Abstract
CSP41 is a ubiquitous chloroplast endoribonuclease belonging to the short chain dehydrogenase/reductase (SDR) superfamily. To help elucidate the role of CSP41 in chloroplast gene regulation, the mechanisms that determine its substrate recognition and catalytic activity were investigated. A divalent metal is required for catalysis, most probably to provide a nucleophile for cleavage 5′ to the phosphodiester bond, and may also participate in cleavage site selection. This requirement distinguishes CSP41 from other Rossman fold-containing proteins from the SDR superfamily, including several RNA-binding proteins and endonucleases. CSP41 is active only in the presence of MgCl2 and CaCl2. Although Mg2+- and Ca2+-activated CSP41 cleave at identical sites in the single-stranded regions of a stem–loop-containing substrate, Mg2+-activated CSP41 was also able to cleave within the double-stranded region of the stem–loop. Mixed metal experiments with Mg2+ and Ca2+ suggest that CSP41 contains a single divalent metal-binding site which is non-selective, since Mn2+, Co2+ and Zn2+ compete with Mg2+ for binding, although there is no activity in their presence. Using site-directed mutagenesis, we identified three residues, Asn71, Asp89 and Asp103, which may form the divalent metal-binding pocket. The activation constant for Mg2+ (KA,Mg = 2.1 ± 0.4 mM) is of the same order of magnitude as the stromal Mg2+ concentrations, which fluctuate between 0.5 and 10 mM as a function of light and of leaf development. These changes in stromal Mg2+ concentration may regulate CSP41 activity, and thus cpRNA stability, during plant development.
INTRODUCTION
In chloroplasts, gene expression is often regulated at the post-transcriptional level, particularly through mechanisms influencing RNA stability. These mechanisms are regulated by light and during leaf development. Most chloroplast mRNAs contain an inverted repeat in their 3′ untranslated region (UTR), which can fold into a stable stem–loop structure. These stem–loops constitute recognition sites for nucleus-encoded RNA-binding proteins (1–3). After removal or disruption of the stem–loop by endonucleases, transcripts become unstable as they enter a degradation pathway that includes polyadenylation and subsequent 3′ to 5′ exonucleolytic degradation (4). In light of the importance of the initial endonucleolytic cleavage of the stem–loop as a committed and potentially regulatory step in this pathway, understanding the specificities and activities of chloroplast endoribonucleases has become of increasing importance. Several endoribonuclease activities that cleave within the coding and/or untranslated regions have been observed in plant chloroplasts. However, the identities and specific catalytic properties of these enzymes mostly remain to be elucidated (5–8).
The most well studied endoribonuclease in chloroplasts has been CSP41 (9–12). CSP41 lacks known ribonuclease motifs, but belongs to the short chain dehydrogenase/reductase (SDR) superfamily (11). As such, CSP41 is predicted to contain an SDR Rossman fold domain. Three other members of the dehydrogenase family, glyceraldehyde phosphate dehydrogenase (GAPDH), and two dehydrogenases from the archaeon Solfolobus solfataricus, have been shown to have endoribonucleolytic activity and to cleave within the secondary structure of phage T7 R1.1 RNA (13). However, unlike CSP41, these enzymes do not require a divalent metal for their RNase activity (10). Two other Rossman fold-containing proteins are the regulatory proteins CheY and mSlo1. Both of these proteins contain low affinity carboxylate cluster-type binding sites, which have dissociation constants for Mg2+ that are close to the physiological Mg2+ concentration (14,15). In chloroplasts, recent attention has been directed at the variation in stromal Mg2+ concentration as a function of light and of leaf development (16,17). The results suggest that Mg2+ may play a regulatory role in the chloroplast, and probably in RNA stability, by modulating the activity of enzymes such as CSP41.
CSP41 was shown to cleave primarily within the stem–loop structures of several chloroplast RNA 3′ UTR substrates in vitro (10,12). Because stem–loop structures are known to be important for chloroplast mRNA stability, CSP41 was hypothesized to play a role in the initiation of RNA degradation. In order to characterize the divalent metal-binding site on CSP41 and divalent metal-dependent catalysis by the SDR Rossman fold, and to begin to understand how Mg2+ activation of CSP41 may play a regulatory role in its function in vivo, we sought to understand the mechanistic details of RNA cleavage by this new type of endoribonuclease.
MATERIALS AND METHODS
Purification and CSP41 assay
Recombinant CSP41 was purified to >95% homogeneity from Escherichia coli SG13009 as previously described (9). The enzyme was assayed according to Yang and Stern (10).
Preparation of synthetic RNA substrates
Templates encoding the petD 3′ UTR RNA substrates were contained between the SacI and EcoRV sites of pBluescript KS (–). Synthetic RNAs were prepared according to Stern and Gruissem (18) after linearizing templates with HindIII. Substrates for CSP41 assays were synthesized with 2.5 µM [α-32P]UTP (3000 Ci/mmol) and 25 µM cold UTP. Substrates for footprinting were synthesized in the presence of 1 mM cold rNTPs and end labeled with [γ-32P]ATP according to Yang and Stern (10). Lengths of RNAs were petDΔ3 224 nt, and petDΔ18 179 nt. Secondary structures for petDΔ18 and petDΔ3 are based on predictions by energy minimization using RNA Structure 3.7 (19) and experimental mapping experiments described elsewhere (12).
Divalent metal activation
Cleavage of the petDΔ3 RNA substrate was measured in the presence of 20 mM MgCl2, CaCl2, MnCl2, CoCl2 or ZnCl2. To measure the rate as a function of Mg2+ concentration, CSP41 was incubated in varying concentrations of MgCl2 and aliquots were taken at 0.5 and 10 min. Each data point was replicated and the average rate was plotted. The rate of reaction versus Mg2+ concentration was plotted using Kaleidagraph (Synergy Software, Perkionen, PA) and fit to the Michaelis–Menten equation:
where v is the initial rate of the CSP41-catalyzed reaction in units per milligram of enzyme (U/mg). One unit of CSP41 activity is defined as 1 nmol petD RNA cleaved/ml/min/mg of CSP41. The concentration of metal is represented by [S], KA,Mg is the concentration of Mg2+ giving a half-maximal rate of petD RNA degradation. Vmax is the maximal rate of petD RNA degradation.
Divalent metal competition
CSP41 was assayed as described above, except that the concentration of MgCl2 was held at 2 mM. The assay was supplemented with 20 mM CaCl2, MnCl2, CoCl2 or ZnCl2.
RNA binding assays
Gel mobility shift assays were performed in a 20 µl sample containing 20 mM HEPES pH 7.5, 10% glycerol, 5 µg of CSP41 and varying concentrations of uniformly labeled petDΔ3. The reaction was incubated at room temperature for 10 min, treated with 40 U of RNase T1 at room temperature for 15 min, and electrophoresed in a 5% Tris–glycine native polyacrylamide gel. The gel was then visualized on a Storm Scanner (Molecular Dynamics). The resulting data were fit to a hyperbolic binding isotherm:
In this equation, I is the intensity of the shifted band as measured on the Storm Scanner, Imax is the intensity of the shifted band at saturating ligand concentrations, [L] is the concentration of ligand and Kd is the concentration of ligand giving 50% maximum intensity of the shifted band.
Mechanism of cleavage
CSP41 was incubated in the presence of 20 mM MgCl2 and petDΔ3 RNA, which was either unlabeled or uniformly labeled with [α-32P]UTP. Following the reaction, each sample was phenol/chloroform extracted and ethanol precipitated. The samples were then resuspended in water, treated with 1 U of calf intestinal alkaline phosphatase for 10 min at 37°C, and boiled to inactivate the enzyme. The samples were labeled with 1 U of polynucleotide kinase and [γ-32P]ATP for 30 min at 37°C. The samples were then extracted with phenol/chloroform, ethanol precipitated, and analyzed in 5% denaturing polyacrylamide gels.
Site-directed mutagenesis
CSP41 mutants were created using the Quickchange kit according to the manufacturer’s instructions (Stratagene, La Jolla, CA), using the following oligonucleotides (5′ to 3′): aagacggtctggggaGCTcctgcagacattggg (N71A), tggggaaatcctgcaGCTattgggaatgta (D74A), aatgtagttggaggaGCTgcctttGCTgtggtgctagataac (E82A/D85A), tttgatgtggtgctaGCTaac aacgggaaagac (D89A), gataacaacgggaaaGCTcttgctactgtcagc (D94A), gtcagccctgtggtaGCTtgggcaaagagttca (D103A), gaacctcctcatattGCTgggGCTgctgttaaatctagt (E132A/D134A) and tcccacgttgcagtaGCTgactacattgctaaa (146A), with mutated codons indicated by uppercase letters. Proteins were expressed in E.coli SG13009 and purified as described above.
RESULTS
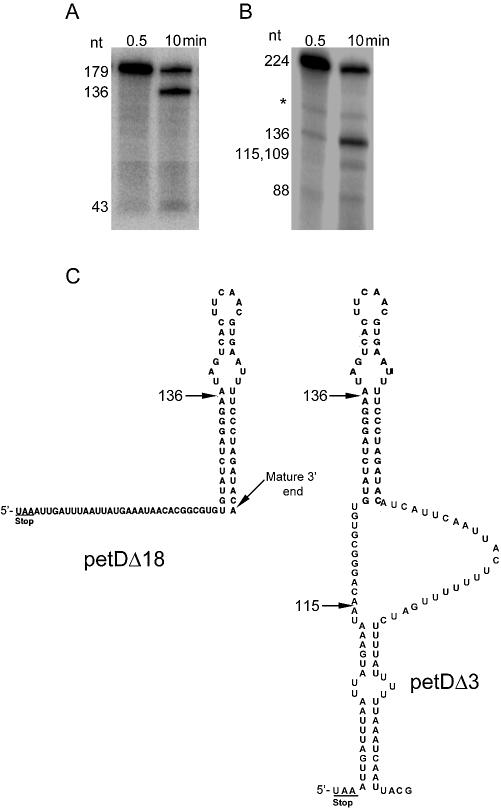
CSP41 cleaves primarily within RNA stem–loops, such as that of the petD 3′ UTR. petD is a chloroplast gene encoding subunit IV of the cytochrome b6/f complex, which is essential for photosynthesis. To establish CSP41 cleavage activity, the enzyme was assayed in the presence of either petDΔ18 or petDΔ3 RNA and 20 mM MgCl2 (Fig. 1A and B, respectively). petDΔ18 corresponds to the mature form of the petD mRNA 3′ UTR, whereas petDΔ3 corresponds to the petD precursor RNA 3′ UTR (Fig. 1C). All cleavage positions will be referred to relative to the 5′ end of the synthetic petD transcript.
Figure 1.
(A and B) Cleavage of petDΔ18 (A) or petDΔ3 RNA (B) by CSP41. A 0.5 µg aliquot of recombinant CSP41 was incubated with 20 fmol petD RNA in buffer containing 20 mM MgCl2. Samples were removed at the indicated times and analyzed in a 5% denaturing polyacrylamide gel. (C) The spinach petD 3′ UTR substrates used in this study. Numbering begins at the 5′ end of the in vitro transcript. The petD stop codon, major cleavage sites of CSP41 and the mature petD 3′ end are indicated. Secondary structures are based on folding predictions (RNA Structure 3.7) and structural mapping experiments.
Following a 10 min reaction, several bands are visible. The 179 nt band in Figure 1A corresponds to the petDΔ18 substrate, and the primary cleavage at position 136 generates 136 and 43 nt products. In Figure 1B, the 224 nt band corresponds to the petDΔ3 substrate. With petDΔ3, there are two sets of cleavage products. The primary cleavage at position 136 generates 136 and 88 nt products. A secondary cleavage at position 115 generates 115 and 109 nt products, which are not resolved in Figure 1B. A band of ∼165 nt was often present in the reaction products, but was also present in controls lacking CSP41. Therefore, some or all of this is a non-enzymatically generated fragment. The positions of CSP41 cleavage are indicated on the theoretically and experimentally determined structures shown in Figure 1C.
Although previous work established the general properties of the endonuclease activity of CSP41 (10), and we were aware that the enzyme was inactive in the absence of divalent metals, the specificity of divalent metal activation had not been formally addressed. Structure–function aspects of CSP41 cannot be understood without dissecting the divalent metal requirement, for which two roles can be envisaged. These are the facilitation of RNA substrate binding to the enzyme, and/or a requirement during catalysis to aid in cleavage of the phosphodiester backbone.
To distinguish between these possibilities, we first tested whether divalent metals were required for binding of petDΔ3 RNA, as shown in Figure 2. CSP41 was incubated with increasing amounts of RNA in the absence of divalent metals, and the formation of a binary CSP41–petDΔ3 complex was monitored by a gel mobility shift assay (inset). The intensity of the band corresponding to the binary CSP41–petDΔ3 complex increased with RNA concentration, and the quantified data were fit to a hyperbolic binding isotherm. The hyperbolic binding curve suggests that the RNA is binding to a specific site on the enzyme. The data in Figure 2 yielded a dissociation constant of 51 ± 0.6 pM for the binary interaction, where the dissociation constant is defined as the concentration of petDΔ3 RNA giving half-maximal saturation of the CSP41 apoenzyme. Thus, CSP41 has a relatively high affinity for the petDΔ3 substrate, comparable with those of other site-specific RNA-binding proteins (20,21). Measurements in the presence of saturating Mg2+ were not done, because the enzyme would have consumed the RNA before complex formation could be analyzed. This RNA binding is most likely to be mediated through the SDR Rossman fold, as observed with other enzymes containing this motif (22). In conclusion, because Mg2+ was not required for RNA binding, the divalent metal must be required for catalysis.
Figure 2.
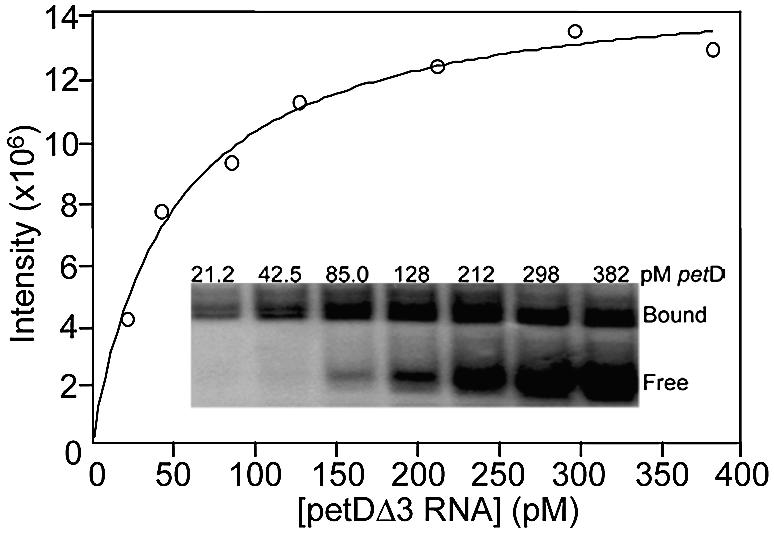
CSP41 binds to petD RNA in the absence of divalent metals. Inset: gel mobility shift assay of petDΔ3 binding by CSP41. Uniformly labeled petDΔ3 RNA at the indicated concentrations was incubated with 5 µg of CSP41. The samples were then treated with 40 U of RNase T1 and analyzed in a 5% native polyacrylamide gel. Reactions were performed in duplicate. The intensity of the shifted band from the gel mobility shift assay was plotted versus the concentration of petDΔ3 RNA. The error in intensity at each data point was typically <5%. The curve represents the best fit to equation 2 (see Materials and Methods).
Although ribonucleases can cleave on either the 5′ or 3′ side of phosphodiester bonds (23), endonucleases that require divalent metals for catalysis generally cleave 5′ to the phosphodiester bond and generate 3′ hydroxyl-terminated products. Because the initiating step in chloroplast mRNA degradation is believed to be endonucleolytic cleavage within the 3′ stem–loop structure and/or the coding region, followed by polyadenylation of the 3′ hydroxyl groups of proximal endonucleolytic cleavage products by polynucleotide phosphorylase (PNPase) (4), it was essential to determine whether CSP41 could generate 3′ hydroxyl groups. If this were the case, it could be argued that CSP41 might initiate cpRNA degradation.
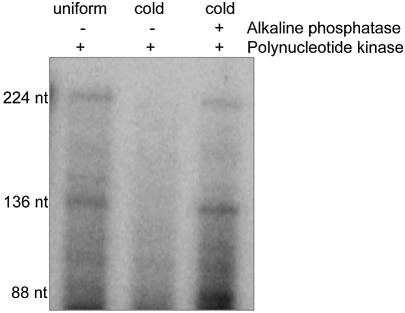
To evaluate the scission products, CSP41 was incubated with unlabeled petDΔ3 RNA and 20 mM MgCl2, and then in the presence or in the absence of alkaline phosphatase. Alkaline phosphatase removes the 5′ phosphate groups of both the triphosphorylated substrate and the cleavage products. Both samples were subsequently incubated with polynucleotide kinase and [γ-32P]ATP. It was assumed that if the products of the CSP41 reaction were 3′ hydroxyl- and 5′ phosphoryl-terminated, they would only be labeled after treatment with alkaline phosphatase. For comparison, a control reaction was run with uniformly labeled petDΔ3 RNA. The results, shown in Figure 3, reveal that labeling of the cleavage products only occurred in the samples treated with alkaline phosphatase (right lane). Therefore, CSP41 cleaves RNA 5′ to the backbone phosphoryl group, consistent with a mechanism requiring a divalent metal and a role in facilitating subsequent polyadenylation.
Figure 3.
CSP41 cleaves petDΔ3 RNA 5′ to the phosphodiester bond. petDΔ3 RNA, either uniformly labeled or unlabeled, was incubated in the presence of 0.5 µg of CSP41 and buffer containing 20 mM MgCl2 for 20 min. The unlabeled reactions were then treated with or without 10 U alkaline phosphatase as indicated. All reactions were subsequently incubated with 10 U of polynucleotide kinase and [γ-32P]ATP. The reactions were then precipitated and analyzed in a 5% denaturing polyacrylamide gel.
The divalent metal specificity of CSP41 was subsequently determined, as shown in Figure 4A. CSP41 was incubated in the presence of 20 fmol petDΔ18 RNA and 20 mM MgCl2, CaCl2, MnCl2, CoCl2 or ZnCl2. Cleavage at position 136 was observed only in the presence of MgCl2. RNase III is known to have a narrow optimum for activation by Mn2+ [e.g. Li et al. (24)]. We therefore assayed CSP41 at Mn2+ concentrations ranging from 50 µM to 1 mM, but no petDΔ18 cleavage activity was observed (data not shown).
Figure 4.
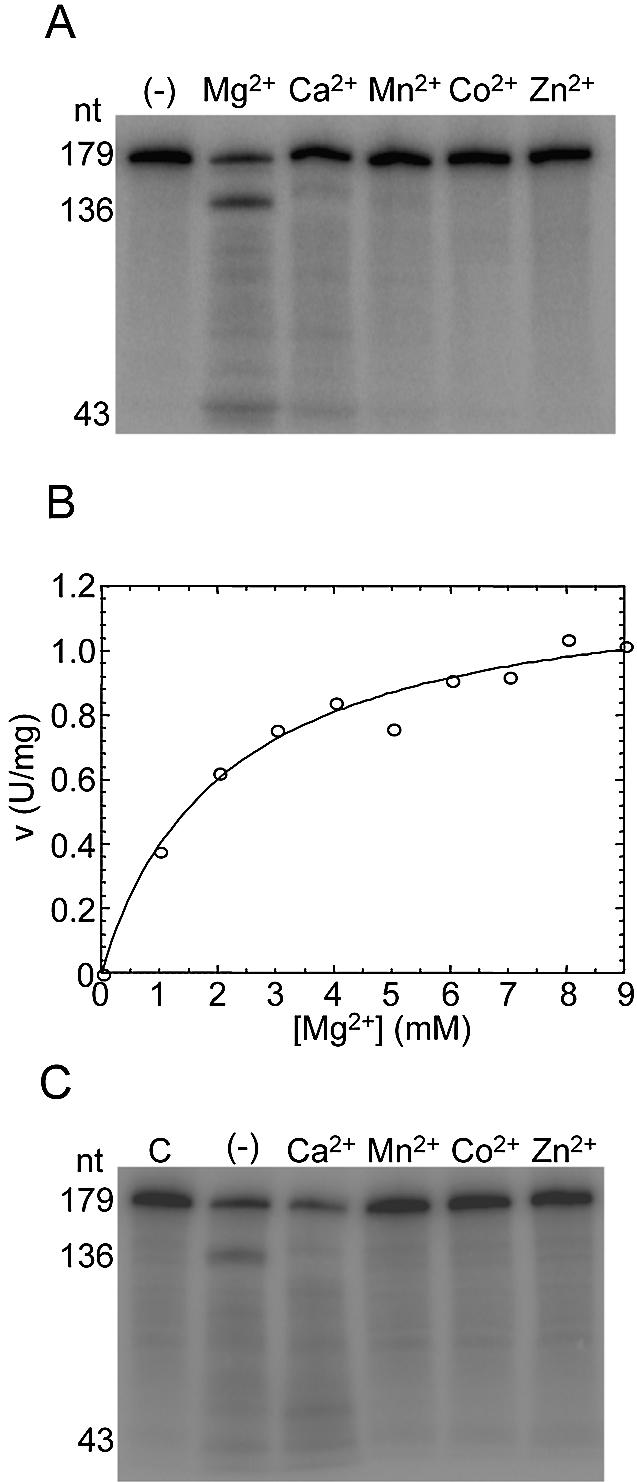
The divalent metal specificity of CSP41. (A) A 0.5 µg aliquot of recombinant CSP41 was incubated with 20 fmol petDΔ18 RNA in buffer containing 20 mM concentrations of the indicated metals. Samples were removed after 10 min and analyzed in a 5% denaturing polyacrylamide gel. Sizes of CSP41 cleavage products are indicated on the left. (B) A 0.5 µg aliquot of recombinant CSP41 was incubated with 20 fmol petDΔ3 RNA in buffers containing varying concentrations of MgCl2 as indicated. Each data point represents the average of duplicate measurements. The variation in the rate at each of the indicated Mg2+ concentrations was typically <10%. The curve represents the best fit to equation 1 (see Materials and Methods). (C) A 0.5 µg aliquot of recombinant CSP41 was incubated with 20 fmol petDΔ18 RNA in buffer containing 2 mM MgCl2 and 20 mM of the indicated divalent metals. The control reaction (C) contained no CSP41. Reactions were stopped after 10 min and analyzed in a 5% denaturing polyacrylamide gel. Sites of CSP41 cleavage are indicated on the left.
To determine whether Ca2+, Mn2+, Co2+ or Zn2+ could bind to CSP41 despite their inability to activate the enzyme catalytically, we conducted competition experiments. First, the KA,Mg, the concentration of Mg2+ giving a half-maximal rate of degradation of petD RNA, was determined, since keeping the concentration of Mg2+ near its KA ensures that measurements of enzyme activity will be most sensitive to the competing cation. The rate of degradation of the petD RNA was measured as a function of Mg2+ concentration (Fig. 4B). It was first determined that under these conditions, the decrease in intensity of the full-length substrate was linear over the course of the reaction at each metal concentration (data not shown). Then, the data measuring activity versus concentration were fit to a hyperbolic rate response, giving KA,Mg, of 2.1 ± 0.4 mM. The KA,Mg for CSP41 is similar to the KA,Mg of 1 mM measured for CheY, a phosphorylation-activated signaling protein from E.coli (15). CheY also contains a metal-binding Rossman fold. This is also similar to the affinity for Mg2+ of mSlo1, a K+ channel protein that contains a low affinity Mg2+-binding site in its intracellular Rossman fold (14).
The results of metal competition experiments are shown in Figure 4C, where CSP41 was incubated in the presence of 2 mM MgCl2 and 20 mM of the indicated divalent metal competitor. As a control, the degradation of petDΔ18 RNA was measured in the presence of 2 mM MgCl2 alone (lane –). The data show that cleavage at position 136 was inhibited by the combination of 2 mM MgCl2 and 20 mM MnCl2. This is not surprising, considering that with most metal-requiring endonucleases, Mg2+ and Mn2+ are interchangeable at the divalent metal-binding site. This is true of other enzymes catalyzing various phosphate chemistries (15); for example, in the case of RNase III, the KA,Mg and KA,Mn are similar (24). petDΔ18 degradation was undetectable in the presence of 2 mM MgCl2 and 20 mM CaCl2, 20 mM CoCl2 or 20 mM ZnCl2. These results suggest that Ca2+, Mn2+, Co2+ and Zn2+ are either direct competitors of Mg2+ for the catalytic metal-binding site, or they bind at a second, inhibitory divalent metal site. The latter type of inhibition has been well characterized for E.coli RNase HI (25).
The activation of CSP41 by Mg2+ was characterized in more detail, with the aim of determining the divalent metal stoichiometry on the enzyme. This could help distinguish between mechanisms that require one and two divalent metals. A two-metal mechanism has been suggested for RNase III (24). We performed a series of experiments in which CSP41 was assayed in the presence of either Mg2+ or Ca2+ alone, or where both Mg2+ and Ca2+ were present at subsaturating concentrations, in order to allow simultaneous binding of both metals. In the case of a single divalent metal-binding site, the reaction rate in the presence of both metals would be the weighted average of the rates in the presence of each metal alone. This rate would depend on the relative affinity of each metal for the binding site, as well as the turnover rate of the enzyme. On the other hand, if two divalent metal sites were present, the reaction rate in the presence of both divalent metals might exceed the sum of individual rates. An example of this response is observed when subsaturating concentrations of Mn2+ are titrated into Archaeoglobus fulgidis RNase HII in the presence of subsaturating concentrations of Mg2+ (26).
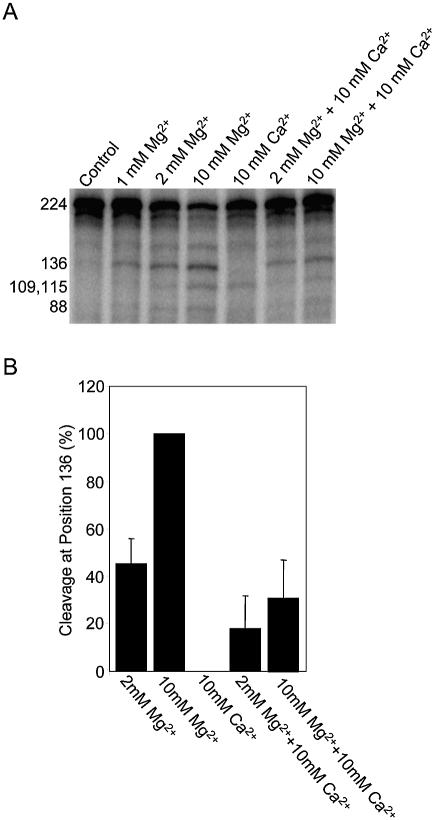
As a reference, the petDΔ3 cleavage reaction was measured in the presence of 1, 2 and 10 mM MgCl2. These experiments were run in triplicate, and a representative gel is shown in Figure 5A. The degradation of petDΔ3 by CSP41 in the presence of 10 mM CaCl2 was inhibited at both 2 and 10 mM MgCl2, as reflected in a decrease in cleavage at positions 136 and 115. This suggests that Mg2+ and Ca2+ compete for the same binding site on CSP41. We found that the specificity of the reaction was different in the presence of Ca2+ and Mg2+. In the presence of Mg2+, two primary 5′ cleavage products are present, corresponding to positions 136 and 115. However, in the presence of Ca2+, CSP41 cleaves only at position 115. In the presence of both Mg2+ and Ca2+, cleavage at position 136 is decreased by >50% relative to the control reactions in the presence of Mg2+ alone (Figure 5B). We did not observe a transient enhancement of activity at lower Ca2+ concentrations (data not shown). These results suggest that Ca2+ and Mg2+ bind at a single divalent metal-binding site on CSP41.
Figure 5.
Mg2+ and Ca2+ bind to the same site on CSP41. (A) CSP41 was incubated for 10 min in the presence of 20 fmol petDΔ3 RNA and divalent metals as indicated. The reaction products were analyzed in a 5% denaturing polyacrylamide gel. Sizes of CSP41 cleavage products are indicated on the left. (B) Relative cleavage rates at position 136 under different mixed metal conditions. Reactions were run in triplicate, the 136 nt band was quantified on a Storm Scanner, and the values were averaged. Cleavage in the presence of 10 mM MgCl2 was adjusted to 100% as a reference.
It is interesting that Mn2+, Co2+ and Zn2+ compete with Mg2+ on CSP41, considering that none of them catalytically activate it. We attempted to measure binding of MgCl2, MnCl2, CoCl2, ZnCl2 or CaCl2 to the CSP41 apoenzyme by monitoring the total intrinsic fluorescence of the enzyme as a function of divalent metal concentration, but none of these divalent metals caused a significant fluorescence change, even at high metal concentrations (data not shown). This suggests either that divalent metal binding does not cause a global conformational change in the apoenzyme, or that the divalent metal can only bind to the CSP41–RNA complex.
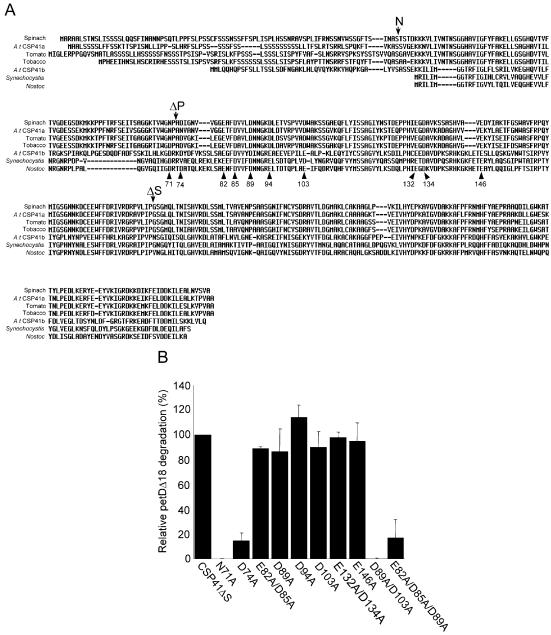
Given the divalent metal specificity and putative structural similarity to dehydrogenases, it was of interest to determine which residues on CSP41 were responsible for metal binding. Previously, we showed that spinach CSP41 could be inactivated by chemical modification of its acidic residues (Glu/Asp) by EDAC and glycine methyl ester (12). Because acidic residues are good candidates for divalent metal-ligating residues, we used multiple sequence alignments of plant, Synechocystis and Nostoc CSP41 proteins to determine which conserved acidic residues were good candidates for mutagenesis. Figure 6A shows the alignment and highlights the N-terminus of the mature protein (N) and the C-termini of two previously characterized spinach deletion mutants (ΔP and ΔS). Previous work showed that while ΔS cleaved petD RNA, ΔP was catalytically inactive, suggesting that the active site of CSP41 was contained within the region between amino acids 72 and 192 of the mature protein (10). Therefore, the residues N71, E82, D85, D89, D94, D103, E132, D134 and E146 of CSP41ΔS were mutated to alanine individually, and in combination (Fig. 6B). Although D74 was not conserved in one sequence, we also mutated it, given its proximity to N71 and because metal-ligating residues are often adjacent.
Figure 6.
Site-directed mutagenesis of conserved acidic residues of CSP41. (A) Sequence alignment of spinach (AAC49424), Arabidopsis CSP41a (NM_116179), tomato (T52071), tobacco (AY324804), Arabidopsis CSP41b (NM_100804), Synechocystis (NP_440784) and Nostoc (NP_488871) CSP41. The sequence is numbered according to the spinach protein, with arrowheads indicating the positions of residues mutated in this study. N indicates the N-terminus of the mature spinach protein, and ΔP and ΔS indicate the C-termini of the CSP41ΔP and CSP41ΔS deletion proteins. (B) Relative rates of degradation of petDΔ18 RNA by CSP41ΔS and site-directed mutants as indicated under each bar. Samples were assayed in duplicate, degradation of the 179 nt fragment was quantified on a Storm Scanner, and the values were averaged. Cleavage by CSP41ΔS was adjusted to 100% as a reference.
Initially, we assayed CSP41ΔS and the mutant enzymes with petDΔ18. The relative activities of the mutants are shown in Figure 6B. Cleavage by N71A was undetectable under the conditions of our experiment, representing a >100-fold reduction in activity. The E82A/D85A, D89A, D94A, D103A, E132A/D134A and E146A mutations had no significant effect on activity, but the petDΔ18 cleavage activity of D74A and the E82A/D85A/D89A triple mutant was reduced by >5-fold relative to CSP41ΔS. D89A/D103A had no detectable activity. Therefore, while single mutations at positions E82/D85, D89 and D103 had no significant effect on enzyme activity, concomitant mutation of these residues did. To determine whether the decrease in petDΔ18 cleavage activity was due to an increase in the KA,Mg, and to determine whether the divalent metal specificity had changed as a result of the mutations, CSP41ΔS and the mutant enzymes were assayed in the presence of up to 100 mM MgCl2 or in the presence of MnCl2, but no effect on activity was observed (data not shown). These data demonstrate that N71, D74, E82, D85, D89, D94 and D103 are important for activity, but do not give direct evidence that they are ligands to the divalent metal.
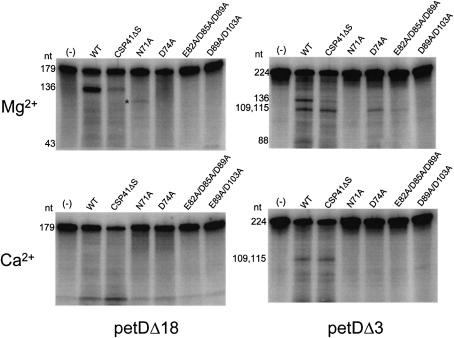
We compared cleavage of petDΔ18 and petDΔ3 with various mutant enzymes, to determine whether the cleavage specificity at positions 136 and 115 with petDΔ3 was retained; the results are shown in Figure 7. Full-length (WT) CSP41 and CSP41ΔS cleave petDΔ18 at position 136, although accumulation of the 136 nt band is lower with CSP41ΔS. None of the site-directed mutants cleaved petDΔ18, even when the assay was repeated at higher enzyme concentrations and for longer incubation times (data not shown). The band marked by an asterisk in the N71A lane was not reproducible. Both WT and CSP41ΔS cleaved petDΔ3 at positions 136 and 115. While accumulation of the 115 nt band was similar with WT and CSP41ΔS in this experiment, accumulation of the 136 nt band was much lower with CSP41ΔS, similar to what was seen with petDΔ18 (upper left panel). None of the site-directed mutants cleaved petDΔ3 at position 136, but D74A and E82A/D85A/D89A showed cleavage at position 115, although the rate was much lower than for CSP41ΔS.
Figure 7.
Cleavage of petD RNA by CSP41, CSP41ΔS and mutants. The top row represents cleavage of petDΔ18 (left) and petDΔ3 (right) in the presence of Mg2+, and the bottom row in the presence of Ca2+. CSP41 was incubated with either 20 fmol petDΔ18 or 20 fmol petDΔ3 and either 20 mM MgCl2 or 20 mM CaCl2 as indicated. The enzyme used is indicated at the top of each lane. The sizes of substrates and cleavage products are indicated to the left of each gel. (–) indicates the non-enzymatic control. The band marked with an asterisk is discussed in the text.
The assays were repeated in the presence of Ca2+. WT CSP41 did not cleave petDΔ18 in the presence of Ca2+. In contrast, CSP41ΔS had significant activity as measured by disappearance of the 179 nt precursor, although no cleavage fragments accumulated, suggesting that the activity of CSP41ΔS under these conditions was non-specific. None of the site-directed mutants digested petDΔ18 in the presence of Ca2+. As observed before, both WT and CSP41ΔS cleaved petDΔ3 exclusively at position 115, but none of the site-directed mutants had detectable activity. The data suggest that the efficiency of cleavage at position 136, but not position 115, is dependent on the C-terminal region of the protein and on the divalent metal. Although the D74A and E82A/D85A/D89A mutations had a significant effect on cleavage at position 136, these mutants still possess cleavage activity at position 115 of petDΔ3, and, therefore, are still capable of binding divalent metal and cleaving RNA. However, these mutants appear to have an altered divalent metal specificity compared with CSP41ΔS, since they were not activated by Ca2+. The reason for this is unclear, although these proteins may have a slightly altered structure at the active site that modifies their ability to recognize and cleave petD RNA at position 136.
DISCUSSION
This report describes the role of divalent metal cations in the reaction catalyzed by CSP41, a ubiquitous endoribonuclease found in plant chloroplasts. In vitro, CSP41 cleaves petD 3′ UTR RNA substrates primarily in the stem–loop, upstream of the stem–bulge junction, and eventually produces short oligoribonucleotides (9,10). Although Rossman fold proteins have been described as RNA-binding proteins and as endonucleases (13), this is the first endonuclease activity described as part of this family that has a metal ion requirement. Therefore, it was of interest to determine the metal ion requirements of the ribonuclease activity of CSP41.
Monovalent cations neither are required for activity nor enhance the activity of CSP41, although KCl has been shown to be inhibitory at concentrations >100 mM in the presence of Mg2+ (10). In contrast, enzyme activity has an absolute requirement for Mg2+. Based on our results, the divalent metal probably participates in RNA cleavage site selection, and in the catalytic mechanism of phosphodiester bond scission. In general, the degradative, low molecular weight RNases cleave RNA substrates such that 3′ phosphoryl-terminated products are generated, via the formation of a 2′,3′-cyclic intermediate, and are active in the absence of divalent metals. In contrast, highly specific endonucleases that participate in processing or turnover of RNA have an absolute requirement for divalent cations and generate 3′ hydroxyl-terminated products. Divalent metal requirement has been observed with endonucleases such as RNase III, RNase H, RNase E and human ARD1 (an RNase E homolog) (27–30). In the case of E.coli RNase H, the role of the divalent metal is thought to be to stabilize the negatively charged pentacovalent intermediate during the phosphoryl transfer reaction (30). RNase III, RNase H, RNase E and ARD1 have been shown to require a divalent metal for catalysis, and all cleave on the 5′ side of the phosphodiester bond. Based on the chemistry of non-enzymatic RNA cleavage, and mechanistic studies of RNases that are known to cleave on the 5′ side of the phosphodiester bond, the role of the divalent metal in these enzymes is to provide a solvent water molecule for attack at the phosphate with the 3′ oxygen as the leaving group (29). This is probably the role of the divalent metal ion in the CSP41-catalyzed reaction.
Mn2+, Co2+ and Zn2+ did not activate CSP41, although they were competitors of Mg2+. It is probable that these metals can occupy the Mg2+-binding site on the enzyme, but are inefficient at activating CSP41 to catalyze bond scission. In the presence of Zn2+, binding of Mg2+ to RNase III, and therefore its RNase activity, are inhibited, although the RNase III–Zn2+ complex still binds RNA (24). RNase III contains a single divalent metal site. Similarly, Mg2+, Ca2+, Ba2+ and Co2+ are able to occupy the same divalent metal-binding site on E.coli RNase H, but the enzyme requires Mg2+ for optimal catalytic activity (31).
Ca2+-activated CSP41 cleaves petDΔ3 at position 115. With Mg2+-activated CSP41, there is an additional cleavage at position 136, generating a 136 nt proximal fragment and an 88 nt distal fragment (Fig. 5A). Cleavage at position 136 occurs at a higher rate than cleavage at position 115. We suggest here that the divalent metal binding plays a role in cleavage site selection by CSP41, since Ca2+- and Mg2+-activated CSP41 have different cleavage specificities. This type of metal-dependent cleavage specificity has been observed with RNase III and also with the restriction endonucleases EcoRI and HindIII (24,32). In the case of EcoRI and HindIII, it has been proposed that the larger ionic radius and altered ligand affinity of Mn2+ confer the ability of the enzyme to cleave at non-canonical sequences compared with those cleaved in the presence of Mg2+. Therefore, Mg2+-activated CSP41 can be viewed as an enzyme with intrinsically broadened RNA cleavage capability over that of the Ca2+-activated enzyme. It is possible that Mg2+ and Ca2+ stabilize different conformations of CSP41, allowing recognition and cleavage at different sites. With RNase III, a divalent metal is required for both RNA cleavage and recognition of at least one of its double-stranded RNA substrates (33).
Two pieces of evidence suggest that N71, D89 and D103 are ligands to the divalent metal. Mutation of N71 to alanine completely abolished activity in the presence of either Mg2+ or Ca2+. Because this side chain has no ionizable group capable of proton transfer, and therefore cannot participate directly in catalysis, particularly in proton transfer, this side chain may be a ligand to the metal. Also, because only the double D89A/D103A mutation abolished activity while the single D89A and D103A mutations had no effect, these side chains may be metal ligands. It is possible that in the single mutants, remaining residues in the metal-binding cluster are sufficient to allow metal binding and catalysis. Similar results have been seen with CheY, where mutation of either D13 or D57 of the Mg2+-binding carboxylate cluster abolishes Mg2+ binding, whereas mutation of D12, the third member of the cluster, does not (34). Therefore, N71, D89 and D103 may form a divalent metal-binding site on CSP41.
CSP41 is not unique in its divalent metal requirement among the proteins containing a Rossman fold. Two other examples are CheY and mSlo1, both of which contain a carboxylate cluster Mg2+-binding site (15). This type of binding site consists of a cluster of side chain carboxylates and is often found in enzymes catalyzing phosphochemistry. These sites have a preference for Mg2+ binding, exclude monovalent cations, and are flexible enough to accommodate various divalent metals (15). The magnesium-binding sites of mSlo1 and CheY are similar in that they consist of three aspartate side chains and a backbone carbonyl coordinating the divalent metal. The positions of the metal-binding sites on the three-dimensional structures of CheY and mSlo1 are very different, however, suggesting that there is no common metal-binding fold or motif within the active sites of metal-binding Rossman fold proteins.
Only Mg2+ activates CSP41 toward endonucleolytic cleavage of petDΔ18 RNA, the predominant form of the petD 3′ UTR in the chloroplast (35). Although Ca2+ activates CSP41 to cleave petDΔ3, which represents the petD pre-mRNA 3′ UTR, the petD pre-mRNA does not accumulate to significant levels in the chloroplast, and would only be a transient substrate prior to pre-mRNA processing. Furthermore, the concentration of free Ca2+ in the chloroplast has been reported to be 150 nM (36), which is too low to activate CSP41 catalytically. Based on relative abundance, Mg2+ is most likely to be the physiological divalent metal activator of CSP41 (16,36). The concentration of Mg2+ in the chloroplast varies from ∼0.5 mM in etiolated spinach leaves to 2–3 mM in young light-grown seedlings and 10 mM in mature light-grown seedlings (16,17). Given that the KA,Mg, the concentration of Mg2+ giving half-maximal catalytic activity of CSP41, is ∼2 mM (Fig. 4B), the in vivo concentration range gives maximum responsiveness for CSP41 endonucleolytic activity, consistent with Mg2+ concentration regulating CSP41 activity in vivo. Several other chloroplast enzymes, including fructose- and sedoheptulosebisphosphatase and inorganic pyrophosphatase, are also regulated by Mg2+ (37,38). Taken together, the activation of CSP41 by Mg2+, and its ability to generate 3′ hydroxyl groups for polyadenylation, suggest that this enzyme may play a major role in regulating chloroplast RNA degradation.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the DOE Energy Biosciences Program, award DE-FG02-90ER20015.
REFERENCES
- 1.Stern D.B., Jones,H. and Gruissem,W. (1989) Function of plastid mRNA 3′ inverted repeats: RNA stabilization and gene-specific protein binding. J. Biol. Chem., 264, 18742–18750. [PubMed] [Google Scholar]
- 2.Chen H.C. and Stern,D.B. (1991) Specific binding of chloroplast proteins in vitro to the 3′ untranslated region of spinach chloroplast petD messenger RNA. Mol. Cell. Biol., 11, 4380–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memon A.R., Meng,B. and Mullet,J.E. (1996) RNA-binding proteins of 37/38 kDa bind specifically to the barley chloroplast psbA 3′-end untranslated RNA. Plant Mol. Biol., 30, 1195–1205. [DOI] [PubMed] [Google Scholar]
- 4.Monde R.A., Schuster,G. and Stern,D.B. (2000) Processing and degradation of chloroplast mRNA. Biochimie, 82, 573–582. [DOI] [PubMed] [Google Scholar]
- 5.Klaff P. (1995) mRNA decay in spinach chloroplasts: psbA mRNA degradation is initiated by endonucleolytic cleavages within the coding region. Nucleic Acids Res., 23, 4885–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudla J., Hayes,R. and Gruissem,W. (1996) Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J., 15, 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H. and Stern,D.B. (1991) Specific ribonuclease activities in spinach chloroplasts promote mRNA maturation and degradation. J. Biol. Chem., 266, 24205–24211. [PubMed] [Google Scholar]
- 8.Nickelsen J. and Link,G. (1993) The 54 kDa RNA-binding protein from mustard chloroplasts mediates endonucleolytic transcript 3′ end formation in vitro. Plant J., 3, 537–544. [DOI] [PubMed] [Google Scholar]
- 9.Yang J., Schuster,G. and Stern,D.B. (1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell, 8, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J. and Stern,D.B. (1997) The spinach chloroplast endoribonuclease CSP41 cleaves the 3′ untranslated region of petD mRNA primarily within its terminal stem–loop structure. J. Biol. Chem., 272, 12784–12880. [DOI] [PubMed] [Google Scholar]
- 11.Baker M.E., Grundy,W.N. and Elkan,C.P. (1998) Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide–sugar epimerases and hydroxysteriod dehydrogenases. Biochem. Biophys. Res. Commun., 248, 250–254. [DOI] [PubMed] [Google Scholar]
- 12.Bollenbach T.J. and Stern,D.B. (2003) Secondary structures common to chloroplast mRNA 3′ UTRs direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J. Biol. Chem., 278, in press. [DOI] [PubMed] [Google Scholar]
- 13.Evguenieva-Hackenberg E., Schiltz,E. and Klug,G. (2002) Dehydrogenases from all three domains of life cleave RNA. J. Biol. Chem., 277, 46145–46150. [DOI] [PubMed] [Google Scholar]
- 14.Shi J., Krishnamoorthy,G., Yang,Y., Hu,L., Chaturvedi,N., Harilal,D., Qin,J. and Cui,J. (2002) Mechanism of magnesium activation of calcium-activated potassium channels. Nature, 418, 876–880. [DOI] [PubMed] [Google Scholar]
- 15.Needham J.V., Chen,T.Y. and Falke,J.J. (1993) Novel ion specificity of a carboxylate cluster Mg(II) binding site: strong charge selectivity and weak size selectivity. Biochemistry, 32, 3363–3367. [DOI] [PubMed] [Google Scholar]
- 16.Horlitz M. and Klaff,P. (2000) Gene-specific trans-regulatory functions of magnesium for chloroplast mRNA stability in higher plants. J. Biol. Chem., 275, 35638–35645. [DOI] [PubMed] [Google Scholar]
- 17.Ishijima S., Uchibori,A., Takagi,H., Maki,R. and Ohnishi,M. (2003) Light-induced increase in free Mg2+ concentration in spinach chloroplasts: measurement of free Mg2+ by using a fluorescent probe and necessity of stromal alkalinization. Arch. Biochem. Biophys., 412, 126–132. [DOI] [PubMed] [Google Scholar]
- 18.Stern D.B. and Gruissem,W. (1987) Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell, 51, 1145–1157. [DOI] [PubMed] [Google Scholar]
- 19.Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 20.Zamore P.D., Patton,J.G. and Green,M.R. (1992) Cloning and domain structure of the mammalian splicing factor U2AF. Nature, 355, 609–614. [DOI] [PubMed] [Google Scholar]
- 21.Bentley R.C. and Keene,J.D. (1991) Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B” protein and through interactions with U2 snRNP-A′ protein. Mol. Cell. Biol., 11, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagy E., Henics,T., Eckert,M., Miseta,A., Lightowlers,R.N. and Kellermayer,M. (2000) Identification of the NAD(+)-binding fold of glyceraldehyde-3-phosphate dehydrogenase as a novel RNA-binding domain. Biochem. Biophys. Res. Commun., 275, 253–260. [DOI] [PubMed] [Google Scholar]
- 23.Deutscher M.P. (1985) E.coli RNases: making sense of alphabet soup. Cell, 40, 731–732. [DOI] [PubMed] [Google Scholar]
- 24.Li H.L., Chelladurai,B.S., Zhang,K. and Nicholson,A.W. (1993) Ribonuclease III cleavage of a bacteriophage T7 processing signal. Divalent cation specificity and specific anion effects. Nucleic Acids Res., 21, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keck J.L., Goedken,E.R. and Marqusee,S. (1998) Activation/attenuation model for RNase H. A one-metal mechanism with second-metal inhibition. J. Biol. Chem., 273, 34128–34133. [DOI] [PubMed] [Google Scholar]
- 26.Chai Q., Qiu,J., Chapados,B.R. and Shen,B. (2001) Archaeoglobus fulgidus RNase HII in DNA replication: enzymological functions and activity regulation via metal cofactors. Biochem. Biophys. Res. Commun., 286, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 27.Claverie-Martin F., Wang,M. and Cohen,S.N. (1997) ARD-1 cDNA from human cells encodes a site-specific single-strand endoribonuclease that functionally resembles Escherichia coli RNase E. J. Biol. Chem., 272, 13823–13828. [DOI] [PubMed] [Google Scholar]
- 28.Szeberenyi J., Roy,M.K. and Apirion,D. (1983) 7 S RNA: a single site substrate for the RNA processing enzyme ribonuclease E of Escherichia coli. Biochim. Biophys. Acta, 740, 282–290. [DOI] [PubMed] [Google Scholar]
- 29.Campbell F.E. Jr, Cassano,A.G., Anderson,V.E. and Harris,M.E. (2002) Pre-steady-state and stopped-flow fluorescence analysis of Escherichia coli ribonuclease III: insights into mechanism and conformational changes associated with binding and catalysis. J. Mol. Biol., 317, 21–40. [DOI] [PubMed] [Google Scholar]
- 30.Goedken E.R. and Marqusee,S. (2001) Co-crystal of Escherichia coli RNase HI with Mn2+ ions reveals two divalent metals bound in the active site. J. Biol. Chem., 276, 7266–7271. [DOI] [PubMed] [Google Scholar]
- 31.Katayanagi K., Miyagawa,M., Matsushima,M., Ishikawa,M., Kanaya,S., Ikehara,M., Matsuzaki,T. and Morikawa,K. (1990) Three-dimensional structure of ribonuclease H from E.coli. Nature, 347, 306–309. [DOI] [PubMed] [Google Scholar]
- 32.Hsu M. and Berg,P. (1978) Altering the specificity of restriction endonuclease: effect of replacing Mg2+ with Mn2+. Biochemistry, 17, 131–138. [DOI] [PubMed] [Google Scholar]
- 33.Chelladurai B., Li,H., Zhang,K. and Nicholson,A.W. (1993) Mutational analysis of a ribonuclease III processing signal. Biochemistry, 32, 7549–7558. [DOI] [PubMed] [Google Scholar]
- 34.Sola M., Lopez-Hernandez,E., Cronet,P., Lacroix,E., Serrano,L., Coll,M. and Parraga,A. (2000) Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY. J. Mol. Biol., 303, 213–225. [DOI] [PubMed] [Google Scholar]
- 35.Monde R.A., Greene,J.C. and Stern,D.B. (2000) The sequence and secondary structure of the 3′-UTR affect 3′-end maturation, RNA accumulation and translation in tobacco chloroplasts. Plant Mol. Biol., 44, 529–542. [DOI] [PubMed] [Google Scholar]
- 36.Johnson C.H., Knight,M.R., Kondo,T., Masson,P., Sedbrook,J., Haley,A. and Trewavas,A. (1995) Circadian oscillations of cytosolic and chloroplastic free calcium in plants. Science, 269, 1863–1865. [DOI] [PubMed] [Google Scholar]
- 37.Portis A.R. Jr, Chon,C.J., Mosbach,A. and Heldt,H.W. (1977) Fructose- and sedoheptulosebisphosphatase. The sites of a possible control of CO2 fixation by light dependent changes of the stromal Mg2+ concentration. Biochim. Biophys Acta, 461, 313–325. [DOI] [PubMed] [Google Scholar]
- 38.Krishnan V.A. and Gnanam,A. (1988) Properties and regulation of Mg2+-dependent chloroplast inorganic pyrophosphatase from Sorghum vulgare leaves. Arch. Biochem. Biophys., 260, 277–284. [DOI] [PubMed] [Google Scholar]