Abstract
The LSF/Grainyhead transcription factor family is involved in many important biological processes, including cell cycle, cell growth and development. In order to investigate the evolutionary conservation of these biological roles, we have characterized two new family members in Caenorhabditis elegans and Xenopus laevis. The C.elegans member, Ce-GRH-1, groups with the Grainyhead subfamily, while the X.laevis member, Xl-LSF, groups with the LSF subfamily. Ce-GRH-1 binds DNA in a sequence-specific manner identical to that of Drosophila melanogaster Grainyhead. In addition, Ce-GRH-1 binds to sequences upstream of the C.elegans gene encoding aromatic l-amino-acid decarboxylase and genes involved in post-embryonic development, mab-5 and dbl-1. All three C.elegans genes are homologs of D.melanogaster Grainyhead-regulated genes. RNA-mediated interference of Ce-grh-1 results in embryonic lethality in worms, accompanied by soft, defective cuticles. These phenotypes are strikingly similar to those observed previously in D.melanogaster grainyhead mutants, suggesting conservation of the developmental role of these family members over the course of evolution. Our phylogenetic analysis of the expanded LSF/GRH family (including other previously unrecognized proteins/ESTs) suggests that the structural and functional dichotomy of this family dates back more than 700 million years, i.e. to the time when the first multicellular organisms are thought to have arisen.
INTRODUCTION
The LSF/Grainyhead (GRH) transcription factor family initially included LSF, LBP-1a, LBP-9 and LBP-32 in Homo sapiens, CP2, NF2d9 and CRTR-1 in Mus musculus, Grainyhead in Drosophila melanogaster and cCP2 in Gallus gallus (Table 1). While D.melanogaster Grainyhead predominantly regulates genes involved in various stages of fruit fly development (1–4), H.sapiens and M.musculus LSF/CP2 members regulate expression of genes involved in a variety of cell cycle, growth and differentiation processes in non-developmental contexts (5–7). The precise basis of division of various functions among different family members is, however, unclear. The recent discovery and characterization of two new mammalian members, MGR (mammalian grainyhead) and BOM (brother of mammalian grainyhead), plus a new D.melanogaster member, dCP2 (considered to be the fruit fly ortholog of LSF), has provided initial evidence for the existence of two distinct divisions in this transcription factor family (8) (referred to as the LSF and GRH subfamilies in Table 1 and hereafter). Still, it is unclear if physiological functions among members within each subfamily are conserved and, if so, to what extent. The taxonomic spread across which physiological functions may be conserved, if at all, also remains to be determined.
Table 1. List of the various previously known LSF/GRH family members.
| Homo sapiens | Mus musculus | Gallus gallus | Drosophila melanogaster |
|---|---|---|---|
| LSF subfamily members | |||
| 1 LSF (34,35), CP2 (36), LBP-1c (37) | CP2 (36) | cCP2 (32) | dCP2 (8) |
| 2 LBP-1a (37) | NF2d9 (38) | ||
| 3 LBP-9 (39) | CRTR-1 (40) | ||
| Grainyhead subfamily members | |||
| 4 LBP-32 (39), MGR (8) | MGR (8) | Grainyhead (2), Elf-1 (41), NTF-1 (1) | |
| 5 BOM (8) | BOM (8) |
Orthologs are listed on the same row, along with literature references (in parentheses) and alternative names, if any.
We report the sequencing of new members in Caenorhabditis elegans and Xenopus laevis and investigate their placement in the family phylogeny. As part of our analysis, we have constructed protein sequence profiles and corresponding multi-alignments of the family members to address two key points: first, assignments of the new sequences from other species (including some ESTs) to the suggested family dichotomy; second, an identification of the distinctive as well as shared features of the two family divisions. In addition, the profiles provided a basis for constructing a phylogenetic tree that exploits the positional sequence variation across the entire family, rather than using just individual, pairwise similarities. This is the first comprehensive comparative analysis of the entire LSF/GRH family, including proteins/ESTs identified from additional genomes such as C.elegans, X.laevis, Danio rerio, Anopheles gambiae and Balanus amphitrite. Our analysis suggests that the family is ancient and that there is a distinct dichotomy among family members in the DNA binding structure and the type of DNA sequences bound, as well as in the type of physiological function. To this end, we have systematically characterized the DNA binding site preference of the C.elegans member, Ce-GRH-1, in vitro and have shown that it binds to sites upstream of the following C.elegans homologs of Grainyhead-regulated genes: mab-5, dbl-1 [both genes involved in post-embryonic development (9,10)] and the gene encoding aromatic l-amino acid decarboxylase. Using RNAi analysis, we have shown that Ce-grh-1 is an essential gene for embryonic development and cuticle morphogenesis therein and that this developmental role is conserved across evolution between C.elegans and D.melanogaster.
MATERIALS AND METHODS
GenBank accession nos of sequences analyzed
Ce-grh-1 mRNA, AY323527; Xl-LSF mRNA, AY323528; A.gambiae CP2, EAA11971; A.gambiae GRH, EAA03941; B.amphitrite BCS-3, BAA99545; Y48G8AR genomic clone, AC024797; Y48G8AR.1 (ORF), AAF60703; Xl-5prime EST, AW765842; Xl-3prime EST, AW640817; D.rerio EST 1, AL721647; D.rerio EST 2, AI794123; X.laevis EST 1, BJ066002; X.laevis EST 2, BJ060916; G.gallus EST 1, BU469673, Strongyloides stercoralis EST, BE581124; Conidiobolus coronatus EST, BQ621910.
Construction of protein sequence profiles
Bayesian prior-based PIMA profiles (11) of the protein sequences were constructed for the LSF and GRH subfamilies using local dynamic programming. Mus musculus and G.gallus LSF/GRH sequences were excluded from the defining set of the profile since they were nearly identical to the corresponding H.sapiens orthologs. Each position in the profile matrix has an estimated probability of being occupied by each of the 20 amino acids. The profile can also be represented as a regular expression pattern of amino acids or classes of amino acids based on physico-chemical side chain similarity (12). Profile searches were done using short-in-long dynamic programming and sequences with z-scores > 10 were considered significant. Division of the family based on similarity scores from BLAST analysis (BLASTP of the set of LSF/GRH proteins used in profile construction, against itself) yielded the same two subfamilies.
Identification of ESTs corresponding to LSF/GRH family members
Initial BLAST searches were performed using the H.sapiens LSF protein sequence as query against the translated dbEST database (TBLASTN) at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/dbEST) with an expectation value cut-off of 1e–10 and all other parameters as default. Selected ESTs, as indicated, were used as queries to search all identified LSF/GRH family protein sequences (BLASTX) using default parameters, in order to identify the protein with closest similarity.
Amplification of cDNAs by RT–PCR
Caenorhabditis elegans Bristol N2 strain worms were grown and staged as described previously (13). Five picomoles of the appropriate primer were annealed to total RNA (2 µg) from worms (13) or X.laevis oocytes (14) and then extended using 200 U Superscript Reverse Transcriptase II (Life Technologies) according to the manufacturer’s instructions. One-tenth of this cDNA was amplified by PCR using 400 nM forward and reverse primers, 200 µM dNTPs and 2.5 U LA Taq (Takara-Shuzo) in the presence of 1.5 mM MgCl2, 20 mM Tris–HCl (pH 8.0), 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol (DTT), 0.5% Tween 20, 0.5% Nonidet P-40 and 50% glycerol. PCR products were visualized by staining with ethidium bromide after electrophoresis through a 0.8% agarose gel. The following primers were used in the RT reactions: Ce-GRH-1 cDNA, CAGTAGAGTCGGAGCATTTCGTC myosin light chain cDNA–oligo(dT) (Fig. 2A); TGAGTTGGAATACGAGTTTGGAT (Fig. 2B); GTAAACGGTAAGCCTTGG (Fig. 2C). The following primers were used in the PCR reactions: Ce-GRH-1 cDNA, forward, GACAAAGTCCCATTCCACAGG, reverse, GTCAACTTTTACGGTGATGCC; myosin light chain cDNA, forward, CCGCCAAGAAGAAGTCCTCA, reverse, GTGGTAATGAGGTGAGCGAAGG (Fig. 2A); forward primer SL1, GTTTAATTACCCAAGTTTGA; forward primer SL2, GGTTTTAACCCAGTTACT; M13125 HindIII primer, gcggaaagcttATGTCATTCCAACTTGACC (bases in lower case are in addition to the cDNA sequence and contain a HindIII restriction site); M12705 primer, ATGCCATCACCAGTGGAT; M12615 primer, ATGCTGGAAAGAAGTAGTG; reverse XbaI primer, tgctgtctagatcaCGAGTTTGGATTGGT GGG (bases in lower case are in addition to the cDNA sequence and contain an XbaI restriction site) (Fig. 2B); forward, ATGAGCGATGTGCTTGCCTTG, reverse, GCCATCAGCAGGACCACAG (Fig. 2C). RT–PCR products were sequenced by Davis Sequencing (Davis, CA).
Figure 2.
Experimental amplification of mRNAs of two newly predicted LSF/GRH family members. (A) RT–PCR of Ce-GRH-1 mRNA using total RNA extracted from C.elegans N2 strain in five developmental stages: egg (E, lane 2), larval stages L2–L4 (lanes 3–5, respectively) and adult (A, lane 6). Upper lanes correspond to Ce-GRH-1 primers and lower lanes correspond to myosin light chain primers (positive control). Lane 1 indicates DNA size markers. (B) RT–PCR analysis of Ce-GRH-1 mRNA to determine the translation initiation site of the coding region mRNA, using total RNA from mixed stage N2 strain worms. Amplifications by PCR were performed in duplicate using forward primers for splice leaders SL1 (lanes 2 and 3) or primers in the region of three different in-frame methionine codons located upstream of the computationally predicted translation initiation site in the putative first exon (lanes 4 and 5, primer M13125, i.e. a primer in the region of the methionine codon at position 13125 on the parent genomic clone, Y48G8AR; lanes 6 and 7, primer M12705; lanes 8 and 9, primer M12615). Lane 1 indicates DNA size markers. The asterisk (*) denotes products from lanes 2–5 that were purified and sequenced. Additional shorter bands observable in lanes 2 and 3 apparently result from other SL1-primed mRNAs containing similarity to the Ce-GRH-1 mRNA in the region of the gene-specific RT primer. (C) RT–PCR of Xl-LSF mRNA using two preparations of total RNA extracted from X.laevis oocytes (lanes 2 and 3). Lane 1 indicates DNA size markers.
Construction of phylogenetic trees
A phylogenetic tree was built using beta version 4.0b10 of PAUP (15) on the basis of variations in each position (total of 189 variable positions) of the alignable region across all identified proteins in the family. Protein sequence profiles (above) were used as the basis for the multi-alignment. These alignments were refined by hand. Regions that could not be aligned across all sequences were discarded. Parsimony was used as the optimality criterion for building the tree. Bootstrap trees were built for 400 replications and values ≥60% were incorporated into the phylogenetic tree. Translated EST sequences were not directly used for the analysis since they did not represent the full-length protein and did not extend over the entire multi-alignment. Rather, their placements in the tree were deduced from BLAST sequence similarity scores to other full-length LSF/GRH family proteins.
Cloning and in vitro transcription/translation
Near full-length Ce-GRH-1 cDNA amplified by RT–PCR using the M13125 HindIII and reverse XbaI primers (above) was cloned into the HindIII and XbaI sites of pBluescript (SK–); this plasmid was named CeGRHpBS. Proteins were synthesized in vitro from 2 µg CeGRHpBS (Ce-GRH-1) or 2 µg pTβStuNTF-1 (GRH) (16) in 25 µl reaction volumes using the TNT T7 Quick Coupled Transcription/Translation System (Promega).
Determination of C.elegans homologs
Caenorhabditis elegans homologs for Ubx, Ddc, PCNA and dpp were assigned by BLASTP of proteins corresponding to these genes against Wormpep 79 (http://www.wormbase.org) with an e-value cut-off of e–20. Reciprocal BLASTP of the most similar Wormpep sequences obtained above against the D.melanogaster protein database (release 2, http://www.flybase.org) correctly yielded back proteins corresponding to Ddc, PCNA and dpp. A one-to-one homolog assignment was not possible for Ubx owing to the presence of other similar homeodomain proteins in these genomes. However, phylogenetic analyses of various nematode HOX genes suggest that mab-5, the top-scoring BLAST match to Ubx, is one of the C.elegans Antp group genes (17), probably an ortholog of ftz (18). Since both Ubx and ftz in the Antp group are regulated by Grainyhead (1), we included mab-5 in our analysis.
Assignment of putative promoter regions of C.elegans genes
cDNAs corresponding to the coding regions of each of the C.elegans genes (above) were used as queries for TBLASTX searches against C.elegans ESTs in dbEST with a threshold of e–50. Genomic locations of those ESTs (if any) with sequences extending 5′ of the translation start site were determined using SIM4 (19) alignment to C.elegans genomic sequence. ESTs mapping to genomic locations distinct from that of the cDNA or mapping to ambiguous genomic locations (less than 85% identity over less than 85% of the length) were discarded. The 5′-most position thus obtained from among the cDNAs and ESTs was defined to be the approximate transcription start site and 1200 bases upstream were chosen for promoter analysis. Promoter regions were scanned for identity to the core GRH binding site, C(C/T)(T/G)G.
Electrophoretic mobility shift assays
One hundred femtomoles of radiolabeled DNA and 6.3 µl of Ce-GRH-1 or 3 µl of GRH protein (quantitated to be equimolar amounts) from in vitro translation reactions were added to a buffer containing 5 mM MgCl2, 16 mM KCl, 165 ng/µl bovine serum albumin (BSA), 10% glycerol, 20 mM HEPES (pH 7.9), 10 mM EDTA and 10 mM DTT along with 13 ng/µl salmon sperm carrier DNA. Non-radiolabeled competitor DNA (where indicated) was added to the reactions in 10- or 40-fold molar excess over radiolabeled DNA. Proteins were incubated with DNA for 25 min at 23–25°C and electrophoresed through a 5% polyacrylamide gel containing 0.2× TBE at 4°C. Gels were dried and analyzed using a phosphorimager (Molecular Dynamics) and ImageQuant software. The following oligonucleotide sequences (annealed to their respective complementary sequences) were used in EMSAs (nucleotides in lower case indicate changes in sequence of the four UbxMt DNAs with respect to the wild-type Ubx DNA): dpp-DREB, 5′-CTTTTACCTGCTCTTCCG-3′; Ubx, 5′-GATCAAACAATCTGGTTTTGAGCGTTA-3′; UbxMt1, 5′-GATCAAACAATtaGGTTTTGAGCG TTA-3′; UbxMt2, 5′-GATCAAACAATCTGGacgTGAGCGTTA-3′; UbxMt3, 5′-GATCAAACctaCTGGTTTTGAGCGTTA-3′; UbxMt4, 5′-GATCAAACAATCTacTTTTGAGCG TTA-3′; Ddc (be-2), 5′-CTAGAGCGATTGAACCGGTCCTGCGGT-3′; PCNA, 5′-TGCCAACTGGTTTGATTGTTCACACTTTTT-3′; dbl-1 I, 5′-TTTCATACTGGTTGCT TGA-3′; dbl-1 II, 5′-AAACATCTGGAACATTTT-3′; mab-5, 5′-AAACAAACCTGATATATT-3′; CeDdc (gene encoding aromatic l-amino acid decarboxylase) I, 5′-ACTTTTCCCTGGGCTAATG-3′; CeDdc II, 5′-CCAAGTTCC CTGATAAATA-3′; CeDdc III, 5′-ACCAATACTGGGAGTTTGC-3′; CeDdc IV, 5′-TAAACGACTTGAAAAATA-3′; CeDdc V, 5′-GTCTACACACCTGTTTTAACA-3′; pcn-1 I, 5′-AAAATCGCTGGTAAATTC-3′; pcn-1 II, 5′-AAATGCCTGGTACGCAAT-3′.
RNAi analysis
Ce-GRH-1 sense and antisense RNA were transcribed from linearized CeGRHpBS plasmid DNA. Transcription reactions were carried out separately using T3 or T7 RNA polymerase Ambion MEGAscript™ High Yield Transcription kits according to the manufacturer’s instructions. Product purity was verified by agarose/formaldehyde gel electrophoresis. Equimolar amounts of sense and antisense RNA were annealed in injection buffer (2% polyethylene glycol 8000, 20 mM potassium phosphate, 3 mM potassium citrate, pH 7.5) to yield double-stranded RNA (dsRNA) at a concentration of 2.5 µg/µl. L4 stage larvae (N2 strain) were injected with dsRNA as described in Fire et al. (20) and were monitored for a period of 3 days post-injection. Injected parent generation (P0) animals were transferred to fresh plates every 12 h. The numbers of live eggs, dead eggs, larvae and adult animals on all plates that had contained an injected animal (P0) were determined.
RESULTS
Identification of new LSF/GRH family members
In order to better understand the evolution and functional divisions of the LSF/GRH protein family, we undertook a comparative analysis of known family members. Starting with the two subfamilies suggested previously (8), we generated two Bayesian prior-based protein sequence profiles (11) which are diagnostic of this subfamily division. These profiles were used to search the database of all annotated proteins to identify new LSF/GRH homologs. Two predicted ORFs in C.elegans (one a 167 amino acid predicted protein, Y48G8AR.a, and the other a 154 amino acid predicted protein, Y48G8AR.b, in Wormpep 19), two predicted proteins in A.gambiae (mosquito) and one protein, BCS-3, in B.amphitrite (barnacle) were identified. BLAST (21) searches using human LSF as the query against the six frame translated dbEST database identified ESTs corresponding to the expected H.sapiens, M.musculus, G.gallus, D.melanogaster and A.gambiae proteins, along with ESTs from several other species.
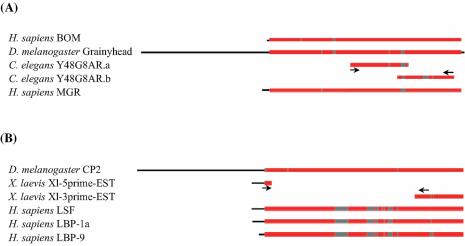
The two C.elegans ORFs aligned to adjacent, partially overlapping regions in the GRH subfamily profile (Fig. 1A) as well as in the parent genomic clone, Y48G8AR. This suggested that these ORFs had been incorrectly predicted and in fact code for a single protein. Further, since the combined span of the two sequences was only around 300 amino acids, rather than the expected 500 amino acids or longer in other family members, we re-analyzed the Y48G8AR genomic clone sequence around these ORFs. The gene prediction program GeneID (22) predicted somewhat different exon boundaries compared to those in the Y48G8AR.a/b sequences and predicted additional N- and C-terminal coding exons. The extended coding sequence showed significantly greater similarity to the profile than did the originally annotated ORFs.
Figure 1.
Bar representation of protein sequence profile-induced multiple alignments of the GRH and LSF subfamilies. (A) GRH profile, including the two original Wormpep 19 ORFs. (B) LSF profile, including two translated X.laevis ESTs [from the Harland stage 19–23 library and Blackshear/Soares normalized X.laevis egg library (25)]. Regions of the sequences in red indicate alignment to the profile, those in gray indicate alignment gaps and those in black indicate non-homologous overhangs. The arrows indicate the location of oligonucleotide primers chosen for RT–PCR analyses shown in Figure 2.
Experimental determination of Ce-grh-1 and Xl-LSF gene structure
We tested our computational prediction of the gene structure of the C.elegans member (hereafter referred to as Ce-grh-1, corresponding to the new gene class ‘grh’ as submitted to the Caenorhabditis Genetics Center) by performing RT–PCR assays (Fig. 2A) using RNA extracted from N2 strain worms in different developmental stages. The positions of the primers are indicated by the arrows in Figure 1A. Sequencing of purified amplified products yielded exon boundaries similar to the GeneID predicted gene structure. In order to determine the translation initiation site, we took advantage of the phenomenon of trans-splicing (23) that occurs to generate mRNAs of an estimated 70% (24) of the genes in C.elegans. In these genes, 5′ untranslated regions of pre-mRNAs are trans-spliced by splice leader 1 (SL1) RNA and internal sites upstream of coding regions in polycistronic transcripts are trans-spliced by SL2 RNA. RT–PCR analysis of C.elegans total RNA was performed using sequence complementary either to SL1, to SL2 or to regions of in-frame methionine codons upstream of the computationally predicted translation start site in the putative first exon as a forward primer (Fig. 2B). No major products were obtained in amplification reactions using the SL2 primer (data not shown). The RT–PCR product corresponding to the forward primer around M13125 (methionine at position 13125 in Y48G8AR) was approximately the same size as the largest product amplified using the SL1 primer (lanes 2–5), suggesting that the Ce-GRH-1 pre-mRNA is trans-spliced to SL1 near the codon for M13125 and that this codon represents the translation initiation site. We confirmed that the junction for SL1 trans-splicing is one base upstream of the codon for M13125 by sequencing the indicated products (Fig. 2B). Therefore, the predicted, full-length Ce-GRH-1 protein is 563 amino acids long, and comprises eight exons. (The ORFs predicted by Wormpep19, Y48G8AR.a and Y48G8AR.b, stand partly corrected in Wormpep 96 as Y48G8AR.1; this predicted ORF has nine exons with a conceptual translated product of 584 amino acids. It still differs from our experimentally verified gene structure in some exon–intron boundaries and in the presence of an additional exon.) Ce-GRH-1 mRNA is expressed in all five developmental stages tested, and at apparently comparable levels.
Two X.laevis ESTs, when translated, aligned to N- and C-terminal regions of the LSF subfamily profile (Xl-5prime EST and Xl-3prime EST in Fig. 1B). In order to determine if they indeed represented two regions of the same transcript and, if so, to determine the full-length protein sequence, we performed RT–PCR assays using RNA extracted from X.laevis oocytes (Fig. 2C) and primers positioned as indicated in Figure 1B. The complete X.laevis member (hereafter referred to as Xl-LSF) sequence was then reconstructed using the sequence from the purified RT–PCR product plus the flanking EST sequences. Given the 87% identity over the alignment between Xl-LSF (506 amino acids) and human-LSF (502 amino acids) and the fact that the 3′ X.laevis EST was generated from poly(A)-selected mRNAs (25), it is likely that the predicted Xl-LSF protein is full length.
Protein sequence profile and phylogenetic analyses of the expanded LSF/GRH family
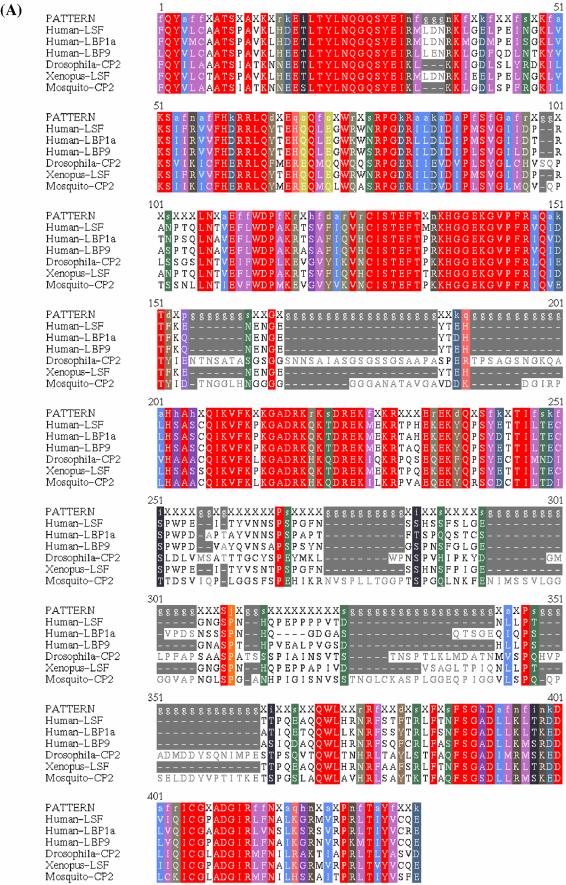
The protein sequence profile for each subfamily was expanded to include the additional members from C.elegans, X.laevis, B.amphitrite and A.gambiae. Ce-GRH-1, BCS-3 (B.amphitrite) and one of the A.gambiae members (referred to as A.gambiae GRH) aligned to the GRH profile. Xl-LSF and the other A.gambiae member (referred to as A.gambiae CP2) aligned to the LSF profile. Profile-induced multi-alignments based on the expanded LSF and GRH subfamilies show conservation within, as well as between, subfamilies in the regions required for DNA binding (26,27) (Fig. 3). However, the regions required for oligomerization (26,27) are conserved only within each subfamily (Fig. 3). It is useful to note that the region required for DNA binding of LSF as mapped by deletion analysis (26,27) partially overlaps the oligomerization-associated region and is consistent with the fact that tetramerization of LSF is a prerequisite for DNA binding. The region directly interacting with DNA, although unknown, is likely to be within the region depicted in Figure 3D.
Figure 3.
(Above and previous two pages) Protein sequence profile analysis of LSF and GRH subfamilies. (A) Profile-induced multi-alignment of protein sequences within the LSF subfamily, spanning the regions associated with DNA binding and oligomerization. (B) Profile-induced multi-alignment of protein sequences within the GRH subfamily in the region associated with DNA binding. (C) Profile-induced multi-alignment of protein sequences within the GRH subfamily in the region associated with oligomerization. (D) Profile–profile alignment of the two LSF and GRH profiles in the region associated with DNA binding. Asterisks (*) in the alignment indicate conserved amino acid/amino acid class identities and dashes (–) indicate similarities based on amino acid classes (below). Profile positions that are identical or similar to each other are indicated in bold. (E) Regions associated with oligomerization that apparently cannot be aligned between the two profiles. Gaps in the profiles are not shown in (D) and (E). Profiles are represented as regular expressions. Lower case letters in the regular expression indicate amino acid classes based on physico-chemical properties (a: I, L and V; d: F,W and Y; e: F, W, Y and H; f: A, I, L, V, M, F, W, Y and C; h: A, G and S; i: S and T; j: G, N and P; k: D and E; l: D and N; n: K and R; o: E and Q; p: D, E, N and Q; q: H, K and R; r: D, E, N, Q, H, K and R; s: D, E, N, Q, H, K, R, S and T; x, any amino acid) (details of the coloring scheme are available at http://bmerc-www.bu.edu/description/aaclasses.html) (12). DNA binding and oligomerization-associated regions are based on mapping by deletion analysis (26,27).
We then constructed a phylogenetic tree exploiting the positional amino acid variations across the entire family (including the new sequences in C.elegans, X.laevis, A.gambiae and B.amphitrite) using PAUP (15). We restricted our analysis to regions that could be aligned across all sequences. The tree has two distinct divisions corresponding to the two subfamily profiles (Fig. 4). The time scales estimated in the tree suggest that this division occurred more than 700 million years ago. It is important to note that in such a reconstruction, the longer branches are more informative. Also, the implied variation in the length of terminal branches suggests some variation in the rates of evolution, particularly between vertebrates and invertebrates in the GRH subfamily.
Figure 4.
Phylogenetic tree based on positional variation within regions of sequence that aligned across all identified LSF/GRH family members. Bootstrap values ≥60% are indicated near the nodes and are based on 400 replications. The scale bar corresponds to 10 substitutions per 100 positions per unit branch length. Dotted lines indicate estimated placements of G.gallus, D.rerio and X.laevis translated ESTs based on BLAST sequence similarity scores. Time scales are based on current estimates of the mammalian radiation at 175 million years and the vertebrate–invertebrate separation at 450 million years (42). Double-headed arrows indicate the temporal uncertainty based on apparent spread in extant branch termini.
Ce-GRH-1 binds DNA sequences upstream of genes involved in post-embryonic development with binding site preference identical to that of Grainyhead
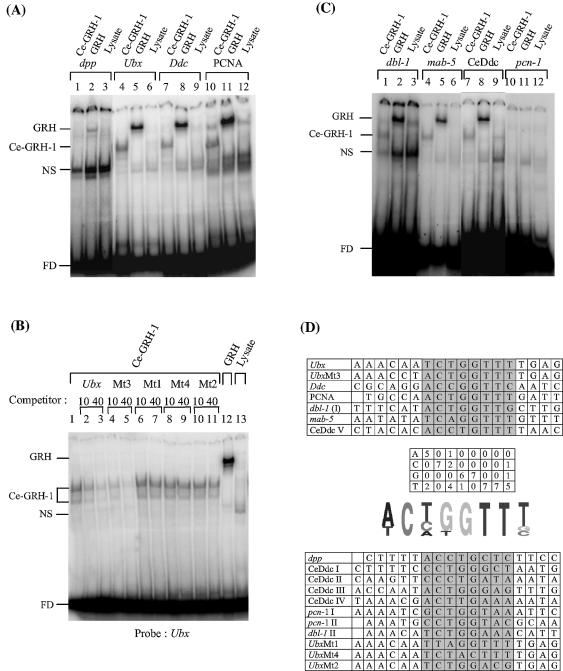
The phylogenetic placement of Ce-grh-1 suggests that it is a member of the GRH subfamily. To test this experimentally and to characterize the DNA binding characteristics of Ce-GRH-1 protein, we performed electrophoretic mobility shift assays using in vitro translated Ce-GRH-1 and four different known Grainyhead binding sites upstream of the following D.melanogaster genes: decapentaplegic (dpp) (4), Ultrabithorax (Ubx) (1), dopa decarboxylase (Ddc) (2) and PCNA (28) (Fig. 5A). Both Ce-GRH-1 and Grainyhead bind to the Ubx (lanes 4 and 5), Ddc (lanes 7 and 8) and PCNA (lanes 10 and 11) sites, although Ce-GRH-1 has an apparent lower binding affinity. Grainyhead only weakly binds the dpp site (lane 2), where no binding is observable with Ce-GRH-1 (lane 1). This is not surprising given the relative affinities for binding of the two proteins to the other sites tested. In a competition binding assay of Ce-GRH-1 with wild-type or mutant Ubx DNAs (Fig. 5B), 10- and 40-fold molar excess of non-radiolabeled wild-type or Mt3 DNA competed effectively for binding (lanes 2 –5), while a 40-fold excess of Mt1, Mt4 or Mt2 DNA offered little competition (lanes 6–11), demonstrating the sequence specificity of the central C(A/C/T)(T/G)G as well as flanking bases in the site required for DNA binding (Fig. 5D). Similar results were obtained for competition assays with Grainyhead (data not shown). Thus, Ce-GRH-1 binds DNA in a sequence-specific manner identical to that of Grainyhead.
Figure 5.
Ce-GRH-1 and Grainyhead have identical DNA binding site preferences. (A) Ce-GRH-1 and Grainyhead have identical binding preferences to known D.melanogaster Grainyhead DNA binding sites. Binding reactions of Ce-GRH-1 and Grainyhead with D.melanogaster DNA binding sites upstream of the following genes: dpp (lanes 1 and 2), Ubx (lanes 4 and 5), Ddc (lanes 7 and 8) and PCNA (lanes 10 and 11) are shown. Lanes 3, 6, 9 and 12 display binding reactions of mock-translated rabbit reticulocyte lysate (negative control). Lanes 1–3 are scanned at higher exposure for ease of visualization. (B) Ce-GRH-1 binds the Grainyhead Ubx site in a sequence-specific manner. Competition assays were performed with 10- or 40-fold molar excess of non- radiolabeled wild-type (lanes 2 and 3) or mutant (4–11) Ubx DNAs and were compared to binding of radiolabeled wild-type Ubx DNA alone (lane 1). Lane 12 displays the binding of D.melanogaster Grainyhead to the Ubx binding site and lane 13 displays the reaction with mock-translated rabbit reticulocyte lysate. (C) Ce-GRH-1 and Grainyhead have identical binding preferences to C.elegans sequences upstream of genes homologous to Grainyhead-regulated genes. Binding reactions of Ce-GRH-1 and Grainyhead with C.elegans sequences upstream of the following genes: dbl-1 (lanes 1 and 2), mab-5 (lanes 4 and 5), the gene encoding aromatic l-amino acid decarboxylase (abbreviated CeDdc, lanes 7 and 8) and pcn-1 (lanes 10 and 11) are shown. Lanes 3, 6, 9 and 12 display binding reactions of mock-translated rabbit reticulocyte lysate (negative control). Lanes 7–9 are scanned at higher exposure for ease of visualization. EMSAs were performed with in vitro translated proteins. FD indicates free DNA and NS indicates the non-specific protein–DNA complex formed by proteins in the rabbit reticulocyte lysate. (D) (Upper) Alignment of sequences bound by Ce-GRH-1. (Middle) Count matrix of the number of occurrences of the four nucleotides in each of the positions deduced to be important for interaction with Ce-GRH-1 and a Pictogram (http://genes.mit.edu/pictogram.html) visualization of these nucleotide frequencies. (Lower) Alignment of sequences not bound by Ce-GRH-1 (under our binding conditions). Positions deduced to be important for binding are shaded in gray.
To test whether there is functional conservation between the two proteins, the upstream regions of C.elegans homologs of the four genes, dpp (C.elegans dbl-1), Ubx (C.elegans mab-5), Ddc (C.elegans gene encoding aromatic l-amino acid decarboxylase) and PCNA (C.elegans pcn-1) were scanned for potential Ce-GRH-1 binding sites. Binding assays of Ce-GRH-1 and Grainyhead with DNAs containing these sites were performed (Fig. 5C and data not shown). Both Ce-GRH-1 and Grainyhead bound to sites upstream of dbl-1 (lanes 1 and 2), mab-5 (lanes 4 and 5) and the gene encoding aromatic l-amino acid decarboxylase (‘CeDdc’, lanes 7 and 8), again with Ce-GRH-1 binding with apparently lower affinity. The various Ce-GRH-1 binding sites (Fig. 5D, upper panel) were then aligned and compared to an alignment of DNAs that were not bound (Fig. 5D, lower panel), at least under our experimental conditions. Based on this small data set, we deduced that at least eight contiguous base positions are important for Ce-GRH-1 binding. A matrix was generated with the number of occurrences of each of the four nucleotides in each of these eight positions (Fig. 5D, middle panels indicating the matrix and the derived pictogram visualization). The preference for an adenine immediately upstream and three thymidines immediately downstream of the central C(A/C/T)(T/G)G is consistent with the lack of binding in DNAs where these positions exhibit transversions.
Ce-grh-1 RNAi worms are embryonic lethal with defective cuticles
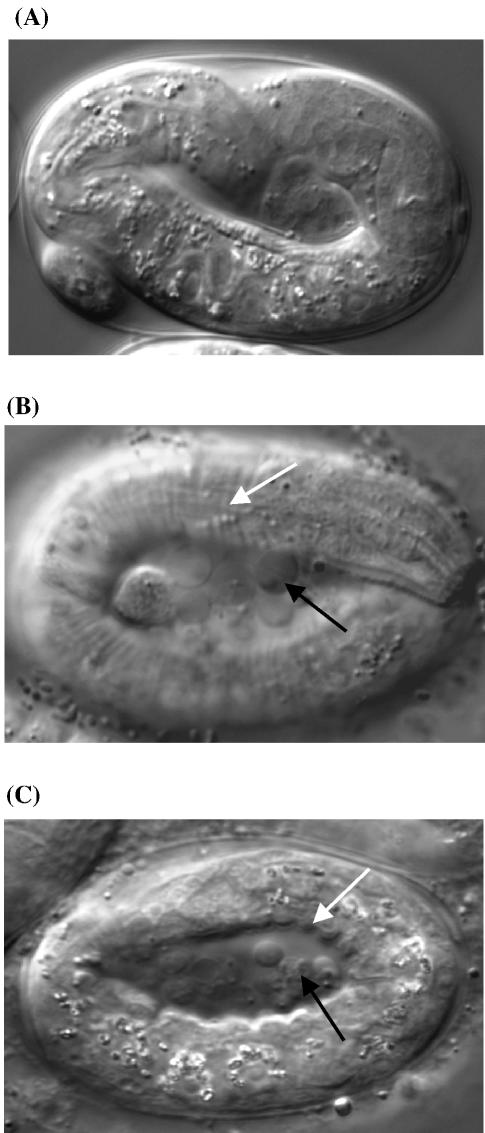
We examined the function of Ce-GRH-1 in vivo using RNAi analysis in C.elegans. A 1.6 kb region of double-stranded RNA corresponding to the full-length Ce-GRH-1 coding region was prepared and injected into N2 strain C.elegans L4 stage larvae. Upon reaching adulthood, these injected animals produced apparently normal numbers of embryos, however, greater than 95% of these embryos failed to hatch. The remainder hatched and then died as L1 stage larvae. The embryos arrested development at the three-fold stage with well differentiated tissues and apparently normal motility within the egg. However, muscle contractions that would normally cause the body and cuticle of the animal to bend smoothly were instead observed to induce constrictions or puckering in the external cuticle (compare Fig. 6A with B and C). All of the arrested embryos observed (n > 500) exhibited this phenotype, suggesting that the cuticles of the Ce-grh-1 (RNAi) embryos are malformed and may lack the rigidity necessary for normal motility and hatching. However, further ultrastructural studies will be required to determine what, if any, specific defects exist in the cuticle architecture. Also consistent with the idea that a cuticle defect underlies the Ce-grh-1 phenotype, we found that ∼8% of the Ce-grh-1 (RNAi) embryos exhibit ruptures in their cuticles resulting in the extrusion of cells from the bodies of the animals (Fig. 6B and C and data not shown).
Figure 6.
Nomarski images of three-fold stage C.elegans embryos. (A) Wild-type animal. (B and C) Ce-GRH-1 RNAi embryos. The embryos in (B) and (C) exhibit abnormal puckering in the cuticle (white arrows) and extruded cells (black arrows).
DISCUSSION
We have undertaken the first comprehensive comparative genome analysis of the LSF/GRH transcription factor family, members of which play an important role in the cell cycle, growth, differentiation and development. We have sequenced two new family members, Ce-GRH-1 and Xl-LSF, and identified several other previously unrecognized members. mRNA of Ce-GRH-1, the novel C.elegans member, is expressed in at least five developmental stages and its mRNA is SL1 trans-spliced one base upstream of the codon representing the translation initiation site. Our protein sequence profile and phylogenetic analysis both reflect the dichotomy in the LSF/GRH family (8). The DNA binding site preference of Ce-GRH-1 is consistent with its phylogenetic placement in the Grainyhead subfamily. Our findings are that (i) Ce-GRH-1 binds to the promoters of three genes involved in post-embryonic development that are homologous to Grainyhead-regulated genes and (ii) inhibition of Ce-grh-1 by RNAi results in embryonic lethality. These strongly support the hypothesis that the GRH subfamily members are involved in development.
Our computational analysis of the expanded LSF/GRH family, together with the results of Wilanowski et al., suggest that the phylogenetic division into two subfamilies reflects significant differences in structure and function between them. Functionally, Grainyhead, the best studied protein in its subfamily, is mainly a regulator of developmental control genes in D.melanogaster, such as Ultrabithorax (1), dopa decarboxylase (2), tailless (3) and decapentaplegic (4). Fruit flies carrying grainyhead mutations display an embryonic lethal phenotype (2). Similarly, H.sapiens MGR (presumably the H.sapiens ortholog of grainyhead) binds to and can activate the promoter of engrailed, a gene involved in development (8). Ce-GRH-1, the putative C.elegans ortholog, binds the promoters of the gene encoding aromatic l-amino acid decarboxylase and genes involved in post-embryonic development, mab-5 and dbl-1 (9,10) (Fig. 5C). These genes are all homologs of D.melanogaster Grainyhead-regulated genes. It remains to be seen if Ce-GRH-1 regulates the transcription of these genes in vivo. Ce-grh-1 phenotypic knockout worms generated by our RNAi analysis are late embryonic lethal and have cuticles apparently defective in rigidity (Fig. 6). The observed cuticle rupturing in these RNAi worms suggests that their cuticles are too weak to maintain structural integrity. These phenotypes are strikingly similar to the phenotypes of grainyhead mutant fruit flies, which also die at the end of embryogenesis, have granular head skeletal structures and misshapen, weak cuticles that manifest distended bulges and rupture easily (2). In addition, Ce-GRH-1 binds to the promoter of the gene encoding aromatic l-amino acid decarboxylase (Fig. 5C), the D.melanogaster homolog of which is involved in cuticle hardening and is regulated by Grainyhead (2). These findings suggest that Grainyhead and Ce-GRH-1 regulate genes in the embryonic epidermis involved in cuticular morphogenesis pathways and that these developmental pathways have been conserved during the course of evolution. Such conservation of developmental gene regulatory networks across evolution has been found previously (29). Specifically, there is phylogenetic evidence for the existence of a monophyletic clade (Ecdysozoa) of moulting animals including arthropods and nematodes (30), suggesting that the moulting process arose once and that related molecular mechanisms are common to these species.
Homo sapiens and M.musculus LSF/CP2, on the other hand, bind to promoters of genes such as DNA polymerase β (31), thymidylate synthase (7), c-fos (R.Misra, H.-C.Huang, M.Greenberg and U.Hansen, unpublished data), ornithine decarboxylase (J.Volker, A.P.Butler and U.Hansen, unpublished data), α-globin (5) and IL-4 (6), involved in a wide variety of processes, including the cell cycle, growth and differentiation.
Although the protein sequence profiles representing the two family divisions show conservation in the DNA binding-associated region as mapped by deletion analysis, they show no commonality in the oligomerization-associated region (Fig. 3). Further, there is no protein–protein interaction observed between proteins belonging to different subfamilies (8,26). Structurally, Grainyhead binds to a ‘single’ DNA site as a dimer while LSF binds two direct repeat DNA sites as a tetramer (16,27,32). Taken together, these observations suggest fundamental differences in the quaternary DNA binding protein structure between the two subfamilies.
Our analysis of Ce-grh-1 is consistent with this family dichotomy, based both on phylogenetic and experimental data. We have demonstrated that the binding site preference for Ce-GRH-1 is identical to that of Grainyhead, although our data set is statistically small. In addition to competition analyses using mutated DNA binding sites, comparison of sequences of DNAs that bound Ce-GRH-1 versus those that did not (at least under our binding conditions) indicated contiguous base positions important for interaction with Ce-GRH-1; these agree with the base preferences estimated in the DNA binding site count matrix.
Among the known full-length proteins in the family, the H.sapiens, M.musculus, D.melanogaster and A.gambiae genomes have evolved members in both the GRH and LSF subfamilies (Fig. 4). Although there is only a single identified member each in X.laevis and G.gallus (both in the LSF subfamily), the dbEST database contains additional X.laevis EST sequences with greater than 85% identity over their entire length to LBP-9 in the LSF subfamily and MGR in the GRH subfamily and a similar G.gallus EST corresponding to MGR (dotted lines in tree, Fig. 4). There is also EST evidence for the existence of genes similar to LSF and MGR in D.rerio (zebrafish) (dotted lines in tree, Fig. 4), reflecting a similar gene duplication event. The B.amphitrite BCS-3 protein groups with the GRH subfamily in the tree, and its cDNA is selectively expressed in the larval stage (33). Given its phylogenetic placement, we anticipate that there is at least one additional member belonging to the LSF subfamily in this genome.
Finally, the C.elegans genome apparently has a single member, which clusters with the GRH subfamily. Similarly, there is evidence for GRH subfamily members in other nematodes such as Caenorhabditis briggsae (contig FPC2032 from assembly cb25.agp8, derived from BLAST analysis of C.briggsae genomic contigs) and S.stercoralis (BLAST analysis of dbEST). However, there is no evidence for a nematode protein in the LSF subfamily in terms of additional products in our RT–PCR assays performed across C.elegans developmental stages (Fig. 2A), sequence similarities to nematode ESTs or sequence similarities to either the complete genome sequence of C.elegans or the available genome sequence of C.briggsae. Thus, the nematodes alone appear to have a representative from only one of these two subfamilies. The function of the other gene(s) may have been lost through the course of evolution in these genomes. There is precedent for gene loss in nematodes in the case of other gene families, including the HOX gene cluster (18). An alternative hypothesis is that the LSF subfamily functions may have been subsumed by the GRH member in nematodes.
Our phylogenetic analysis, along with the known functions and quaternary structures of the LSF/GRH family members, suggests that the family underwent a major gene duplication event more than 700 million years ago (Fig. 4), when the first multicellular organisms are thought to have evolved. (Consistent with this estimated time line, the only EST from fungi with even weak sequence similarity to proteins in the LSF/GRH family is an EST from C.coronatus, a multicellular fungus.) This gene duplication event may have resulted in a distinct functional and structural division among its members.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Laura Attardi and Robert Tjian for the pTβStuNTF-1 plasmid, Modular Genetics Inc. for the generous gift of oligonucleotides used in the EMSAs, Chris Li for providing reagents and C.elegans RNA samples, Kyuhyung Kim for help with RNAi injections, Yanxia Bei for help with manipulating embryos, Jim Deshler for providing X.laevis oocytes and for facilitating searches of X.laevis ESTs and Ying-Bing Zhou, John Finnerty, Scott Mohr and John Spieth for helpful discussions. K.V. and T.F.S. were supported by NSF grant DBI-98097993. K.V., U.H. and cost of supplies were supported by NIH grant CA81157. H.R.M. participated in the Federal Work-study Program.
REFERENCES
- 1.Dynlacht B.D., Attardi,L.D., Admon,A., Freeman,M. and Tjian,R. (1989) Functional analysis of NTF-1, a developmentally regulated Drosophila transcription factor that binds neuronal cis elements. Genes Dev., 3, 1677–1688. [DOI] [PubMed] [Google Scholar]
- 2.Bray S.J. and Kafatos,F.C. (1991) Developmental function of Elf-1: an essential transcription factor during embryogenesis in Drosophila. Genes Dev., 5, 1672–1683. [DOI] [PubMed] [Google Scholar]
- 3.Huang J.-D., Dubnicoff,T., Liaw,G.-J., Bai,Y., Valentine,S.A., Shirokawa,J.M., Lengyel,J.A. and Courey,A.J. (1995) Binding sites for transcription factor NTF-1/Elf-1 contribute to the ventral repression of decapentaplegic. Genes Dev., 9, 3177–3189. [DOI] [PubMed] [Google Scholar]
- 4.Liaw G.-J., Rudolph,K.M., Huang,J.-D., Dubnicoff,T., Courey,A.J. and Lengyel,J.A. (1995) The torso response element binds GAGA and NTF-1/Elf-1, and regulates tailless by relief of repression. Genes Dev., 9, 3163–3176. [DOI] [PubMed] [Google Scholar]
- 5.Lim L.C., Fang,L., Swendeman,S.L. and Sheffery,M. (1993) Characterization of the molecularly cloned murine α-globin transcription factor CP2. J. Biol. Chem., 268, 18008–18017. [PubMed] [Google Scholar]
- 6.Casolaro V., Keane-Myers,A.M., Swendeman,S.L., Steindler,C., Zhong,F., Sheffery,M., Georas,S.N. and Ono,S.J. (2000) Identification and characterization of a critical CP-2 binding element in the human interleukin-4 promoter. J. Biol. Chem., 275, 36605–36611. [DOI] [PubMed] [Google Scholar]
- 7.Powell C.M.H., Rudge,T.L., Zhu,Q., Johnson,L.F. and Hansen,U. (2000) Inhibition of the mammalian transcription factor LSF induces S-phase dependent apoptosis by down regulating thymidylate synthase expression. EMBO J., 19, 4665–4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilanowski T., Tuckfield,A., Cerruti,L., O’Connell,S., Saint,R., Parekh,V., Tao,J., Cunningham,J.M. and Jane,S.M. (2002) A highly conserved novel family of mammalian developmental transcription factors related to Drosophila grainyhead. Mech. Dev., 114, 37–50. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki Y., Yandell,M.D., Roy,P.J., Krishna,S., Savage-Dunn,C., Ross,R.M., Padgett,R.W. and Wood,W.B. (1999) A BMP homolog acts as a dose-dependent regulator of body size and mail tail patterning in Caenorhabditis elegans. Development, 126, 241–250. [DOI] [PubMed] [Google Scholar]
- 10.Liu J. and Fire,A. (2000) Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development, 127, 5179–5190. [DOI] [PubMed] [Google Scholar]
- 11.Das S. and Smith,T.F. (2000) Identifying nature’s protein lego set. In Bork,P. (ed.), Advances in Protein Chemistry. Academic Press, San Diego, CA, Vol. 54, pp. 159–183. [DOI] [PubMed] [Google Scholar]
- 12.Smith R.F. and Smith,T.F. (1992) Pattern-induced multiple sequence alignment (PIMA) algorithm employing secondary structure-dependent gap penalties for use in comparative protein modeling. Protein Eng., 5, 35–41. [DOI] [PubMed] [Google Scholar]
- 13.Nelson L., Kim,K., Memmott,J. and Li,C. (1998) FMRFamide-related gene family in the nematode, Caenorhabditis elegans. Brain Res. Mol. Brain Res., 58, 103–111. [DOI] [PubMed] [Google Scholar]
- 14.Evans J.P. and Kay,B.K. (1991) Biochemical fractionation of oocytes. In Kay,B.K. and Peng,H.B. (eds), Methods in Cell Biol. Academic Press, San Diego, CA, Vol. 36, pp. 133–148. [DOI] [PubMed] [Google Scholar]
- 15.Swofford D.L. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods), Version 4. Sinauer Associates, Sunderland, MA. [Google Scholar]
- 16.Attardi L.D. and Tjian,R. (1993) Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev., 7, 1341–1353. [DOI] [PubMed] [Google Scholar]
- 17.Ruvkun G. and Hobert,O. (1998) The taxonomy of developmental control in Caenorhabditis elegans. Science, 282, 2033–2041. [DOI] [PubMed] [Google Scholar]
- 18.Aboobaker A.A. and Blaxter,M.L. (2003) Hox gene loss during dynamic evolution of the nematode cluster. Curr. Biol., 13, 37–40. [DOI] [PubMed] [Google Scholar]
- 19.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Guigo R., Knudsen,S., Drake,N. and Smith,T.F. (1992) Prediction of gene structure. J. Mol. Biol., 226, 141–157. [DOI] [PubMed] [Google Scholar]
- 23.Krause M. and Hirsh,D. (1987) A trans-spliced leader sequence on actin mRNA in C. elegans. Cell, 49, 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blumenthal T. (1995) Trans-splicing and poly-cistronic transcription in Caenorhabditis elegans. Trends Genet., 11, 132–136. [DOI] [PubMed] [Google Scholar]
- 25.Blackshear P.J., Lai,W.S., Thorn,J.M., Kennington,E.A., Staffa,N.G., Moore,D.T., Bouffard,G.G., Beckstrom-Sternberg,S.M., Touchman,J.W., Bonaldo,M.F. and Soares,M.B. (2001) The NIEHS Xenopus maternal EST project: interim analysis of the first 13,879 ESTs from unfertilized eggs. Gene, 267, 71–87. [DOI] [PubMed] [Google Scholar]
- 26.Uv A.E., Thompson,C.R.L. and Bray,S.J. (1994) The Drosophila tissue-specific factor grainyhead contains novel DNA-binding and dimerization domains which are conserved in human protein CP2. Mol. Cell. Biol., 14, 4020–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirra M.K. and Hansen,U. (1998) LSF and NTF-1 share a conserved DNA-recognition motif yet require different oligomerization states to form a stable protein-DNA complex. J. Biol. Chem., 273, 19260–19268. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi Y., Yamagishi,M., Nishimoto,Y., Taguchi,O., Matsukage,A. and Yamaguchi,M. (1999) A binding site for the transcription factor Grainyhead/Nuclear Transcription Factor-1 contributes to regulation of the Drosophila proliferating cell nuclear antigen promoter. J. Biol. Chem., 274, 35080–35088. [DOI] [PubMed] [Google Scholar]
- 29.Holland P.W.H. (1999) The future of evolutionary developmental biology. Nature, 402, C41–C44. [DOI] [PubMed] [Google Scholar]
- 30.Aguinaldo A.A., Turbeville,J.M., Linford,L.S., Rivera,M.C., Garey,J.R., Raff,R.A. and Lake,J.A. (1997) Evidence for a clade of nematodes, arthropods and other moulting animals. Nature, 387, 489–493. [DOI] [PubMed] [Google Scholar]
- 31.Weis L. and Reinberg,D. (1992) Transcription by RNA polymerase II: initiator-directed formation of transcription-competent complexes. FASEB J., 6, 3300–3309. [DOI] [PubMed] [Google Scholar]
- 32.Murata T., Nitta,M. and Yasuda,K. (1998) Transcription factor CP2 is essential for lens-specific expression of the chicken alphaA-crystallin gene. Genes Cells, 3, 443–457. [DOI] [PubMed] [Google Scholar]
- 33.Okazaki Y. and Shizuri,Y. (2000) Structures of six cDNAs expressed specifically at cypris larvae of barnacles, Balanus amphitrite. Gene, 250, 127–135. [DOI] [PubMed] [Google Scholar]
- 34.Kim C.H., Heath,C., Bertuch,A. and Hansen,U. (1987) Specific stimulation of simian virus 40 late transcription in vitro by a cellular factor binding the simian virus 40 21-base-pair repeat promoter element. Proc. Natl Acad. Sci. USA, 84, 6025–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shirra M.K., Zhu,Q., Huang,H.-C., Pallas,D. and Hansen,U. (1994) One exon of the human LSF gene includes conserved regions involved in novel DNA-binding and dimerization motifs. Mol. Cell. Biol., 14, 5076–5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim L.C., Swendeman,S.L. and Sheffery,M. (1992) Molecular cloning of the α-globin transcription factor CP2. Mol. Cell. Biol., 12, 828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoon J.-B., Li,G. and Roeder,R.G. (1994) Characterization of a family of related cellular transcription factors which can modulate human immunodeficiency virus type I transcription in vitro. Mol. Cell. Biol., 14, 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sueyoshi T., Kobayashi,R., Nishio,K., Aida,K., Moore,R., Wada,T., Handa,H. and Negishi,M. (1995) A nuclear factor (NF2d9) that binds to the male-specific P450 (Cyp 2d-9) in mouse liver. Mol. Cell. Biol., 15, 4158–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang N. and Miller,W.L. (2000) Cloning of factors related to HIV-inducible LBP proteins that regulate steroidogenic factor-1-independent human placental transcription of the cholesterol side-chain cleavage enzyme, P450scc. J. Biol. Chem., 275, 2852–2858. [DOI] [PubMed] [Google Scholar]
- 40.Rodda S., Sharma,S., Scherer,M., Chapman,G. and Rathjen,P. (2001) CRTR-1, a developmentally regulated transcriptional repressor related to the CP2 family of transcription factors. J. Biol. Chem., 276, 3324–3332. [DOI] [PubMed] [Google Scholar]
- 41.Bray S.J., Burke,B., Brown,N.H. and Hirsh,J. (1989) Embryonic expression pattern of a family of Drosophila proteins that interact with a central nervous system regulatory element. Genes Dev., 3, 1130–1145. [DOI] [PubMed] [Google Scholar]
- 42.Kumar S. and Hedges,B. (1998) A molecular timescale for vertebrate evolution. Nature, 392, 917–920. [DOI] [PubMed] [Google Scholar]