Abstract
An international effort is underway to generate a comprehensive haplotype map (HapMap) of the human genome represented by an estimated 300 000 to 1 million ‘tag’ single nucleotide polymorphisms (SNPs). Our analysis indicates that the current human SNP map is not sufficiently dense to support the HapMap project. For example, 24.6% of the genome currently lacks SNPs at the minimal density and spacing that would be required to construct even a conservative tag SNP map containing 300 000 SNPs. In an effort to improve the human SNP map, we identified 140 696 additional SNP candidates using a new bioinformatics pipeline. Over 51 000 of these SNPs mapped to the largest gaps in the human SNP map, leading to significant improvements in these regions. Our SNPs will be immediately useful for the HapMap project, and will allow for the inclusion of many additional genomic intervals in the final HapMap. Nevertheless, our results also indicate that additional SNP discovery projects will be required both to define the haplotype architecture of the human genome and to construct comprehensive tag SNP maps that will be useful for genetic linkage studies in humans.
INTRODUCTION
With the nearing completion of the human genome sequence, a number of studies have been initiated to identify natural genetic variation in human populations (1,2). Several large-scale projects have been conducted to identify single nucleotide polymorphisms (SNPs), including studies involving specific genes (3–6), chromosomes (7,8) and the whole genome (1,2). Two basic experimental strategies have been used to identify SNPs on a genome-wide scale. In the first approach, DNA from 24 diverse humans was pooled together and shotgun sequenced. The traces generated were compared with each other, or to finished genomic sequence, to identify SNPs. In the second approach, base substitutions were identified within the overlap regions of adjacent bacterial artificial chromosomes (BACs) that were used to sequence the human genome. Using a combination of these two methods, The SNP Consortium (TSC) developed an initial map of human genome variation containing 1.4 million SNPs (2). The human SNP map now has grown to ∼2.2 million non-redundant polymorphisms due to contributions from a number of laboratories (www.ncbi.nlm.nih.gov/SNP).
In addition to serving as a repository for human genetic variation, this collection of human SNPs will be useful for genetic linkage studies in humans. In fact, an international effort currently is underway to develop a comprehensive haplotype map (HapMap) of the human genome using these SNPs (9–11). The HapMap will greatly facilitate genetic linkage studies in humans by providing a genome-wide map of SNPs that are commonly inherited together as ‘haplotype blocks’. Tag SNPs representing these haplotype blocks then will be used to map traits to specific genomic intervals.
Haplotype architecture varies greatly across the human genome, with haplotype blocks ranging in size from <1 kb in length to >100 kb (9–14). Thus, it will be necessary to have very high SNP densities in some regions of the genome, but lower densities in others, in order to identify all of the common haplotype blocks in humans. Since we cannot determine in advance which regions of the genome will require high or low densities, it will be necessary to have uniformly high distributions across the genome at the onset of the project in order to identify all of the haplotype blocks. Conservative estimates indicate that 300 000 tag SNPs will be necessary to represent all of the common haplotype blocks in the human genome, corresponding to an average density of one tag SNP per 10 kb. Higher estimates indicate that as many as 1 million tag SNPs (or one tag SNP per 3 kb) may be required (11–14). These tag SNPs will be selected from a larger set of SNPs that define the haplotype architecture of the genome.
Although the current human SNP map (build 110) contains 2.2 million non-redundant SNPs, it is presently unclear as to whether the density and spacing of these SNPs across the human genome is sufficient to define the haplotype architecture of the genome. It is also unclear whether a sufficient number of tag SNPs could be selected from this collection to adequately represent all of the underlying haplotypes. We have studied the distribution of SNPs in the human SNP map (build 110), and have determined that there are 38 497 inter-SNP intervals that are >10 kb in length and 221 511 inter-SNP intervals that are >3 kb in length. Since even the most minimal tag SNP map will require an average spacing of one SNP per 10 kb, we conclude that the current SNP map is not sufficiently dense to support the HapMap project. In an effort to generate higher SNP densities in the largest gaps of the current SNP map, we conducted a new SNP discovery project, which has led to significant improvements in these regions. Nevertheless, our results indicate that additional SNP discovery projects will be required both to define the haplotype architecture of the human genome and to construct comprehensive tag SNP maps that will be useful for genetic linkage studies in humans.
MATERIALS AND METHODS
Computational pipeline for SNP identification
The 7.1 million trace files generated by TSC were obtained from Cold Spring Harbor Laboratory (2). The traces were trimmed using the VecScreen system developed by the National Center for Biotechnology Information (NCBI) together with a custom Perl trimming program that used the Phred quality scores (15) to trim traces upon encountering five bases in a row with quality scores below 25. The single longest, high quality interval from each trace then was selected for further analysis and all other data were set aside. We required that the average Phred score for the trimmed trace exceed a minimum of 25. The minimum trace length after trimming was 100 bases (due to our imposed limit), and the maximum was 905 bases, with an average of 346 bases. After the trimming step, the number of useful traces was reduced from 7 120 020 to 4 293 807 (60.3% of the original traces).
These trimmed traces were masked for known repeats using Repeatmasker and Maskeraid (16). The single longest unmasked interval within the trace then was used to map the trace with MegaBLAST (NCBI) and the Golden Path (June 2002 release) (17). We required a minimum of a 50 base match at 100% identity to a single location in the genome for a successful trace mapping. Traces that matched to more than one location, or that lacked at least a 50 base match at 100% identity, were eliminated from the analysis due to potential segmental duplications (18). Using this approach, 2 759 010 traces were successfully mapped to unique genomic locations (64.3% of the trimmed traces, equivalent to 980 655 789 bases, or ∼33% of the genome). The traces then were unmasked and aligned with their assigned genomic locations by pair-wise alignment, allowing for insertions and deletions of up to 16 bases in length using the program BL2Seq (NCBI). Sixty-five percent of the traces were completely identical to their assigned genomic locations within the Golden Path after unmasking and alignment, whereas another 35% of the traces contained sequence variations. Ninety-nine percent of these ‘variant’ traces had <10% variation from their assigned genomic locations. We have found that this collection of variant traces is an excellent resource for identifying a variety of DNA sequence polymorphisms, including SNPs, insertion/deletion polymorphisms and transposon polymorphisms (Fig. 2 and unpublished data).
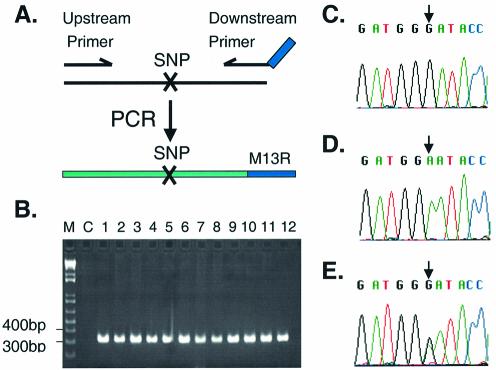
Figure 2.
Strategy for SNP validation. (A) The diagram shows the strategy used to amplify each SNP candidate by PCR. A recombinant PCR product is generated by introducing the M13R sequence into the downstream primer. (B) A gel showing PCR products generated for a typical SNP (SNP 9 is shown, generating a PCR product of 319 bp in length). Note the robust PCR products in lanes 1–12, and the absence of a band in the negative control lane (C). A 1 kb ladder marker is shown on the left (M). (C–E) Examples of chromatograms from SNP 9 (a ‘C’ is present at position 67 671 151 of chromosome 8 according to our predictions, whereas a ‘T’ is present in the Golden Path. The sequences shown are of the complementary strand. Arrows show the single nucleotide of interest. (C) Homozygous for our prediction based on the TSC trace sequence (human sample 3). (D) Homozygous for the Golden Path (human sample 6). (E) Heterozygous for our prediction and the Golden Path sequence (human sample 5).
A total of 709 492 SNPs were identified from the 7.1 million traces analyzed with this pipeline. We used a neighborhood quality assessment to call the final base differences. The SNP itself was required to have a minimum Phred score of 25, and all five bases on each side flanking the SNP were required to have minimum quality scores of 20. Of the 709 492 SNPs identified in our analysis, 568 796 (80.2%) were present in dbSNP (build 110) and, therefore, had been identified previously (2). An additional 140 696 SNPs were identified only by our analysis. These new SNPs were deposited into build 111 of dbSNP under accession numbers ss7844264 through ss7984919. TSC identified ∼1 million SNPs using the same 7.1 million traces (compared with 709 492 here). We did not expect to find all of the TSC SNPs because we aggressively trimmed the traces and discarded over half of the sequence information from those traces.
The distribution of our SNPs also was examined relative to genes in the human genome using the classification system established by dbSNP (www.ncbi.nlm.nih.gov/SNP). Build 113 of dbSNP, which included our SNPs, was used for this analysis. Overall, 70 046 of our 140 696 SNPs (49.7%) fell within ‘locus regions’ according to the dbSNP definition (i.e. fell within 3 kb of a gene or predicted gene in the upstream direction, or within 500 bp of a gene or predicted gene in the downstream direction). In comparison, 45.2% of all SNPs in the same build of dbSNP (113) fell within such regions. Our 70 046 ‘locus SNPs’ could be broken down further according to the locations of these SNPs within the genes. A total of 29 316 of our SNPs (or 40.5% of our locus SNPs) were located within 3 kb (upstream) or 500 bp (downstream) of a gene but were not located within the transcribed portion of the gene. In comparison, 36.9% of all locus SNPs in build 113 of dbSNP were found within this category. A total of 343 of our locus SNPs (0.47%) were predicted to cause synonomous changes within coding regions (compared with 1.0% of all locus SNPs in build 113 of dbSNP). A total of 513 of our locus SNPs (0.71%) were predicted to cause non-synonomous changes in coding regions (compared with 1.2% for all locus SNPs in build 113 of dbSNP). A total of 6676 (9.3%) of our locus SNPs were found within predicted untranslated regions (compared with 16.5% of all locus SNPs in build 113 of dbSNP). A total of 34 440 of our locus SNPs (47.7%) were located within predicted introns (compared with 47.7% for all locus SNPs in build 113 of dbSNP). Finally, a total of 13 of our locus SNPs (0.02%) fell within splice sites (compared with 0.02% for all locus SNPs within build 113 of dbSNP). Thus, the overall distribution of our SNPs relative to genes and other genomic features was remarkably similar to the distribution of all SNPs in the same build of dbSNP
Analysis of the SNP map
We examined the 2.2 million SNPs of dbSNP (build 110) that could be mapped to unique locations within the Golden Path (June 2002 release) (17). The SNP TSC (random read) and SNP NIH (overlap) tables were downloaded from the UC Santa Cruz website (www.genome.ucsc.edu) and combined into a single table. These tables then were merged with a third table containing the locations of all gaps in the genome (also obtained from the UC Santa Cruz website). A custom Perl program then was used to measure the intervals between all adjacent SNPs, excluding intervals caused by genomic gaps. The results were deposited into an Oracle database to assign intervals to size classes and then count each class (Table 1). Similar results were obtained using equivalent SNP tables obtained from dbSNP (www.ncbi.nlm.nih.gov/SNP/). Finally, this process was repeated after adding our new SNPs to assess the impact of our SNPs on the intervals (Table 2).
Table 1. Distribution of SNP intervals (intervals between adjacent SNPs) based on dbSNP (build 110).
| Class | Interval size (in kb) | No. of intervals | Total length (bp) | % of genome covered by this class |
|---|---|---|---|---|
| 0 | ≤0.1 | 540 376 | 22 982 827 | 0.76 |
| 1 | >0.1 and ≤0.5 | 728 703 | 187 697 440 | 6.17 |
| 2 | >0.5 and ≤1 | 330 788 | 237 817 929 | 7.82 |
| 3 | >1 and ≤3 | 399 275 | 689 502 930 | 22.67 |
| 4 | >3 and ≤5 | 105 666 | 405 154 007 | 13.32 |
| 5 | >5 and ≤10 | 77 348 | 533 016 204 | 17.52 |
| 6 | >10 and ≤50 | 37 046 | 618 629 083 | 20.34 |
| 7 | >50 and ≤500 | 1451 | 113 191 946 | 3.72 |
| Total | 2 220 653 | 2 807 992 366 | 92.32 |
Table 2. Distribution of 10–500 kb SNP intervals (intervals between adjacent SNPs) by chromosome.
| Chr | Current distribution | With our newly discovered SNPs | % Change | ||
|---|---|---|---|---|---|
| Number | Total length (bp) | Number | Total length (bp) | ||
| SNP Intervals of length >50 kb and ≤500 kb | |||||
| 1 | 78 | 5 585 535 | 12 | 844 883 | –84.87 |
| 2 | 106 | 7 505 978 | 15 | 1 125 383 | –85.01 |
| 3 | 76 | 5 537 160 | 13 | 1 161 678 | –79.02 |
| 4 | 146 | 11 323 424 | 16 | 1 217 843 | –89.24 |
| 5 | 115 | 8 779 105 | 17 | 1 272 405 | –85.51 |
| 6 | 110 | 9 053 122 | 13 | 911 330 | –89.93 |
| 7 | 63 | 4 910 285 | 12 | 1 343 660 | –72.64 |
| 8 | 136 | 10 621 236 | 31 | 2 306 466 | –78.28 |
| 9 | 30 | 2 223 391 | 10 | 900 842 | –59.48 |
| 10 | 37 | 2 606 920 | 8 | 632 838 | –75.72 |
| 11 | 42 | 3 025 765 | 9 | 706 986 | –76.63 |
| 12 | 48 | 3 804 568 | 11 | 979 539 | –74.25 |
| 13 | 22 | 1 601 924 | 3 | 186 953 | –88.33 |
| 14 | 8 | 516 358 | 1 | 50 299 | –90.26 |
| 15 | 27 | 1 866 030 | 7 | 467 171 | –74.96 |
| 16 | 39 | 3 143 940 | 13 | 1 067 630 | –66.04 |
| 17 | 25 | 1 634 730 | 8 | 494 328 | –69.76 |
| 18 | 30 | 2 421 414 | 4 | 375 489 | –84.49 |
| 19 | 9 | 743 099 | 8 | 665 629 | –10.43 |
| 20 | 0 | 0 | 0 | 0 | 0 |
| 21 | 0 | 0 | 0 | 0 | 0 |
| 22 | 0 | 0 | 0 | 0 | 0 |
| X | 202 | 15 497 366 | 111 | 7 670 345 | –50.51 |
| Y | 102 | 10 790 596 | 100 | 10 457 071 | –3.09 |
| Total | 1451 | 113 191 946 | 422 | 34 838 768 | –69.22 |
| SNP intervals of length >10 kb and ≤50 kb | |||||
| 1 | 2477 | 40 672 204 | 1 676 | 25 252 780 | –37.91 |
| 2 | 3453 | 56 757 325 | 2 514 | 37 698 436 | –33.58 |
| 3 | 2519 | 42 062 241 | 1 692 | 25 473 302 | –39.44 |
| 4 | 2822 | 47 874 122 | 1 828 | 27 109 984 | –43.37 |
| 5 | 1886 | 32 513 067 | 1 162 | 17 455 100 | –46.31 |
| 6 | 2008 | 33 560 739 | 1 374 | 20 408 992 | –39.19 |
| 7 | 2041 | 32 910 970 | 1 552 | 23 218 090 | –29.45 |
| 8 | 1835 | 32 010 697 | 1 093 | 16 755 825 | –47.66 |
| 9 | 1569 | 25 556 884 | 1 164 | 18 030 068 | –29.45 |
| 10 | 1688 | 27 772 540 | 1 158 | 17 701 341 | –36.26 |
| 11 | 1332 | 21 844 811 | 871 | 13 007 496 | –40.45 |
| 12 | 1634 | 26 718 713 | 1 233 | 18 639 999 | –30.24 |
| 13 | 1152 | 18 687 944 | 783 | 11 638 775 | –37.72 |
| 14 | 1148 | 17 969 446 | 950 | 14 359 234 | –20.09 |
| 15 | 1079 | 17 676 577 | 727 | 10 902 358 | –38.32 |
| 16 | 1018 | 17 378 709 | 683 | 10 903 977 | –37.26 |
| 17 | 1064 | 16 989 104 | 765 | 11 554 814 | –31.99 |
| 18 | 696 | 11 233 681 | 416 | 5 808 116 | –48.30 |
| 19 | 807 | 12 880 609 | 667 | 10 406 368 | –19.21 |
| 20 | 766 | 11 268 333 | 653 | 9 430 374 | –16.31 |
| 21 | 133 | 1 767 854 | 118 | 1 571 726 | –11.09 |
| 22 | 332 | 5 040 570 | 276 | 4 201 143 | –16.65 |
| X | 3216 | 60 143 734 | 2 758 | 49 583 696 | –17.56 |
| Y | 371 | 7 338 209 | 367 | 7 196 772 | –1.93 |
| Total | 37 046 | 618 629 083 | 26 480 | 408 308 766 | –34.00 |
Analysis of SNPs by PCR and DNA sequencing
Primer pairs were designed to amplify each SNP using the flanking DNA sequences. In each case, primers were designed to have melting temperatures above 64°C. The M13 Reverse (M13R) primer sequence (5′-CAGGAAACAGCTATGACC-3′) was added to the 5′-end of the downstream primer such that this sequence was incorporated into the PCR product to generate a recombinant template for sequencing. PCR products were amplified from the first 12 human DNAs from the Coriell diversity panel (19), and these 12 PCR products were sequenced using M13R BigDye Primer kits from Applied Biosystems (version 1.0 and version 3.0). The PCR products were diluted in water 1:4 before sequencing, and 1.0 µl of DNA was used as a template. Upon completion of the cycling protocol recommended by Applied Biosystems, the four dye primer sequencing reactions were pooled together and precipitated with ethanol. The dried precipitates were resuspended in formamide and run on an ABI 3100 Genetic Analyzer. If the SNP was not verified in the first 12 samples, then the remaining 12 samples were sequenced.
RESULTS
In order to examine the distribution and spacing of human SNPs across the genome, we measured the distances between all adjacent SNPs in the human SNP map (build 110), and then classified the observed intervals by size (Table 1). We found a wide degree of variation in the distances between adjacent SNPs, ranging from 1 to 427 780 bases. A large number of intervals were greater than the 10 kb average spacing that will be necessary to construct even a minimal tag SNP map containing 300 000 SNPs (Table 1). For example, 1451 SNP intervals were 50–500 kb in length, and 37 046 SNP intervals were 10–50 kb in length (Table 1). In fact, we found a total of 38 497 intervals (occupying 24.6% of the human genome) that currently lack SNPs at the minimal 10 kb average spacing required for a tag SNP map of 300 000 SNPs. Moreover, 221 511 intervals (occupying 54.9% of the genome) currently fall below a density of one SNP per 3 kb (the density required for a tag SNP map of 1 million SNPs; Table 1). Therefore, between 24.6 and 54.9% of the human genome currently lacks SNPs at the minimal densities that are estimated to be required to construct tag SNP maps containing 300 000 and 1 million SNPs, respectively.
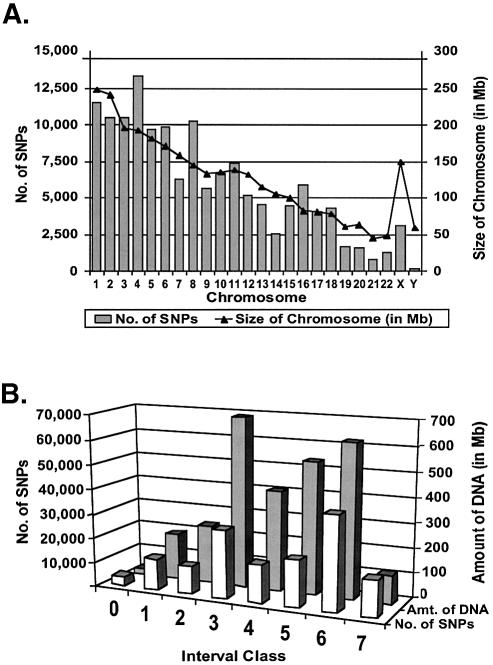
In an effort to generate higher SNP densities in the largest gaps of the current SNP map, we re-mined the original 7 million traces that were generated by The SNP Consortium (TSC) using a new computational pipeline (see Materials and Methods), and identified 140 696 additional SNP candidates that had not been identified previously. These SNP candidates were distributed on all 24 chromosomes (Fig. 1A), and also fell within genes at a frequency that was very similar to the frequency that all SNPs in dbSNP fell within genes (Fig. 1A and Materials and Methods). A significant fraction of these SNPs were located within the largest SNP gaps of the genome (Fig. 1B). For example, 14 253 of our SNP candidates mapped to the 1451 largest SNP ‘deserts’ of the human genome (ranging from 50 to 500 kb in length; Fig. 1B, class 7). As a result of adding our SNP candidates to the map, 1029 (69.2%) of these largest gaps were reduced to intervals that were smaller than 50 kb in length, and this group now has an average size that is <10 kb (Fig. 1 and Table 2). In fact, 80–90% of the largest gaps were eliminated on chromosomes 1, 2, 4, 5, 6, 13, 14 and 18 (Table 2). Another 37 517 of our SNP candidates (20.3%) mapped to the second-largest class of intervals (10–50 kb in length; Fig. 1B, class 6). The addition of these SNPs to the map resulted in a ∼48% reduction of the 10–50 kb intervals on chromosomes 8 and 18, and led to a 34% genome-wide reduction in 10–50 kb intervals overall (Table 2). Therefore, over 51 000 of our SNPs mapped to the two largest interval groups (50–500 and 10–50 kb), and yielded significant reductions in the amount of DNA contained within these groups.
Figure 1.
Analysis of 140 696 new SNP candidates. (A) The bar graph depicts the genomic distribution of our 140 696 SNP candidates by chromosome. Note that the SNPs are distributed on all 24 chromosomes proportional to the amount of DNA (indicated by a line above the bar graph). The possible exceptions are the X and Y chromosomes, which have fewer SNPs than average per kb of DNA. The distribution of our SNPs also was examined relative to genes (see Materials and Methods). Overall, 49.7% of our 140 696 SNPs fell within or near genes (defined as being within 3 kb of a gene or predicted gene in the upstream direction, or within 500 bp of a gene or predicted gene in the downstream direction). In comparison, 45.2% of all SNPs in the equivalent build of dbSNP (build 113) fell within such regions using the same criteria. Thus, the overall distribution of our SNPs relative to genes was highly similar to the overall distribution of all SNPs in the same build of dbSNP. (B) The row of white bars (front) shows the number of SNPs from our collection in interval classes 0 through to 7 (defined in Table 1). The row of black bars (back) depicts the amount of DNA (in Mb) contained within each class before adding our SNPs to the map (listed in Table 1 under ‘Total length’ column). Note that our SNPs occur in all classes, and are generally proportional to the amount of DNA in each class. The 51 770 SNPs in classes 6 and 7 close many of the largest gaps in the SNP map, and the 85 757 SNPs in classes 4–7 are likely to be immediately useful for construction of the HapMap. The SNPs in classes 0–3 may be useful in cases where the allelic frequencies of existing SNPs are unfavorable.
It is noteworthy that chromosomes 20, 21 and 22 lacked 50–500 kb gaps altogether, presumably due to SNP discovery projects that were focused on these chromosomes (7,8,12). In contrast, both the X and Y chromosomes had many more gaps than the average chromosome, and also had a significant number of gaps that were refractory to closure by our SNPs (Table 2). This also was the trend with SNPs identified by TSC (2) and others (11), presumably due to the smaller effective population sizes and different mutation rates of these chromosomes. Natural selection (both positive and negative), and the lower recombination rates of these chromosomes, also may have contributed to the creation of SNP deserts on these chromosomes.
Although the two largest interval groups discussed above (50–500 and 10–50 kb) are the most important with respect to closing gaps in the SNP map, we also identified SNPs that will be useful for identifying smaller haplotype blocks in the genome (<10 kb). Indeed, up to half of the haplotype blocks identified in previous studies were found to be <3 kb in length, and a large number of blocks were in the 3–10 kb range as well (12). Many of our SNPs mapped to these smaller intervals. For example, 18 760 of our SNPs mapped to 5–10 kb intervals (Fig. 1B, class 5), and another 15 197 mapped to 3–5 kb intervals (Fig. 1B, class 4). A total of 85 757 (61.0%) of our 140 696 SNP candidates mapped to intervals that were >3 kb in length and thus are likely to be immediately useful for the construction of the HapMap. The remaining SNP candidates (54 969 or 39.1%) mapped to genomic regions containing SNP densities more than one SNP per 3 kb. Although these more densely populated regions already contain a large number of SNPs, our SNPs may be useful in cases where other SNPs in the region have low allelic frequencies.
In order to measure the accuracy of our SNP predictions, 30 SNP candidates were chosen randomly from the collection of 140 696 and examined further. If our predictions were accurate for a given SNP, then we expected to be able to identify that SNP in at least one of the original 24 people that were used to generate the 7.1 million TSC traces (2). If, on the other hand, the SNP was not confirmed in at least one of the 24 individuals, then we would know with certainty that our bioinformatics prediction was incorrect. To perform these validation studies, each SNP candidate was amplified by PCR in 12–24 of the original 24 humans and individually sequenced. Twenty-nine of the thirty SNP candidates examined were confirmed by this analysis for a confirmation rate of 96.7% (Fig. 2 and Table 3). This is comparable with the overall verification rate of 95% obtained by TSC (2), indicating that our pipeline is highly accurate.
Table 3. SNP verification by PCR and sequencing.
| SNP | Genomic location | Trace base | Golden path base | Alleles sequenced | % Trace | % Golden path |
|---|---|---|---|---|---|---|
| 1 | chr8: 138543564 | t | g | 22 | 95.0 | 5.0 |
| 2 | chr11: 2786593 | g | a | 24 | 83.0 | 17.0 |
| 3 | chr4: 173806221 | c | g | 24 | 37.5 | 62.5 |
| 4 | chr4: 140987424 | a | g | 18 | 94.0 | 6.0 |
| 5 | chr8: 117177669 | t | c | 22 | 77.0 | 23.0 |
| 6 | chr18: 39493166 | g | t | 44 | 100.0 | 0.0 |
| 7 | chr6: 85928202 | a | g | 22 | 9.0 | 91.0 |
| 8 | chr3: 51223303 | t | c | 22 | 91.0 | 9.0 |
| 9 | chr8: 67671151 | c | t | 20 | 50.0 | 50.0 |
| 10 | chr8: 27015545 | t | c | 24 | 67.0 | 33.0 |
| 11 | chr4: 52910312 | a | g | 24 | 75.0 | 25.0 |
| 12 | chr4: 44538473 | c | t | 42 | 100.0 | 0.0 |
| 13 | chr5: 177158144 | a | g | 24 | 21.0 | 79.0 |
| 14 | chr15: 84691055 | c | t | 22 | 23.0 | 77.0 |
| 15 | chr3: 186142005 | g | a | 22 | 59.0 | 41.0 |
| 16 | chr3: 99873851 | g | a | 22 | 32.0 | 68.0 |
| 17 | chr4: 3649237 | a | g | 24 | 54.0 | 46.0 |
| 18 | chr15: 55487474 | t | c | 24 | 96.0 | 4.0 |
| 19 | chr1: 178793436 | g | a | 10 | 40.0 | 60.0 |
| 20 | chr11: 124193407 | t | g | 24 | 37.5 | 62.5 |
| 21 | chr16: 16356684 | g | a | 22 | 0.0 | 100.0 |
| 22 | chr20: 11220172 | g | t | 22 | 27.0 | 73.0 |
| 23 | chr2: 19825504 | a | t | 18 | 28.0 | 72.0 |
| 24 | chr18: 38060888 | g | a | 18 | 83.0 | 17.0 |
| 25 | chr19: 36651599 | g | a | 22 | 32.0 | 68.0 |
| 26 | chr1: 12919864 | c | t | 14 | 21.0 | 79.0 |
| 27 | chr5: 58480456 | g | a | 24 | 17.0 | 83.0 |
| 28 | chr1: 186077564 | g | a | 22 | 100.0 | 0.0 |
| 29 | chr2: 29123555 | g | a | 22 | 100.0 | 0.0 |
| 30 | chr10: 56311213 | t | c | 24 | 58.0 | 42.0 |
A range of allelic frequencies was observed among the SNPs sequenced in our validation study (Table 3). Interestingly, four of the SNPs in the study were present in all individuals examined except for the person(s) represented by the Golden Path sequence (Table 3). The most likely explanation for these results is that the person(s) represented by the Golden Path sequence had rare ‘private’ alleles at those positions that were absent from the majority of humans. Overall, 83% of the SNPs examined in our validation study had minor allelic frequencies that were >5% (Table 3). This fraction is similar to estimates obtained previously by TSC and others (2), indicating that our SNPs will be equally useful for mapping studies.
DISCUSSION
Since the HapMap project is rapidly ramping up to meet the goal of completion within 3 years, it is desirable at this stage to identify all of the SNPs that will be necessary to meet the demands of the project. Our SNP discovery pipeline, which has several notable differences from previous pipelines (see below), has allowed us to discover a novel collection of human SNPs that will be immediately useful for the HapMap project. The addition of our SNP candidates to the current SNP map has improved the most problematic areas of the map, and has been particularly useful in the 24% of the genome that (prior to our study) contained SNP intervals of 10–500 kb in length. We have contributed over 51 000 SNPs to these largest intervals and, as a consequence, have reduced the amount of DNA contained in these gaps by 34 (10–50 kb gaps) to 69% (50– 500 kb gaps) genome-wide. Therefore, these SNPs will allow for the inclusion of many additional genomic intervals in the final HapMap.
Nevertheless, our efforts did not completely close all of the gaps in the SNP map, and our analysis indicates that additional SNP discovery projects should be launched now to meet the demands of the HapMap project (Table 2). Ideally, a uniformly dense SNP map would be generated now in order to ensure an adequate supply of SNPs as each region of the HapMap is developed. For example, global SNP discovery methods such as shotgun re-sequencing could be extended such that every region of the genome exceeds a minimum SNP density (one SNP per 1–3 kb?). Such densities may not be required throughout the genome; however, up to half of the haplotype blocks in the genome will be missed at lower SNP densities (12). Having more SNPs at the beginning of the project also will ensure that the final HapMap contains SNPs that have more desirable allelic frequencies and are technically easier to genotype. An alternative approach would be to construct an initial draft of the HapMap using all currently available SNPs, followed by a gap closure phase in which additional SNPs are identified as necessary. Irrespective of the specific strategy that is chosen, however, additional SNPs will be required to complete the goals of the project.
There are several key differences between our pipeline and previous pipelines that allowed us to discover new SNPs in the TSC traces. First, 13 different methods were used previously to identify SNPs in the TSC traces, and no single method was applied to all of the traces uniformly (2). In contrast, we processed all 7.1 million traces using only a single pipeline (see Materials and Methods). Since some of the 13 methods used previously compared only the traces to each other, our analysis represents the first time that some of these traces were compared with the draft sequence for the purpose of cataloging SNPs. Secondly, we used only the largest, high quality segment from each trace, which allowed us to include insertions and deletions (INDELs) in our analysis. The inclusion of INDELs facilitated SNP discovery in traces that otherwise would have been set aside (TSC discarded traces with <99% match). Thirdly, our procedure for mapping traces was different from methods employed previously and allowed accurate mapping while still retaining flexibility in the subsequent pair-wise alignment step. We required a minimum of a 50 base match at 100% identity to a single location in the genome for a successful trace mapping. In contrast, many of the previous methods used the entire trace for mapping and required a match of >99%. Some of the methods also set aside traces that contained >50% repetitive sequences; however, our mapping methods allowed us to utilize many of these traces successfully. Fourthly, because a number of gaps in the human genome sequence have been closed recently, we were able to map traces for the first time to these segments of the genome. Since our collection of SNPs mapped to all SNP interval sizes, however, only a fraction of the SNPs discovered in our study can be attributed to such gaps (Table 2). Therefore, the success of our pipeline is due to a combination of the factors listed above.
In conclusion, the current human SNP map will serve as an excellent resource to fuel the construction of an initial ‘draft’ of the human HapMap. Although the addition of our new SNPs to the human SNP map led to significant improvements in the largest gaps of the map, additional SNP discovery projects will be required to fully support the HapMap project. Higher SNP densities will be necessary to fully discover the haplotype architecture of the genome and to construct comprehensive tag SNP maps that will be useful for genetic linkage studies in humans.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by Research Scholar Grant RSG-01-173-01-MBC from the American Cancer Society (S.E.D.), the Woodruff Foundation (S.E.D.), and the Emory University Research Council (S.E.D.). We thank Shari Corin for helpful discussions and critical review of the manuscript. We thank the people at TSC, dbSNP and UC Santa Cruz for the use of their data and for helpful advice.
REFERENCES
- 1.Altshuler D., Pollara,V.J., Cowles,C.R., Van Etten,W.J., Baldwin,J., Linton,L. and Lander,E.S. (2000) An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature, 407, 513–516. [DOI] [PubMed] [Google Scholar]
- 2.The International SNP Map Working Group (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 3.Nickerson D.A., Taylor,S.L., Weiss,K.M., Clark,A.G., Hutchinson,R.G., Stengard,J., Salomaa,V., Vartiainen,E., Boerwinkle,E. and Sing,C.F. (1998) DNA sequence diversity in a 9.7-kb region of the human lipoprotein lipase gene. Nature Genet., 19, 216–217. [DOI] [PubMed] [Google Scholar]
- 4.Rieder M.J., Taylor,S.L., Clark,A.G. and Nickerson,D.A. (1999) Sequence variation in the human angiotensin converting enzyme. Nature Genet., 22, 59–62. [DOI] [PubMed] [Google Scholar]
- 5.Taillon-Miller P., Piernot,E.E. and Kwok,P.Y. (1999) Efficient approach to unique single-nucleotide polymorphism discovery. Genome Res., 9, 499–505. [PMC free article] [PubMed] [Google Scholar]
- 6.Taillon-Miller P. and Kwok,P.Y. (2000) A high-density single-nucleotide polymorphism map of Xq25-q28. Genomics, 65, 195–202. [DOI] [PubMed] [Google Scholar]
- 7.Mullikin J.C., Hunt,S.E., Cole,C.G., Mortimore,B.J., Rice,C.M., Burton,J., Matthews,L.H., Pavitt,R., Plumb,R.W., Sims,S.K. et al. (2000) An SNP map of human chromosome 22. Nature, 407, 516–520. [DOI] [PubMed] [Google Scholar]
- 8.Dawson E., Chen,Y., Hunt,S., Smink,L.J., Hunt,A., Rice,K., Livingston,S., Bumpstead,S., Bruskiewich,R., Sham,P. et al. (2001) A SNP resource for human chromosome 22: extracting dense clusters of SNPs from the genomic sequence. Genome Res., 11, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly M.J., Rioux,J.D., Schaffner,S.F., Hudson,T.J. and Lander,E.S. (2001) High resolution haplotype structure in the human genome. Nature Genet., 29, 229–232. [DOI] [PubMed] [Google Scholar]
- 10.Reich D.E., Cargill,M., Bolk,S., Ireland,J., Sabeti,P.C., Richter,D.J., Lavery,T., Kouyoumjian,R., Farhadian,S.F., Ward,R. et al. (2001) Linkage disequilibrium in the human genome. Nature, 411, 199–204. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel S.B., Schaffner,S.F., Nguyen,H., Moore,J.M., Roy,J., Blumenstiel,B., Higgins,J., DeFelice,M., Lochner,A., Faggart,M., et al. (2002) The structure of haplotype blocks in the human genome. Science, 296, 2225–2229. [DOI] [PubMed] [Google Scholar]
- 12.Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,H.H., Marjoribands,C., McDonough,D.P. et al. (2001) Blocks of limited haplotype diversity revealed by high resolution scanning of human chromosome 21. Science, 294, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 13.Stephens J.C., Schneider,J.A., Tanguay,D.A., Choi,J., Acharya,T., Stanley,S.E. et al. (2001) Haplotype variation and linkage disequilibrium in 313 human genes. Science, 293, 489–493. [DOI] [PubMed] [Google Scholar]
- 14.Judson R., Salisbury,B., Schneider,J., Windemuth,A. and Stephens,J.C. (2002) How many SNPs does a genome-wide haplotype map require? Pharmacogenomics, 3, 279–391. [DOI] [PubMed] [Google Scholar]
- 15.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res., 8, 186–194. [PubMed] [Google Scholar]
- 16.Bedell J.A., Korf,I. and Gish,W. (2000) MaskerAid: a performance enhancement to repeatMasker. Bioinformatics, 16, 1040–1041. [DOI] [PubMed] [Google Scholar]
- 17.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey J.A., Gu,Z., Clark,R.A., Reinert,K., Samonte,R.V., Schwartz,S., Adams,M.D., Myers,E.W., Li,P.W. and Eichler,E.E. (2002) Recent segmental duplications in the human genome. Science, 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 19.Collins F.S., Brooks,L.D. and Chakravarti,A. (1999) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res., 8, 1229–1231. [DOI] [PubMed] [Google Scholar]