Membrane proteins, encoded by ~20% of genes in almost all organisms, including humans, are critical for cellular communication, electrical and ion balances, structural integrity of the cells and their adhesions, and other functions. Atomic-resolution structures of these proteins furnish important information for understanding their molecular organization and constitute major breakthroughs in our understanding of how they participate in physiological processes. However, obtaining structural information about these proteins has progressed slowly (1, 2), mostly because of technical difficulties in the purification and handling of integral membrane proteins. Instability of the proteins in environments lacking phospholipids, the tendency for them to aggregate and precipitate, and/or difficulties with highly heterogeneous preparations of these proteins isolated from heterologous expression systems have hindered application of standard structure determination techniques to these molecules.
Among membrane proteins, G-protein-coupled receptors (GPCRs)1 are of special importance because they form one of the largest and most diverse groups of receptor proteins. More than 400 nonsensory receptors identified in the human genome are involved in the regulation of virtually all physiological processes. Drug addiction, mood control, and memory (via 5-HT6 or neuropeptide receptors) are just a short list of processes in which GPCRs are critically implicated. Another even larger group of GPCRs consist of sensory receptors involved in the fundamental process of translation of light energy (rhodopsin and cone pigments), the detection of chemoattractant molecules, or the detection of compounds stimulating the taste buds (3, 4). The activity of GPCRs comes about when binding of diffusable extracellular ligands causes them to switch from quiescent forms to an active conformation capable of interaction with hundreds of G-proteins. Their roles as extracellular ligand-binding proteins make them attractive targets for drug design. GPCRs account for ~40% of all therapeutic intervention, and major GPCR research projects are found throughout the pharmaceutical industry (5, 6).
A paucity of structural data is available for GPCRs. The crystal structure of a member of the largest subgroup (I) of GPCRs, rhodopsin (7), and a ligand-binding domain of the metabotropic glutamate receptor with and without the ligand (8) have been determined recently. The data allow models, firmly based on the atomic-resolution structural information, to be further tested as to the conformational changes that these receptors undergo in going from the quiescent to the signaling state. In this article, we describe the further refinement of rhodopsin (7) and provide some clues about how the receptor could be activated by light.
RHODOPSIN
Rhodopsin is involved in the molecular transformation of light energy into a neuronal signal transmitted to the secondary neurons of the retina and ultimately to the brain. The role of rhodopsin in this physiologically interesting and important process is that of a classic G-protein-coupled receptor. These receptors interact with environmental signals and initiate, via G-proteins, intracellular responses to those signals. Rhodopsin is also a member of the largest subfamily of the membrane receptors, constituting ~90% of all GPCRs. The family includes cone pigments and adrenergic and other ligand receptors (9–15). Most GPCRs respond to binding of a ligand. In the case of rhodopsin, the signal is made up of two components: the bound chromophore, which undergoes cis → trans photoisomerization, and a photon. In fact, the binding site for retinal is viewed as representative of the binding mode of an inverse agonist in other GPCRs. Indeed, a rhodopsin mutant lacking Lys296, the residue to which 11-cis-retinal binds via a protonated Schiff base linkage (16, 17), can bind and is inactivated by a noncovalently bound 11-cis-retinal analogue (18).
Rhodopsin is an integral membrane protein located in the outer segments of rod cells. It provides an environment in which its 11-cis-retinal chromophore can undergo a cis → trans conformational switch in response to absorption of a photon with a very high quantum yield of 0.67 (19). This process is completed in <200 fs (20). Due to this fast photochemical process, one of the fastest chemical reactions known, it is believed that a large fraction of the energy of a photon, 32 kcal/mol, is first stored in the chromophore–rhodopsin complex (21, 22). Further structural changes in photoactivated rhodopsin lead to a well-defined set of photostates and are well-characterized spectroscopically by low-temperature trapping experiments (for example, see refs 23 and 24). Interestingly, other GPCRs may also undergo multiple conformational changes upon agonist binding (25). In milliseconds, the formation of the active species of rhodopsin, so-called metarhodopsin II, allows photoactivated rhodopsin to interact with a G-protein, transducin (Gt), and activate it (26, 27). The resulting signal transduction cascade involves activation of cGMP phosphodiesterase, reduction in levels of cGMP, closure of cGMP cation channels in the cellular membrane, hyperpolarization of the cells, and synaptic signaling (28–33).
The crystal structure of rhodopsin (7) provided the first detailed three-dimensional structural model for a GPCR. Earlier low-resolution work on rhodopsin had revealed the organization of this receptor at 7.5 Å resolution in the plane of the membrane and 16.5 Å resolution perpendicular to the membrane (34–39). This low-resolution three-dimensional study of rhodopsin predicted the location of the seven rods of density that correspond to transmembrane helices and was our first view of rhodopsin. The electron density map also allowed an estimation of the tilt angles for these seven helices and showed clear differences between rhodopsin, bacteriorhodopsin, and other retinylidene-binding proteins (39, 40; for theoretical models, see refs 41–44). Further refinement of the structural model reported at 2.8 Å resolution (7) has resulted in an improved model for ground-state bovine rhodopsin with many structural details that were not seen via cryoelectron microscopy.
REFINEMENT OF THE RHODOPSIN STRUCTURE
The crystals of rhodopsin are twinned. Two refinements of rhodopsin were carried out to account for the twinning. In one refinement protocol, the twin law relating the indices of the overlapping reflections was used in conjunction with a twinning parameter. Alternatively, once a twinning parameter was available, the intensities of the overlapping reflections could be calculated and used in a standard refinement. Since this latter procedure involves fitting of derived data (de-twinned) rather than the observations themselves, we consider it a less desirable procedure.
The data set used for the refinements was part of the same set published previously (APS 19-ID) (7). The starting model was PDB entry 1F88. The entire refinement was carried out using CNS (45), with weak noncrystallographic symmetry restraints applied to the two molecules in the asymmetric unit (molecules A and B). For the refinement, reflections with F < 2σ(F) were omitted. This was done because the reflections in the last shell are rather weak with an 〈I/σ(I)〉 of <2.
Palmitoyl groups, heptanetriol, and β-nonylglucoside topology and parameter files were taken from the HIC-Up server (46) and modified as necessary. The bond angles and bond lengths of retinal, which were used in this refinement, differ slightly from those previously used (7). They were taken from Cambridge Structural Database entry cretal10 (47). XtalView (48) was used for the model building stages. For the twinned data, the twin fraction was repeatedly determined on the basis of the model and this fraction used for the next steps in refinement. The twin fraction initially was close to 0.29 and decreased throughout the refinement to a final value of 0.277. Additionally, the statistical method of Yeates as implemented in CNS was used to de-twin the data; the value determined using the average value of H was 0.252 (49). This value appeared to give cleaner maps than the value of 0.242 determined from 〈H2〉.
The R-values and statistics for these two refinements are shown in Table 1. The R-values for refinement using the de-twinned data differ from those from the refinement against the twinned data by the expected ratio of 1/√2 (50). The two resulting refined models differ slightly, but likely not significantly. Accordingly, the model obtained using twinned data will be the focus of the following discussion. This model has been deposited in the Protein Data Bank (1HZX).
Table 1.
Refinement Statistics
| twinneda refinement | “de-twinned”b refinement | |
|---|---|---|
| resolution range (Å) | 30–2.8 | 30–2.8 |
| no. of reflections in working set | 42036 | 39588 |
| no. of reflections in test set | 2026 | 1913 |
| Rcryst | 0.175c | 0.248d |
| Rfree | 0.212c | 0.277d |
| no. of atoms | 5552 | 5552 |
| rms deviation in bond lengths (Å) | 0.010 | 0.011 |
| rms deviation in bond angles (deg) | 1.47 | 1.65 |
| rms deviation in dihedral angles (deg) | 20.1 | 19.8 |
| rms deviation in improper angles (deg) | 0.85 | 1.27 |
| average B-value (Å2) | 45.3 | 39.0 |
The twinned data set is part of the APS 19-ID set as reported in ref 7. The data were collected at λ = 1.03320 Å. Friedel mates were retained and used in refinement.
The de-twinned data set was obtained by applying a twin fraction of 0.252 and the twinning law within CNS (45).
When this model is tested against the de-twinned data, Rcryst = 0.236 and Rfree = 0.266.
When this model is tested against the twinned data, Rcryst = 0.197 and Rfree = 0.227 using a twin fraction of 0.277.
DOMAINS OF RHODOPSIN
The overall shape of the various parts of the rhodopsin molecule indicates the dimensions of the cytoplasmic, transmembrane, and extracellular regions and their footprint of the molecule in the plane of the membrane. These regions, however, are not domains in the protein folding sense.
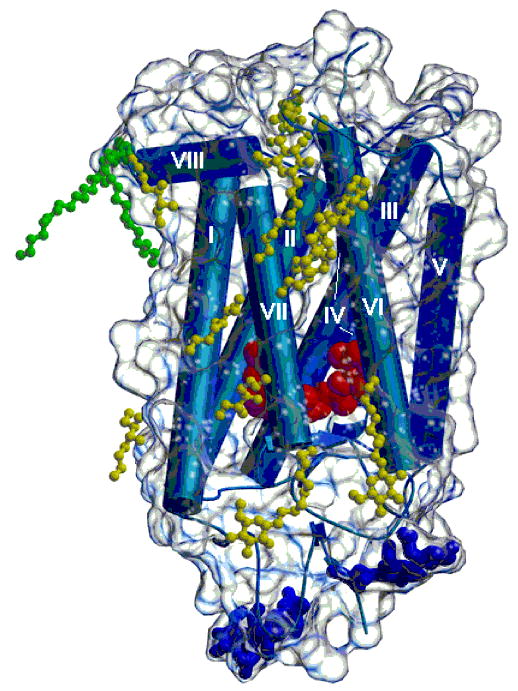
Examination of the figures in the “standard view” (Figure 1) shows an ellipsoidal shape. The dimensions of the ellipsoid are ~75 Å perpendicular to the membrane, ~48 Å wide in the standard view, and ~35 Å thick in this view.
Figure 1.
Three-dimensional crystal structure of rhodopsin with bound detergent and amphiphile molecules. Helical portions of the protein, including the seven transmembrane helices, are shown as blue rods, and β-strands are shown as blue arrows. The polypeptide connecting the helices appears as blue coils. A transparent envelope around the protein represents the molecular surface. The dark blue ball-and-stick groups at the bottom of the figure denote carbohydrate groups attached to the protein. Two palmitoyl groups covalently attached to the protein are shown in green. Nonylglucoside and heptanetriol molecules located near the hydrophobic surface of the protein are shown in yellow. The figure was drawn using Molscript (116) and Raster3d (117).
The transmembrane domain was determined by bisecting helix VIII as the cytoplasmic limit. The extracellular limit of the transmembrane region was determined by locating a plane parallel to the membrane where the external residues of rhodopsin shift from hydrophobic to polar. The shape of this transmembrane domain is that of an elliptic cylinder ~41 Å in height. The length and width of the elliptic footprint on the plane at the middle of the membrane are roughly 45 and 37 Å, respectively.
The cytoplasmic domain was taken simply as the residues above the transmembrane domain, and it is reasonably well fitted by an ellipsoid of axes 44 Å × 30 Å × 18 Å along x, y, and z, respectively. Similarly, the extracellular domain was determined as an ellipsoid of axes 38 Å × 32 Å × 27 Å. These two ellipsoids encompass about 95% of the atoms of the respective domains.
It is of some interest to project the molecule into the membrane plane and determine the area. This area is connected to the density of the molecules in the membrane and their closest approaches. The area is close to 1500 Å2. The molecule has a strong dipole perpendicular to the membrane, and one would anticipate that these parallel dipoles would repel one another due to the electrostatic repulsion at the two ends.
Due to the elongation of the cytoplasmic region by inclusion of helix VIII, the area of the cytoplasmic domain is sufficient to dock a single transducin Gt trimer (α-, β-, and γ-subunits) on the surface. However, the C-terminal residues must move aside to allow all the residues implicated in binding to be on the surface. A reasonable but speculative model of this complex has been built taking into account the known interactions of Gt and rhodopsin (data not shown). In such a complex, Gt would form a 1:1 complex with photoactivated rhodopsin as proposed previously (51).
MODEL OF RHODOPSIN
Bovine rhodopsin contains 348 amino acids, has a molecular mass of ~40 kDa, and folds into seven transmembrane helices varying in length from 19 to 34 residues and one cytoplasmic helix (Figure 1, blue rods encapsulating α-helices; Figure 2). These helices differ in their length and are irregular, and they tilt at various angles with respect to the expected membrane surface (Figure 1). They contain a mix of α- and 310-helices, and they possess a large number of kinks, twists, and bends (52). Further refinement has not reduced the number of these conformational anomalies. Many of the bends and twists were also seen in the two-dimensional cryoelectron microscopy study of frog rhodopsin (36), and the helical axes from that study can be readily superposed with the helices in our refined model. This indicates that the irregular helices are not artifacts of data set resolution or the methods used for crystallographic refinement.
Figure 2.
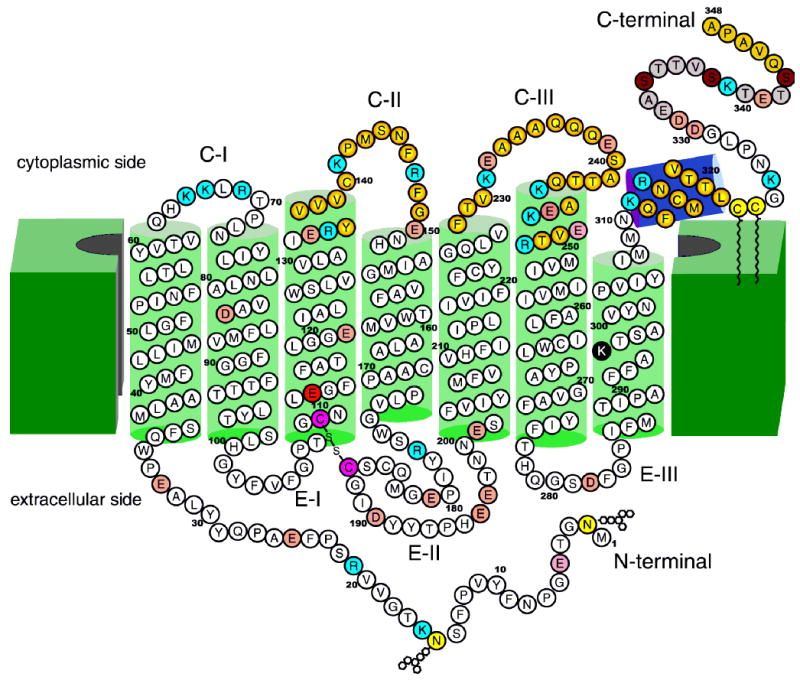
Two-dimensional model of bovine rhodopsin adopted after that of Hargrave (9). About ¼ of the polypeptide chain of opsins has cytoplasm exposure; ¼ is sequestered in the intradiscal space, and ½ forms a core of transmembrane helices. Green cylinders represent helices that in large part are imbedded in the disk membranes (19–34 residues each). The violet cylinder represents the amphiphilic helix (H-VIII) that runs parallel to the membranes. The color of rhodopsin results from the protonated Schiff base linkage of the 11-cis-retinal chromophore (100). Lys296 forms this protonated Schiff base with retinal and is shown as a black filled circle. Glu113 is a counterion of the Schiff base and is shown as a red filled circle. The disulfide bridge conserved among GPCRs is shown as purple filled circles (60). Two carbohydrate moieties at Asn2 and Asp15 and two palmitoyl groups at Cys322 and Cys323 (15, 64–71) are shown as light yellow filled circles (32, 72, 73). Light red filled circles represent acidic residues, and blue filled circles represent basic residues. Photoisomerization of the 11-cis-retinal chromophore of rhodopsin to all-trans-retinal leads to a conformational change in the protein, including the cytoplasmic surface (9–14, 32, 118, 119) leading to activation of Gt. Loops CII and CIII and helix VIII are involved in the recognition of Gt and are marked with filled yellow circles at the center, changing to red filled circles in the periphery. These residues are also important in binding of two regulatory proteins: rhodopsin kinase and arrestin. The functional phosphorylation occurs in the C-terminal region (represented by light brown filled circles) at phosphorylation sites denoted by dark brown filled circles (86–91, 96). The last five C-terminal residues were postulated to be involved in the vectorial transport within highly differentiated rod photoreceptors (92, 93, 120). The carbohydrate chains and the disulfide linkage (in pink filled circles) are oriented toward the lumen (extracellular) face of rhodopsin, and the C-terminal domain is cytoplasmic. To date, more than 100 mutations in the human rhodopsin gene have been associated with recessive and dominant retinitis pigmentosa (RP), as well as congenital stationary night blindness (CSNB).
To appraise the tilt, the membrane plane was assumed to be perpendicular to the long axis of rhodopsin molecule A. The helix tilt angles are the angles between the helical axes and the long axis of the molecule. Thus, a helix at 0° is perpendicular to the membrane, while a helix (such as helix VIII) with a tilt angle of 90° lies parallel to the membrane plane. The second method was to measure the bending of the helices from an ideal straight helix. Helix I (Figure 2) is 44 Å long and tilted from the plane of the membrane by 25° and contains a 12° bend within it (see Table 2 in the Supporting Information), mostly due to the presence of Pro53. This helix forms multiple hydrogen bond interactions with helix II and helix VII, but it appears that it does not interact by hydrogen bonding with helices III–VI (Scheme 1). Furthermore, the interactions of helices I and VII are strengthened by hydrophobic interactions between Leu40 (helix I) and Phe293 (helix VII).
Scheme 1.
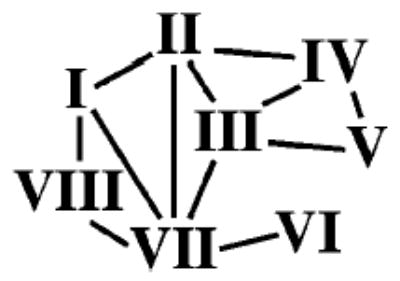
Connections between Helices via Hydrogen Bondinga
a The helix–helix hydrogen bond interactions parallel the loss of accessible surface.
Helix II is tilted from the plane of the membrane about the same as helix I, 25°, and deviates 30° from an ideal helix (Table 2 in the Supporting Information), in the region of Gly89 and Gly90. Because of this bending, Gly90 is in the vicinity of Glu113, the counterion for the protonated Schiff base of the chromophore. This helix forms multiple hydrogen bond interactions with helices I, III, IV, and VII (Scheme 1). Helix III is the longest helix, most tilted from the plane of the membranes, and has two small internal bends. The helix begins at Pro107, near Cys110 which is engaged in a disulfide bridge with Cys187 and bends first at residues Gly120 and Gly121. The second bend occurs at Ser127, but this bend has no apparent correlation with the sequence composition. It seems to be related to the packing of the helix among the others. The tilt of this 48 Å helix is 33°, and the two bends are 12° and 11°. The 33° tilt results in running the helix across the helical bundle and making contacts with four helices (II, IV, V, and VII) (Scheme 1). The C-terminal end of helix III is particularly important, because it contains the E(D)RY motif implicated in the regulation of the receptor’s interaction with its G-protein. This motif is in a very hydrophobic environment formed among residues from helices II and IV–VI, and there is a salt bridge between Glu134 and Arg135. Reversing these residues through mutagenesis leads to formation of metarhodopsin II, and abolishes binding and activation of transducin (53). During photoactivation, it is possible that this Glu residue becomes protonated (12, 54, 55). This salt bridge disruption would be one of the constraints abolished as rhodopsin assumes the metarhodopsin II conformation. In the current model, Glu247 faces the solvent, but it is in a position such that it could hydrogen bond to Arg135 by rotation about χ1 and possibly trigger disruption of the salt bridge. Helix IV is the shortest one, and runs almost perpendicular to the membranes, but it is significantly bent from an ideal helix due to Pro170 and Pro171 (Table 2 in the Supporting Information). We hypothesize that it is involved in the stabilization of the dark-state rhodopsin through additional hydrophobic interactions with helices II, III, and V.
Helix V is 35 Å long and tilted from the membrane normal by 26°. It has two internal kinks of 25° and 15° (Table 2 of the Supporting Information). The bends occur at residues Phe203 and His211 with no apparent correlation with the sequence, apart from a striking number of aromatic residues in the region of the bend. The helix forms multiple hydrogen bond interactions with helix III and helix IV (Scheme 1). Helix VI requires special considerations, as movement of this helix could be a clue to receptor activation (56, 121). It is the second longest and most bent helix (36°) because of the presence of Pro267, one of the most conserved residues among GPCRs. Overall, it is almost perpendicular to the membrane plane. The presence of specific hydrogen bond interactions with only helix VII would allow movement of VI relative to the rest of the helices (Scheme 1). In contrast to the rest of the helices, helix VI interacts with helices II, III, and V only through van der Waals interactions. Helix VII shows a considerable distortion and elongation in the region around the retinal attachment site Lys296 (shown in red in Figure 1) and contains Pro291 and Pro303, a part of the highly conserved NPXXY motif. The NPXXY motif might be involved in the formation of a structural domain that would allow interaction with helix VI (e.g., Met253 is involved). These interactions could be one of the critical constraints holding rhodopsin in the inactive state. Helix VII (containing the chromophore) is nicely situated in the bundle of helices and interacts with all of them, except helices IV and V.
In addition to these transmembrane helices, another short helix in the cytoplasmic surface, termed helix VIII, is located at the extension of helix VII. This helical region, in addition to loops CII and CIII (reviewed in ref 26) of rhodopsin, is a part of the binding sites for the Gt α-subunit and plays a role in the regulation of Gtγ binding (57). Helix VIII is straight and amphipathic. The helix starts at Lys311 and contains Arg314, whose side chains hydrogen bond to surrounding residues. Cys322 is also on the cytoplasmic face of the helix, but the attached palmitoyl moiety makes a U-turn to lie adjacent to the transmembrane portion of the structure (Figure 1). Cys323 is the C-terminal residue of the helix, and its palmitoyl portion associates with molecule B in the crystals of rhodopsin. This particular arrangement may distort the C-terminus of helix VIII.
The cytoplasmic loops are poorly determined in the structure presented here. This is the region of the protein with the highest B-factors, and these loops are probably mobile in solution. Residues missing from the current model are residues 236–240 in cytoplasmic loop II and residues 331–333 in the C-terminal region. C-Terminal residues 334–348 possess electron density, but it is of poor quality. Diffuse density between molecule A and a crystallographically symmetric molecule is sufficiently good to build the structure, but its reliability is probably poor. In molecule B, more residues are missing, including the C-terminus beyond residue Asn326 (Figure 2). Almost certainly, the C-terminal residues of rhodopsin are flexible and mobile in the physiological milieu, lacking a definite single conformation (58, 59).
The extracellular region of rhodopsin consists of the globular N-terminus from residue 1 to 33, the short loop of residues 101–105, “plug” residues 173–198 between helices IV and V, and residues 277–285 between helices VI and VII. The current model does not differ significantly from the previous report (7) in this region. Residues 1–33 form a compact, glycosylated unit which overlays the rest of the loops. Prior to the determination of the rhodopsin structure, we had thought of this region as unstructured because it was not part of the helices, so its observation as a globular unit came somewhat as a surprise. The extended structure of residues 173–198 was another aspect that had not been anticipated. This sequence forms a twisted β-hairpin from residue 177 to 190 and contains the Cys187 portion of the disulfide bridge. As previously noted, this structure forms a plug upon which the retinal lies. The carbonyl oxygen of Cys187 approaches retinal C12 at only 3.00 Å (see Table 2 of the Supporting Information), so the retinal appears to lie on the plug. Mutation of Cys187 gives rise to proteins that are abnormally glycosylated and cannot bind retinal (60). The plug structure also serves to bring Glu181 into proximity (4.4 Å) of retinal at the C12 position. This residue is highly conserved in rhodopsins and short- and middle-wavelength visual pigments (15), and a corresponding Glu residue in this position serves as a counterion in retinochrome, a member of the rhodopsin family (61). In long-wavelength visual pigments, such as red visual pigments, a His residue occupies this position. The His residue and a Lys residue present three residues toward the C-terminus, and form a chloride ion-binding site (62). Chloride ion, when bound, further causes a bathochromic shift by ~40 nm for these cone visual pigments (63).
MOLECULES BOUND TO RHODOPSIN
In addition to the chromophore, discussed below, rhodopsin is modified by two palmitoyl groups at Cys322 and Cys323 in the cytoplasmic region (Figure 2) (15, 64–71). Improved electron density for three of the four palmitoyl groups covalently attached to the two protein molecules in the asymmetric unit has allowed their identification during refinement (Figure 1, in green). Two palmitoyl side chains are attached in molecule A of the model. The palmitoyl group attached to Cys322 is roughly aligned with the transmembrane helices in an orientation where it could interact with the hydrophobic region of the membrane bilayer (Figure 1). The palmitoyl bound to Cys322 in molecule B also takes on this orientation. The second palmitoyl bound to molecule A at Cys323 interacts with a neighboring protein molecule in this crystal form, is extended away from the protein, and is likely not mimicking the hydrophobic parts of the membrane. No density is observed for the palmitoyl bound to Cys323 in molecule B, presumably due to the mobility of the hydrocarbon tail.
Rhodopsin is also modified by two carbohydrate moieties at Asn2 and Asn15 that are oriented toward the lumen (extracellular) face (Figure 1, blue) (32, 72, 73). Two major species identified as Man3GlcNAc3 are attached to Asn2 and Asn15 in bovine rhodopsin (74). Additional mannose groups lead to formation of a minor component. This earlier analysis has been confirmed using mass spectrometry (75). Only partial three-dimensional structural models have been assembled for these groups. In the latest model of rhodopsin, we have been able to build three additional carbohydrate residues, extending the original model (7).
All the carbohydrate chains are in contact with protein residues of crystallographically related molecules; carbohydrates of molecule A contact crystallographically related molecules of B and vice versa. The intermolecular contacts appear to be mediated by water molecules or Zn2+. Superposition of the carbohydrate chains shows significant deviations, however. It is not possible to say at present if this is real or due to disorder; the B-factors of the most deviant parts of the chains are quite large.
Noncrystallographic symmetry was imposed on the protein residues where the carbohydrate is attached, but was not imposed for the carbohydrate. Superposition of the NAG-NAG-MAN residues attached to Asn15 gave a root-mean-square deviation of 2.18 Å. The main discrepancy between the two chains is that the mannose rings are flipped. For the carbohydrate chains attached to residue 2, the two NAG residues of molecule A fit to those of molecule B with a root-mean-square deviation of 1.24 Å. The major deviation in this case is between the N-acetyl moieties of the terminal NAG residues.
At position Asn15, we have built a mannose residue as an addition to the NAG-NAG residues previously reported on molecule A (7). Two mannose residues have also been added to the NAG-NAG chain of molecule B. Asn15 is close to a local 2-fold axis between molecule A and symmetry-related molecule B. This particular NAG-NAG-MAN chain is aligned adjacent to the Lys16 and Thr17 of molecule B in a neighboring cell. The interaction of the two molecules appears to be mediated by a water molecule or Zn2+ ion with low occupancy. Protein linkages to the carbohydrate are in the β-configuration. All linkages are β-1,4 for this NAG-NAG-MAN chain. For the carbohydrate attached to residue Asn2, there is also a water- or Zn2+-mediated association with a B molecule in the crystal. In this case, only two residues of the carbohydrate can be observed and these connect through a probable Zn2+ (Zn2+ 956, occupancy = 0.5) to His195. Glu197 makes a long contact (3.4 Å) with NAG; this may be a hydrogen bond. This carbohydrate chain also contacts one of the BNG (β-nonylglucoside) ligands with a long contact of 4.2 Å between NAG O7 and O3 and O4 of BNG (BNG 1503). For molecule B and the carbohydrate chain attached to Asn15, one additional mannose residue was included in the model. This residue has an α-1,3 linkage to the remainder of the chain according to ref 74, and we have used that configuration in the model. The interactions of this carbohydrate with molecule A are with residues Lys16 and Thr17 due to the 2-fold axis. Like the carbohydrate molecules that interact with Asn2 of molecule A, those attached to residue 2 of molecule B interact through a water or Zn2+ with a crystallographically related molecule. A water molecule forms a hydrogen bond between the first NAG and His195. Glu197 makes a long (4.0 Å) contact with the second NAG. No density was observed for carbohydrate beyond these two residues.
Interactions of rhodopsin with phospholipid could be very important during the photoactivation of rhodopsin and movement of the helices (76, 77). When rhodopsin is reconstituted into saturated phospholipids, the formation of metarhodopsin II is inhibited or does not occur (78–82). There is also lipid restructuring during the photoactivation process (83). Lipids are not removed in the purification and crystallization steps used here (84, 85), but no electron density due to phosphorus atoms could be found. However, six heptanetriol molecules and seven β-nonylglucosides are now part of the structural model (Figure 1, yellow). One heptanetriol and one nonylglucoside are located on the cytoplasmic face of molecule A and are probably artifacts of crystallization. The other additive and detergent molecules are found near the hydrophobic surfaces of the transmembrane helices (Figure 1). The hydrophobic surface of molecules A and B are not completely masked by these amphipathic molecules. About half of the surface is not covered by ordered detergent or additives. Crystal packing interactions among the rhodopsin molecules in the unit cell do not entirely account for the unmasked hydrophobic surfaces.
Other significant additives in the crystals of rhodopsin are the metal ions. Zn2+ ions are an integral component of the purification and crystallization protocols (84), and Hg2+ compounds were bound to the protein to provide the anomalous scatterers used in obtaining phases for the structure determination. Seven Zn2+ ions (occupancy of 0.4–1.0) and six Hg2+ ions (occupancy of 0.5–1.0) have been included in the structural model. Identification of electron density peaks as Zn2+ or Hg2+ ions has been based on the nature of the protein ligands. The occupancies and B-factors were adjusted using the B-factors of liganding atoms and |Fo| − |Fc| difference electron density maps. All of the Hg2+ ions are associated with cysteine residues at positions 222, 264, and 316. Cys140 resides on the cytoplasmic surface but does not appear to bind ions. Zn2+ ions are found near Cys167, but the Zn2+ does not appear to bind to the sulfur atom.
Small improvements in the protein portion of the model are distributed across the entire polypeptide chain. While amino acid side chains have been added for the residues at the C-terminus of molecule A, the refinement has not improved the electron density for the C-terminal tail of molecule B or for the missing residues of the cytoplasmic loops.
The cytoplasmic C-terminal region has two important functions (Figure 2). In photoactivated rhodopsin, the C-terminal is a substrate for rhodopsin kinase at the C-terminal Ser and Thr residues (86–91). The last five amino acids are part of the vectorial transport machinery involved in exporting rhodopsin to outer segments in highly differentiated rod cells of the retina (92, 93). The structural work reported here adds another interesting twist to this physiologically important region. The C-terminal structure of rhodopsin does not have any structural elements that would stabilize this region in a rigid conformation. As described previously (7), the C-terminal region from Gly324 to Asp330 folds back over helix VIII so that Asp330 is located close to Lys311 at the beginning of helix VIII. Residues 331–333 have no discernible electron density. At Thr335, the backbone forms hydrogen bonds to Gln312 of helix VIII, but diverges until Ser338. At Ser338, the backbone forms hydrogen bonds to the carbonyl O of His65 at the end of helix I. The chain makes a loop from Ser338 to Glu341 and then associates with another molecule A in the crystal at the C-terminal Ala348. In molecule B, the electron density cannot be fitted beyond residue Asn326.
Our rhodopsin structure represents the inactive form of the receptor, but in conjunction with other biochemical data from previous studies, it provides us with a better understanding of allowed changes upon activation. The sites of light-dependent multiple phosphorylations are six to seven Ser/Thr residues at the C-terminal end. These sites were discovered by specific cleavage of rhodopsin by Asp-N endoproteinase (94). The resultant 19-amino acid C-terminal peptide contained all of the phosphorylation sites. Heterogeneity and multiple phosphorylation of rhodopsin in vitro has been well documented (87–89, 91, 95, 96). In vivo there are three sites, which were found to be phosphorylated by direct and quantitative methods after 20–40% bleaching of the protein (97, 98). Recently, Mendez and colleagues used a combination of transgenic mice lacking selective phosphorylation sites and electrophysiological recordings to test the effect of mutation on the physiological responses. They concluded that multiple phosphorylation events are needed to shut off photoactivated rhodopsin (90). An abundance of Ser/Thr residues at this region of rhodopsin could speed phosphorylation, and removal of these sites could slow it. It is also known that arrestin binding depends on the presence of hydroxyl groups for high affinity (99).
RHODOPSIN CHROMOPHORE
Rhodopsin is a red-colored protein due to a prosthetic group chromophore, 11-cis-retinal (100, 101) (Figure 1, chromophore in red). The chromophore is bound in the hydrophobic core of the molecule, causing its absorption maximum at approximately 380 nm to be shifted bathochromically to that characteristic of intact rhodopsin at 500 nm (102). The chromophore is covalently linked to Lys296 on helix VII in bovine rhodopsin (Figure 2) (16, 17) through a protonated Schiff base. When extracted into detergent, rhodopsin maintains its chromophore (103, 104). The counterion for the protonated Schiff base is provided by Glu113, which is highly conserved among all known vertebrate visual pigments (105, 106). The counterion has two important functions. (1) It stabilizes the protonated Schiff base by increasing the Ka for this group by up to 107 (e.g., ref 107) and preventing its spontaneous hydrolysis. (2). It also causes a bathochromic shift in the maximum absorption for visual pigments to make them more sensitive to longer wavelengths as UV light is filtered by the front of the eye in most animals.
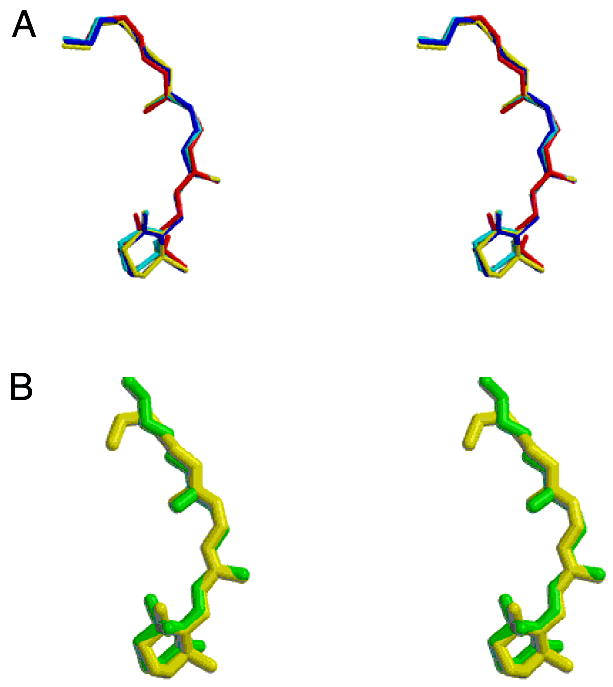
The 11-cis-retinylidene moieties in the molecular model have been refined independently. Aligning these structures gives an estimate of the uncertainty in positions, bond angles, and bond lengths. The chromophore, albeit a conjugated system, is not planar. It could be speculated that the visual cone opsins might make use of this lack of planarity in modulating the absorption maxima of their chromophore. Table 2 (Supporting Information) contains the torsion angles found for the chromophore. It should be noted that the torsion angle restraints for the chromophore were removed from our refinement except for the cis restraint for the C11–C12 bond. This relieves the torsion angles of the chromophore from any computational artifact due to the refinement method or restraints.
Another interesting conformational issue concerns the orientation of the β-ionone ring with respect to the rest of the chromophore. The diffraction data at 2.8 Å resolution are sufficient to identify and orient the ring, although there is a possibility of partial occupancy of rings rotated by 180° about the C6–C7 bond. Solid-state NMR measurements have shown the orientation of various C–C vectors in the ring with respect to the membrane normal (108). The NMR results are consistent with a ring rotated 180° from the ring found in the crystal structure; see Figure 3B. However, other studies indicate that the retinal chromophore is in the twisted 6-s-cis conformation in rhodopsin, in contrast to the planar 6-s-trans conformation found in bacteriorhodopsin (109). The inconsistency between the X-ray crystallographic and solid-state NMR studies requires further study, but it should be pointed out that the binding site for the β-ionone ring was demonstrated to accommodate a variety of substituents (110, 111). Thus, it can be oriented as either 6s-cis or -trans in the ground state (K. Palczewski, unpublished).
Figure 3.
Conformation of the retinylidine chromophore. (A) Stereoview of the chromophores from the refined rhodopsin models showing the small variation in the conformation and orientation of the group in the four refined subunits. The rms deviation among the four molecules is 0.26 Å for all atoms. (B) Stereoview showing the chromophore from molecule A of the crystallographic model refined against twinned data (yellow) and the chromophore structure obtained by Watts and colleagues (108) (green).
CAVITIES WITHIN THE RHODOPSIN MOLECULE
An accessible surface calculation for rhodopsin reveals several cavities within the molecule (Figure 4). This might be expected for proteins containing helices since the divergence of the helices will provide spaces between them that might not be fully occupied by amino acid side chains. The cavities could play a role in the conformational change leading to activation of rhodopsin. In particular, several cavities are near the cytoplasmic end of helix III, and wholesale motion of the helix to occupy them seems possible. Likewise, residues from helix III contribute to the binding site for the ionone ring and conjugated system of the chromophore. What is not clear is how, or whether, the cis–trans conformation change of the chromophore would be coupled to movement of helix III.
Figure 4.
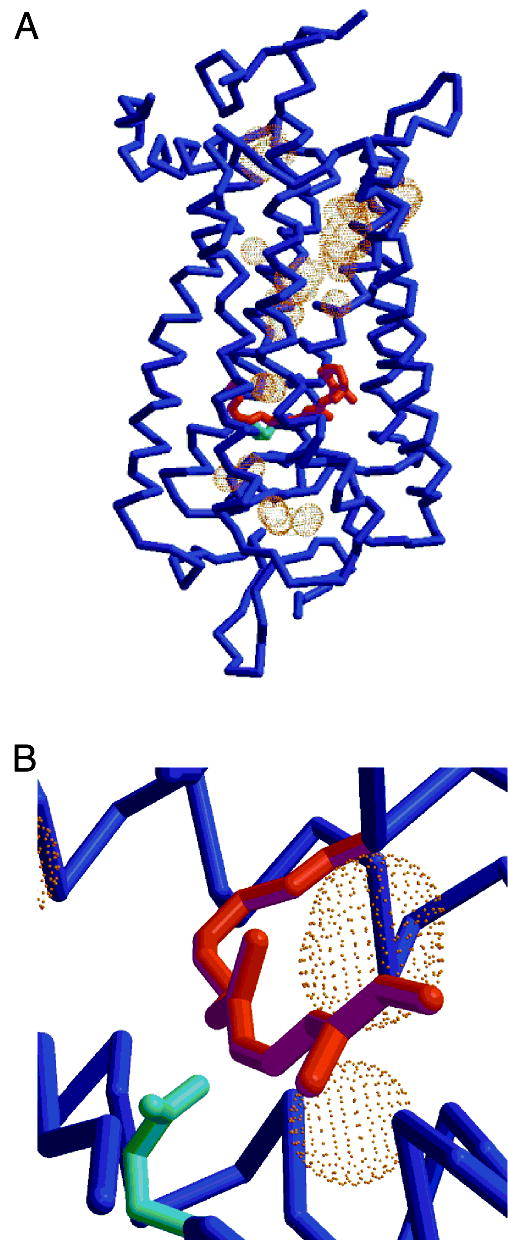
Cavities within the rhodopsin molecule. (A) Cα tracing (blue) with cavities shown by molecular surface dots around the test probes (1.4 Å radius). Glu113 is depicted in cyan and is about 3.6 Å from the Lys296 NZ. The cavities are of sufficient size to hold one or more water molecules. (B) Closeup of two cavities adjacent to the protonated Schiff base portion of the chromophore.
A large connected cavity is found lying parallel to the two β-strands of the plug. On the other side of the cavity, the polypeptide chain runs roughly parallel to the β-strands. Clearly, some packing interaction keeps the two β-strands from adding another to make a three-stranded sheet. One question arising from the structure of rhodopsin concerns how the chromophore makes its way to the binding pocket when the protein is reconstituted. This question is of great importance when considering the binding of ligands to other GPCRs. If ligands pass directly to the binding site from the extracellular space and do not pass into the site from the hydrophobic center of the bilayer, large portions of the structure (including the plug) must move. This cavity might be part of a “loosened” structure that can easily move for ligand binding.
Two of the cavities are located near the nitrogen atom of the Schiff base. Water molecules have been implicated in the hydrolysis of the Schiff base, but no electron density appropriate for bound water molecules near 296 NZ has been seen in our crystallographic studies. The current molecular model contains very few bound water molecules, but this is appropriate for a 2.8 Å resolution study. Series termination effects and experimental and computational noise complicate the identification of water at this resolution. At this point, we have chosen to add water molecules to the model only when electron density is found for both the twinned and de-twinned refined models. In this case, the only water molecule near the retinal chromophore is located near C13 and hydrogen bonded to Glu181 OE2 and Ser186 OG. It is found in only one molecule in the asymmetric unit, molecule A. This water molecule is 4.4 Å from the carboxylate of Glu113 and also 4.4 Å from the Schiff base nitrogen atom. The lack of electron density in molecule B argues for caution in ascribing biochemical significance to this water molecule. A small cavity is found at this site in molecule B, consistent with the possibility of water binding at this site.
On the other hand, the two cavities near the Schiff base are more likely candidates for water molecules involved in the chemistry of the chromophore since the cavities exist in all views of the molecular structure available to us at this time. One of the cavities is close to Glu113, and the other is located on the other side of the Schiff base nitrogen atom away from Glu113. Both cavities are buried in the protein with no access to the surface of the protein, but dynamical motions of the structure should be sufficient to allow water to diffuse into these sites.
RHODOPSIN VERSUS BACTERIORHODOPSIN
It is interesting to compare the rhodopsin helices with those in bacteriorhodopsin (PDB entry 1C3W) (112). Panels A and B of Figure 5 show a superposition of rhodopsin molecule A with bacteriorhodopsin. The models were superimposed using structural alignments only. No amino acid sequence information was used in the superposition. After an initial alignment, residues from each of the molecules were used for superposition if they were located within 3.8 Å of each other. Iteration of the superposition procedure eventually led to a superposition using 79 Cα atoms and resulted in a root-mean-square difference between the molecules of 2.13 Å based on these 79 Cα atoms alone.
Figure 5.
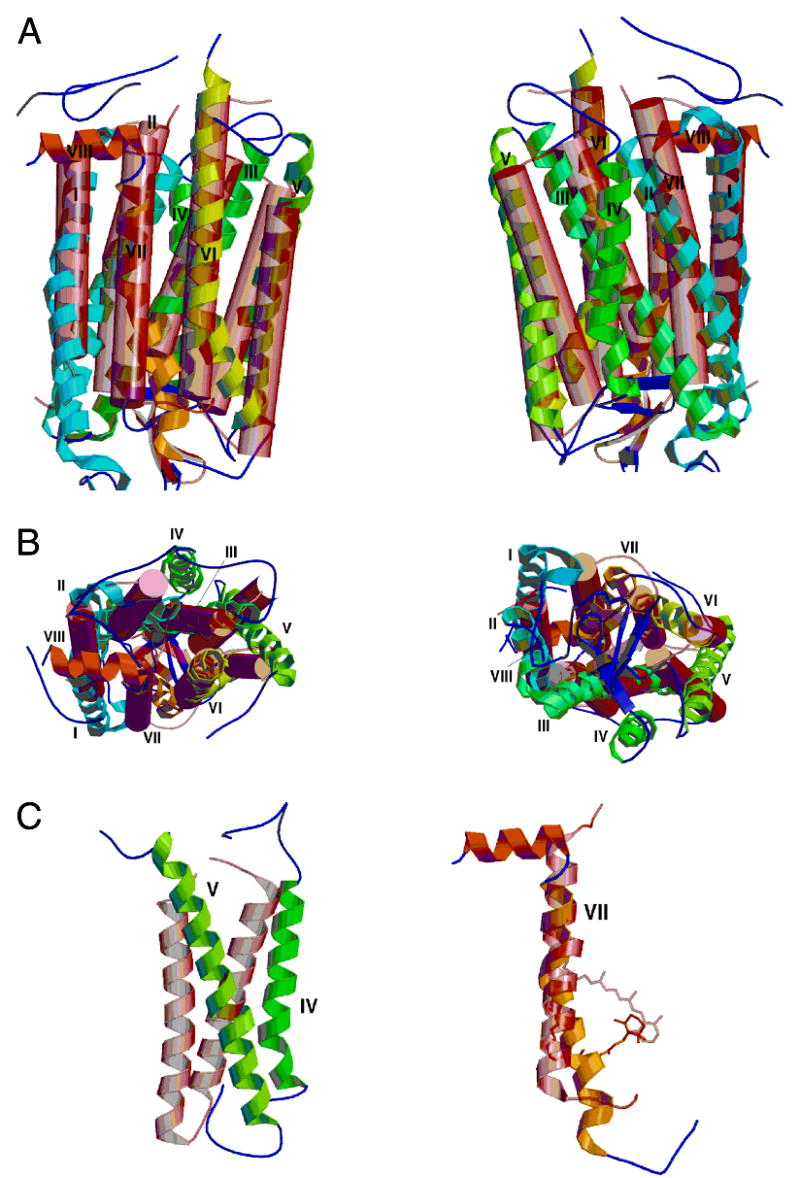
Superposition of bacteriorhodopsin (pink transparent cylinders and connecting coil) on molecule A of rhodopsin (colored helical ribbons and connecting coils). (A) At the left are molecules in the same orientation as in Figure 1. At the right is shown a similar view, with the molecules rotated 180° about the vertical axis. (B) At the left is a view of the top surface of the molecules. This is the cytoplasmic surface of rhodopsin. Note the substantial differences between helices IV and V in the two molecules. At the right is a bottom view of the molecules. (C) At the left, helices IV and V in the two molecules do not overlap significantly. At the right is helix VII. Note the irregular and kinked helix in rhodopsin as well as the differences in the location of the Schiff base attachment and orientation of the retinal chromophore.
Rhodopsin and bacteriorhodopsin have the same overall topology of their polypeptide fold as seen in Figure 5A. The helices in rhodopsin are slightly longer than those in bacteriorhodopsin. We have compared the individual helix positions by maintaining the same superposition as shown in Figure 5A but isolating the individual helices. In this comparison, helices I–III superimpose reasonably well. Pairwise comparison of the remaining helices (Figure 5C, left) shows that helices IV and V do not superimpose. Additionally, the twists and kinks in the helices create substantial differences between the two molecules. These differences are clearly large enough to affect homology modeling efforts for other GPCRs which are based on the bacteriorhodopsin structure.
The major function of rhodopsin is to couple the conformational change of a retinal chromophore (caused by absorption of a photon) with a structural change in the protein generating a signal on the cytoplasmic face of the molecule. Accordingly, there is great interest in the retinal environment and conformation within the protein. The comparison between the rhodopsin model and the high-resolution structure of bacteriorhodopsin (113–115) demonstrates how uniquely nature adapted each protein to amplify the light signal and to pump protons across plasma membranes, respectively. However, when the activation process is dissected into fine details at atomic resolution, it is possible that both mechanisms for rhodopsin and bacteriorhodopsin may share many similarities, such as movement of protons within the hydrophobic core of both proteins. In Figure 5C, on the right is shown a comparison of helix VII of bacteriorhodopsin and rhodopsin. The Schiff base attachment is in a different position relative to the membrane plane, but surprisingly, the ionone rings are rather close. The significance of this, if any, is unknown.
CONCLUSIONS
Due to the importance of GPCRs in vast numbers of physiological processes, understanding how rhodopsin is activated, as well as other GPCRs, is one of the most fundamental problems currently unsolved in neuroscience. Our refined model, in conjunction with many biochemical studies, including our most recent work using rhodopsin regenerated with ring-constrained 11-cis-retinal analogues, suggests that cis–trans isomerization is merely a mechanism for repositioning the β-ionone ring that ultimately determines receptor activation (K. Palczewski, unpublished). However, more molecular information is needed to understand how rhodopsin and other GPCRs are activated. Now with the first fundamental step taken by determination of the rhodopsin structure, further investigations will fill our gaps in understanding how this and other GPCRs switch into the signaling state.
Acknowledgments
We thank Drs. Klaus Peter Hofmann and Paul A. Hargrave for their comments on the manuscript.
Footnotes
This research was supported by NIH Grant EY-09339, awards from Research to Prevent Blindness, Inc. (RPB), to the Department of Ophthalmology at the University of Washington, the Foundation Fighting Blindness, the Ruth and Milton Steinbach Fund, and the E. K. Bishop Foundation. K.P. is a RPB Senior Investigator.
The coordinates have been deposited in the Protein Data Bank as entry 1HZX.
Abbreviations: GPCR, G-protein-coupled receptor; MAN, β-d-mannose; NAG, 2-N-acetyl-β-d-glucose; BNG, β-nonylglucoside; |Fo|, observed structure factor; |Fc|, calculated structure factor. R = ∑||Fo| − |Fc||/∑|Fo|. Rcryst is the value for the working set of |Fo|, while Rfree is that calculated for the 5% of reflections set aside for cross validation.
SUPPORTING INFORMATION AVAILABLE
Tabular listing of the following topics: tilt of the helices, bends within helices, possible hydrogen bonds between helices, closest atoms to the 11-cis-retinal, retinal atoms closest to these protein atoms, and torsion angles for the chromophore. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kuhlbrandt W, Gouaux E. Curr Opin Struct Biol. 1999;9:445–7. doi: 10.1016/S0959-440X(99)80062-1. [DOI] [PubMed] [Google Scholar]
- 2.Caffrey M. Curr Opin Struct Biol. 2000;10:486–97. doi: 10.1016/s0959-440x(00)00119-6. [DOI] [PubMed] [Google Scholar]
- 3.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. Cell. 2000;100:693–702. doi: 10.1016/s0092-8674(00)80705-9. [DOI] [PubMed] [Google Scholar]
- 4.Mombaerts P. Science. 1999;286:707–11. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 5.Milligan G, Rees S. Trends Pharmacol Sci. 1999;20:118–24. doi: 10.1016/s0165-6147(99)01320-6. [DOI] [PubMed] [Google Scholar]
- 6.Marchese A, George SR, Kolakowski LF, Jr, Lynch KR, O’Dowd BF. Trends Pharmacol Sci. 1999;20:370–5. doi: 10.1016/s0165-6147(99)01366-8. [DOI] [PubMed] [Google Scholar]
- 7.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 8.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature. 2000;407:971–7. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 9.Hargrave PA, McDowell JH, Curtis DR, Wang JK, Juszczak E, Fong SL, Rao JK, Argos P. Biophys Struct Mech. 1983;9:235–44. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- 10.Hargrave PA, McDowell JH. FASEB J. 1992;6:2323–31. doi: 10.1096/fasebj.6.6.1544542. [DOI] [PubMed] [Google Scholar]
- 11.Khorana HG. J Biol Chem. 1992;267:1–4. [PubMed] [Google Scholar]
- 12.Sakmar TP. Prog Nucleic Acid Res Mol Biol. 1998;59:1–34. doi: 10.1016/s0079-6603(08)61027-2. [DOI] [PubMed] [Google Scholar]
- 13.Fahmy K, Siebert F, Sakmar TP. Biophys Chem. 1995;56:171–81. doi: 10.1016/0301-4622(95)00030-2. [DOI] [PubMed] [Google Scholar]
- 14.Rao VR, Oprian DD. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- 15.Ebrey T, Koutalos Y. Prog Retina Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- 16.Hargrave PA, Bownds D, Wang JK, McDowell JH. Methods Enzymol. 1982;81:211–4. doi: 10.1016/s0076-6879(82)81033-1. [DOI] [PubMed] [Google Scholar]
- 17.Bownds D. Nature. 1967;216:1178–81. doi: 10.1038/2161178a0. [DOI] [PubMed] [Google Scholar]
- 18.Zhukovsky EA, Robinson PR, Oprian DD. Science. 1991;251:558–60. doi: 10.1126/science.1990431. [DOI] [PubMed] [Google Scholar]
- 19.Birge RR, Einterz CM, Knapp HM, Murray LP. Biophys J. 1988;53:367–85. doi: 10.1016/S0006-3495(88)83114-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenlein RW, Peteanu LA, Mathies RA, Shank CV. Science. 1991;254:412–5. doi: 10.1126/science.1925597. [DOI] [PubMed] [Google Scholar]
- 21.Honig B, Ebrey T, Callender RH, Dinur U, Ottolenghi M. Proc Natl Acad Sci USA. 1979;76:2503–7. doi: 10.1073/pnas.76.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper A. FEBS Lett. 1981;123:324–6. doi: 10.1016/0014-5793(81)80319-5. [DOI] [PubMed] [Google Scholar]
- 23.Shichida Y, Imai H. Novartis Found Symp. 1999;224:142–57. doi: 10.1002/9780470515693.ch9. [DOI] [PubMed] [Google Scholar]
- 24.Shichida Y, Tachibanaki S, Mizukami T, Imai H, Terakita A. Methods Enzymol. 2000;315:347–63. doi: 10.1016/s0076-6879(00)15853-7. [DOI] [PubMed] [Google Scholar]
- 25.Gether U, Lin S, Ghanouni P, Ballesteros JA, Weinstein H, Kobilka BK. EMBO J. 1997;16:6737–47. doi: 10.1093/emboj/16.22.6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann KP. Novartis Found Symp. 1999;224:158–80. doi: 10.1002/9780470515693.ch10. [DOI] [PubMed] [Google Scholar]
- 27.Okada, T., Ernst, O. P., Palczewski, K., and Hofmann, K. P. (2001) Trends Biochem. Sci. (in press). [DOI] [PubMed]
- 28.Fain GL, Matthews HR, Cornwall MC, Koutalos Y. Physiol Rev. 2001;81:117–51. doi: 10.1152/physrev.2001.81.1.117. [DOI] [PubMed] [Google Scholar]
- 29.Polans A, Baehr W, Palczewski K. Trends Neurosci. 1996;19:547–54. doi: 10.1016/s0166-2236(96)10059-x. [DOI] [PubMed] [Google Scholar]
- 30.Koutalos Y, Yau KW. Curr Opin Neurobiol. 1993;3:513–9. doi: 10.1016/0959-4388(93)90049-5. [DOI] [PubMed] [Google Scholar]
- 31.Baylor DA, Burns ME. Eye. 1998;12:521–5. doi: 10.1038/eye.1998.140. [DOI] [PubMed] [Google Scholar]
- 32.Hargrave PA, McDowell JH. Int Rev Cytol. 1992;137b:49–97. doi: 10.1016/s0074-7696(08)62600-5. [DOI] [PubMed] [Google Scholar]
- 33.Stryer L. Proc Natl Acad Sci USA. 1996;93:557–9. [Google Scholar]
- 34.Schertler GF, Hargrave PA. Methods Enzymol. 2000;315:91–107. doi: 10.1016/s0076-6879(00)15837-9. [DOI] [PubMed] [Google Scholar]
- 35.Unger VM, Hargrave PA, Baldwin JM, Schertler GF. Nature. 1997;389:203–6. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]
- 36.Schertler GF, Hargrave PA. Proc Natl Acad Sci USA. 1995;92:11578–82. doi: 10.1073/pnas.92.25.11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schertler GF. Novartis Found Symp. 1999;224:54–66. [PubMed] [Google Scholar]
- 38.Unger VM, Schertler GF. Biophys J. 1995;68:1776–86. doi: 10.1016/S0006-3495(95)80354-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Henderson R, Schertler GF. Philos Trans R Soc London, Ser B. 1990;326:379–89. doi: 10.1098/rstb.1990.0019. [DOI] [PubMed] [Google Scholar]
- 40.Schertler GF. Eye. 1998;12:504–10. doi: 10.1038/eye.1998.138. [DOI] [PubMed] [Google Scholar]
- 41.Baldwin JM, Schertler GF, Unger VM. J Mol Biol. 1997;272:144–64. doi: 10.1006/jmbi.1997.1240. [DOI] [PubMed] [Google Scholar]
- 42.Herzyk P, Hubbard RE. J Mol Biol. 1998;281:741–54. doi: 10.1006/jmbi.1998.1981. [DOI] [PubMed] [Google Scholar]
- 43.Pogozheva ID, Lomize AL, Mosberg HI. Biophys J. 1997;72:1963–85. doi: 10.1016/S0006-3495(97)78842-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neumuller M, Jahnig F. Proteins. 1996;26:146–56. doi: 10.1002/(SICI)1097-0134(199610)26:2<146::AID-PROT4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 45.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. 1998;D54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 46.Kleywegt GJ, Jones TA. Acta Crystallogr. 1998;D54:1119–31. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 47.Gilardi R, Karle I, Karle J, Sperling W. Nature. 1971;232:187–8. doi: 10.1038/232187c0. [DOI] [PubMed] [Google Scholar]
- 48.McRee DE. J Struct Biol. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 49.Yeates TO. Methods Enzymol. 1997;276:344–58. [PubMed] [Google Scholar]
- 50.Redinbo MR, Yeates TO. Acta Crystallogr. 1993;D49:375–80. doi: 10.1107/S090744499300294X. [DOI] [PubMed] [Google Scholar]
- 51.Kisselev OG, Meyer CK, Heck M, Ernst OP, Hofmann KP. Proc Natl Acad Sci USA. 1999;96:4898–903. doi: 10.1073/pnas.96.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riek RP, Rigoutsos I, Novotny J, Graham RM. J Mol Biol. 2001;306:349–62. doi: 10.1006/jmbi.2000.4402. [DOI] [PubMed] [Google Scholar]
- 53.Franke RR, Konig B, Sakmar TP, Khorana HG, Hofmann KP. Science. 1990;250:123–5. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 54.Fahmy K, Sakmar TP, Siebert F. Biochemistry. 2000;39:10607–12. doi: 10.1021/bi000912d. [DOI] [PubMed] [Google Scholar]
- 55.Arnis S, Fahmy K, Hofmann KP, Sakmar TP. J Biol Chem. 1994;269:23879–81. [PubMed] [Google Scholar]
- 56.Gerber BO, Meng EC, Dotsch V, Baranski TJ, Bourne HR. J Biol Chem. 2001;276:3394–400. doi: 10.1074/jbc.M007748200. [DOI] [PubMed] [Google Scholar]
- 57.Ernst OP, Meyer CK, Marin EP, Henklein P, Fu WY, Sakmar TP, Hofmann KP. J Biol Chem. 2000;275:1937–43. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 58.Langen R, Cai K, Altenbach C, Khorana HG, Hubbell WL. Biochemistry. 1999;38:7918–24. doi: 10.1021/bi990010g. [DOI] [PubMed] [Google Scholar]
- 59.Cai K, Langen R, Hubbell WL, Khorana HG. Proc Natl Acad Sci USA. 1997;94:14267–72. doi: 10.1073/pnas.94.26.14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karnik SS, Sakmar TP, Chen HB, Khorana HG. Proc Natl Acad Sci USA. 1988;85:8459–63. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terakita A, Yamashita T, Shichida Y. Proc Natl Acad Sci USA. 2000;97:14263–7. doi: 10.1073/pnas.260349597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Asenjo AB, Oprian DD. Biochemistry. 1993;32:2125–30. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 63.Crescitelli, F. (1977) Handbook of Sensory Physiology Vol. VII/5, Springer-Verlag, Berlin.
- 64.Papac DI, Thornburg KR, Bullesbach EE, Crouch RK, Knapp DR. J Biol Chem. 1992;267:16889–94. [PubMed] [Google Scholar]
- 65.Morrison DF, O’Brien PJ, Pepperberg DR. J Biol Chem. 1991;266:20118–23. [PubMed] [Google Scholar]
- 66.Ovchinnikov YA, Abdulaev NG, Bogachuk AS. FEBS Lett. 1988;230:1–5. doi: 10.1016/0014-5793(88)80628-8. [DOI] [PubMed] [Google Scholar]
- 67.Veit M, Sachs K, Heckelmann M, Maretzki D, Hofmann KP, Schmidt MF. Biochim Biophys Acta. 1998;1394:90–8. doi: 10.1016/s0005-2760(98)00097-6. [DOI] [PubMed] [Google Scholar]
- 68.Pepperberg DR, Morrison DF, O’Brien PJ. Methods Enzymol. 1995;250:348–61. doi: 10.1016/0076-6879(95)50084-7. [DOI] [PubMed] [Google Scholar]
- 69.Traxler KW, Dewey TG. Biochemistry. 1994;33:1718–23. doi: 10.1021/bi00173a014. [DOI] [PubMed] [Google Scholar]
- 70.Karnik SS, Ridge KD, Bhattacharya S, Khorana HG. Proc Natl Acad Sci USA. 1993;90:40–4. doi: 10.1073/pnas.90.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.O’Brien PJ, Zatz M. J Biol Chem. 1984;259:5054–7. [PubMed] [Google Scholar]
- 72.Fukuda MN, Papermaster DS, Hargrave PA. J Biol Chem. 1979;254:8201–7. [PubMed] [Google Scholar]
- 73.Hargrave PA. Biochim Biophys Acta. 1977;492:83–94. doi: 10.1016/0005-2795(77)90216-1. [DOI] [PubMed] [Google Scholar]
- 74.Hargrave PA, McDowell JH, Feldmann RJ, Atkinson PH, Rao JK, Argos P. Vision Res. 1984;24:1487–99. doi: 10.1016/0042-6989(84)90311-0. [DOI] [PubMed] [Google Scholar]
- 75.Ball LE, Oatis JE, Jr, Dharmasiri K, Busman M, Wang J, Cowden LB, Galijatovic A, Chen N, Crouch RK, Knapp DR. Protein Sci. 1998;7:758–64. doi: 10.1002/pro.5560070325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isele J, Sakmar TP, Siebert F. Biophys J. 2000;79:3063–71. doi: 10.1016/S0006-3495(00)76541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beck M, Siebert F, Sakmar TP. FEBS Lett. 1998;436:304–8. doi: 10.1016/s0014-5793(98)01156-9. [DOI] [PubMed] [Google Scholar]
- 78.Zorn M, Futterman S. J Biol Chem. 1971;246:881–6. [PubMed] [Google Scholar]
- 79.Bonting SL, van Breugel PJ, Daemen FJ. Adv Exp Med Biol. 1977;83:175–89. doi: 10.1007/978-1-4684-3276-3_16. [DOI] [PubMed] [Google Scholar]
- 80.Williams TP, Baker BN, McDowell JH. Exp Eye Res. 1974;18:69–75. doi: 10.1016/0014-4835(74)90044-x. [DOI] [PubMed] [Google Scholar]
- 81.Litman BJ, Mitchell DC. Lipids. 1996;31(Suppl):S193–7. doi: 10.1007/BF02637075. [DOI] [PubMed] [Google Scholar]
- 82.Mitchell DC, Straume M, Litman BJ. Biochemistry. 1992;31:662–70. doi: 10.1021/bi00118a005. [DOI] [PubMed] [Google Scholar]
- 83.Hessel E, Muller A, Herrmann A, Hofmann KP. J Biol Chem. 2001;276:2538–43. doi: 10.1074/jbc.M009061200. [DOI] [PubMed] [Google Scholar]
- 84.Okada T, Le Trong I, Fox BA, Behnke CA, Stenkamp RE, Palczewski K. J Struct Biol. 2000;130:73–80. doi: 10.1006/jsbi.1999.4209. [DOI] [PubMed] [Google Scholar]
- 85.Okada T, Takeda K, Kouyama T. Photochem Photobiol. 1998;67:495–9. [PubMed] [Google Scholar]
- 86.Buczylko J, Saari JC, Crouch RK, Palczewski K. J Biol Chem. 1996;271:20621–30. doi: 10.1074/jbc.271.34.20621. [DOI] [PubMed] [Google Scholar]
- 87.Ohguro H, Rudnicka Nawrot M, Buczylko J, Zhao X, Taylor JA, Walsh KA, Palczewski K. J Biol Chem. 1996;271:5215–24. doi: 10.1074/jbc.271.9.5215. [DOI] [PubMed] [Google Scholar]
- 88.Ohguro H, Palczewski K, Ericsson LH, Walsh KA, Johnson RS. Biochemistry. 1993;32:5718–24. doi: 10.1021/bi00072a030. [DOI] [PubMed] [Google Scholar]
- 89.McDowell JH, Nawrocki JP, Hargrave PA. Biochemistry. 1993;32:4968–74. doi: 10.1021/bi00069a036. [DOI] [PubMed] [Google Scholar]
- 90.Chen J, Makino CL, Peachey NS, Baylor DA, Simon MI. Science. 1995;267:374–7. doi: 10.1126/science.7824934. [DOI] [PubMed] [Google Scholar]
- 91.Papac DI, Oatis JE, Jr, Crouch RK, Knapp DR. Biochemistry. 1993;32:5930–4. doi: 10.1021/bi00074a002. [DOI] [PubMed] [Google Scholar]
- 92.Wolfrum U, Schmitt A. Cell Motil Cytoskeleton. 2000;46:95–107. doi: 10.1002/1097-0169(200006)46:2<95::AID-CM2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 93.Deretic D, Schmerl S, Hargrave PA, Arendt A, McDowell JH. Proc Natl Acad Sci USA. 1998;95:10620–5. doi: 10.1073/pnas.95.18.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Palczewski K, Buczylko J, Kaplan MW, Polans AS, Crabb JW. J Biol Chem. 1991;266:12949–55. [PubMed] [Google Scholar]
- 95.Ohguro H, Yoshida N, Shindou H, Crabb JW, Palczewski K, Tsuda M. Photochem Photobiol. 1998;68:824–8. [PubMed] [Google Scholar]
- 96.Ohguro H, Johnson RS, Ericsson LH, Walsh KA, Palczewski K. Biochemistry. 1994;33:1023–8. doi: 10.1021/bi00170a022. [DOI] [PubMed] [Google Scholar]
- 97.Ohguro H, Van Hooser JP, Milam AH, Palczewski K. J Biol Chem. 1995;270:14259–62. doi: 10.1074/jbc.270.24.14259. [DOI] [PubMed] [Google Scholar]
- 98.Ohguro H, Palczewski K. FEBS Lett. 1995;368:452–4. doi: 10.1016/0014-5793(95)00710-q. [DOI] [PubMed] [Google Scholar]
- 99.Brannock MT, Weng K, Robinson PR. Biochemistry. 1999;38:3770–7. doi: 10.1021/bi982419w. [DOI] [PubMed] [Google Scholar]
- 100.Wald G. Nature. 1968;219:800–7. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 101.Wald G. Science. 1968;162:230–9. doi: 10.1126/science.162.3850.230. [DOI] [PubMed] [Google Scholar]
- 102.Kosower EM. Proc Natl Acad Sci USA. 1988;85:1076–80. doi: 10.1073/pnas.85.4.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kühne W. Vision Res. 1977;17:1273–316. doi: 10.1016/0042-6989(77)90114-6. [DOI] [PubMed] [Google Scholar]
- 104.Kühne, W. (1879) Chemische vorgange in der netzhaut, Vol. 3, F. C. W. Vogel, Leipzig, Germany.
- 105.Sakmar TP, Franke RR, Khorana HG. Proc Natl Acad Sci USA. 1989;86:8309–13. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhukovsky EA, Oprian DD. Science. 1989;246:928–30. doi: 10.1126/science.2573154. [DOI] [PubMed] [Google Scholar]
- 107.Ebrey TG. Methods Enzymol. 2000;315:196–207. doi: 10.1016/s0076-6879(00)15844-6. [DOI] [PubMed] [Google Scholar]
- 108.Grobner G, Burnett IJ, Glaubitz C, Choi G, Mason AJ, Watts A. Nature. 2000;405:810–3. doi: 10.1038/35015604. [DOI] [PubMed] [Google Scholar]
- 109.Smith SO, Palings I, Copie V, Raleigh DP, Courtin J, Pardoen JA, Lugtenburg J, Mathies RA, Griffin RG. Biochemistry. 1987;26:1606–11. doi: 10.1021/bi00380a018. [DOI] [PubMed] [Google Scholar]
- 110.Okada T, Kandori H, Shichida Y, Yoshizawa T, Denny M, Zhang BW, Asato AE, Liu RS. Biochemistry. 1991;30:4796–802. doi: 10.1021/bi00233a022. [DOI] [PubMed] [Google Scholar]
- 111.Imamoto Y, Sakai M, Katsuta Y, Wada A, Ito M, Shichida Y. Biochemistry. 1996;35:6257–62. doi: 10.1021/bi9519359. [DOI] [PubMed] [Google Scholar]
- 112.Luecke H, Richter HT, Lanyi JK. Science. 1998;280:1934–7. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 113.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 114.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Science. 1999;286:255–61. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 115.Lanyi JK. J Phys Chem B. 2000;104:11441–8. [Google Scholar]
- 116.Kraulis PJ. J Appl Crystallogr. 1991;24:946–50. [Google Scholar]
- 117.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–24. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 118.Khorana HG. Ann NY Acad Sci. 1986;471:272–88. doi: 10.1111/j.1749-6632.1986.tb48043.x. [DOI] [PubMed] [Google Scholar]
- 119.Khorana, H. G. (1993) Nucleic Acids Symp. Ser, 219. [PubMed]
- 120.Deretic D. Methods Enzymol. 2000;315:77–88. doi: 10.1016/s0076-6879(00)15836-7. [DOI] [PubMed] [Google Scholar]
- 121.Jensen AD, Guarnieri F, Rasmussen SG, Asmar F, Ballesteros JA, Gether U. J Biol Chem. 2001;276:9279–90. doi: 10.1074/jbc.M004871200. [DOI] [PubMed] [Google Scholar]