Abstract
Frameshift mutations generally result in loss-of-function changes since they drastically alter the protein sequence downstream of the frameshift site, besides creating premature stop codons. Here we present data suggesting that frameshift mutations in the C-terminal domain of specific ancestral MADS-box genes may have contributed to the structural and functional divergence of the MADS-box gene family. We have identified putative frameshift mutations in the conserved C-terminal motifs of the B-function DEF/AP3 subfamily, the A-function SQUA/AP1 subfamily and the E-function AGL2 subfamily, which are all involved in the specification of organ identity during flower development. The newly evolved C-terminal motifs are highly conserved, suggesting a de novo generation of functionality. Interestingly, since the new C-terminal motifs in the A- and B-function subfamilies are only found in higher eudicotyledonous flowering plants, the emergence of these two C-terminal changes coincides with the origin of a highly standardized floral structure. We speculate that the frameshift mutations described here are examples of co-evolution of the different components of a single transcription factor complex. 3′ terminal frameshift mutations might provide an important but so far unrecognized mechanism to generate novel functional C-terminal motifs instrumental to the functional diversification of transcription factor families.
INTRODUCTION
Plants exhibit a wide range of ornamental and functional differences in number and appearance of the organs that constitute their flowers. In general, such differences may be ascribed to variations in a basic set of key developmental regulators (called homeotic selector genes). These variations may simply represent differences in the expression patterns of an otherwise standard set of genes that determine the underlying morphogenetic processes. On the other hand, changes in the coding sequence might also lead to changes in gene function. Extensive analysis of plant floral developmental mutants during the last decade has revealed the importance of the MADS-box transcription factor family in flower development and plant architecture. The identity of the floral organs has been shown to be governed by the combined activity of specific MADS-box floral homeotic genes and it has been suggested that gene duplications followed by functional diversification within the MADS-box gene family must have been key processes in floral evolution (1–3). Phylogenetic studies of the MADS-box gene family thus have the potential to correlate differences in floral organ morphology with molecular and functional changes in MADS-box genes. The best-known subfamilies are the A (SQUA/AP1), B (DEF/AP3 and GLO/PI) and C function (AG) MADS-box subfamilies, representing the basic players in the historical ABC model of flower organ identity.
Recent progress by reverse genetics strategies has uncovered redundant functions (4,5) that obviously have been missed by classical forward genetics approaches (6–13). Combined with the elucidation of protein–protein interactions between the different MADS-box genes, these results have led to extensions of the ABC model towards models with a higher complexity (14–18). All data together presently suggest a quartet model (14) in which the identity of the four different floral organs, sepals, petals, stamens and carpels, is specified by four different protein complexes consisting of various combinations of MADS-box proteins and yet unknown factors.
All MADS-box genes discussed here belong to the Type II class MADS-box genes; the proteins encoded by these genes share a conserved modular organization, called the MIKC type domain structure, consisting of a MADS (M), intervening (I), keratin-like (K) and C-terminal domain (2,19–21). The MADS-domain is responsible for DNA binding, but it is also involved in dimerization and accessory factor-binding functions (21). The K-domain seems to be plant-specific (2) and is involved in protein dimerization (19,21). Several lines of evidence demonstrate the functional importance of the C-terminal domain. Loss-of-function alleles may carry mutations in the C-terminus and dominant-negative phenotypes can be generated by overexpressing MADS-box genes lacking the C-terminus (summarized in 16). The first half of the C-terminal domain of DEF and GLO proteins appears to be essential for ternary complex formation between SQUA (A-function) and DEF and GLO (B-function) MADS-domain proteins in vitro (16). Several reports suggest the presence of a C-terminal transcriptional activation domain in proteins encoded by genes belonging to different MADS-box subfamilies (18,22–24). Recently, it was demonstrated that truncated versions of the Arabidopsis B-function genes AP3 and PI, only lacking the characteristic C-terminal euAP3 and Pi motif, respectively, were unable to rescue the corresponding ap3 and pi mutants (25). This implies that the C-terminal motifs are essential for the full function of these proteins. Finally, although the C-terminus is overall the most divergent region among the different MADS-domain proteins, members of the same subfamily usually contain highly conserved C-terminal motifs (26). This suggests that the C-terminus may have played an important role in the functional diversification of the major MADS-box gene subfamilies. Because a less-conserved region of variable length often precedes these highly conserved motifs, the C-terminal region has mostly been excluded from phylogenetic analyses. While the high sequence similarity in the MADS- and K-domains of all MIKC type MADS-domain proteins strongly suggests that they are derived from a common ancestor, and differences in the MIK domains between the different subfamilies can be attributed to mutational events like single amino acid substitutions in combination with small in-frame insertions or deletions, the origin of the highly divergent C-terminal motifs remains obscure. The goal of the present study was to obtain a better understanding of how these putatively functionally important C-terminal motifs may have originated at the DNA level.
MATERIALS AND METHODS
Assembling the MADS-box sequence dataset
We screened the available nucleotide (non-redundant and EST) and protein databases with a diverged set of sequences containing representatives of all known MIKC type MADS-box gene subfamilies, resulting in a collection of over 400 unique plant MIKC type MADS-box sequences from over 100 plant species. More details about the pursued approach are provided in the Supplementary Material. For expressed sequence tag (EST) sequences included in the phylogenetic analysis, consensus sequences covering the full coding sequence were derived from several overlapping ESTs (indicated with ‘merge’ in Figure 2).
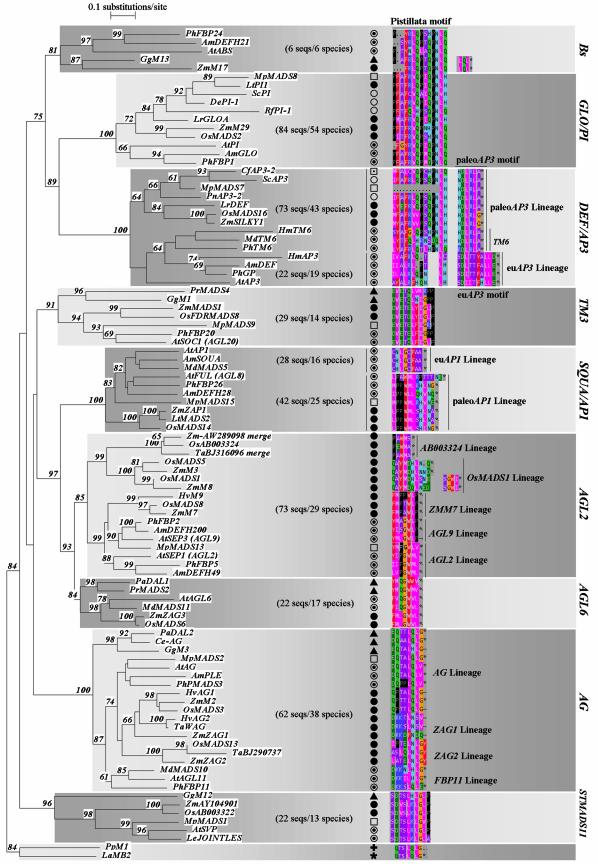
Figure 2.
(Opposite) Neighbor-joining tree of the MIKC type MADS-box gene family. The Neighbor-joining tree has been constructed using the MIK domains of a representative subset of 97 sequences from the total collection of available plant MIKC type MADS-box sequences (see Supplementary Material). These 97 sequences have been selected as follows: subclasses within subfamilies were determined based on the presence of deviating but conserved C-terminal motifs. For each subclass, one to three representative sequences from each major plant group (when available) were selected. The tree was rooted with two MIKC type MADS-box genes from the moss Physcomitrella patens and the clubmoss Lycopodium annotinum. To assess support for the inferred relationships, 1000 bootstrap samples were generated. In a final step, we mapped C-terminal conserved epitopes on the tree. Local bootstrap probabilities are indicated for branches supported with more than 60%. Asterisks behind protein motifs represent stop codons. Motifs not terminating with an asterisk are followed by a variable number of non-conserved residues (not shown). A two-letter code preceding the gene names as found in the database indicates the species involved. Species names and taxa are indicated as follows. Angiosperms: Higher eudicots (open circles with inner filled circles): Am: Antirrhinum majus; At: Arabidopsis thaliana; Hm: Hydrangea macrophylla; Le: Lycopersicon esculentum; Md: Malus domestica; Ph: Petunia hybrida; Basal eudicots (open circles): De: Dicentra eximia; Pn: Papaver nudicaule; Rf: Ranunculus ficaria; Sc: Sanguinaria canadensis; Monocotyledons (filled circles): Hv: Hordeum vulgare; Lr: Lilium regale; Lt: Lolium temulentum; Os: Oryza sativa; Ta: Triticum aestivum; Zm: Zea mays; Others: Mp: Magnolia praecocissima (Magnoliales) (open squares), Cf: Calycanthus floridus (Laurales) (open square with inner filled square). Gymnosperms (filled triangles): Pa: Picea abies (Coniferales); Pr: Pinus radiata (Coniferales); Gg: Gnetum gnemon (Gnetales); Ce: Cycas edentata (Cycadales). Outgroup: La: Lycopodium annotinum (Lycopodiophyta) (filled star); Pp: Physcomitrella patens (Bryophyta) (plus sign). For each subfamily, the total number of analyzed sequences and different species is indicated in parentheses (no. sequences/no. species).
Sequence alignments
Full-length sequences were aligned using the PILEUP function, followed by a manual alignment of the C-terminal regions using the Seqlab Editor of the GCG software package [Wisconsin Package Version 10.0, Genetics Computer Group (GCG), Madison, WI, USA]. For each gene, the cDNA sequence and the corresponding putative protein sequence were coupled and for both, the C-terminal domains were aligned manually.
Phylogenetic analysis
For simplicity reasons, the Neighbor Joining tree (Fig. 2) has been constructed using a representative subset of 97 sequences from the total collection of available plant MIKC type MADS-box sequences. These sequences have been selected as follows: subclasses within subfamilies were determined based on the presence of deviating but conserved C-terminal motifs. For each subclass, one to three representative sequences from each major plant group (when available) were selected. The MIK domains of the selected MADS-box genes were aligned using ClustalW (27) and subjected to a phylogenetic analysis. Phylogenetic trees were computed using the TREECON program (28) according to the neighbor-joining algorithm (29), based on Poisson and Tajima and Nei (30) corrected evolutionary distances.
RESULTS
The DEFICIENS (DEF)/AP3 subfamily
So far, only for B-function MADS-box genes has a detailed sequence analysis of the C-terminus been performed for a diverged set of species (31,32). Although protein sequences belonging to the DEF/AP3 subfamily share extensive similarity, two lineages can clearly be distinguished on the basis of their completely different C-terminal motifs (31). The first motif is referred to as the paleoAP3 motif and is found in DEF/AP3 proteins from lower eudicots, magnoliid dicots, monocots and basal angiosperms, while a second type, named the euAP3 motif is uniquely present in DEF/AP3 proteins from higher eudicots. In addition, some higher eudicots possess both the euAP3 and paleoAP3 type (TM6 lineage). Recently, Lamb and Irish published data on C-terminal motif swapping experiments involving euAP3 and paleoAP3 motifs, and demonstrating that these two motifs clearly encode a diverged function (25): a chimeric construct in which the euAP3 motif of the Arabidopsis AP3 gene was replaced by a paleoAP3 motif displayed differential rescue of the second and third whorls of the ap3-3 mutant: second whorl organs remained fully sepaloid while stamen formation was partially rescued. These results indicate that the C-terminal motif of paleoAP3 proteins promote stamen but not petal formation in higher eudicots. Our own attention was initially drawn to paleoAP3 B-function MADS-box genes while analyzing the Petunia B-function family (manuscript in preparation). The paleoAP3 motif containing PhTM6 gene of Petunia exhibits some atypical characteristics compared to the classical euAP3 B-function MADS-box genes. During later stages of floral development, PhTM6 mRNAs are abundantly present in carpels [similar to the tomato TM6 gene (33)], to a lesser extent in stamens and to even lower levels in petals and sepals. Also, the Petunia Green Petals (GP) mutant (a null mutant for the euAP3 Pmads1 gene) displays a homeotic conversion of petals to sepals, while the formation of stamens remains unaffected (34), suggesting that PhTM6 cannot substitute the euAP3 Pmads1 gene in petal formation, but most likely can complement its function in stamen development. These findings suggest that sequence diversification at the C-terminus may be responsible for differences in function between the AP3 genes in higher eudicots as compared to other angiosperms and thus reflect part of the species diversification at the level of floral organ determining genes.
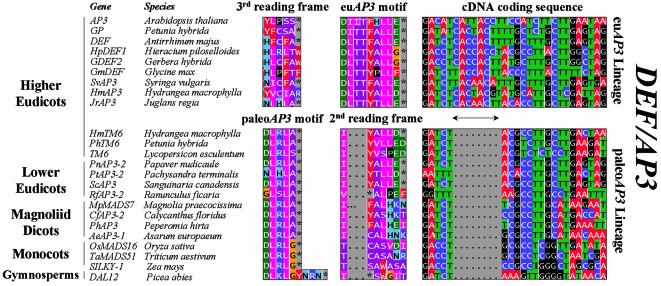
To understand how these different peptide motifs may have arisen at the molecular level during evolution, we compared the coding sequences of paleoAP3 and euAP3 motif-encoding MADS-box genes in detail. To our surprise, we discovered that the C-terminal euAP3 motif can simply be explained by an eight base pair insertion in the C-terminus of paleoAP3 genes, thus causing a frameshift mutation beyond the insertion site in euAP3 genes, when compared to the original reading frame of paleoAP3 genes. A subset of the alignment of paleoAP3 and euAP3 genes is shown in Figure 1. In a number of cases, translation of the C-terminus of paleoAP3 genes according to the second reading frame indeed yields a motif that closely resembles the euAP3 motif (Fig. 1). It is interesting to note that although the paleoAP3 motif is highly conserved among paleoAP3 members, frameshift translations of the lower eudicot and TM6 members resemble the euAP3 motif most, in contrast to frameshift translations of monocot paleoAP3 genes, thus reflecting the phylogenetic relationships of the host species involved. Furthermore, the majority of paleoAP3 members contain a clearly recognizable internal PI motif, while in euAP3 proteins this motif is degenerating (Fig. 2), suggesting that recruitment of the novel euAP3 motif may have been accompanied by a subsequent loss of the internal PI motif in euAP3 B-function proteins. The fact that both paleoAP3 (TM6 lineage) and euAP3 genes have been isolated from several higher eudicots suggests that euAP3 genes have originated after duplication of a paleoAP3 ancestral gene, followed by a frameshift mutation in one of the copies. Species such as Petunia, tomato and Hydrangea macrophylla have retained both copies, while Arabidopsis apparently has lost the paleoAP3 copy. Although the overall sequence analysis clearly points towards an eight base pair insertion in the euAP3 lineage, its exact origin remains elusive, because most likely it may have evolved further. We can presently envisage two putative mechanisms for this event: the insertion can be the result of a footprint left behind upon transposon excision or it may result from DNA polymerase slippage.
Figure 1.
Alignment of paleoAP3 and euAP3 C-terminal motifs present within the DEF/AP3 subfamily. Although protein sequences belonging to the DEF/AP3 subfamily display extensive homology almost along their entire length (not shown), two lineages can be distinguished on the basis of their completely different C-terminal motifs (columns indicated with paleoAP3 and euAP3 motifs). In contrast, the cDNA fragments encoding the conserved motifs align very well (right column) upon the introduction of a gap of eight base pairs in the coding sequences of paleoAP3 lineage members. The euAP3 motif, which is uniquely present in DEF/AP3 subfamily members isolated from higher eudicots, may thus have originated by a frameshift mutation caused by the eight base pair insertion (indicated by a double headed arrow) into a paleoAP3 ancestral gene. This is illustrated by the second reading frame translation of paleoAP3 members (indicated with 2nd reading frame), which resembles the euAP3 motif. For details on the 3rd reading frame of the euAP3 motif, we refer to the text. A full set of analyzed sequences is presented in the Supplementary Material.
It is quite remarkable that a frameshift mutation just upstream of a highly conserved motif would yield a new, equally highly conserved motif. However, the data presented here are based on MADS-box sequences isolated from different species by different laboratories, rendering the possibility of sequencing mistakes unlikely. In addition, paleoAP3 and euAP3 genes have been aligned in two different classes, solely based on the comparison of the non-C-terminal sequences (31). Finally, evolutionary conservation of the newly evolved frameshifted motif at the amino acid level changes the position of degenerate nucleotides compared to the original codon triplets. As a consequence, nucleotide substitutions, which may be silent in the new motif, may hamper the recognition of the original protein motif when translated according to the progenitor reading frame. This is in accordance with our observations that translating euAP3 genes according to the progenitor reading frame (third reading frame of the euAP3 Lineage in Figure 1) yields in the best cases only a highly diverged paleoAP3 motif. If paleoAP3 and euAP3 motifs had originated artificially from simple sequencing errors, the asymmetry in degree of conservation between correct and alternative reading frames would not be observed.
Intrigued by such a simple frameshifting mechanism, we were curious to find indications for a similar scenario in other subfamilies of the MADS-box gene family. Since only the C-terminus of the B-function subfamily has been analyzed in greater detail in a wide range of species (31,32,35), we first determined whether conserved C-terminal motifs existed in other major subfamilies as well. Therefore, we analyzed over 400 MIKC type MADS-box genes covering a wide range of species and representing all major subfamilies. Sequences were first grouped in subfamilies based on sequence homology in the MIK region. Once grouped, the C-terminal regions were aligned manually to determine C-terminal motifs. To illustrate this, we selected a representative set of sequences from each subclass for a diverged set of species, and performed a phylogenetic analysis to map the corresponding C-terminal motifs on the tree (Fig. 2). For simplicity reasons, the complexity of Figure 2 has been reduced in several ways. A number of subfamilies contain several conserved motifs separated by less conserved patches in the C-domain; we only show the conserved residues closest to the C-terminus. Monophyletic clades (e.g. the AGL12, AGL15 and AGL17 subfamilies) for which only a limited set of family members has been isolated, or that contain sequences from just a few species, were not included in the analysis. For these clades, sample numbers and/or species diversity were too low to allow a reliable identification of C-terminal conserved motifs.
For the majority of the subfamilies, we could identify subfamily specific C-terminal motifs. In an increasing level of detail, a number of subfamilies (e.g. the AGAMOUS subfamily) can be further divided into subclasses displaying distinct but related C-terminal motifs of which the differences can be attributed to normal nucleotide substitutions. On the other hand, we found that some subfamilies (e.g. the SQUA/AP1 and AGL2 subfamilies) could be further divided into subclasses displaying completely different but highly conserved C-terminal motifs, comparable to the situation found in the DEF/AP3 subfamily. Other clades (e.g. TM3 and STMADS11 subfamilies) display C-terminal motifs that are highly conserved among protein sequences isolated from distantly related species such as angiosperms versus gymnosperms, suggesting that these C-terminal motifs were already fixed in the ancestors preceding the split of angiosperms and gymnosperms. It has been estimated that the lineages that led to extant gymnosperms and angiosperms probably separated about 300 million years ago, while the lineages that led to extant monocots probably separated from the lineage that led to extant eudicots about 160–200 million years ago (1 and references therein). A number of these small C-terminal peptide motifs thus have been preserved for several hundreds of millions of years. Similarly, it is remarkable that the AGAMOUS (AG) type C-terminal motif can be clearly recognized in the two MADS-box genes PpM1 and LaMB2 isolated from the moss Physcomitrella patens and clubmoss Lycopodium annotinum. C-terminal motifs of the full MADS-box gene dataset have been added in the Supplementary Material. In Figure 2, we have indicated the total number of analyzed sequences and the number of species from which genes belonging to a particular class have been isolated (in parentheses).
A minority of the analysed sequences did not exhibit the C-terminal peptide motif(s) as identified in the majority of the members of that subfamily or subclass. With the currently available data, we cannot exclude that at least some of these aberrant proteins represent the first isolated members of new classes of variants, perhaps only present in a subset of species of the plant kingdom.
However, for a substantial part of the sequences that did not exhibit the sub(class)family-specific motif, we were able to demonstrate extensive homology and the appearance of the sub(class)family-specific motif in either one of the three different reading frames downstream of the K-region, often beyond the proposed stop codon. Thus, the latter sequences presumably contain sequencing mistakes. Alternatively they might represent degenerating copies of recently duplicated genes. Besides a complete loss of the conserved C-terminal epitope, we also found pairs of recently duplicated paralogs of which one copy contained the consensus C-terminal motif, while the second copy displayed a more diverged motif. A clear example of such a case is the Arabidopsis AGL13 gene, a member of the AGL6 subfamily. The putative AGL13 protein terminates prematurely after only the first three amino acid residues of the AGL6 motif, but still displays homology beyond the stopcodon (see Supplementary Material).
Having defined C-terminal motifs for the major subfamilies, we specifically searched for further examples of putative frameshift mutations in these regions. Much to our surprise, we found additional examples in the SQUAMOSA/AP1 and AGL2 subfamilies.
The SQUAMOSA (SQUA)/AP1 subfamily
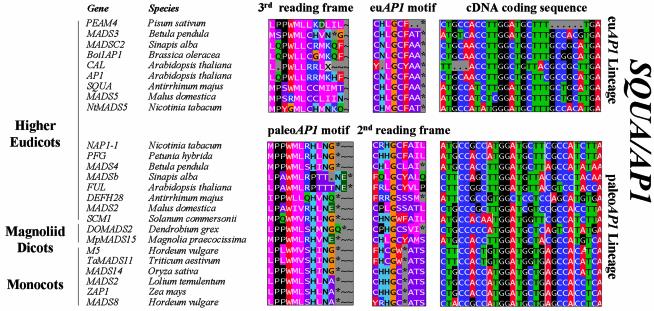
The majority of protein sequences belonging to the SQUA/AP1 subfamily display either one of two highly conserved C-terminal motifs (Figs 2 and 3). We have designated these motifs the paleoAP1 and euAP1 motifs, respectively. The highly conserved paleoAP1 motif is present in AP1 homologs from magnoliid dicots, monocots and higher eudicots. So far, no paleoAP1 like proteins have been isolated from gymnosperm species. Note that the Arabidopsis FRUITFULL gene and SAMADSB from white mustard display a quite diverged paleoAP1 motif compared to the other paleoAP1 genes. The C-terminal euAP1 motif as found in the Arabidopsis AP1 and Antirrhinum SQUA proteins seems to be restricted to the higher eudicots, since we extensively screened the available monocot EST databases without finding them. However, we also found higher eudicot sequences that displayed a more diverged euAP1 motif (e.g. the pea protein PEAM4 in Fig. 3). Although the two AP1 subclasses exhibit a divergent C-terminal peptide motif, cDNA sequences encoding the terminal euAP1 and paleoAP1 motifs align very well. Indeed, translation of the C-terminal part of paleoAP1 genes according to the second reading frame yields motifs that closely resemble the euAP1 motif, and translation of the C-terminal part of euAP1 genes according to the third reading frame yields motifs that closely resemble the paleoAP1 motif (Fig. 3). Similar to the situation in the DEF/AP3 subfamily, frameshift translations of paleoAP1 genes from dicot origin resemble the euAP1 motif most, which reflects the phylogenetic origin of the euAP1 genes. Also, correct reading frame translations yield motifs that are more rigidly conserved than frameshift translations, suggesting that the presence of these two different motifs have not originated from sequencing errors. Because the coding sequence preceding the terminal motifs appeared to be too divergent between paleoAP1 and euAP1 genes to align, we could not determine the nature or the exact position of the putative frameshift mutation. The restriction of euAP1 type genes to the higher eudicots suggests that euAP1 type genes have originated after duplication of a paleoAP1 type gene followed by a mutational event creating a frameshift in the C-terminus of one of two copies. In addition, higher eudicots such as Arabidopsis, snapdragon, tobacco, apple, birch and cauliflower have retained both variants. The taxonomic distribution suggests that the gene duplication happened close to or at the base of the higher eudicots.
Figure 3.
Alignment of paleoAP1 and euAP1 C-terminal motifs present within the SQUA/AP1 subfamily. Within the SQUA/AP1 subfamily, two distinct lineages (euAP1 and paleoAP1 lineages) can be distinguished, each displaying highly conserved but completely different C-terminal motifs (columns indicated with paleoAP1 and euAP1 motifs). Representatives of both lineages have been isolated from a number of higher eudicot species, while magnoliid dicot and monocot species appear to yield only the paleoAP1 type. Although these two types of C-terminal motifs are totally unrelated at the protein level, the cDNA fragments encoding these conserved motifs align surprisingly well (right column). This suggests that the euAP1 motif may have originated by a frameshift mutation in a paleoAP1 ancestral gene at a position upstream of the paleoAP1 motif. To illustrate this, we have shown frameshift translations of paleoAP1 members (column indicated with 2nd reading frame) and of euAP1 members (column indicated with 3rd reading frame), which resemble the euAP1 motif and the ancestral paleoAP1 motif, respectively. A full set of analyzed sequences is presented in the Supplementary Material.
The AGL2 subfamily
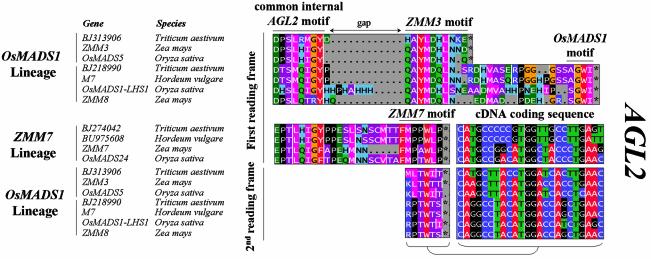
In higher eudicots, two closely related types of AGL2 genes can be distinguished, each displaying a distinct but related C-terminal motif, represented by AGL2(SEP1) and AGL9 (SEP3) types, respectively (Fig. 2). For monocots, we have identified three clearly divergent types of AGL2-like genes (Fig. 2). A first group, the ZMM7 type, has a C-domain that is closely related to the AGL9 type from higher eudicots. The other two groups exhibit very divergent C-domains. For the first of these, designated the AB003324 type, only the rice cDNA sequence AB003324 was found in the nucleotide database at the time of analysis; a search in EST databases with this gene revealed highly homologous copies from maize and wheat, indicating that this type of AGL2-like gene may be functionally conserved among monocots (Fig. 2). Recently, the maize ZMM24 and ZMM31 MADS-box genes have been published (36) that correspond with the identified EST sequences. A second group with a deviating but conserved C-domain, named the OsMADS1 lineage, contains two homologous types of MADS-box genes, exhibiting a difference in length. Both types contain a ZMM3 motif; the long variant in addition contains a conserved C-terminal motif named the OsMADS1 motif. Short and long version sequences have been isolated from both maize and rice, and searching the EST databases with these sequences showed the presence of both types in wheat. This indicates that both forms are conserved among monocots. To investigate the molecular origin of the divergent C-terminus of AB003324 and OsMADS1 types, we aligned these sequences with the ZMM7 type AGL2-like genes. For the AB003324 type, we were unable to find convincing C-terminal homologies with any of the other monocot AGL2-like genes. For the OsMADS1 type genes, the sequences encoding the ZMM3 motif clearly align with the C-terminus of ZMM7 type genes by introducing an internal gap causing a downstream frameshift in the coding sequence of the OsMADS1 type genes compared to the reading frame of the ZMM7 type (Fig. 4). These results suggest that the C-terminal ZMM3 motif of the OsMADS1 type genes has originated after duplication of a monocot ZMM7 type ancestral gene followed by a small deletion immediately downstream of the common internal motif. The C-terminus of the long versions may have been recruited from a sequence beyond the original stop codon of the ZMM7 type genes.
Figure 4.
Alignment of C-terminal motifs of monocot OsMADS1 and ZMM7 type AGL2 like subfamily members. In monocot species, we have identified three distinct types of AGL2 like subfamily members, each displaying different C-terminal motifs (Fig. 2). Here we show part of the C-terminal domain alignment of OsMADS1 and ZMM7 type sequences. Both types have an internal motif in common (indicated with common internal AGL2 motif), while their C-termini have fully diverged at the protein level. Sequences belonging to the OsMADS1 type can be further subdivided into two classes: a short version terminating with a ZMM3 motif, and a longer version with a C-terminal extension terminating with a short conserved OsMADS1 motif. As in the previous cases, we found that the cDNA fragments encoding the ZMM3 motif of the OsMADS1 type align with those encoding the ZMM7 motif by introducing a gap representing a frameshift mutation in the OsMADS1 type sequences. The alignment of the cDNA fragments encoding these ZMM3 and ZMM7 motifs is shown on the right and the 2nd reading frame translation of the ZMM3 motif is shown below the ZMM7 motif.
DISCUSSION
Towards a model for neo-functionalization by C-terminal motif selection
While basic features such as DNA binding domains and motifs necessary for protein–protein interactions must be rigidly conserved in order to maintain the basic capacity to function as a transcription factor, generation of novel C-terminal motifs may have been of major importance for functional diversification. C-terminal domains may play a key role in determining partner specificity in higher order complex formation, may contain activation domains, or may be subject to post-translational modifications that may influence DNA binding specificity, subcellular localization or the ability to attract interacting partners. Although the exact role of these small peptide motifs residing in the C-domain largely remains to be determined (25), the fact that a number of these motifs has been preserved for hundreds of million of years of evolution (see above) strongly suggests that they may have been instrumental in the functional diversification of the MADS-box gene family. Furthermore, we found a comparable C-terminal domain conservation in the WUSCHEL, the NAM and the AP2 transcription factor families. Members of these transcription factor families all possess a highly conserved DNA binding domain while their C-terminus is strongly divergent between different subfamilies. Within subfamilies however, small motifs of variable length occur that are highly conserved even between proteins isolated from distantly related species (unpublished results).
Based on these observations and the results presented here, we propose a model for the functional diversification of duplicated members of transcription factor families (Fig. 5). After duplication of an ancestral gene X, one of the copies (Y) may accumulate mutations in the C-terminus, while retaining features such as DNA binding, essential for its function as a transcription factor, in the upstream coding regions. Apart from in frame insertions/deletions and single nucleotide substitutions, mutations in the coding sequence at the 3′ end will also induce frameshifts, as such masking the ancestral origin of the motif at the protein level. While most frameshift mutations will be deleterious for the existing function, in specific cases they may yield novel functional C-terminal motifs. The three cases we have described are perfect examples of such a neo-functionalization process. This widens the emerging view that plant transcription factors evolve mainly by changes in cis-regulatory elements that affect their expression pattern (37,38), and that after gene duplication, mainly degeneration and selection of complementary functioning, i.e. sub-functionalization occurs (39,40). At first sight, it may seem extraordinary that in all three cases, frameshift mutations of highly conserved motifs yielded novel highly conserved motifs. However, this specific situation is the only type of motif generation that can still be recognized after millions of years of independent evolution of both copies. If the new motif had been recruited from a sequence in a non-conserved (Y3 and Y4, Fig. 5) or less conserved region of the C-terminus (e.g. Y2), it would be impossible to trace back the ancestral motif. Equally important, either the new or the ancestral motif must contain amino acid residues that are not too highly degenerate in order to be able to recognize the related motif after frameshifting. Thus, the only cases of frameshift mutations that we still can recognize are those in highly conserved motifs that yield novel highly conserved motifs. Finally, novel motifs may be acquired in an additive way downstream of existing motifs as an extra feature, with retention of the ancestral motif that in such a case becomes internal (e.g. Y3); or with subsequent loss of the ancestral motif (all other cases).
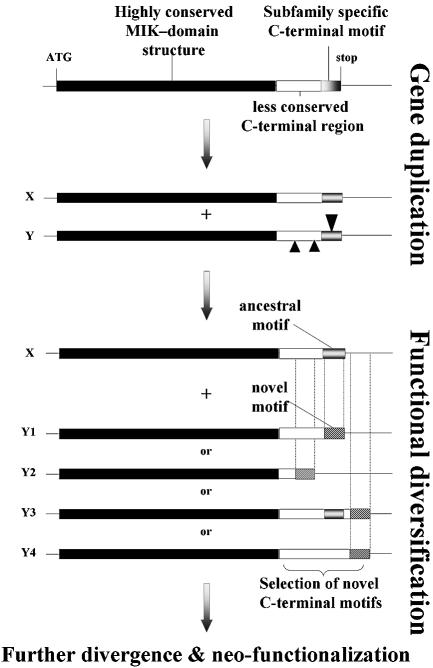
Figure 5.
Model for the generation of novel C-terminal motifs within the MADS-box gene family. After duplication of an ancestral gene X, the Y copy accumulates mutations in the C-terminal domain, while retaining the essential MIK domain. Insertions or deletions will cause a frameshift in the coding sequence. Rarely, these frameshift mutations may yield novel functional motifs that consequently will be conserved. In cases where the novel motif is recruited from poorly conserved regions (e.g. Y 2–4) in the ancestral sequence, the sequence relation with the ancestral gene X will become unclear after a period of independent evolution. In the Y copy, new motifs may be added downstream of the ancestral motif as an extra feature, with retention of the ancestral motif which in this case becomes internal (e.g. Y3); or with subsequent loss of the ancestral motif (all other cases).
Higher order complex formation and importance for flower evolution
We have identified drastic changes in the conserved C-terminal motifs of the core eudicot B-function subfamily (DEF/AP3), the SQUA/AP1 subfamily and the AGL2 subfamily. These mutations appear to be associated with changes in gene function. The apparent coincidence between the origin of euAP1 (A) and euAP3 (B) motifs, and the origin of the higher eudicots is remarkable. Higher eudicots show a characteristic canalization of floral development and thus a standardization of floral architecture (41 and references cited therein). Moreover, although petaloid organs may have evolved several times independently during evolution, the higher eudicot petals seem to be homologous organs that trace back to a single origin at the base of higher eudicots (30,31,33,36). Strikingly, higher eudicot petal identity is specified by A+B function genes encoding euAP1 and euAP3 motifs, respectively. It is conceivable, therefore, that there is a causal relationship between the parallel frameshift mutations described here and both the canalization of floral structure and the origin of a certain type of petals at the base of higher eudicots.
Recently, it has been demonstrated that B-function MADS-box proteins may form higher order complexes with SQUA in Anthirrinum and with SEP3 and AP1, and, alternatively, with SEP3 and AG in Arabidopsis (16,18). We speculate therefore, that the frameshift mutations represent an example of co-evolution between different components of a single transcription factor complex and that these mutations may have modulated the function. Clearly, in a next step, complex formation and function of complexes consisting of paleoAP3 and paleoAP1 proteins have to be studied in monocot and basal angiosperm species in comparison to eudicots.
CONCLUSIONS
The data presented here indicate an excitingly rapid mode of protein evolution: novel, highly conserved motifs at the C-terminus may originate by frameshift mutations in the existing coding sequence. This phenomenon may explain a substantial part of the high sequence divergence in the C-terminal region between and within the different MADS-box gene subfamilies. It will be interesting to see how general the mechanism of protein evolution by novel motif selection (whether or not induced by frameshift mutations) at the C-terminus will appear to be. There is evidence, however, that at least some aspects of it apply not only to plants.
The Ultrabithorax protein acquired a poly-ala tail in the lineage that led to Drosophila, but only after Crustaceans had branched off (42,43). This poly-ala tail is involved in suppressing abdominal leg development. It thus appears that a change in a C-terminal sequence motif of a homeodomain transcription factor can be correlated with a neo-functionalization event affecting the arthropod body plan.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
REFERENCES
- 1.Theissen G., Becker,A., Di Rosa,A., Kanno,A., Kim,J.T., Munster,T., Winter,K.U. and Saedler,H. (2000) A short history of MADS-box genes in plants. Plant Mol. Biol., 42, 115–149. [PubMed] [Google Scholar]
- 2.Theissen G., Kim,J.T. and Saedler,H. (1996) Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol., 43, 484–516. [DOI] [PubMed] [Google Scholar]
- 3.Theissen G. (2001) Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol., 4, 75–85. [DOI] [PubMed] [Google Scholar]
- 4.Pelaz S., Ditta,G.S., Baumann,E., Wisman,E. and Yanofsky,M.F. (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature, 405, 200–203. [DOI] [PubMed] [Google Scholar]
- 5.Liljegren S.J., Ditta,G.S., Eshed,Y., Savidge,B., Bowman,J.L. and Yanofsky,M.F. (2000) SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature, 404, 766–770. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D., Carpenter,R., Sommer,H., Hartley,N. and Coen,E. (1993) Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell, 72, 85–95. [DOI] [PubMed] [Google Scholar]
- 7.Carpenter R. and Coen,E.S. (1990) Floral homeotic mutations produced by transposon-mutagenesis in Antirrhinum majus. Genes Dev., 4, 1483–1493. [DOI] [PubMed] [Google Scholar]
- 8.Coen E.S. (1992) Flower development. Curr. Opin. Cell Biol., 4, 929–933. [DOI] [PubMed] [Google Scholar]
- 9.Coen E.S. and Meyerowitz,E.M. (1991) The war of the whorls: genetic interactions controlling flower development. Nature, 353, 31–37. [DOI] [PubMed] [Google Scholar]
- 10.Jack T., Brockman,L.L. and Meyerowitz,E.M. (1992) The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell, 68, 683–697. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz-Sommer Z., Hue,I., Huijser,P., Flor,P.J., Hansen,R., Tetens,F., Lonnig,W.E., Saedler,H. and Sommer,H. (1992) Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J., 11, 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwarz-Sommer Z., Huijser,P., Nacken,W., Saedler,H. and Sommer,H. (1990) Genetic control of flower development by homeotic genes in Antirrhinum majus. Science, 250, 931–936. [DOI] [PubMed] [Google Scholar]
- 13.Yanofsky M.F., Ma,H., Bowman,J.L., Drews,G.N., Feldmann,K.A. and Meyerowitz,E.M. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature, 346, 35–39. [DOI] [PubMed] [Google Scholar]
- 14.Theissen G. and Saedler,H. (2001) Plant biology. Floral quartets. Nature, 409, 469–471. [DOI] [PubMed] [Google Scholar]
- 15.Theissen G. (2001) Genetics of identity. Nature, 414, 491. [DOI] [PubMed] [Google Scholar]
- 16.Egea-Cortines M., Saedler,H. and Sommer,H. (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J., 18, 5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jack T. (2001) Relearning our ABCs: new twists on an old model. Trends Plant Sci., 6, 310–316. [DOI] [PubMed] [Google Scholar]
- 18.Honma T. and Goto,K. (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature, 409, 525–529. [DOI] [PubMed] [Google Scholar]
- 19.Ma H., Yanofsky,M.F. and Meyerowitz,E.M. (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev., 5, 484–495. [DOI] [PubMed] [Google Scholar]
- 20.Munster T., Pahnke,J., Di Rosa,A., Kim,J.T., Martin,W., Saedler,H. and Theissen,G. (1997) Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc. Natl Acad. Sci. USA, 94, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shore P. and Sharrocks,A.D. (1995) The MADS-box family of transcription factors. Eur. J. Biochem., 229, 1–13. [DOI] [PubMed] [Google Scholar]
- 22.Cho S., Jang,S., Chae,S., Chung,K.M., Moon,Y.H., An,G. and Jang,S.K. (1999) Analysis of the C-terminal region of Arabidopsis thaliana APETALA1 as a transcription activation domain. Plant Mol. Biol., 40, 419–429. [DOI] [PubMed] [Google Scholar]
- 23.Moon Y.H., Jung,J.Y., Kang,H.G. and An,G. (1999) Identification of a rice APETALA3 homologue by yeast two-hybrid screening. Plant Mol. Biol., 40, 167–177. [DOI] [PubMed] [Google Scholar]
- 24.Lim J., Moon,Y.H., An,G. and Jang,S.K. (2000) Two rice MADS domain proteins interact with OsMADS1. Plant Mol. Biol., 44, 513–527. [DOI] [PubMed] [Google Scholar]
- 25.Lamb R.S. and Irish,V.F. (2003) Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl Acad. Sci. USA, 100, 6558–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies B. and Schwarz-Sommer,Z. (1994) Control of floral organ identity by homeotic MADS-box transcription factors. Results Probl. Cell Differ., 20, 235–258. [DOI] [PubMed] [Google Scholar]
- 27.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van De Peer Y. and De Wachter,R. (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci., 10, 569–570. [DOI] [PubMed] [Google Scholar]
- 29.Saitou N. and Nei,M. (1987) The neighbour-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol., 4, 406–425. [DOI] [PubMed] [Google Scholar]
- 30.Tajima F. and Nei,M. (1984) Estimation of evolutionary distance between nucleotide sequences. Mol. Biol. Evol., 1, 269–285. [DOI] [PubMed] [Google Scholar]
- 31.Kramer E.M., Dorit,R.L. and Irish,V.F. (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics, 149, 765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer E.M. and Irish,V.E. (2000) Evolution of the petal and stamen developmental programs: evidence from comparative studies of the lower eudicots and basal angiosperms. Int. J. Plant Sci., 16, 29–30. [Google Scholar]
- 33.Pnueli L., Abu-Abeid,M., Zamir,D., Nacken,W., Schwarz-Sommer,Z. and Lifschitz,E. (1991) The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J., 1, 255–266. [PubMed] [Google Scholar]
- 34.Van Der Krol A.R., Brunelle,A., Tsuchimoto,S. and Chua,N.H. (1993) Functional analysis of petunia floral homeotic MADS box gene pMADS1. Genes Dev., 7, 1214–1228. [DOI] [PubMed] [Google Scholar]
- 35.Kramer E.M., Di Stilio,V.S. and Schluter,P.M. (2003) Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci., 164, 1–11. [Google Scholar]
- 36.Munster T., Deleu,W., Wingen,L.U., Cacharrón,J., Ouzunova,M., Faigl,W., Werth,S., Kim,J.T.T., Saedler,H. and Theissen,G. (2002) Maize MADS-box genes galore. Maydica, 47, 287–301. [Google Scholar]
- 37.Doebley J. and Lukens,L. (1998) Transcriptional regulators and the evolution of plant form. Plant Cell, 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R.L., Stec,A., Hey,J., Lukens,L. and Doebley,J. (1999) The limits of selection during maize domestication. Nature, 398, 236–239. [DOI] [PubMed] [Google Scholar]
- 39.Force A., Lynch,M., Pickett,F.B., Amores,A., Yan,Y.L. and Postlethwait,J. (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics, 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prince V.E. and Pickett,F.B. (2002) Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet., 3, 827–837. [DOI] [PubMed] [Google Scholar]
- 41.Winter K.U., Weiser,C., Kaufmann,K., Bohne,A., Kirchner,C., Kanno,A., Saedler,H. and Theissen,G. (2002) Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol., 19, 587–596. [DOI] [PubMed] [Google Scholar]
- 42.Galant R. and Carroll,S.B. (2002) Evolution of a transcriptional repression domain in an insect Hox protein. Nature, 415, 910–913. [DOI] [PubMed] [Google Scholar]
- 43.Levine M. (2002) How insects lose their limbs. Nature, 415, 848–849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.