Abstract
Cyclophilins are a family of proteins that share a common, highly conserved sequence motif. Cyclophilins bind transiently to other proteins and facilitate their folding. One member of the family, hCypH, is part of the human spliceosomal [U4/U6·U5] tri-snRNP complex; it associates specifically and stably with the U4/U6-specific protein 60K. Here, we demonstrate that recombinant hCypH exhibits peptidyl–prolyl isomerase (PPIase) activity, and describe mutagenesis studies demonstrating that it shares the catalytic pocket with other members of the cyclophilin family. However, neither the PPIase activity nor the catalytic pocket is required for binding of protein 60K. Rather, hCypH contains a small insertion in a loop of the otherwise conserved cyclophilin backbone, and this minor change creates a highly specific binding site that is responsible for the association of this cyclophilin, but not others, with protein 60K. hCypH is thus the first small cyclophilin shown to have a second protein–protein interaction site and the ability to bind stably to another protein. Since the catalytic pocket and the second binding site are located on opposite sides of the cyclophilin structure, this opens up the interesting possibility that hCypH may serve as a bridge mediating interactions between protein 60K of the U4/U6 snRNP and other as yet unknown factors.
INTRODUCTION
Cyclophilins form a large family of proteins that share a common sequence domain (‘cyclophilin domain’) and are found in all organisms from bacteria to eukaryotes. Some cyclophilins consist entirely of this domain, while others contain additional domains [see, for example, Schiene-Fischer and Yu (1)]. In vitro, cyclophylins exhibit peptidyl–prolyl isomerase (PPIase) activity (2). Since peptidyl–prolyl isomerisation is a rate-limiting step in protein folding, cyclophylins accelerate protein folding in vitro (3). Their role in vivo is not clear, and two possible functions, which are not mutually exclusive, are currently under discussion. First, cyclophylins may participate in protein folding in vivo (4). The effect of the cyclophylins on protein folding, however, is only incremental, arguing against an essential role for them in protein maturation. This argument is strengthened by the finding that all deletions of cyclophylin genes in yeast produce strains that are viable, as is the mutant that lacks all cyclophylins (5). Secondly, PPIases (like the cyclophylins), can bind at hydrophobic patches on their target proteins without necessarily isomerising a peptidyl–prolyl bond, thereby preventing misfolding (6–8) and/or modulating activity of the target protein (9–13). No matter which of the two functions is fulfilled by the cyclophylin, the interaction with the target protein is transient. However, some of the cyclophylins associate stably with other proteins. In this case, the stable interaction is mediated by the additional domains present on the cyclophylin, such as the Ran-binding or the TPR domain (1,7).
One member of the cyclophylin family associates stably with the human [U4/U6·U5] tri-snRNP complex involved in pre-mRNA splicing. This protein consists of a single domain with clear sequence homology (14,15) and structural resemblance (16) to the known cyclophylins, and was therefore denoted hCypH (formerly also named Snu-Cyp-20 or USA-Cyp). Biochemical fractionation showed that hCypH forms a very stable complex with the U4/U6-specific 60K/90K dimer (14,15), binding primarily to the N-terminus of protein 60K (17). This binding appears to be highly specific, since at present only one other protein, the non-snRNP splicing factor hPrp18, is known to be recognised by hCypH (17). The specificity and stability of the 60K–CypH interaction is particularly remarkable, since on the one hand hCypH contains no additional protein domains that could mediate this interaction, while, on the other hand, a cyclophylin domain does not usually mediate stable protein–protein interactions. Knowledge of how the cyclophylin is stably integrated into this spliceosomal 60K/90K complex will thus reveal important insights into the function of cyclophylins in general and hCypH in particular.
In the work described here, we mutagenised hCypH and found that mutations in the presumed catalytic pocket suppressed both the PPIase activity and the binding of cyclosporin A (CsA), confirming that hCypH functions as a typical cyclophylin. However, the same mutations hardly affected hCypH’s binding of protein 60K; instead, we identified a specific binding site for protein 60K, which is expressed on the conserved cyclophylin domain structure but distinct from the catalytic pocket. hCypH is thus the first small cyclophylin shown to contain a second protein–protein interaction site.
MATERIALS AND METHODS
Expression of hCypH
The expressed sequence tag (accession no. T53949) encoding full-length hCypH was subcloned into the BamHI and NotI sites of the pBluescript SK– vector (Stratagene). The protein was expressed in vitro by coupled T7 transcription and reticulocyte lysate translation (Promega). Mutagenesis was performed according to a PCR-based protocol (QuikChange, Stratagene). The sequence of the mutated open reading frames (ORFs) was verified by dideoxy sequencing.
For overexpression, the ORF was subcloned into the pGEX-4T-2 vector (Amersham Pharmacia), inserting an artificial glycine hinge (GSGGGS) between the GST and the hCypH to enhance solubility of the fusion protein. Expression was induced in Escherichia coli BL21(DE3) by 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After 4 h at 37°C, cells were harvested and lysed by sonification. The fusion protein was loaded onto a 2 ml glutathione–Sepharose 4B column, washed and eluted as recommended by the manufacturer (Amersham Pharmacia). The purity of the protein was checked by SDS–PAGE.
PPIase assay
The enzymatic assay was performed in 35 mM HEPES buffer pH 7.8 at 11°C using the substrate peptide succ-Ala-Ala-Pro-Phe-pNA (in which the Ala–Pro bond is in equilibrium between the cis and trans conformations) and α-chymotrypsin to cleave the chromophore p-nitroaniline (pNA) from the all-trans peptide (18). Release of the chromophore was monitored by measuring the absorption at 390 nm. Since the Michaelis constant KM for the cyclophylin-catalysed isomerisation of such peptides is very high (KM>>[S]), the Michaelis–Menten equation can be simplified to a second-order kinetic equation yielding the overall apparent first-order rate constant kapp = kcat [Enz]/KM + ksp, where Enz is the catalyst (here, cyclophylin) and ksp is the first-order rate constant of the spontaneous (uncatalysed) isomerisation. The rate constant kapp was determined by iteratively fitting the data to the equation [P] = [P]∞ (1 – e–k*t), where [P]∞ is the product concentration at the end of the reaction. Fitting was carried out by the Marquardt method (GraphPad Software).
CsA precipitation assay
For each precipitation, we used 10 µl (settled volume) of glutathione–Sepharose beads, 10 µg (217 pmol) of GST–hCypH or the equivalent molar amounts of GST and GST–hCypA. [3H]CsA (0.2 µCi; 25 pmol; New England Nuclear) and 0, 25 or 125 pmol of non-radioactive CsA were incubated for 1 h in 300 µl of precipitation buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.05% NP-40). The beads were pelleted and washed three times with 1 ml of precipitation buffer, and bound protein was eluted with 10 mM glutathione. Co-purified CsA was quantified in a scintillation counter.
hCypH precipitation assays
[35S]Methionine-labelled hCypH was prepared by expression in vitro (see above); it was mixed with protein 60K (14) and precipitated with polyclonal anti-60K antibodies as described elsewhere (14): briefly, antibodies from 20 µl of serum in 250 µl of precipitation buffer (see above) containing 1% bovine serum albumin (BSA) were immobilised on 10 µl of protein A–Sepharose beads. The beads were washed and incubated with the respective in vitro expression mixtures in 300 µl of precipitation buffer for 1 h at 4°C. The beads were washed three times, and bound proteins were released by boiling the beads in SDS loading buffer. The proteins were separated on SDS–12% acrylamide gels. hCypH and protein 60K were detected by autoradiography and quantified on a densitometer.
RESULTS AND DISCUSSION
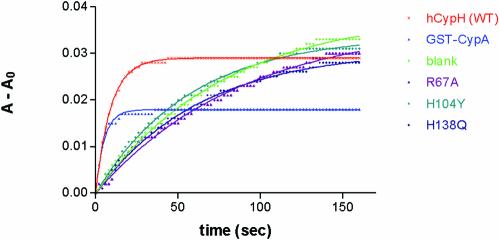
Purified human [U4/U6·U5] tri-snRNP particles have previously been shown to possess PPIase activity (14). To verify that hCypH carries this activity, a GST–hCypH fusion protein was overexpressed in E.coli and purified by affinity chromatography. PPIase activity was measured in a coupled isomerisation/proteolysis assay, with a model proline- containing tetrapeptide as substrate: an initial, rate-limiting cis→trans isomerisation step generates an all-trans peptide from which fast chymotryptic cleavage liberates a chromophore (18). As shown in Figure 1, this reaction occurred, albeit slowly, in the absence of PPIase owing to background thermal isomerisation. Upon addition of hCypA or wild-type hCypH, the reaction proceeded much more rapidly and reached equilibrium after 15–30 s. The first-order decay constant kapp = kcat [Enz]/KM + ksp was estimated by fitting the data iteratively to a pseudo first-order kinetic equation (see Materials and Methods). As can be seen in the figure, the best-fit exponential curves agree well with the measurements. With the conditions and concentration chosen here, hCypA catalyses the isomerisation of the peptide with a kapp of 0.195 –1. Subtraction of the background value (0.012 s–1) gave a contribution of 0.183 s–1 due to the catalysis by CypA, representing a kcat/KM value of 8.8 × 106 M–1s–1. For hCypH, the corresponding kapp was 0.109 s–1 (catalytic contribution 0.097 s–1, kcat/KM 1.3 × 106 M–1s–1). Thus, hCypH exhibits significant PPIase activity, while being only ∼15% as active as hCypA. Recently, a very similar kcat/KM value has been reported for hCypH (19). The variability in the catalytic activity of hCypA and hCypH may be a consequence of different intrinsic catalytic activities, different substrate affinities or both. For comparison, a study of all cyclophylins in the Caenorhabditis elegans genome (20) has shown that their PPIase activity towards a model peptide varies by two orders of magnitude.
Figure 1.
Wild-type hCypH, but not its mutants, exhibit PPIase activity. hCypA (25 pmol) or GST–hCypH (wild-type or mutant; 87.5 pmol) were assayed for PPIase activity by mixing with α-chymotrypsin and 38 nmol substrate peptide in a 1.2 ml assay (see Materials and Methods). Cleavage of the chromophore from the substrate peptide was monitored by measuring the absorbance (A) at 390 nm every other second over a period of 160 s. Non-linear regression to a single-exponential curve gave a best-fit curve for each set of data points, as shown overlaid with the data points in the graph. The height of the plateau reflects the amount of substrate reacted between mixing the reactants and starting the measurement, so the plateau is lower for the faster reactions.
To probe the catalytic pocket of hCypH, we introduced mutations designed to inhibit its PPIase activity. Sequence comparison showed that all residues implicated in substrate binding and PPIase activity of hCypA are conserved in the hCypH protein (14,15). We selected three mutations corresponding to ones previously reported to inhibit PPIase activity of other cyclophylins (21,22) and performed the corresponding amino acid exchanges in hCypH. Thus, Arg67 was changed to alanine (R67A, to imitate the effect of R55A in hCypA), His104 to tyrosine (H104Y, cf. H90Y in the yeast Cpr1p) and His138 to glutamine (H138Q, cf. H126Q in hCypA). The three mutated proteins were expressed as before and assayed for PPIase activity. As shown in Figure 1, each of the three mutations greatly impaired the PPIase activity. Respective kapp values were 0.011, 0.017 and 0.016 s–1; thus the mutant R67A exhibits no catalytic activity (kapp ≈ background), and the other two mutants showed an activity of ∼5% of that of the wild-type protein (respective kcat/KM = 7 × 104 and 5 × 104 M–1s–1). We infer that all three mutated residues are either part of the catalytic site in the hCypH protein, or else are required for proper folding of this part of the molecule.
In order to determine to what extent the mutations affect substrate binding by hCypH, we measured the ability of the wild-type and mutant hCypH to bind CsA. CsA is known to form a tight dimeric complex with hCypA that interferes with the calcineurin signalling cascade; this is the origin of the drug’s immunosuppresive effect. CsA fits exactly into the catalytic pocket of cyclophilins and inhibits their PPIase activity competitively (23). Therefore, the affinity of hCypH and its mutants for CsA will reflect their affinity for the substrate. We performed pull-down assays using the GST–hCypH fusion proteins. These proteins were incubated with tritiated CsA and then precipitated with glutathione–Sepharose. After washing, the GST fusion proteins were eluted with glutathione, and the amount of CsA co-precipitated was determined by scintillation counting. Unlabelled CsA was added to the binding reaction mixtures to give respective total amounts of 25, 50 and 125 pmol CsA (equivalent to 80, 160 and 400 nM). For each CsA concentration, the background precipitation obtained with GST alone was defined as 0%, and the precipitation obtained with a GST–hCypA control employing the same amount of CsA was defined as 100% (this count was always at least 100 times the background level). At high CsA concentrations, hCypH binds 50–70% of the CsA precipitated by the hCypA control (Fig. 2, first three bars). The difference between the two proteins may be due to a larger amount of active hCypA immobilised on the beads. At the lower CsA concentration, however, the amount of CsA bound to hCypH is only 20% of that bound to hCypA, which probably reflects inherently weaker binding. Note that the lowest CsA concentration is below the dissociation constant KD of the [CsA·hCypH] complex, 120 nM (19), so binding will be sensitive to changes in CsA concentration. The hCypH mutants, in contrast to the wild type, show a drastically reduced CsA precipitation, with R67A and H104Y precipitating only background amounts of CsA, and H138Q showing no more than 5% of the binding yield of hCypA.
Figure 2.
Wild-type hCypH protein binds CsA, whereas the mutant proteins do not. The CsA binding assay was conducted as described in Materials and Methods with 25, 50 and 125 pmol CsA and 10 µg (217 pmol) of wild-type GST fusion proteins. For each value, background precipitation by GST alone was subtracted, and the result was then normalized to the amount of CsA precipitated by GST–hCypA under the same conditions.
In conclusion, all three mutations of hCypH inhibit its PPIase activity and suppress CsA binding. This indicates that the residues are located in or around the catalytic site and the CsA-binding pocket. The alternative possibility, that these mutations inhibit proper folding of hCypH, rendering it non-functional, is virtually excluded by the fact that R67 and H104 are surface residues and do not make significant intramolecular contacts (16), so their replacement should not disturb the overall folding of the protein. H138 may contribute to local folding of the catalytic pocket, but it does not influence the folding of the core (16). The assertion that the substitutions do not lead to a general misfolding of the mutants is further supported by the observation that all three mutants are recognised by protein 60K (see below), indicating that at least this interaction surface remains intact. Therefore, we conclude that the residues R67, H104 and H138 are required for substrate binding and/or PPIase catalysis. This is in good agreement with recent crystallographic results, which show both R67 and H104 in direct contact with a proline residue from a neighbouring molecule protruding into the catalytic pocket of hCypH (29).
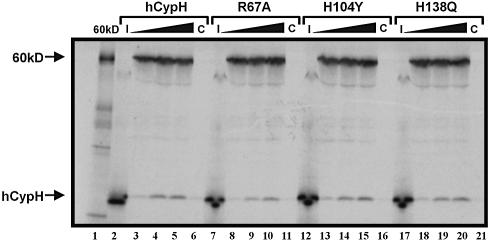
To investigate whether the interaction of hCypH with the U4/U6-specific protein 60K involves the catalytic pocket, we performed co-immunoprecipitation experiments. 35S-labelled hCypH and protein 60K were mixed and precipitated with antibodies against protein 60K. The antibody-bound proteins were then released with SDS and analysed by SDS–PAGE. As demonstrated previously (14), wild-type hCypH binds to protein 60K; it was therefore co-precipitated with anti-60K antibodies only in the presence of this protein (Fig. 3, compare lanes 3–5 with lane 6). Furthermore, when increasing concentrations of hCypH translate were incubated with constant amounts of 60K translate, the yield of co-precipitating hCypH reached a plateau, indicating that there is a limited number of saturable binding sites for hCypH on protein 60K. Intriguingly, the substitutions R67A (lanes 8–10), H104Y (lanes 13–15) and H138Q (lanes 18–20) did not have a significant effect on binding to protein 60K. Quantification of the intensities by densitometry shows a 9% reduction in binding yield for R67A and H104Y compared with the wild type, and a 6% reduction for H138Q; both changes are within the error of this experiment. Therefore, mutations that drastically reduce both the PPIase activity of hCypH and the binding of CsA to its catalytic site have only a minor effect, if any at all, on 60K binding.
Figure 3.
Mutations in the catalytic pocket do not affect 60K binding. A 2 µl aliquot of 60K translate (translation reaction mixture, without subsequent purification) was incubated with increasing amounts (3, 6 and 9 µl) of corresponding translates of wild-type hCypH (lanes 3–5), the R67A mutant (lanes 8–10), the H104Y mutant (lanes 13–15) and the H138Q mutant (lanes 18–20). Complexes were precipitated with anti-60K antibodies, eluted, and separated by SDS–PAGE. An autoradiograph of the gel is shown. As control, 9 µl of the hCypH and mutant translation mixtures were precipitated in the absence of protein 60K (lanes 6, 11, 16 and 21). The loading control shows 2 µl of the 60K translation mixture (lane 1), and 6 µl of the hCypH and mutant translation mixtures (lanes 2, 7, 12 and 17).
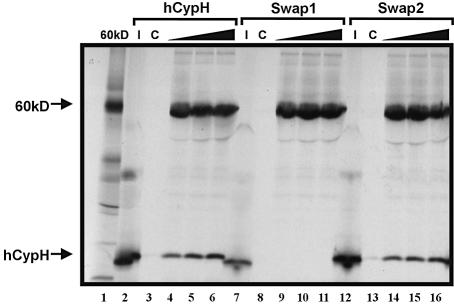
It is well established that cyclophilin proteins can bind to other proteins either through the catalytic pocket of its cyclophylin domain or, if it contains additional domains, through these extra domains (see Introduction). hCypH appears to consist of the cyclophylin domain only, with no other distinguishable domain (14,15); still, the catalytic pocket is not essential for stable binding of protein 60K. It was therefore of considerable interest to find the 60K interaction site on the hCypH protein. To obtain a first clue about the location of this binding site, we made use of the fact that hCypA does not bind to protein 60K (14). The two proteins share extensive sequence and structural homology (16), but there are two structural differences that might be important for the specific recognition of hCypH by protein 60K. First, the loop connecting helix α1 with strand β3 of hCypH is enlarged by a five amino acid insertion. Secondly, proline is inserted at position 158 of hCypH, leading to considerable conformational change of the loop connecting helix α2 with strand β8 (16). To find out whether one of these features is necessary for binding protein 60K, we constructed hCypH mutants in which the α1–β3 loop (51FRKDGVPI58→KGF, termed ‘swap1’) or the α2–β8 loop (156NVPTGPNN163→RFGSRNG, ‘swap2’) was replaced by the corresponding hCypA sequence. Binding assays carried out as in Figure 3 demonstrated that the swap2 mutant exhibited wild-type binding of protein 60K (Fig. 4, compare lanes 2–6 with lanes 12–16). Therefore, the α2–β8 loop is not crucial for 60K binding. The swap1 mutation, in contrast, led to a complete loss of 60K binding (Fig. 4, compare lanes 2–6 with lanes 7–11). Importantly, both swap mutants bound CsA with the same yield as wild-type hCypH (data not shown), indicating that these mutations did not compromise the overall folding of the protein. Therefore, we conclude that the α1–β3 loop is directly involved in the binding of protein 60K.
Figure 4.
Mutagenesis of a specific loop sequence inhibits 60K binding. hCypH and the swap mutants which have the loops α1–β3 (swap1) and α2–β8 loop (swap2) exchanged for hCypA sequences, respectively (see text) were assayed for 60K binding exactly as described in Figure 3.
According to the three-dimensional structure determined by crystallography (16), hCypH is a small, round molecule, on which the catalytic site and the hydrophilic lobe comprising the α1–β3 loop are located roughly on opposite sides. This lobe contributes to the binding of protein 60K, since two mutations in the loop (K53E and V56E) each showed a small reduction in the binding of protein 60K. However, the loop is not sufficient for recognition, because (i) neither point mutation abolished 60K binding, and (ii) insertion of the α1–β3 loop into the hCypA structure did not create a 60K-binding site (data not shown). Thus, structural details surrounding the loop must also contribute to the binding site. In fact, the lobe creates a wide cleft with predominantly hydrophobic character that is well suited for protein–protein interactions, and protein 60K probably recognises both the cleft and the lobe. The importance of this binding site is underlined by its strong evolutionary conservation: homologues of hCypH can be identified in the genomes of all orders, from mammals, insects and plants down to the slime mould. All CypH homologues contain an α1–β3 loop of precisely the same size and the conserved sequence (K/R)-x-(D/A)-G-x-P. A cyclophylin identified in the yeast Schizosaccharomyces pombe (SpCyp-3) also contains an extended α1–β3 loop, albeit with different length and sequence. Despite this divergence, its nuclear localisation suggests that SpCyp-3 is the functional homologue of hCypH (19). The yeast Saccharomyces cerevisiae, in contrast, has no similar cyclophylin. This, however, does not contradict the hypothesis that this motif is required for 60K binding, since no cyclophilin has been found in purified [U4/U6·U5] tri-snRNPs from S.cerevisiae (24,25).
Thus, a minor modification of the cyclophylin backbone creates a new, specific protein-binding site that is located on the opposite side of the protein from the catalytic pocket. The catalytic pocket is thus freely accessible in the hCypH·60K dimer, as borne out by the observations that native, intact tri-snRNP particles exhibit PPIase activity (14) and that hCypH can simultaneously bind CsA and protein 60K (17). Therefore, hCypH is able to mediate a binding interaction between protein 60K and another, as yet unidentified protein target. In this model, hCypH can function in simply mediating a protein–protein interaction with the U4/U6-specific protein 60K, or it can induce a conformational change in the target protein. Whatever its identity, the target protein appears to be directly involved in splicing, since blocking the hCypH protein significantly reduces splicing efficiency (17); however, splicing is not completely inhibited, arguing against an essential role for the target protein. Furthermore, it is remarkable that hCypH specifically binds a second splice factor, hPrp18 (see Introduction). Since the binding sites in hPrp18 and protein 60K are homologous, hCypH presumably binds both proteins in a similar mode. This suggests that two splice factors, which enter the spliceosome at different steps of the spliceosomal cycle, associate with hCypH in such a way that its catalytic pocket is accessible. Moreover, two other cyclophylins are known (SR-Cyp and Matrin-Cyp) that contain an Arg/Ser-rich domain (26,27); this domain is typical of splicing factors, and these cyclophylins are therefore believed to take part in splicing too. As members of the cyclophylin family, they possess the same catalytic binding pocket; in particular, the residues R67, H104 and H138—identified as being important for the PPIase activity and substrate binding of hCypH—are also found in these cyclophilins. Since SR domain proteins interact with each other, these proteins could mediate protein–protein interactions between the network of SR proteins (28) and a target for the cyclophylin domain. It will be interesting to see whether all these spliceosomal cyclophylins, the two copies of hCypH, SR-Cyp and Matrin-Cyp, all interact with similar, or even the same, target proteins.
CONCLUSIONS
Like other cyclophylins, the U4/U6 snRNP-associated hCypH exhibits PPIase activity. However, specific binding of the U4/U6-specific protein 60K does not require this activity, or even the integrity of the catalytic site. Instead, a small insertion in the otherwise conserved cyclophylin backbone creates a site that is specifically recognised by protein 60K. hCypH is thus the first small cyclophylin shown to possess two protein–protein interaction sites and, in consequence, the ability to mediate interactions between two proteins.
Acknowledgments
ACKNOWLEDGEMENT
R.L. and M.A.M. were supported by the Deutsche Forschungs gemeinschaft (SFB 532 and SFB 395, respectively).
REFERENCES
- 1.Schiene-Fischer C. and Yu,C. (2001) Receptor accessory folding helper enzymes: the functional role of peptidyl prolyl cis/trans isomerases. FEBS Lett., 495, 1–6. [DOI] [PubMed] [Google Scholar]
- 2.Fischer G., Wittmann-Liebold,B., Lang,K., Kiefhaber,T. and Schmid,F.X. (1989) Cyclophilin and peptidyl-prolyl cis–trans isomerase are probably identical proteins. Nature, 337, 476–478. [DOI] [PubMed] [Google Scholar]
- 3.Schmid F.X. (1995) Protein folding. Prolyl isomerases join the fold. Curr. Biol., 5, 993–994. [DOI] [PubMed] [Google Scholar]
- 4.Schiene C. and Fischer,G. (2000) Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol., 10, 40–45. [DOI] [PubMed] [Google Scholar]
- 5.Dolinski K., Muir,S., Cardenas,M. and Heitman,J. (1997) All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 13093–13098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker E.K., Colley,N.J. and Zuker,C.S. (1994) The cyclophilin homolog NinaA functions as a chaperone, forming a stable complex in vivo with its protein target rhodopsin. EMBO J., 13, 4886–4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira P.A., Nakayama,T.A., Pak,W.L. and Travis,G.H. (1996) Cyclophilin-related protein RanBP2 acts as chaperone for red/green opsin. Nature, 383, 637–640. [DOI] [PubMed] [Google Scholar]
- 8.Arié J.P., Sassoon,N. and Betton,J.M. (2001) Chaperone function of FkpA, a heat shock prolyl isomerase, in the periplasm of Escherichia coli. Mol. Microbiol., 39, 199–210. [DOI] [PubMed] [Google Scholar]
- 9.Braaten D., Ansari,H. and Luban,J. (1997) The hydrophobic pocket of cyclophilin is the binding site for the human immunodeficiency virus type 1 Gag polyprotein. J. Virol., 71, 2107–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streblow D.N., Kitabwalla,M., Malkovsky,M. and Pauza,C.D. (1998) Cyclophilin A modulates processing of human immunodeficiency virus type 1 p55Gag: mechanism for antiviral effects of cyclosporin A. Virology, 245, 197–202. [DOI] [PubMed] [Google Scholar]
- 11.Shen M., Stukenberg,P.T., Kirschner,M.W. and Lu,K.P. (1998) The essential mitotic peptidyl–prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev., 12, 706–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crenshaw D.G., Yang,J., Means,A.R. and Kornbluth,S. (1998) The mitotic peptidyl–prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J., 17, 1315–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X., Wilcox,C.B., Devasahayam,G., Hackett,R.L., Arevalo-Rodriguez,M., Cardenas,M.E., Heitman,J. and Hanes,S.D. (2000) The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery. EMBO J., 19, 3727–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teigelkamp S., Achsel,T., Mundt,C., Gothel,S.F., Cronshagen,U., Lane,W.S., Marahiel,M. and Lührmann,R. (1998) The 20kD protein of human [U4/U6·U5] tri-snRNPs is a novel cyclophilin that forms a complex with the U4/U6-specific 60kD and 90kD proteins. RNA, 4, 127–141. [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz D.S., Kobayashi,R. and Krainer,A.R. (1997) A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA, 3, 1374–1387. [PMC free article] [PubMed] [Google Scholar]
- 16.Reidt U., Reuter,K., Achsel,T., Ingelfinger,D., Lührmann,R. and Ficner,R. (2000) Crystal structure of the human U4/U6 small nuclear ribonucleoprotein particle-specific SnuCyp-20, a nuclear cyclophilin. J. Biol. Chem., 275, 7439–7442. [DOI] [PubMed] [Google Scholar]
- 17.Horowitz D.S., Lee,E.J., Mabon,S.A. and Misteli,T. (2002) A cyclophilin functions in pre-mRNA splicing. EMBO J., 21, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer G., Bang,H. and Mech,C. (1984) Determination of enzymatic catalysis for the cis–trans-isomerization of peptide binding in proline-containing peptides. Biochim. Biophys Acta, 43, 1101–1111. [PubMed] [Google Scholar]
- 19.Pemberton T.J., Rulten,S.L. and Kay,J.E. (2003) Identification and characterisation of Schizosaccharomyces pombe cyclophilin 3, a cyclosporin A insensitive orthologue of human USA-CyP. J. Chromatogr. B Anal. Technol. Biomed. Life Sci., 786, 81–91. [DOI] [PubMed] [Google Scholar]
- 20.Page A.P., MacNiven,K. and Hengartner,M.O. (1996) Cloning and biochemical characterization of the cyclophilin homologues from the free-living nematode Caenorhabditis elegans. Biochem. J., 317, 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zydowsky L.D., Etzkorn,F.A., Chang,H.Y., Ferguson,S.B., Stolz,L.A., Ho,S.I. and Walsh,C.T. (1992) Active site mutants of human cyclophilin A separate peptidyl–prolyl isomerase activity from cyclosporin A binding and calcineurin inhibition. Protein Sci., 1, 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cardenas M.E., Lim,E. and Heitman,J. (1995) Mutations that perturb cyclophilin A ligand binding pocket confer cyclosporin A resistance in Saccharomyces cerevisiae. J. Biol. Chem., 270, 20997–21002. [DOI] [PubMed] [Google Scholar]
- 23.Kunz J. and Hall,M.N. (1993) Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem. Sci., 18, 334–338. [DOI] [PubMed] [Google Scholar]
- 24.Gottschalk A., Neubauer,G., Banroques,J., Mann,M., Lührmann,R. and Fabrizio,P. (1999) Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6·U5] tri-snRNP. EMBO J., 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens S.W. and Abelson,J. (1999) Purification of the yeast U4/U6·U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA, 96, 7226–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourquin J.P., Stagljar,I., Meier,P., Moosmann,P., Silke,J., Baechi,T., Georgiev,O. and Schaffner,W. (1997) A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res., 25, 2055–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mortillaro M.J. and Berezney,R. (1998) Matrin CYP, an SR-rich cyclophilin that associates with the nuclear matrix and splicing factors. J. Biol. Chem., 273, 8183–8192. [DOI] [PubMed] [Google Scholar]
- 28.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reidt U., Wahl,M.C., Fasshauer,D., Horowitz,D.S., Lührmann,R. and Ficner,R. (2003) Crystal structure of a complex between human spliceosomal cyclophilin H and a U4/U6 snRNP-60K peptide. J. Mol. Biol., in press. [DOI] [PubMed] [Google Scholar]