Abstract
The Erm family of adenine-N6 methyltransferases (MTases) is responsible for the development of resistance to macrolide–lincosamide–streptogramin B antibiotics through the methylation of 23S ribosomal RNA. Hence, these proteins are important potential drug targets. Despite the availability of the NMR and crystal structures of two members of the family (ErmAM and ErmC′, respectively) and extensive studies on the RNA substrate, the substrate-binding site and the amino acids involved in RNA recognition by the Erm MTases remain unknown. It has been proposed that the small C-terminal domain functions as a target-binding module, but this prediction has not been tested experimentally. We have undertaken structure-based mutational analysis of 13 charged or polar residues located on the predicted rRNA-binding surface of ErmC′ with the aim to identify the area of protein–RNA interactions. The results of in vivo and in vitro analyses of mutant protein suggest that the key RNA-binding residues are located not in the small domain, but in the large catalytic domain, facing the cleft between the two domains. Based on the mutagenesis data, a preliminary three-dimensional model of ErmC′ complexed with the minimal substrate was constructed. The identification of the RNA-binding site of ErmC′ may be useful for structure-based design of novel drugs that do not necessarily bind to the cofactor-binding site common to many S-adenosyl-l- methionine-dependent MTases, but specifically block the substrate-binding site of MTases from the Erm family.
INTRODUCTION
A major form of resistance against the widely used macrolide–lincosamide–streptogramin B (MLS) antibiotics (such as erythromycin) in pathogenic bacteria results from the methylation of a specific adenine in the peptidyl transferase loop of 23S ribosomal RNA (A2058 in Escherichia coli, A2085 in Bacillus subtilis) (1). As a result, the MLS antibiotics lose their ability to bind to the ribosome and no longer exhibit antibacterial activity. The specific mono- or dimethylation of adenine at the N6 position (generating m6A and m62A, respectively) is carried out by methyltransferases (MTases) from the Erm (erythromycin resistance MTase) family, using S-adenosyl-l-methionine (AdoMet) as a methyl group donor. Erm MTases are found in a wide variety of bacteria, including antibiotic-producing actinomycetes and clinical pathogens, both Gram-positive and Gram-negative (2).
The structures of two Erm MTases, ErmAM determined by NMR (3) and ErmC′ determined by X-ray crystallography (4,5), are nearly identical (reviewed in 6). They are comprised of two structural domains. The larger N-terminal catalytic domain exhibits a typical α/β/α sandwich architecture common to the ‘classical’ Rossmann-fold MTases that modify DNA, RNA, proteins and small molecules (7,8). The catalytic domain is relatively well conserved; it contains nine (X and I-VIII) motifs typically found in this family of enzymes. The smaller C-terminal domain consists of three α-helices and is unique to this family of MTases and is unrelated to known RNA-binding domains. The conservation of the Erm MTases on the sequence and structural levels is paralleled by the conservation of the nucleic acid substrate across vast evolutionary distances (9–11). In accordance with this, Erm MTases can modify rRNAs from phylogenetically distinct species (12). The smallest RNA that shows methyl-accepting activity is a 27 nt stem–loop, corresponding to the 23S rRNA sequences 2048–2063 and 2610–2620, with the substrate adenosine situated as an unpaired base within the loop (13).
Analysis of the electrostatic potential distribution suggested that the small domain and the cleft at the junction between the two domains form a positively charged rRNA-binding site (3,4). However, to our knowledge this presumption has not been validated experimentally and no specific residues that participate in key protein–RNA interactions have been identified. On the other hand, the recognition elements have been relatively well studied on the RNA level. The unique RNA motif recognized by the MTase ErmE appears to be formed by a moderately conserved sequence of bases (aNNNcgGAHAg; A is methylated, N is any nucleotide, H is ‘not G’, lowercase is preference, uppercase is essential) that is displayed in a particular secondary structure (14,15). It was shown that an irregular stem is required immediately 5′ to the substrate adenosine (A2058 in E.coli), with an unpaired nucleotide, preferably a cytidine residue, at position 2055. Minor preferences in the identities of nucleotides 2051–2055 (5′ side of the helix) were reported, while there was little or no restriction on the identities of individual nucleotides 2611–2616 (3′ side of the helix). It was concluded that the main role of these residues is in maintaining the irregular secondary structure rather than in making specific base-mediated contacts with the MTase (16).
The compounds, which inhibit Erm MTases can sensitize MLS-resistant bacteria to macrolide antibiotics, as demonstrated both in vivo and in vitro for ErmC′ inhibitors (17). Erm inhibitors used in combination with a broad-spectrum macrolide antibiotic could be potentially useful for the treatment of infections caused by MLS-resistant pathogens. Using the NMR-based screen and parallel synthesis, new lead compounds have been generated that bind to the AdoMet-binding site on the ErmAM and ErmC′ MTases and can reverse Erm-mediated MLS antibiotic resistance (18). Nevertheless, inhibitors that block the AdoMet-binding site may be poisonous for the humans, because they could act against many essential AdoMet-dependent DNA, RNA, protein, lipid and small-molecule MTases (19). Hence, MTase inhibitors with potential medicinal use will require improved selectivity among different classes of MTases and greater potency for the targeted Erm MTases. Another possible means of combating the drug resistance by inhibiting the A2058 methylation would be to develop drugs that block the unique RNA-binding site of the Erm MTases. Precise delineation of the RNA-binding site on the surface of the Erm crystal structure would be an essential step towards this goal. With the aim of identification of essential protein–RNA interactions we carried out systematic replacement of charged side chains on the predicted target-binding surface of ErmC′ with alanine and studied the function of the single- and multiple-site mutants in vitro and in vivo.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacillus subtilis BD1167 carrying naturally occurring plasmid pIM13 with ermC′ gene (20) was kindly provided by Dr David Dubnau, New York University School of Medicine. Escherichia coli DH5α and BL21(DE3)pLysS and the expression vector pET-25b(+) were obtained from Novagen. Cloning vector pUC18 was from Amersham Biosciences.
Gene cloning and site-directed mutagenesis
The ermC′ gene together with its native promoter and terminator were amplified from the B.subtilis plasmid pIM13. Simultaneously, four new restriction sites were introduced using the PCR-overlapping method. PstI and BamHI sites were introduced at the 5′ and 3′ end of the construct, respectively, while NdeI and XhoI sites were introduced to flank the ermC′ coding sequence. Additionally, codons for His6-Tag were introduced immediately before the STOP codon to facilitate protein purification. The construct obtained was cloned into PstI and BamHI sites of pUC18 vector. Site-directed mutagenesis was carried out according to either the PCR-overlapping method or QuikChange protocol (Stratagene). All introduced changes as well as the absence of unwanted mutations were confirmed by DNA sequencing. For protein expression and purification, coding sequences of mutant genes were recloned into NdeI and XhoI sites of the expression vector pET-25b(+). The oligonucleotides used in this work are listed in Table 1.
Table 1. Deoxyoligonucleotides employed.
| Deoxyoligonucleotide | Description | Sequence |
|---|---|---|
| Oligo-1 | Sense PCR primer for ermC′ cloning | AAAACTGCAGTATAAATTTAACGATCAC |
| PstI | ||
| Oligo-2 | Antisense PCR primer for ermC′ cloning | CGCGGATCCCCCTACGAGGTTGTCG |
| BamHI | ||
| Oligo-3 | Sense PCR primer for introduction of NdeI site | AAGAGGGTTCATATGAACGAGAAAA |
| NdeI | ||
| Oligo-4 | Antisense PCR primer for introduction of NdeI site | CTCGTTCATATGAACCCTCTTTATTT |
| NdeI | ||
| Oligo-5 | Sense PCR primer for introduction of XhoI site and His6-Tag | CACCATCACCATCACCATTAACTCGAGGTTAAGGGATGCATAAACTGC |
| His6-Tag XhoI | ||
| Oligo-6 | Antisense PCR primer for introduction of XhoI site and His6-Tag | CTCGAGTTAATGGTGATGGTGATGGTGCTTATTAAATAATTTATAGCTATTG |
| XhoIHis6-Tag | ||
| Oligo-7 | Sense PCR primer for T108A | TAACATAAGTGCGGATATAATACG |
| Oligo-8 | Antisense PCR primer for T108A | CGTATTATATCCGCACTTATGTTAT |
| Oligo-9 | Sense PCR primer for R112A | GTACGGATATAATAGCCAAAATTGTTT |
| Oligo-10 | Antisense PCR primer for R112A | AAACAATTTTGGCTATTATATCCGTAC |
| Oligo-11 | Sense PCR primer for R112D | GTATTGTACGGATATAATAGACAAAATTGTTTAAC |
| Oligo-12 | Antisense PCR primer for R112D | GTTAAACAATTTTGTCTATTATATCCGTACAATAC |
| Oligo-13 | Sense PCR primer for K133A | TACGGGTTTGCTGCAAGATTATTA |
| Oligo-14 | Antisense PCR primer for K133A | TAATAATCTTGCAGCAAACCCGTA |
| Oligo-15 | Sense PCR primer for R134A | GGTTTGCTAAAGCATTATTAAATACAA |
| Oligo-16 | Antisense PCR primer for R134A | ATTTAATAATGCTTTAGCAAACCCGT |
| Oligo-17 | Sense PCR primer for R140A | ATACAAAAGCCTCATTGGCATTATT |
| Oligo-18 | Antisense PCR primer for R140A | TAATGCCAATGAGGCTTTTGTATTT |
| Oligo-19 | Sense PCR primer for N192A | GATAAACAGAAGTATGCTTATTTCGTTATGAAATGG |
| Oligo-20 | Antisense PCR primer for N192A | CCATTTCATAACGAAATAAGCATACTTCTGTTTATC |
| Oligo-21 | Sense PCR primer for M196A | AATTATTTCGTTGCGAAATGGGTTAACAAAGAATAC |
| Oligo-22 | Antisense PCR primer for M196A | GTATTCTTTGTTAACCCATTTCGCAACGAAATAATT |
| Oligo-23 | Sense PCR primer for K197A | CGTTATGGCATGGGTTAACAAAGA |
| Oligo-24 | Antisense PCR primer for K197A | TTGTTAACCCATGCCATAACGAAAT |
| Oligo-25 | Sense PCR primer for N200A | TGAAATGGGTTGCCAAAGAATACA |
| Oligo-26 | Antisense PCR primer for N200A | TGTATTCTTTGGCAACCCATTTCA |
| Oligo-27 | Sense PCR primer for E202A | GGGTTAACAAAGCATACAAGAAAA |
| Oligo-28 | Antisense PCR primer for E202A | AATATTTTCTTGTATGCTTTGTTA |
| Oligo-29 | Sense PCR primer for K204A | GTTAACAAAGAATACGCGAAAATA |
| Oligo-30 | Antisense PCR primer for K204A | TAAATATTTTCGCGTATTCTTTGTTAA |
| Oligo-31 | Sense PCR primer for K205A | CAAAGAATACAAGGCAATATTTACAAA |
| Oligo-32 | Antisense PCR primer for K205A | TTTGTAAATATTGCCTTGTATTCTTTG |
| Oligo-33 | Sense PCR primer for K209A | AATATTTACAGCAAATCAATTTAACAAT |
| Oligo-34 | Antisense PCR primer for K209A | ATTGTTAAATTGATTTGCTGTAAATATT |
| Oligo-35 | Sense PCR primer for clustera | GGCATGGGTTGCCAAAGCATACGCGGCAATATTTACAAAAAATC |
| Oligo-36 | Antisense PCR primer for clustera | TTGTAAATATTGCCGCGTATGCTTTGGCAACCCATGCCATAACGAA |
| Oligo-37 | Sense PCR primer for Δ193–203 | TAAACAGAAGAAGAAAATATTTACAAAAAATCAATT |
| Oligo-38 | Antisense PCR primer for Δ193–203 | ATATTTTCTTCTTCTGTTTATCTTTGTGTGAT |
| Oligo-39 | Sense PCR primer for Δ207–219 | CAAGAAAATAGGAATTGACGATTTAAACAATAT |
| Oligo-40 | Antisense PCR primer for Δ207–219 | CGTCAATTCCTATTTTCTTGTATTCTTTGTTAAC |
| Oligo-41 | Antisense PCR primer for Δ182–244 | CCGCTCGAGTTAATGGTGATGGTGATGGTGTGATTTTTTTCTATTTAATCTGAT |
Δ, indicated deletion.
aCluster, K197A/N200A/E202A/K204A/K205A.
Protein expression and purification
Escherichia coli BL21(DE3)pLysS cells carrying mutant genes in the pET-25b(+) vector were grown at 30°C in Luria–Bertani medium supplemented with 100 µg/ml ampicillin and 50 µg/ml chloramphenicol. At OD600 of 1.0, the expression was induced with 1 mM IPTG and carried out for 5 h at 30°C. Proteins were purified in a two-step chromatographic procedure. Affinity chromatography was carried out on a HiTrap Chelating column (Amersham Biosciences) according to the manufacturer’s instructions. Mutant proteins were eluted with the linear gradient of imidazole (0.1–0.3 M imidazole in 50 mM phosphate buffer, pH 8.0, containing 0.3 M NaCl). Fractions containing partially purified proteins were pooled and diluted with TDGM buffer (50 mM Tris–Cl, pH 8.0, 5 mM DTT, 10% glycerol, 10 mM MgCl2) to reduce the ionic strength and then applied to a cation exchange HiTrap SP column (Amersham Biosciences) equilibrated with TDGM buffer. ErmC′ variants were eluted with the linear NaCl gradient (0.2–0.6 M NaCl in TDGM buffer). The pure proteins were concentrated on the YM-10 (Amicon) membrane and stored at –80°C in TDGM buffer containing 150 mM NaCl.
Determination of erythromycin minimal inhibitory concentrations (MICs)
Erythromycin MICs were determined in E.coli DH5α essentially as described previously (21). Briefly, overnight cultures of DH5α cells carrying mutant genes in pUC18 vector were diluted 1:25 in fresh 2YT medium supplemented with 100 µg/ml ampicillin and grown until OD500 of 0.7–0.8. Following 100-fold dilution, a 5 µl aliquot was applied to 2YT plates which contained 100 µg/ml ampicillin and various concentrations of erythromycin (80, 160, 320, 640, 1280 and 2560 mg/l). Plates were grown for 18 h at 37°C and the MIC was determined as a minimal concentration of erythromycin that inhibits confluent growth.
Filter-binding assay
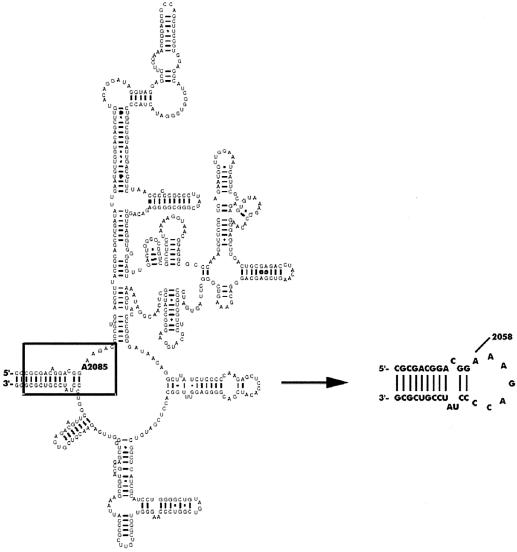
The RNA-binding properties of mutant proteins were determined using synthetic RNA oligonucleotide (32mer CGCGACGGACGGA2085AAGACCCCUAUCCGUCGCG, hairpin structure) designed to mimic the adenine loop in domain V of B.subtilis 23 S rRNA (residues 2073–2090 and 2638–2651) and used previously in studies on the wild-type (wt) enzyme (Fig. 1) (5). The oligonucleotide substrate was labeled radioactively using adenosine-5′ [γ-33P]triphosphate (Amersham Biosciences) and T4 polynucleotide kinase (New England Biolabs). RNA (10 nM) was titrated with increasing concentrations (50–1500 nM) of ErmC′ variants in binding buffer [40 mM Tris–Cl, pH 7.6, 40 mM KCl, 4 mM Mg(OAc)2, 10 mM DTT, 1 mM EDTA, 0.2 mg/l BSA] with the addition of 1 U of RNasin (Promega) per reaction mixture. Binding reactions were carried out in a reaction volume of 20 µl for 25 min at 37°C. Nitrocellulose filter sheets Optitran BA-S 83 (pore size 0.22 µm) from Schleicher and Schuell were preincubated for 2 h in binding buffer. Presoaked filters were placed in a dot-blot apparatus (96 wells; Schleicher and Schuell). Wells were washed with 100 µl of binding buffer immediately before the samples were applied. To minimize non-specific background retention of RNA, only six samples were applied to the wells at a time. Fifteen microliters of reaction mixture were vacuum filtered and the wells were immediately flushed with 100 µl of binding buffer. After drying, the filters were exposed overnight to the intensifying screen and the amounts of bound complexes were determined using Cyclon Phosphoimager System (Canberra Packard). Binding curves and determination of apparent dissociation constants were done using the program SigmaPlot, version 8.0 (SPSS, Inc.). Experiments were repeated at least three times in duplicate.
Figure 1.
Schematic diagrams of the secondary structure of domain V of B.subtilis 23S rRNA and the synthetic oligonucleotide used in this work and in previous studies of the wt ErmC′ (5). Domain V is redrawn from the Gutell Lab’s Comparative RNA Website (http://www.rna.icmb.utexas.edu). A2085 (B.subtilis numbering) indicates the target for the ErmC′ MTase.
Methylation assay
In vitro methylation of RNA was done according to previously described procedures (4,22) with some modifications. RNA oligo was denatured at 90°C for 1 min and renatured by cooling down slowly to room temperature. The reaction was carried out in methylation buffer (50 mM Tris–Cl, pH 7.5, 40 mM KCl, 4 mM MgCl2, 10 mM DTT) containing 1.1 µM RNA, 0.2 µM MTase ErmC′, 0.13 µM [3H]-S-adenosyl-l-methionine ([methyl-3H]AdoMet, 16.5 Ci/mmol) and 1 U of RNasin in a total reaction volume of 50 µl. [methyl-3H]AdoMet (82 Ci/mmol) was from Amersham Biosciences, non-radioactive AdoMet was from Sigma. All additions were performed at 0°C and the reaction mixtures were then transferred to a 25°C water bath for 40 min.
In kinetic experiments, reaction mixtures were preincubated at room temperature before the addition of the enzyme. Kinetic parameters for AdoMet were determined using AdoMet concentrations from 1 to 10 µM. RNA kinetic parameters were determined using an RNA concentration range from 0.1 to 1.5 µM.
Reactions were stopped by the addition of 0.5 ml of 10% (w/v) tri-chloro-acetic acid (TCA; Sigma) and the carrier RNA was added to facilitate the precipitation. RNA pellets were washed with 1 ml of 10% TCA, dried and counted for radioactivity. RNA-free and enzyme-free blanks yielded 30–60 c.p.m. under these conditions. All experiments were done at least three times in duplicate. Kinetic parameters were calculated from double reciprocal plots using the Kinetic module of the program SigmaPlot, version 8.0 (SPSS, Inc.).
Modeling of ErmC′–RNA interactions
Modeling of the substrate RNA (residues 2073–2090 and 2638–2651 of B.subtilis 23S rRNA) was carried out using HyperChem 7 (Hypercube Inc). Manual manipulations with protein and RNA structures, including docking, were carried out using SwissPDBViewer (23). Optimization of electrostatic and van der Waals interactions between the protein and the RNA in the preliminary docked complex was carried out for ‘frozen’ protein and ‘thawed’ RNA using SCULPT 3.0 (24). Geometry optimization was carried out in vacuo using the CHARMM force-field (25) and the Steepest Descent and Conjugate Gradients (Polak-Ribiere) molecular mechanics optimization methods implemented in HyperChem, with default parameters, until convergence. The protein and the target adenine were not allowed to change the conformation during optimization.
RESULTS AND DISCUSSION
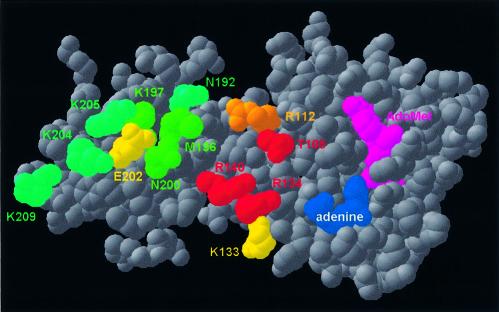
The crystal structure of ErmC′ was solved in the presence of the AdoMet cofactor, but without the RNA substrate. The distribution of the electrostatic potential on the protein surface has been analyzed, and a large positively charged region was identified in the concave cleft between the two domains and on the small domain (3,4). Based on this finding, the small domain was termed the ‘substrate-binding domain’ (3) or the ‘RNA-recognition domain’ (4). It was also predicted that the target adenine would bind to the catalytic domain, in a manner similar to the ‘base-flipping’ mechanism reported for DNA MTases (5). However, to date these predictions have not been tested experimentally and the true substrate-binding site of ErmC′ remains unknown. To identify individual amino acids required for the RNA recognition and binding, we systematically substituted 13 residues of the positively charged region with alanine and characterized the single-site mutants, as well as one multiple mutant in vivo and in vitro (Figs 2 and 3).
Figure 2.
Multiple sequence alignment of the Erm family; only representative sequences are shown (members of the major subfamilies). Conserved residues are shaded. Residues targeted by mutagenesis in this work are indicated by ‘X’.
Figure 3.
ErmC′ structure (5) shown in the ‘spacefill’ representation, reflecting van der Waals radii of non-hydrogen atoms. Residues analyzed by mutagenesis in this work are colored according to the effect of the mutation: red, in vitro activity (A) < 5%; orange, 5% < A < 20%; yellow, 20% < A < 60%; green, 60% < A < 80%; teal, A > 80%.
In vivo characterization of ErmC′ variants
With the aim to investigate the effect of mutations in vivo, we have determined erythromycin MICs in the erythromycin-sensitive E.coli DH5α cells transformed with pUC18 vectors carrying mutant genes. The MIC value for the wt enzyme was >2560 mg/l, while for the negative control it was 80 mg/l. As indicated in Table 2, mutation R134A exhibited the most severe effect on the ErmC′ ability to generate erythromycin resistance—only this mutant has completely lost the activity in vivo. Mutants T108A, R112A, K133A and R140A showed decreased ability in rendering DH5α cells resistant to erythromycin, suggesting that these amino acids are not absolutely indispensable for the ErmC′ activity, but might be involved in important RNA–protein interactions. Since R112 is not fully conserved in the Erm family but substituted with aspartate in some members, we constructed a R112D mutant to see whether this replacement could be sustained by ErmC′ as well. Unexpectedly, the negative effect was even more pronounced with the R112D, indicating that ErmC′ does not support the negatively charged residue at this position. All the above-mentioned residues are located in the large (AdoMet-binding) domain and their side chains point towards the concave cleft between the two domains. Remarkably, single mutations of all of them resulted in decreased enzyme activity in vivo.
Table 2. Effects of ErmC′ mutations on erythromycin resistance.
| ErmC′ variant | Erythromycin MIC (mg/l) |
|---|---|
| Negative control (empty pUC18) | 80 |
| wt | >2560 |
| T108A | 640–1280 |
| R112A | 1280 |
| R112D | 640–1280 |
| K133A | 1280 |
| R134A | 80 |
| R140A | 640–1280 |
| K197A | >2560 |
| N192A | 2560 |
| M196A | >2560 |
| N200A | >2560 |
| E202A | 2560 |
| K204A | 2560 |
| K205A | 2560 |
| K209A | >2560 |
| Clustera | 2560 |
| Δ182–244 | 80 |
| Δ193–203 | 80 |
| Δ207–219 | 80 |
Δ, indicated deletion.
aCluster, K197A/N200A/E202A/K204A/K205A.
On the other hand, mutants of the residues positioned on the surface of the small domain, N192A, M196A, K197A, N200A, E202A, K204A, K205A and K209A did not display substantial defects in activity compared with the wt enzyme. In order to exclude the possibility that single changes are tolerated by ErmC′ and to see whether the small domain really exhibits its predicted RNA-binding role, we created a multiple mutant of spatially adjacent residues K197A/N200A/E202A/K204A/K205A (hereafter referred to as the ‘cluster’ mutant). Surprisingly, even five mutations together did not show a visible cumulative effect on the ErmC′ activity in vivo. This gave us the first indication that the small domain does not have as important a role in RNA recognition and binding as was suggested previously.
RNA-binding affinity of purified ErmC′ mutants
To explore how selected residues are involved in substrate binding, we have compared the RNA-binding affinities of purified mutant proteins with that of the wt enzyme. We have expressed and purified all the proteins as described in Materials and Methods, with the exception of R134A, which was found in inclusion bodies when expressed at 30°C. The expression temperature was therefore decreased to 25°C, but the yield of the soluble protein was much lower than for the other mutants. We have performed equilibrium binding studies with a 32mer stem–loop RNA oligonucleotide that corresponds to the adenine loop in domain V of 23S rRNA of B.subtilis (Fig. 1); this was used in previous studies of the wt enzyme (5). We determined the apparent dissociation constants for all ErmC′ variants from the binding curves obtained by the filter-binding assay. It should be noted that the nitrocellulose filter retained only 40–60% of the complexes, as presented previously in the binding studies of the native 23 S rRNA and ErmC′ (26).
Table 3 presents the summary of all apparent dissociation constants obtained. Apart from the mutant K133A, which showed no change in affinity towards the RNA substrate, all alanine replacements of residues located in the concave interface between the two domains resulted in decreased binding affinities when compared with the wt enzyme, proving that this area is of a considerable significance for the RNA binding. Among them, the T108A mutation showed the most prominent negative effect (5-fold increased Kdapp), suggesting that this amino acid makes an important contact with the RNA moiety. The R112D mutant showed a more pronounced decrease in RNA-binding affinity compared with R112A (3-fold versus 2-fold), again confirming that ErmC′ tolerates better the alanine over the aspartate at this position. In addition, mutants R134A and R140 showed 2-fold elevated apparent dissociation constants, indicating that these residues also contribute to the RNA binding with their positive charge. On the other hand, all alanine substitutions of the residues placed in the C-terminal domain did not have any notable effect on the RNA binding. Moreover, the M196A and E202A mutants were able to bind RNA even better than the wt. Finally, to our surprise, the cluster mutant exhibited only a minor decrease in affinity towards the RNA. These results clearly showed us that neither individual nor multiple changes influence substrate binding in the small domain. Together with the results obtained in vivo this observation strongly suggests that the C-terminal domain of ErmC′ is of minor importance for the RNA recognition and binding.
Table 3. Summary of the in vitro characterization of ErmC′ variants.
| Mutants | Kdapp (M × 10–7) | AdoMet | RNA | ||||
|---|---|---|---|---|---|---|---|
| |
|
KM (M × 10–6) |
kcat (s–1 × 10–2) |
Relative kcat/KM |
KM (M × 10–6) |
kcat (s–1 × 10–3) |
Relative kcat/KM |
| wt | 1.24 ± 0.29 | 2.82 ± 0.20 | 6.55 ± 0.10 | 1.00 | 0.69 ± 0.17 | 6.37 ± 0.42 | 1.00 |
| T108A | 6.41 ± 1.67 | 25.67 ± 5.24 | 2.29 ± 0.22 | 0.004 | 0.43 ± 0.07 | 0.11 ± 0.004 | 0.03 |
| R112A | 2.03 ± 0.61 | 5.80 ± 1.13 | 1.61 ± 0.09 | 0.12 | 1.70 ± 0.39 | 1.13 ± 0.09 | 0.07 |
| R112D | 4.22 ± 0.99 | 10.76 ± 2.92 | 1.21 ± 0.12 | 0.005 | 3.64 ± 1.92 | 0.49 ± 0.11 | 0.02 |
| K133A | 1.29 ± 0.47 | 6.00 ± 0.68 | 3.80 ± 0.13 | 0.27 | 1.86 ± 0.72 | 2.22 ± 0.33 | 0.13 |
| R134A | 2.46 ± 0.67 | 21.32 ± 7.57 | 1.57 ± 0.25 | 0.0032 | 2.50 ± 1.37 | 0.21 ± 0.05 | 0.01 |
| R140A | 2.98 ± 0.98 | 22.28 ± 10.28 | 1.49 ± 0.31 | 0.014 | 1.41 ± 0.35 | 0.34 ± 0.06 | 0.03 |
| N192A | 1.72 ± 0.42 | 2.00 ± 0.18 | 3.41 ± 0.06 | 0.73 | 0.79 ± 0.09 | 5.44 ± 0.17 | 0.74 |
| M196A | 0.76 ± 0.21 | 3.28 ± 0.38 | 3.18 ± 0.06 | 0.42 | 0.62 ± 0.05 | 3.18 ± 0.12 | 0.56 |
| K197A | 1.23 ± 0.22 | 3.34 ± 0.64 | 5.22 ± 0.23 | 0.67 | 1.59 ± 0.26 | 7.31 ± 0.42 | 0.50 |
| N200A | 1.18 ± 0.33 | 7.09 ± 0.52 | 6.88 ± 0.16 | 0.42 | 0.89 ± 0.18 | 4.39 ± 0.26 | 0.54 |
| E202A | 0.88 ± 0.23 | 9.77 ± 2.33 | 7.35 ± 0.62 | 0.32 | 0.57 ± 0.06 | 2.11 ± 0.05 | 0.40 |
| K204A | 1.27 ± 0.32 | 3.83 ± 0.89 | 6.48 ± 0.37 | 0.73 | 0.89 ± 0.23 | 7.29 ± 0.54 | 0.89 |
| K205A | 1.25 ± 0.29 | 4.43 ± 0.86 | 6.49 ± 0.33 | 0.63 | 0.91 ± 0.23 | 7.66 ± 0.57 | 0.92 |
| K209A | 1.24 ± 0.34 | 4.62 ± 0.72 | 7.75 ± 0.32 | 0.72 | 0.95 ± 0.23 | 6.37 ± 0.47 | 0.73 |
| Clustera | 1.48 ± 0.46 | 6.56 ± 1.60 | 3.14 ± 0.23 | 0.21 | 1.21 ± 0.28 | 1.93 ± 0.14 | 0.17 |
aCluster, K197A/N200A/E202A/K204A/K205A.
Kinetic characterization of the ErmC′ mutants
We have determined the kinetic parameters of the purified mutant proteins for both the RNA substrate and the AdoMet factor to explore in detail the functional properties of the amino acids chosen for mutagenesis. All mutants were catalytically active in vitro, as shown in Table 3. The kinetic analysis of alanine mutants of the residues that create a positively charged line in the proposed binding cleft provided us with interesting results. R112A, K133A, R134A and R140A show a 2–3-fold increase in KM for the substrate, with the catalytic constant reduced 2-fold for R112A and K133A, and even 30-fold in the case of R134A and R140 (Table 3). Similar results are also obtained for the R112D and T108A mutants. Remarkably, mutations affect even the cofactor AdoMet binding, with the affinity decreased 2-fold for R112A and K133A, 3-fold for R112D and 8-fold for R134A, R140A and T108A. Together with the increased apparent dissociation constants, these results strongly suggest that the indicated residues take part in specific RNA recognition; however, they also significantly influence the catalytic machinery. For alanine mutants of residues in the cleft, the catalytic efficiency, represented by the kcat/KM values, is extremely diminished mainly due to the decrease in kcat. This indicates that the energy of the transition state complex is higher than in the wt enzyme. Hence, it appears that these residues may be involved in positioning the core of the RNA substrate in a way that ensures the optimal delivery of the target adenine into the catalytic pocket and hence, formation of a stable ternary complex. Positively charged side chains of Lys and Arg and a hydroxyl group of the Thr may form hydrogen bonds with the bases and/or the phosphate backbone of the RNA substrate, which could then help to stabilize the accurate congregation of the catalytic center. Interestingly, the R134A mutant was completely inactive in vivo, in contrast to the low but still measurable activity in vitro. The mutant protein was isolated from bacteria grown at 25°C, while the in vivo experiments were carried out at 37°C. The R134A mutation may destabilize the protein structure, which could result in disintegration of the functional sites at the elevated temperature. Hence, in addition to the important role in RNA recognition, R134 may also maintain the functional conformation of the ErmC′ enzyme. Since none of the mutations caused the complete loss of the enzyme activity in vitro, we suggest that RNA binding is based on the complex network of many contacts, rather than on a few essential interactions.
The positively charged surface of the small C-terminal domain was predicted by Yu et al. (3) and Bussiere et al. (4) to be involved in contacts with the RNA substrate. We have analyzed eight residues from this area, N192, M196, K197, N200, E202, K204, K205 and K209, which form a cluster rich in charged groups. Only the K197A mutant showed a 2-fold decrease in affinity for the substrate, while most of the individual mutants exhibited little or no difference in KM for the RNA when compared with the wt ErmC′. Furthermore, KM for the E202A was even slightly lowered. Amazingly, even the ‘cluster’ mutant that lacked five side chains (K197A/N200A/E202A/K204A/K205A) showed only a very mild increase in KM for the RNA. These results fully support the data obtained in RNA binding experiments, strongly indicating that the small domain of ErmC′ does not mediate important RNA–protein contacts. It is peculiar though, that we observed slightly decreased affinities towards the cofactor and diminished kcat values for M196A, N200A, E202A and the cluster, while these mutants were rather active in vivo. The possible cause of this effect could be the use of different substrates in the two experiments. In vivo ErmC′ methylates the native 23S rRNA molecule, while in the experiments in vitro we have used a 32mer oligonucleotide. It is possible that the binding of the large 23S rRNA molecule is additionally stabilized by non-specific long-range interactions that rely on electrostatic complementarity between the whole macromolecules and/or binding of other substructures of RNA to distal parts of ErmC′. On the other hand, interactions of ErmC′ with the 32mer oligonucleotide are probably limited to contacts within the cleft and in the active site and can be influenced by a small modification of the electrostatic field generated by residues that do not make direct contacts with the substrate. This hypothesis is strongly supported by the observation that the RNA binding by the E202A mutant is enhanced, while mutations of the positively charged residues reduce the binding.
Modeling of ErmC′–RNA interactions
Identification of amino acid residues of ErmC′ important for the RNA binding, together with the knowledge of essential bases and structures in the RNA substrate characterized previously (13–16), prompted us to propose a model for protein–RNA interactions. We extracted the coordinates of the peptidyl transferase loop in domain V of 23 S rRNA from the crystal structures of the large ribosomal subunit of Haloarcula marismortui (27) and Deinococcus radiodurans (10) and used them to build a homology model of the 32mer substrate. HyperChem 7 was used to superimpose the original coordinates and to ‘mutate’ the bases to obtain the sequence identical to the oligonucleotide used as the substrate in the experimental analysis. Breaks in the chain corresponding to deletions and disrupted hydrogen bonding in ‘mutated’ base pairs were repaired by molecular mechanics optimizations (see Materials and Methods). We carried out automated low-resolution docking of the modeled RNA substrate to the ErmC′ crystal structure using GRAMM (28). However, no solution could be identified, which would simultaneously satisfy the following conditions: (i) the protein–RNA interface formed by >50% of amino acid residues of ErmC′ and bases in the RNA substrate known to be essential for the complex formation; and (ii) the distance between the methyl group of AdoMet and any atom of the target adenine <20 Å. Therefore, we decided to build a manual model to illustrate our interpretation of the available data.
We have manually docked the modeled RNA substrate to the ErmC′ using the M.TaqI-DNA complex structure (29) and the mutagenesis data as a guide. The RNA was docked so as to maximize the contacts between the protein and RNA surfaces (especially the residues known to be important), while minimizing steric clashes between them. Following this crude manual docking, the coordinates of the target adenine were copied into the active site of ErmC′ from the M.TaqI–DNA complex structure (29). The only modification of the ErmC′ structure introduced during modeling was manual reconfiguration of the Y104 rotamer in the catalytic center to mimic the stacking interaction with the target adenine observed for the homologous Y108 in the M.TaqI–DNA complex structure (29). No changes of the ErmC′ structure were allowed during docking and energy minimization. Limited steric clashes between the protein and the RNA were relieved by fast energy minimization of the RNA structure using SCULPT (24). In the final model, the geometry of the bound RNA substrate was refined using HyperChem.
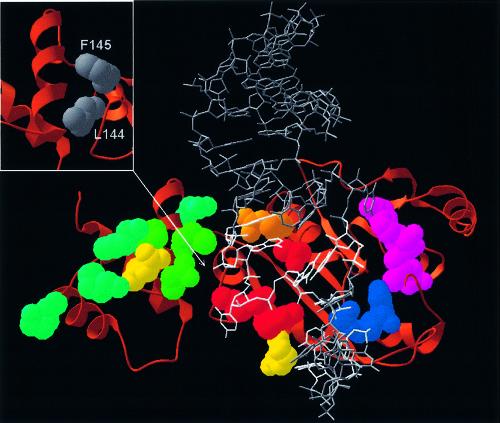
The docking model of the ErmC′–AdoMet–RNA complex is shown in Figure 4. It satisfies all constraints used for its construction (see above). In particular, all amino acids shown to be essential for binding in this work (T108, R112, R134, R140) are buried in the protein–RNA interface and are found spatially close to the bases and phosphate groups of the essential nucleosides (G2053, G2054, C2055, G2056, G2057, A2058, E.coli numbering). Moreover, the model agrees with additional data that were not used during the modeling. The 32mer interacts primarily with the catalytic domain and does not make close interactions with the side chains of the ‘cluster’ residues of the small domain we found to be dispensable for the RNA binding and catalysis. The presence or the lack of contacts between the RNA and the small domain of ErmC′ was not used as a criterion during our docking exercise.
Figure 4.
Speculative model of ErmC′–RNA interactions (the atomic coordinates in the PDB format are available as Supplementary Material and from the website ftp://genesilico.pl/iamb/models/Erm/). The protein is shown in the ‘ribbons’ representation, with experimentally studied residues shown in the ‘spacefill’ representation, colored according to their importance (see the caption to Fig. 2). The model of the RNA substrate 32mer is shown in gray, the target adenosine (A2085) is shown in red, nucleosides important for protein–RNA interactions (13–16) are shown in white. Note that all key RNA-binding residues are located in the large, catalytic (AdoMet-binding) domain. Small panel (upper left corner): hydrophobic residues of the N-terminal domain, which stabilize the structure of the RNA-binding region and may be exposed to the solvent if the C-terminal domain is deleted.
What is the role of the C-terminal domain of ErmC′?
Our study showed unequivocally that the side chains of eight residues on the surface of the C-terminal domain are of little if any importance for RNA binding and catalysis, while four side chains of the cleft, localized in the N-terminal domain, are essential for the MTase activity. This result suggested that the universally conserved small C-terminal domain may be entirely dispensable or required not for binding and catalysis per se, but for maintaining the structural integrity of the large N-terminal domain. Hence, we constructed three deletion mutants, in which either the entire C-terminal domain (amino acids 182–244) was removed, or which lacked the first or the second α-helix (amino acids 193–203 or 207–219, respectively). All these deletion mutants were completely inactive in vivo (Table 3) and could not be purified due to strong aggregation (data not shown). This result demonstrates that the C-terminal domain is essential for the MTase activity of ErmC′ and suggests that it may be required for stability or proper three-dimensional folding of the N-terminal AdoMet-binding/catalytic domain. Indeed, visual analysis of the ErmAM and ErmC′ structures reveals that the ‘RNA-binding region’ of the N-terminal domain (with the key residues T108, R112, R134 and R140) is stabilized by hydrophobic interactions with the C-terminal domain. We speculate that removal or partial deletion of the C-terminal domain destabilizes the structure of the ‘RNA-binding region’, thereby destroying the RNA-binding ability of ErmC′ and leads to aggregation of the mutant protein due to exposure of hydrophobic amino acids such as L144 or F145 (Fig. 4).
CONCLUSIONS
We have undertaken structure-based mutational analysis of 13 residues located on the predicted rRNA-binding surface of ErmC′ with the aim to identify the area of protein–RNA interactions. Our results suggest that the key RNA-binding residues are located not in the small domain (as suggested previously), but in the large catalytic domain, facing the cleft between the two domains. We suggest that the small domain may be most important for structural stabilization of the large domain (especially of the RNA-binding region) and therefore rather indirectly involved in RNA binding. Based on the mutagenesis data, we constructed a preliminary three-dimensional model of ErmC′ complexed with the minimal substrate. We regard this model as purely speculative and far from ‘definite’—specifically, we did not consider any conformational changes on the protein level, which are likely to occur upon RNA binding. Therefore, our prediction must be taken as one of very low resolution, at best at the level of residues, and not individual atoms. Nevertheless, we believe this model will serve as a useful guide for future analyses of ErmC′–RNA interactions as it highlights the regions of protein–RNA contacts that may be studied in more detail using biochemical and biophysical methods. In particular, preliminary characterization of the core of the RNA-binding site of ErmC′ may be useful for structure-based design of novel drugs that do not necessarily bind to the cofactor-binding site common to many AdoMet-dependent MTases, but specifically block the substrate-binding site of MTases from the Erm family.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr David Dubnau for the generous gift of the B.subtilis BD1167. We are indebted to Dr Stephen Douthwaite for the critical reading of the manuscript and useful comments. This work was supported by the ICGEB fellowship to G.M. and by the Ministry of Science of Republic Croatia (grant Nos 006320, 006611). J.M.B. is supported by the Young Investigator award from EMBO and Howard Hughes Medical Institute and by the Young Scholar fellowship from the Foundation for Polish Science. The work at the ICGEB Protein Structure and Function Group was partly supported by EU grant ENGEM QLK3-CT-2001-00448.
REFERENCES
- 1.Weisblum B. (1995) Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother., 39, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M., Brisson-Noel,A. and Courvalin,P. (1987) Origin and evolution of genes specifying resistance to macrolide, lincosamide and streptogramin antibiotics: data and hypotheses. J. Antimicrob. Chemother., 20, 783–802. [DOI] [PubMed] [Google Scholar]
- 3.Yu L., Petros,A.M., Schnuchel,A., Zhong,P., Severin,J.M., Walter,K., Holzman,T.F. and Fesik,S.W. (1997) Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide–lincosamide–streptogramin antibiotic resistance. Nature Struct. Biol., 4, 483–489. [DOI] [PubMed] [Google Scholar]
- 4.Bussiere D.E., Muchmore,S.W., Dealwis,C.G., Schluckebier,G., Nienaber,V.L., Edalji,R.P., Walter,K.A., Ladror,U.S., Holzman,T.F. and Abad-Zapatero,C. (1998) Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry, 37, 7103–7112. [DOI] [PubMed] [Google Scholar]
- 5.Schluckebier G., Zhong,P., Stewart,K.D., Kavanaugh,T.J. and Abad-Zapatero,C. (1999) The 2.2 A structure of the rRNA methyltransferase ErmC′ and its complexes with cofactor and cofactor analogs: implications for the reaction mechanism. J. Mol. Biol., 289, 277–291. [DOI] [PubMed] [Google Scholar]
- 6.Abad-Zapatero C., Zhong,P., Bussiere,D.E., Steward,K. and Muchmore,S.W. (1999) rRNA methyltransferases (ErmC′ and ErmAM) and antibiotic resistance. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Inc., Singapore, pp. 199–226. [Google Scholar]
- 7.Bujnicki J.M. (1999) Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol., 1, 1–8. [PubMed] [Google Scholar]
- 8.Fauman E.B., Blumenthal,R.M. and Cheng,X. (1999) Structure and evolution of AdoMet-dependent methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. World Scientific Inc., Singapore, pp. 1–38. [Google Scholar]
- 9.Noller H.F. (1984) Structure of ribosomal RNA. Annu. Rev. Biochem., 53, 119–162. [DOI] [PubMed] [Google Scholar]
- 10.Harms J., Schluenzen,F., Zarivach,R., Bashan,A., Gat,S., Agmon,I., Bartels,H., Franceschi,F. and Yonath,A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell, 107, 679–688. [DOI] [PubMed] [Google Scholar]
- 11.Hansen J.L., Ippolito,J.A., Ban,N., Nissen,P., Moore,P.B. and Steitz,T.A. (2002) The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell, 10, 117–128. [DOI] [PubMed] [Google Scholar]
- 12.Skinner R., Cundliffe,E. and Schmidt,F.J. (1983) Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem., 258, 12702–12706. [PubMed] [Google Scholar]
- 13.Vester B., Nielsen,A.K., Hansen,L.H. and Douthwaite,S. (1998) ErmE methyltransferase recognition elements in RNA substrates. J. Mol. Biol., 282, 255–264. [DOI] [PubMed] [Google Scholar]
- 14.Hansen L.H., Vester,B. and Douthwaite,S. (1999) Core sequence in the RNA motif recognized by the ErmE methyltransferase revealed by relaxing the fidelity of the enzyme for its target. RNA, 5, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen A.K., Douthwaite,S. and Vester,B. (1999) Negative in vitro selection identifies the rRNA recognition motif for ErmE methyltransferase. RNA, 5, 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villsen I.D., Vester,B. and Douthwaite,S. (1999) ErmE methyltransferase recognizes features of the primary and secondary structure in a motif within domain V of 23 S rRNA. J. Mol. Biol., 286, 365–374. [DOI] [PubMed] [Google Scholar]
- 17.Clancy J., Schmieder,B.J., Petitpas,J.W., Manousos,M., Williams,J.A., Faiella,J.A., Girard,A.E. and McGuirk,P.R. (1995) Assays to detect and characterize synthetic agents that inhibit the ErmC methyltransferase. J. Antibiot., 48, 1273–1279. [DOI] [PubMed] [Google Scholar]
- 18.Hajduk P.J., Dinges,J., Schkeryantz,J.M., Janowick,D., Kaminski,M., Tufano,M., Augeri,D.J., Petros,A., Nienaber,V., Zhong,P. et al. (1999) Novel inhibitors of Erm methyltransferases from NMR and parallel synthesis. J. Med. Chem., 42, 3852–3859. [DOI] [PubMed] [Google Scholar]
- 19.Chiang P.K., Gordon,R.K., Tal,J., Zeng,G.C., Doctor,B.P., Pardhasaradhi,K. and McCann,P.P. (1996) S-adenosylmethionine and methylation. FASEB J., 10, 471–480. [PubMed] [Google Scholar]
- 20.Monod M., Denoya,C. and Dubnau,D. (1986) Sequence and properties of pIM13, a macrolide–lincosamide–streptogramin B resistance plasmid from Bacillus subtilis. J. Bacteriol., 167, 138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrow K.A., Lyras,D., Polekhina,G., Koutsis,K., Parker,M.W. and Rood,J.I. (2002) Identification of essential residues in the ErmB rRNA methyltransferase of Clostridium perfringens. Antimicrob. Agents Chemother., 46, 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong P., Pratt,S.D., Edalji,R.P., Walter,K.A., Holzman,T.F., Shivakumar,A.G. and Katz,L. (1995) Substrate requirements for ErmC′ methyltransferase activity. J. Bacteriol., 177, 4327–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 24.Surles M.C., Richardson,J.S., Richardson,D.C. and Brooks,F.P.,Jr (1994) Sculpting proteins interactively: continual energy minimization embedded in a graphical modeling system. Protein Sci., 3, 198–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brooks B.R., Bruccoleri,R.E., Olafson,B.D., States,D.J., Swaminathan,S. and Karplus,M. (1983) CHARMM: a program for macromolecular energy, minimization and dynamics calculations. J. Comput. Chem., 4, 187–217. [Google Scholar]
- 26.Su S.L. and Dubnau,D. (1990) Binding of Bacillus subtilis ermC′ methyltransferase to 23S rRNA. Biochemistry, 29, 6033–6042. [DOI] [PubMed] [Google Scholar]
- 27.Ban N., Nissen,P., Hansen,J., Moore,P.B. and Steitz,T.A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science, 289, 905–920. [DOI] [PubMed] [Google Scholar]
- 28.Vakser I.A. (1995) Protein docking for low-resolution structures. Protein Eng., 8, 371–377. [DOI] [PubMed] [Google Scholar]
- 29.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M. TaqI in complex with DNA and a cofactor analog. Nature Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.