescents in metaphase NBs (I, J). During telophase, Mira and Pon are segregated into the future GMCs in the majority of the NBs (N, O). In DTRAF1 insc double-mutant embryos, Mira forms extended crescent or cortical in the majority of dividing NBs during metaphase (K) and by telophase Mira is only partially segregated into the future GMCs, leaving a Mira ‘tail' in the large cell (arrow, P). Whereas Pon is still localized normally in the same dividing NB and segregated exclusively into the future GMC (L, Q). Superimposed images of (K), (L) and (P), (Q) (M, R). Apical is up, DNA is in blue. White dots outline the cell body.
DTRAF1 is required for normal Mira/Pros crescent formation at metaphase in insc NBs
We have proposed earlier that two parallel pathways are involved in basal localization of the cell fate determinants (Cai et al, 2001). An Insc-dependent pathway is dominant and functions throughout mitosis. Baz, Insc and Pins are the members of the Insc-dependent pathway. On the other hand an Insc-independent telophase rescue pathway is cryptic (insofar as its function can only be seen in the absence of the Insc-dependent pathway). To investigate whether DTRAF1 might be a member of the proposed telophase rescue pathway, we compared the localization of basal proteins during metaphase and telophase in insc single-mutant NBs with that of DTRAF1 insc double-mutant NBs. We reasoned that if DTRAF1 was indeed involved in telophase rescue, then the segregation of the basal proteins to the future GMCs at telophase should be compromised in the DTRAF1 insc double-mutant NBs. In order to quantitatively score the localization of cell fate determinants at metaphase, we arbitrarily define three categories of protein localizations: normal crescents, extended crescents and uniformly cortical localization (Figure 4A). When basal proteins occupy less than 50% of the NB cortex, we consider them as normal crescents and when they completely occupy the entire NB cortex we score them as cortical localization. NBs with crescents occupying more than 50% of the cortex are scored as extended crescents.
Figure 4.
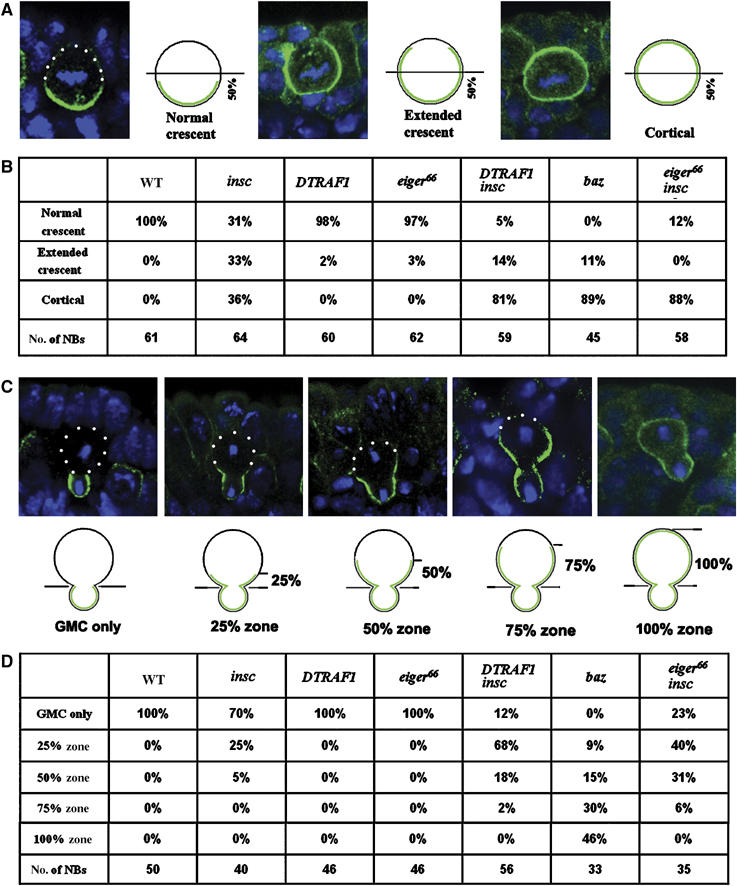
Mira telophase rescue in various mutant backgrounds. The localization of Mira in metaphase NBs is subdivided into three classes: the normal crescents that occupy less than 50% of the NB cortex; the extended crescents that occupy more than 50% of the cortex and uniform cortical distribution, as indicated by the confocal images and the diagrams (A). Quantitation of Mira localization in metaphase NBs according to the standards set in panel A (B). Mira telophase rescue is quantitatively assayed by scoring of the ‘Mira tail' in four arbitrary zones (25, 50, 75 and 100%) according to its distance from the future GMCs as indicated by the confocal images and diagrams (C). The quantitation of Mira localization in telophase NBs following the criterion described in panel C is summarized in (D). NBs from stages 10/11 embryos are used. Apical is up. Mira is in green and DNA is in blue. White dots outline the cell body.
In WT metaphase NBs, both Mira and Pon were colocalized and formed normal crescents (Figure 3A and C). In metaphase insc mutant NBs, Mira formed normal crescents only in one third of the cells (31%, n=64) (Figure 4B) and in the remaining NBs, Mira showed extended crescents (33%, n=64; Figure 3I) or uniformly cortical localization (36%, n=64), consistent with previous observations.
In DTRAF1 insc double-mutant NBs, we saw a dramatic decrease in the frequency of normal crescents (5%, n=59; compared with 31% in insc NBs) and extended crescents (14%, n=59; compared with 33% in insc NBs) of Mira in metaphase cells (Figure 4B). The majority of metaphase DTRAF1 insc mutant NBs showed uniform cortical Mira distribution (Figure 3K) (81%, n=59; compared with 36% in insc NBs). These observations clearly indicate that DTRAF1 plays a role in Mira asymmetric localization in insc NBs as early as in metaphase. In the absence of DTRAF1, Mira tends to be uniformly cortical in most of the metaphase insc NBs.
Mira telophase rescue is compromised in the absence of DTRAF1
We further studied the function of DTRAF1 at telophase in insc mutant NBs. For a quantitative analysis, we arbitrarily divided the large cell of the telophase NB into four equal zones and assigned zones according to its distance from the future GMC as 25, 50, 75 and 100% (Figure 4C). At telophase when the cell fate determinants are exclusively segregated into future GMCs, we score them as complete telophase rescue (GMC only). The higher the percentage of NBs with exclusive segregation, the better the telophase rescue. In mutant NBs where the basal proteins fail to be completely segregated into GMCs at telophase, they are left behind as a cortical ‘tail' in the larger daughter cell (arrow, Figures 3P and 4C). The longer the tail, the weaker the telophase rescue. We assessed telophase rescue potency based on these parameters.
At telophase, Mira and Pon were exclusively deposited into future GMCs in WT NBs (Figure 3E and G). In the absence of Insc, about two-thirds of the NBs were able to redistribute and segregate Mira exclusively into the GMC daughter (70%, n=40; Figures 3N and 4D), and one quarter of the NBs had Mira remaining in the 25% zone (25%, n=40, Figure 4D) of the big daughter cell. We also saw about 5% (n=40) of the NBs containing a Mira ‘tail' in the 50% zone (Figure 4D). These data indicate that, in the absence of Insc, the telophase rescue pathway is able to redistribute and segregate most of Mira/Pros into the future GMCs in NBs.
Telophase rescue was affected when DTRAF1 was further removed from insc NBs: only 12% (n=56; Figure 4D) (compared with 70% in insc NBs) of the NBs segregated Mira exclusively into future GMCs and NBs containing Mira tail extending within the 25% zone increased about 2.5-fold (68%, n=56; Figure 3P) (compared with 25% in insc NBs). The frequency of NBs that contain Mira tails in the 50% zone was about three times as high (18%; n=56; Figure 4D) as that seen in insc NBs. These results demonstrate that telophase rescue of Mira in insc NBs is compromised in the absence of DTRAF1. We also studied telophase rescue of Pon in DTRAF1 insc NBs and found that it was not affected (Figure 3L and Q). Thus, we conclude that DTRAF1 is specifically involved in Mira/Pros telophase rescue.
Loss of DTRAF1 apical localization correlates with defective telophase rescue in baz NBs
In insc mutant NBs, loss of DTRAFl disrupts Mira/Pros telophase rescue. As DTRAF1 retains its apical crescent in insc mutant NBs, we considered the possibility that the apical localization of DTRAF1 might be required for Mira telophase rescue. In baz NBs, not only is the Insc protein level decreased and often undetectable with antibody staining (Wodarz et al, 1999, Schober et al, 1999, Cai et al, 2003, Siegrist and Doe, 2005) but DTRAF1 also loses its apical localization and becomes cytoplasmic (Figure 2D). We should therefore expect to see defective Mira telophase rescue in baz mutant NBs if the apical localization of DTRAF1 is important for the process.
In metaphase baz NBs, Mira localization tended to be uniformly cortical in most NBs (89%, n=45; Figure 5B) and we did not observe any baz NBs with normal crescents (Figure 4B). Similar uniformly cortical Mira localization was also seen in DTRAF1 insc NBs (81%, n=59), whereas in insc NBs only 36% (n=64) of the metaphase NBs showed uniformly cortical Mira (Figure 4B). These observations clearly demonstrate that, similar to DTRAF1 insc NBs, asymmetric Mira localization is disrupted in baz NBs at metaphase.
Figure 5.
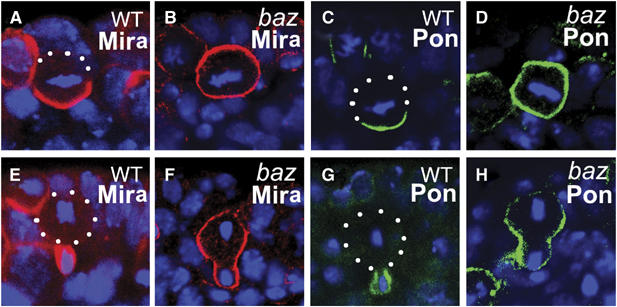
Baz is required for both Mira and Pon telophase rescue. Stage 10 WT and bazGLC embryos are stained with anti-Mira (A, B, E, F; red) or anti-Pon (C, D, G, H; green). Mira and Pon form basal crescents at metaphase (A, C) and segregate into the future GMCs in telophase (E, G) in WT NBs. In bazGLC NBs, Mira and Pon are evenly distributed on the cell cortex in metaphase NBs (B, D) and retained largely on the cortex of the larger cell during telophase (F, H). Apical is up. DNA is in blue. White dots outline the cell body.
At telophase, Mira telophase rescue in baz NBs appeared to be severely impaired; exclusive segregation of Mira into the future GMCs was not observed (0%, n=33; Figure 4D). In addition, about 30% (n=33) of the telophase NBs contained Mira crescents, which extended into the 75% zone and about 46% (n=33) showed cortical Mira (Figure 5F), which was seldom seen in telophase insc or DTRAF1 insc NBs (Figure 4D). However, we noticed the redistribution of Mira even in baz NBs by comparing carefully the Mira distribution patterns at metaphase and at telophase. Mira was uniformly cortical in about 90% of baz NBs (Figure 4B) at metaphase, whereas at telophase, only about half of the NBs originally with cortical Mira showed the same Mira distribution (46%, Figure 4D), indicating that in the other half of the NB population Mira was redistributed, albeit inefficiently, towards the future GMCs. In about a quarter of baz telophase NBs, Mira was found enriched in the future GMCs with ‘tails' extended to the 25–50% zones (Figure 4D). These data indicate that although the redistribution of Mira towards the future GMCs is noticeable in baz telophase NBs, Mira telophase rescue in baz NBs is largely defective and the phenotype is more pronounced than that seen in the DTRAF1 insc double mutant.
We also examined the Pon/Numb telophase rescue in baz NBs. By metaphase, Pon was evenly cortical in most NBs (Figure 5D) and remained in the big cell late in mitosis (Figure 5H), suggesting that the Pon/Numb telophase rescue was also affected in baz mutant NBs. These observations are consistent with our hypothesis that apically localized DTRAF1 is required for telophase rescue of Mira/Pros. The stronger phenotype seen in the baz NBs is most likely due to the disruption of additional players required for the telophase rescue process, consistent with baz acting upstream of DTRAF1 in this process.
Egr, the Drosophila homolog of TNF, is required for DTRAF1 apical localization and Mira telophase rescue
In mammals, TRAFs associate with TNF receptors (TNFRs) and function in the TNF signal transduction pathway. The involvement of DTRAF1 in Mira telophase rescue led us to consider the possibility that other Drosophila homologs in the TNF pathway might also be involved in this process. Two other fly homologs in the mammalian TNF signaling pathway have been identified, Egr, the fly homolog of TNF (Igaki et al, 2002; Moreno et al, 2002; Kauppila et al, 2003) and Wgn, the fly homolog of TNFR (Kanda et al, 2002; Kuranaga et al, 2002; Kauppila et al, 2003). As it had been reported that wgn was not expressed in NBs (Igaki et al, 2002; Kauppila et al, 2003), we focused on Egr.
RNA in situ hybridization data showed that egr was expressed in NBs (Kauppila et al, 2003, data not shown). The egr mutants were generated by mobilization of the P-element in KG02299, which was inserted ∼160 bp 5′ of the gene (Figure 6A). One revertant egr66 contained a deletion of 7 kb, uncovering the entire egr transcription unit. PCR analyses indicated that the deletion did not extend to its neighboring genes (data not shown). The egr66 allele (egr mutant) is homozygous viable and does not show any defect in Mira asymmetric localization (Figure 6B and E). Interestingly, in egr mutant, DTRAF1 lost its apical localization and became cytoplasmic in mitotic NBs (Figure 6C), indicating that DTRAF1 apical localization requires Egr.
Figure 6.
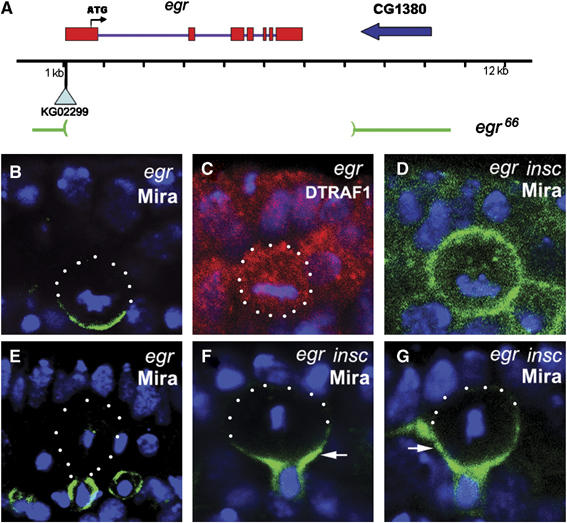
Egr is involved in Mira telophase rescue. A schematic diagram showing the egr genomic locus (A). egr66 deletes the entire coding region of egr. In egr66 NBs, Mira (green) forms basal cortical crescents at metaphase (B) and segregates into the basal daughter cells at telophase (E). The asymmetric apical localization of DTRAF1 (red) seen in WT metaphase NBs (see Figure 1C) is disrupted and becomes cytoplasmic in egr66 metaphase NBs (C). In egr insc NBs, Mira (green) is evenly distributed to the cell cortex of the majority of the metaphase NBs (D) and retained in the large cell at telophase (arrow, F and G). DNA is in blue. Apical is up. White dots outline the cell body.
We further assayed for Mira telophase rescue in egr insc double-mutant embryos. In metaphase egr insc NBs, Mira was cortical (Figure 6D) in 88% (n=58) of the NBs, which was higher than in insc NBs (36%, n=64), but comparable to DTRAF1 insc NBs (81%, n=59, Figure 4B). At telophase, only 23% (n=35) of NBs segregated Mira exclusively to the future GMCs, compared to 70% (n=40, Figure 4D) seen in insc single-mutant NBs. We saw increased populations of telophase NBs with Mira tails retained in the 25% zone (40%, n=35, Figure 6F), 50% zone (31%, n=35, Figure 6G) and 75% zone (6%, n=35, Figure 4D). However, telophase rescue of Pon/Numb remained unaffected in egr insc double-mutant NBs (data not shown).
These results show that Egr is also required for both Mira/Pros crescent formation at metaphase and Mira telophase rescue. Mira telophase rescue is compromised in egr insc double-mutant NBs in a manner similar to DTRAF1 insc. Based on these observations, we conclude that Egr, like DTRAF1, is also involved in the Mira/Pros telophase rescue. As DTRAF1, which is normally apically localized, becomes cytoplasmic in egr insc mutant NBs, it is likely that Egr functions through DTRAF1 and acts upstream of DTRAF1 in the telophase rescue process. Furthermore, the defective Mira telophase rescue in egr insc NBs further supports the view that apical localization of DTRAF1 is required for Mira/Pros telophase rescue.
Discussion
In telophase NBs, segregation of cell fate determinants, such as Pros into future GMCs, is critical for their proper development (Doe et al, 1991; Vaessin et al, 1991; Matsuzaki et al, 1992). Telophase rescue appears to be one of the safeguard mechanisms, which acts to ensure that GMCs inherit the cell fate determinants and adopt the correct cell identity when the mechanisms, which normally operate during NB divisions, fail (e.g., in insc mutant). Telophase rescue is a phenomenon for which the underlying mechanism involved remains largely unknown. Our data demonstrate for the first time that DTRAF1 and Egr are two members of the Insc-independent telophase rescue pathway specific for Mira/Pros.
Apical asymmetric localization of DTRAF1
Although it is apically enriched in mitotic NBs and can directly interact with Baz in vitro, DTRAF1 does not seem to be involved with the functions normally associated with the apical complex proteins. One distinct feature of DTRAF1 that differs from the other known apical proteins is its localization pattern; it is cytoplasmic in interphase and the apical crescent is prominent only at metaphase. In contrast, proteins of the apical complex are largely undetectable during interphase and form distinct apical crescents, starting from late interphase or early prophase. The protein localization difference between DTRAF1 and other apical proteins suggests that DTRAF1 and apical proteins are not always colocalized during mitosis. If DTRAF1 is a bona fide member of the apical complex, we expect to observe the localization defects of other apical proteins in DTRAF1 mutant, as well as mislocalization of basal proteins, which we did not detect. In addition, no spindle orientation or geometry defects were observed in the absence of DTRAF1. Based on these observations, we conclude that DTRAF1 is not involved with the functions normally associated with the apical complex proteins.
The in vitro GST fusion protein pull-down assay suggests that DTRAF1 may physically bind to Baz. This result is consistent with our genetic data, indicating that DTRAF1 acts downstream of baz and that its apical localization requires baz. These observations are consistent with the view that DTRAF1 is recruited to the apical cortex by apical Baz in mitotic NBs. Baz, even at very low levels, can recruit DTRAF1 to the apical cortex of the mitotic NBs. For example, in insc mutant NBs, DTRAF1 remains apical probably owing to the low levels of Baz which remain localized to the apical cortex (Cai et al 2003, Siegrist and Doe, 2005). This speculation is supported by our Mira/Pros telophase rescue data, which clearly demonstrate that the telophase rescue seen in insc mutant NBs is severely damaged in baz mutant, suggesting that the Baz function required for Mira/Pros (and Pon/Numb) telophase rescue is intact in insc mutant NBs.
A recent publication has shown that Pins/Gαi asymmetric cortical localization can be induced at metaphase by the combination of astral microtubules, kinesin Khc-73 and Dlg in the absence of Insc (Siegrist and Doe, 2005), which coincides with our observation that DTRAF1 also only forms tight crescent at metaphase in both WT and insc mutant NBs. Does DTRAF1 apical crescent formation also require the functions of astral microtubules, kinesin Khc-73 and Dlg? Our data do not favor this hypothesis based on the following observations. (1) In TE35BC-3, a small deficiency uncovering sna family genes insc is not expressed but Pins and Gαi are asymmetrically localized (Cai et al, 2001), indicating that the astral microtubules, kinesin Khc-73 and Dlg pathway remain functional. DTRAF1 is delocalized and is uniformly cortical in this deficiency line (data not shown). (2) Similarly, in egr insc NBs, DTRAF1 is cytoplasmic whereas the functions of astral microtubules, kinesin Khc-73 and Dlg are intact. (3) In egr NBs DTRAF1 is cytoplasmic, whereas the apical complex is normal and astral microtubules, kinesin Khc-73 and Dlg are present. (4) DTRAF1 apical localization remains unchanged in dlg mutant NBs (data not shown). Based on these observations, we conclude that DTRAF1 apical localization is unlikely to share similar mechanism with Pins and Gαi and is likely to be independent of astral microtubules, kinesin Khc-73 and Dlg. DTRAF1 apical localization appears to specifically require Egr and Baz.
DTRAF1 is specifically required for Mira/Pros telophase rescue
In DTRAF1 insc double-mutant embryos, the complete segregation of Mir/Pros into future GMCs only occur in about 12% of the total population, and in the remaining NBs, only a fraction of Mira/Pros segregate into future GMCs as indicated by the Mira ‘tail' extending into the future NBs (Figure 4) at telophase. As it is difficult to address the global effect of this partial segregation of Mira/Pros on GMC specification in DTRAF1 insc double mutant, we focused on a well-defined GMC, GMC4-2a in NB4-2 lineage (Doe et al, 1988), to evaluate this issue. We assumed that as long as the RP2 neuron (progeny of GMC4-2a, Even-skipped (Eve)-positive) was identified in a particular hemisegment, the GMC cell fate of GMC4-2a in that hemisegment should have been correctly specified. In insc mutants, almost all hemisegments (99%, n=154) contain RP2s, indicating that GMC4-2a has adopted the correct GMC cell fate in 99% of the total hemisegments (data not shown). When DTRAF1 insc double-mutant embryos were stained with anti-Eve, we found the frequency of loss of Eve-positive RP2 neuron increased (to 8%, n=177) in late embryos (data not shown), suggesting that about 8% of the GMCs in DTRAF1 insc double mutant did not inherit sufficient Pros to specify the GMC fate in these embryos. The relatively low frequency (8%) of mis-specification of GMCs suggests that the threshold amount of Pros protein needed is sufficiently low such that just a partial inheritance of Pros, even when telophase rescue is compromised, is sufficient for most GMCs to be correctly specified.
Although Mira/Pros and Pon/Numb share similar basal localization patterns in insc NBs, further removal of either DTRAF1 or Egr compromised telophase rescue only for Mira/Pros, but not for Pon/Numb. This difference between Mira/Pros and Pon/Numb indicates that the detailed mechanisms of basal localization and segregation of Mira/Pros differ from those of Pon/Numb, which is consistent with the observations that the dynamics of Mira/Pros and Pon/Numb localization early in mitosis are different and the basal localization for Mira/Pros and Pon/Numb requires different regions of the Insc coding sequence (Tio et al, 1999).
Dlg/Lgl/Scrib are required for correct basal localization of Mira/Pros and Pon/Numb in mitotic NBs (Ohshiro et al, 2000; Peng et al, 2000; Albertson and Doe, 2003). Recently, Dlg has been shown to be involved in the Mira telophase rescue (Siegrist and Doe, 2005). In dlg insc double-mutant NBs, not only was spindle geometry symmetric but Mira telophase rescue was also affected. It would be interesting to know if Dlg belongs to the same pathway as DTRAF1 and Egr and if Dlg is also involved in Pon/Numb telophase rescue.
Roles of DTRAF1 and Egr in Mira/Pros telophase rescue
Two other members of the TRAF family have also been identified in Drosophila: DTRAF2 (DTRAF6) and DTRAF3 (Grech et al, 2000). In contrast to the specific and strong expression of DTRAF1 in the embryonic NBs (Preiss et al, 2001), only low levels of ubiquitous signals similar to the control background were seen in the NBs with DTRAF2 and DTRAF3 probes (data not shown). It is likely that DTRAF2 and DTRAF3 are not expressed in NBs and do not play an important role in Mira/Pros telophase rescue pathway as the Mira/Pros telophase rescue is dramatically compromised in DTRAF1 insc and egr insc NBs.
In mammals, the TNF pathway works as a typical receptor-mediated signal transduction pathway. TNFR is a key player in transducing external signal to the cytoplasm. In the Drosophila compound eyes, ectopic Egr, Wgn and DTRAF1 seem to work in a similar receptor-mediated signal pathway to induce apoptosis through the activation of the JNK pathway (Igaki et al, 2002; Kanda et al, 2002; Kuranaga et al, 2002; Moreno et al, 2002). Does the same Egr, Wgn and DTRAF1 receptor-mediated signal pathway play a role in Mira/Pros telophase rescue? If it does, we might expect to see the coexpression of Egr, Wgn and DTRAF1 in dividing NBs and the potential interaction between DTRAF1 and the cytoplasmic domain of Wgn. Three observations argue against this hypothesis: (1) wgn is not expressed in embryonic NBs but in the mesoderm (Igaki et al, 2002; Kauppila et al, 2003). (2) The domain analysis suggests that the Drosophila Wgn cytoplasmic domain is unique with no sequence homology to any mammalian TNFR family members and has neither a TRAF-binding domain nor a death domain (Kanda et al, 2002), which is required for the interaction between TNFR and TRAF in mammals. (3) More informatively, Wgn knockdown by a UAS head-to-head inverted repeat construct of wgn (UAS-wgn-IR) (Kanda et al, 2002) driven by a strong maternal driver, mata-gal4 V32A, in WT embryos did not affect DTRAF1 apical localization (data not shown). These observations are consistent with the view that the receptor Wgn may not be involved in Mira/Pros telophase rescue or is redundant in this pathway. If this is the case, then how do DTRAF1 and Egr function in Mira/Pros telophase rescue? It has been reported that TRAFs associate with numerous receptors other than the TNFR superfamily in mammals (Cao et al, 1996). We speculate that Egr and DTRAF1 may adopt an alternative receptor in NBs for Mira/Pros telophase rescue. However, until an anti-Wgn antibody and wgn mutant alleles are available, we cannot formally exclude the possibility that Wgn is involved in Mira/Pros telophase rescue.
Figure 7 summarizes our current view of a genetic hierarchy that regulates telophase rescue and where DTRAF1 and Egr lie within this hierarchy. The snail family transcription factors Sna, Esg and Wor control the expression of Baz, as well as Insc in the NBs of the ventral cord. Baz functions upstream of DTRAF1 and is required for both Mira/Pros and Pon/Numb telophase rescue. Egr and DTRAF1 are only involved in mediating the basal localization and telophase rescue of Mira/Pros.
Figure 7.
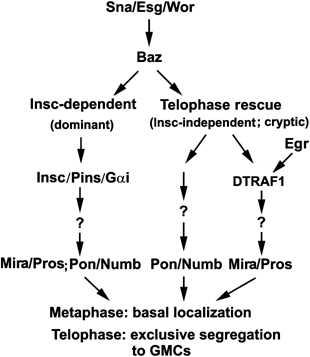
Diagram illustrating where DTRAF1 and Egr lie within a proposed genetic hierarchy that regulates telophase rescue.
Materials and methods
Fly stocks and genetics
Fly stocks are maintained according to (Ashburner, 1989) in either 25 or 22°C fly room. Mutant lines insc22, pins89and bazXi106 were used in the study. The P-element lines EP0578 and KG02299 were from the Szeged Drosophila Stock Center and the Bloomington Stock Center. The standard genetic protocol was used for the mobilization of the P-elements in EF0578 and KG02299. Fifty-four lethal revertants from EP0578 and about 200 independent w− revertants from KG02299 were screened by Southern blot analysis or PCR method to identify the extension of deletions. The female germline clones of baz and DTRAF1 were generated with the standard FLP–DFS technique (Chou and Perrimon, 1992).
Fusion protein and generation of anti-DTRAF1 antisera
A GST fusion construct carrying aa 223–486 of DTRAF1 coding region was expressed in Escherichia coli. The DTRAF1 fusion protein was purified and used to immunize mice according to the standard protocol.
Immunofluorescence staining and confocal microscopy
Embryos were fixed with 4% paraformaldehye according to the standard method (Ashburner, 1989). Antibodies used in this study were raised in this lab unless otherwise stated. It is interesting to note that the two anti-Mira antibodies available (rabbit anti-N-Mira and monoclonal anti-C-Mira, gifts from F Matsuzaki) showed slightly different staining pattern in insc NBs, especially at metaphase. Anti-N-terminal Mira showed overlapped signal with anti-Pros whereas the anti-C-terminal Mira tended to be cortical. For consistent results, only anti-N-terminal Mira was used in the telophase rescue assays. Rabbit anti-Insc (1:1000), rabbit and rat anti-Pins (1:1000), rabbit anti-Baz (1:500; from F Matsuzaki), rabbit anti-Mira (1:1000; anti-N-terminal Mira, from F Matsuzaki), rabbit anti-Pon (1:500;from YN Jan), rabbit anti-Numb (1:500; from YN Jan), rabbit anti-β-gal (1:5000; MP Biomedical), Mouse anti-DTRAF1(1:500), mouse anti-Mira (1:50, monoclonal, anti-C-terminal Mira; from F Matsuzaki), anti-Pros MR1A (1:5; from CQ Doe), mouse anti-β-gal (1:3000; Chemicon) were used in these studies. Cy3- or fluorescein isothiocyanate (FITC)-conjugated secondary antibodies were obtained from Jackson Laboratories. Stained embryos were incubated with ToPro3 (1:10 000, Molecular Probes) for chromosome visualization and subjected to the laser scanning confocal microscopy (Zeiss LSM510). Images were processed with Adobe Photoshop.
In vitro protein-binding assay
PCR fragments containing full-length baz and full length-DTRAF1 were cloned into pGEX vectors (Amersham). The constructs were expressed in BL21 and the fusion proteins were purified by Glutathione–Sepharose-4B beads (Amersham). The full-length baz and -DTRAF1 were inserted into pGBKT7 or pGADT7 (Clontech). In vitro translations were performed with the TNT in vitro transcription and translation kit (Promega) in the presence of 35S methionine. In vitro binding assays were carried out by incubating beads with 35S methionine-labeled proteins for 1 h. A buffer containing 25 mM Tris, pH 7.5, 150 mM NaCl, 25 mM MgCl2, 1% Triton-X 100, 1 mM DTT and protease inhibitor was used. The beads were boiled in SDS sample buffer and proteins were separated by SDS–PAGE. Gels were fixed (50% methanol, 10% acetic acid) for half an hour and soaked for 5 min in the solution containing 7% methanol, 7% acetic acid and 1% glycerol before drying and the exposure to X-ray film.
Acknowledgments
We thank YN Jan, C Doe, F Matsuzaki, K Matthews, A Wodarz, M Miura, the Bloomington Stock Center and the Szeged Drosophila Stock Center for antibodies and flies. We also thank Dr S Bahri and other members of our lab for suggestions and discussions. This work was supported by the IMCB, A*STAR, Singapore.
References
- Albertson R, Doe CQ (2003) Dlg, Scrib and Lgl regulate neuroblast cell size and mitotic spindle asymmetry. Nat Cell Biol 5: 166–170 [DOI] [PubMed] [Google Scholar]
- Ashburner M (1989) Drosophilia: A Laboratory Handbook. Cold Spring Harbour, NY: Cold Spring Harbour Laboratory Press [Google Scholar]
- Betschinger J, Knoblich JA (2004) Dare to be different: asymmetric cell division in Drosophila, C. elegans and Vertebrates. Curr Biol 14: R674–R685 [DOI] [PubMed] [Google Scholar]
- Cai Y, Chia W, Yang X (2001) A family of snail-related zinc finger proteins regulates two distinct and parallel mechanisms that mediate Drosophila neuroblast asymmetric divisions. EMBO J 20: 1704–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu F, Lin S, Chia W, Yang X (2003) Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112: 51–62 [DOI] [PubMed] [Google Scholar]
- Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV (1996) TRAF6 is signal transducer for interleukin-1. Nature 383: 443–446 [DOI] [PubMed] [Google Scholar]
- Chou TB, Perrimon N (1992) Use of a yeast site-specific recombinase to produce female germline chimeras in Drosophila. Genetics 131: 643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP (1991) The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65: 451–464 [DOI] [PubMed] [Google Scholar]
- Doe CQ, Smouse D, Goodman CS (1988) Control of neuronal fate by the Drosophila segmentation gene even-skipped. Nature 333: 376–378 [DOI] [PubMed] [Google Scholar]
- Grech A, Quinn R, Srinivasan D, Badoux X, Brink R (2000) Complete structural characterisation of the mammalian and Drosophila TRAF genes: implications for TRAF evolution and the role of RING finger splice variants. Mol Immunol 37: 721–734 [DOI] [PubMed] [Google Scholar]
- Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, Miura M (2002) Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J 21: 3009–3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F (1997) Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390: 625–629 [DOI] [PubMed] [Google Scholar]
- Kanda H, Igaki T, Kanuka H, Yagi T, Miura M (2002) Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J Biol Chem 277: 28372–28375 [DOI] [PubMed] [Google Scholar]
- Kauppila S, Maaty SAW, Chen P, Tomar SR, Eby TM, Chapo J, Chew S, Rathore N, Zachariah S, Sinha KS, Abrams1 MJ, Chaudhary MP (2003) Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene 33: 4860–4867 [DOI] [PubMed] [Google Scholar]
- Kraut R, Chia W, Jan LY, Jan YN, Knoblich JA (1996) Role of inscuteable in orienting asymmetric cell divisions in Drosophila. Nature 383: 50–55 [DOI] [PubMed] [Google Scholar]
- Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, Miura M (2002) Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol 4: 705–710 [DOI] [PubMed] [Google Scholar]
- Liu H, Su YC, Becker E, Treisman J, Skolnik EY (1999) A Drosophila TNF-receptor-associated factor (TRAF) binds the ste20 kinase Misshapen and activates Jun kinase. Curr Biol 9: 101–104 [DOI] [PubMed] [Google Scholar]
- Lu B, Jan L, Jan YN (2000) Control of cell divisions in the nervous system: symmetry and asymmetry. Annu Rev Neurosci 23: 531–556 [DOI] [PubMed] [Google Scholar]
- Lu B, Rothenberg M, Jan LY, Jan YN (1998) Partner of Numb colocalizes with Numb during mitosis and directs Numb asymmetric localization in Drosophila neural and muscle progenitors. Cell 95: 225–235 [DOI] [PubMed] [Google Scholar]
- Matsuzaki F, Koizumi K, Hama C, Yoshioka T, Nabeshima Y (1992) Cloning of the Drosophila prospero gene and its expression in ganglion mother cells. Biochem Biophys Res Commun 182: 1326–1332 [DOI] [PubMed] [Google Scholar]
- Moreno E, Yan M, Basler K (2002) Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol 12: 1263–1268 [DOI] [PubMed] [Google Scholar]
- Ohshiro T, Yagami T, Zhang C, Matsuzaki F (2000) Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature 408: 593–596 [DOI] [PubMed] [Google Scholar]
- Parmentier ML, Woods D, Greig S, Phan PG, Radovic A, Bryant P, O'Kane CJ (2000) Rapsynoid/partner of inscuteable controls asymmetric division of larval neuroblasts in Drosophila. J Neurosci 20: RC84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Manning L, Albertson R, Doe CQ (2000) The tumoursuppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature 408: 596–600 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Knoblich JA (2001) DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol 3: 43–49 [DOI] [PubMed] [Google Scholar]
- Preiss A, Johannes B, Nagel AC, Maier D, Peters N, Wajant H (2001) Dynamic expression of Drosophila TRAF1 during embryogenesis and larval development. Mech Dev 100: 109–113 [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN (1994) Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell 76: 477–491 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA (2001) Heterotrimeric g proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell 107: 183–194 [DOI] [PubMed] [Google Scholar]
- Schaefer M, Shevchenko A, Knoblich JA (2000) A protein complex containing Inscuteable and the Galpha-binding protein Pins orients asymmetric cell divisions in Drosophila. Curr Biol 10: 353–362 [DOI] [PubMed] [Google Scholar]
- Schober M, Schaefer M, Knoblich JA (1999) Bazooka recruits Inscuteable to orient asymmetric cell divisions in Drosophila neuroblasts. Nature 402: 548–551 [DOI] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN (1997) Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90: 449–458 [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ (2005) Microtubule-induced Pins/G alpha i cortical polarity in Drosophila neuroblasts. Cell 123: 1323–1335 [DOI] [PubMed] [Google Scholar]
- Su YC, Treisman JE, Skolnik EY (1998) The Drosophila Ste20-related kinase misshapen is required for embryonic dorsal closure and acts through a JNK MAPK module on an evolutionarily conserved signaling pathway. Genes Dev 12: 2371–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tio M, Zavortink M, Yang X, Chia W (1999) A functional analysis of inscuteable and its roles during Drosophila asymmetric cell divisions. J Cell Sci 112: 1541–1551 [DOI] [PubMed] [Google Scholar]
- Uemura T, Shepherd S, Ackerman L, Jan LY, Jan YN (1989) Numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 58: 349–360 [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN (1991) Prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67: 942–953 [DOI] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E (2000) Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol 150: 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Kuchinke U, Knust E (1999) Bazooka provides an apical cue for Inscuteable localization in Drosophila neuroblasts. Nature 402: 544–547 [DOI] [PubMed] [Google Scholar]
- Yu F, Cai Y, Kaushik R, Yang X, Chia W (2003) Distinct roles of Galphai and Gbeta13F subunits of the heterotrimeric G protein complex in the mediation of Drosophila neuroblast asymmetric divisions. J Cell Biol 162: 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Morin X, Cai Y, Yang X, Chia W (2000) Analysis of partner of inscuteable, a novel player of Drosophila asymmetric divisions, reveals two distinct steps in inscuteable apical localization. Cell 100: 399–409 [DOI] [PubMed] [Google Scholar]
- Yu F, Wang H, Qian H, Kaushik R, Bownes M, Yang X, Chia W (2005) Locomotion defects, together with Pins, regulates heterotrimeric G-protein signalling during Drosophila neuroblast asymmetric division. Genes Dev 19: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]


