Abstract
Molecular models of six anthracycline antibiotics and their complexes with 32 distinct DNA octamer sequences were created and analyzed using HINT (Hydropathic INTeractions) to describe binding. The averaged binding scores were then used to calculate the free energies of binding for comparison with experimentally determined values. In parsing our results based on specific functional groups of doxorubicin, our calculations predict a free energy contribution of –3.6 ± 1.1 kcal mol–1 (experimental –2.5 ± 0.5 kcal mol–1) from the groove binding daunosamine sugar. The net energetic contribution of removing the hydroxyl at position C9 is –0.7 ± 0.7 kcal mol–1 (–1.1 ± 0.5 kcal mol–1). The energetic contribution of the 3′ amino group in the daunosamine sugar (when replaced with a hydroxyl group) is –3.7 ± 1.1 kcal mol–1 (–0.7 ± 0.5 kcal mol–1). We propose that this large discrepancy may be due to uncertainty in the exact protonation state of the amine. The energetic contribution of the hydroxyl group at C14 is +0.4 ± 0.6 kcal mol–1 (–0.9 ± 0.5 kcal mol–1), largely due to unfavorable hydrophobic interactions between the hydroxyl oxygen and the methylene groups of the phosphate backbone of the DNA. Also, there appears to be considerable conformational uncertainty in this region. This computational procedure calibrates our methodology for future analyses where experimental data are unavailable.
INTRODUCTION
Anthracycline antibiotics, such as doxorubicin and daunorubicin, are some of the most commonly used anticancer agents used today, mainly attributable to their broad spectrum of activity versus human cancers (1). Although the exact mechanism of action is still somewhat unclear, there are several proposed mechanisms, including inhibition of DNA biosynthesis by intercalation, interference with topoisomerase II and induction of DNA double-strand breaks (2–5). Furthermore, there have been quite a few experimental studies as well as molecular modeling and X-ray crystallography studies conducted that provide various explanations for the DNA binding sequence specificity of doxorubicin, yet we have only begun to assemble a model that explains the relationship between doxorubicin binding affinity and DNA sequence specificity (6–14). Applying the methods of rational drug design by exploiting the structure–activity relationships of anthracycline antibiotics may eventually yield more potent anticancer agents, but certainly could assist in the design of more useful molecular tools for understanding DNA structure and anthracycline intercalation.
An initial computational experiment was conducted in which doxorubicin was intercalated into 64 unique DNA base pair quartet sequences, and this led us to an observation that there may be a significant fourth base pair interaction between doxorubicin and DNA (15). This led us to consider a quartet model for the binding of doxorubicin to DNA, which serves as a hypothesis in the analysis of a series of doxorubicin analogs versus a variety of DNA quartet sequences. However, before we are able to determine sequence specificity of compounds analyzed, it is necessary to calibrate our models with respect to available experimental binding data. While there is not an abundance of data of this nature, Chaires et al. have elegantly determined the binding free energy for a series of six anthracycline antibiotics in an effort to parse the free energy contributions of various functional groups present and to determine their overall effect on binding to DNA (16). So, the purpose of our calculations is to determine binding scores for these six doxorubicin analogs (Fig. 1) in a broad sampling of DNA quartet sequences using the HINT (Hydropathic INTeractions) program (17–21) and to explore and understand the interatomic basis for the doxorubicin–DNA interaction that Chaires et al. have looked at experimentally. The binding scores that we calculate with the HINT model (vide infra) can then be converted to approximate free energies (ΔG) (approximately –515 HINT score units per kcal mol–1) (17–21) and these free energies can be compared to the experimental free energies of binding of Chaires et al. (16). The ultimate goal is to develop a rapid computational protocol for the determination of the approximate binding strength of anthracycline analogs as well as to search for and develop novel molecular tools with dynamic sequence specificity. These molecular tools could then be useful in the search for new and powerful antitumor agents.
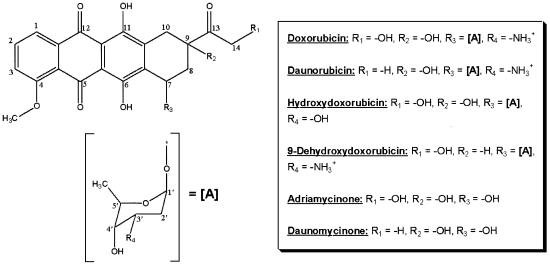
Figure 1.
Structures of the six anthracycline antibiotics used in our studies, showing the differences in functional groups that are deficient in various compounds.
MATERIALS AND METHODS
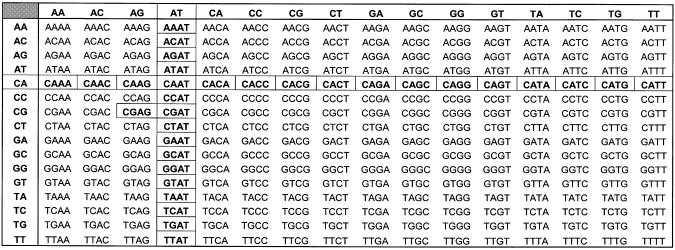
Molecular models were created and minimized using the SYBYL v. 6.7 molecular modeling package (Tripos Inc., St Louis, MO). The starting point was the complex structure of doxorubicin intercalated into the DNA octamer sequence d(CGC|AATCG/CGA|TTGCG). The underlined sequence is the DNA ‘quartet’ hereafter referred to, with the doxorubicin analog intercalated between the first two base pairs in the sequence (indicated by the |). Double helical B-DNA was constructed using the SYBYL Biopolymer module. Doxorubicin, in the same conformation as found in the crystal structure (pdb accession no. 1D12) (22), was placed in the intercalation site, as calculated by the program GRID (GRID v. 17; Molecular Discovery Ltd, London, UK), as previously described (15). The ionization state of the compounds was initially the same as described in previous modeling studies (i.e. the ammonium and all hydroxyls are protonated) (8,13). The structures of the other five compounds were modified from this structure, using SYBYL. The DNA oligomer sequences were then modified using the SYBYL Biopolymer ‘Mutate Monomers’ function. Thirty-two DNA quartet sequences were modeled, representing a sampling of one-eighth of the 256 DNA quartet sequences possible (Table 1). By building the set of all sequences along both the ‘row’ and ‘column’ containing the prototype ‘C|AAT’ sequence, we felt that we would thus examine models representing each type of interaction between DNA quartets and the anthracycline intercalators. Each structure was then solvated using the droplet protocol in SYBYL with a single layer of water molecules. From previous experiments we have found that this water monolayer is sufficient to retard helix unwinding during the complex minimization process without adding significant complexity to the system (15). All structures were then energy minimized using the Tripos Force Field and Gasteiger–Hückel charges with 300 cycles of steepest-descent energy minimization followed by conjugate gradient energy minimization until the energy difference was <0.05 kcal mol–1 as reported previously (15). We earlier showed that this protocol builds model structures with good agreement with available crystal structures (15). In the present study, our model for doxorubicin intercalated with the C|GAT quartet sequence superimposes onto the crystal structure (22), with an RMS deviation of 1.14 Å. We should also highlight, as it is one of our primary assumptions, that the models thus generated are not at the global energy minima, however, as they were all built with consistent protocols, the relative differences between them should be significant.
Table 1. 32 DNA quartet sequences that were modeled are highlighted, out of the 256 DNA quartet sequences possible.
The interactions of each complex were then analyzed using the HINT program v. 2.35S (Tripos Inc., St Louis, MO), as described previously (15,17–21). The HINT model describes specific interactions between two molecules, DNA and the anthracycline in our case, as a double sum over the atoms within each component (equation 1).
where a is the hydrophobic atom constant, S is the solvent accessible surface area, T is a descriptor function and R and r are distance functions. The binding score, bij, describes the specific interaction between two atoms in the complex, i and j, and B describes the total interaction between both molecules (15,17–21).
The hydropathic atom constants (a) are calculated using a partitioning algorithm based on the CLOGP method of Hansch and Leo (23). Positive atom constants indicate a hydrophobic atom and negative atom constants indicate polar or hydrophilic atoms. These calculated atom constants can be added together to give a partition coefficient (logP), and the calculated logP values from the HINT program have been found to be similar to calculated logP values of other programs (15,17–21).
Solvent accessible surface area (S) describes the shape and accessibility of the atom and its tendency for interaction, such that atoms on the surface of the molecule will have a higher S while buried atoms will have a smaller S and are less likely to be involved in interactions (15,17–21). The descriptor function (T) differentiates among the three possibilities for polar interactions (acid–acid, acid–base and base–base). Each atom type is assigned descriptor variables to represent its hydrogen bond acceptor/donor character, charge, Brönsted acid/base character and Lewis acid/base character. These are then used by the T function to calculate a value of +1 or –1 for each atom–atom interaction. So this function is essentially a ‘sign change’ function which maintains the convention that favorable interactions have positive HINT scores while unfavorable interactions have negative HINT scores (15,17–21).
The final components of the HINT model are the distance functions (R and r). R describes the dependence on the hydrophobic atom constant (a) and solvent accessible surface area (S) on the distance between the two atoms, while r is independent of hydropathy and accounts only for variations in distance. For this work, R is a simple exponential (e–r) and r is calculated by a Lennard–Jones 6-12 function (15,17–21).
Equation 1 was then used to calculate an interaction score for each structurally minimized anthracycline/DNA complex. We have found through previous experiments that –515 HINT score units is approximately equal to 1 kcal mol–1 (17–21). This empirical score factor has been determined from the slope of ΔG versus HINT score plots and so is more appropriate for estimating ΔΔG than ΔG (18). Each HINT score was converted to an approximate free energy score in kcal mol–1 by dividing the total HINT score for each sequence by –515. The calculated free energy scores of daunorubicin (14-OH), hydroxydoxorubicin (NH3+), 9-dehydroxydoxorubicin (9-OH), adriamycinone (sugar) and daunomycinone (sugar and 14-OH) were then subtracted from the calculated free energy score of doxorubicin, for all sequences modeled. This allowed us to calculate, or ‘parse’, the approximate free energy contribution of the relevant functional group (in parentheses) for these compounds.
RESULTS AND DISCUSSION
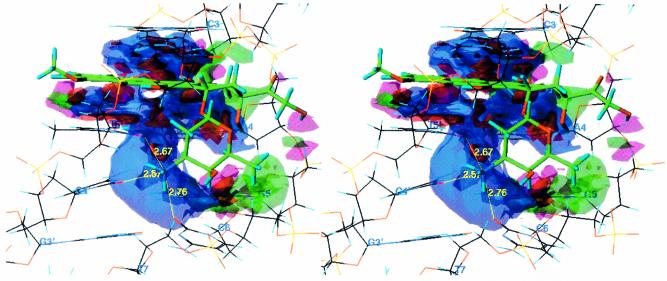
A look at the atomic level interactions of doxorubicin intercalated into DNA identifies some of the important functional groups present that contribute to its binding (Fig. 2). Perhaps the most significant contributor to binding, but also the most difficult to modify, is the basic anthracycline ring scaffold of the molecule, which intercalates between two base pairs of DNA and is stabilized by the stacking interactions of the aromatic anthracycline rings with the DNA base pairs. This is further stabilized latitudinally by the presence of the 4-methoxy group, which finds its optimal position at an angle slightly above that of the anthracycline plane, due to adverse hydrophobic interactions with the methyl group of the T5′ base. Additional latitudinal stabilization comes from the presence of the 9-OH, which forms hydrogen bonds with the aromatic carbon and nitrogen atoms present on the A4 base, as well as the ether oxygen on the ribose of the G5 base on the DNA. The daunosamine sugar also contributes quite significantly to the binding of doxorubicin with DNA. Most notable is the presence of a quaternary ammonium cation at the 3′ position on the ring. If positively charged, this functional group is well adapted for hydrogen bonding, and appears to form three hydrogen bonds with the carbonyl oxygen atoms of the T5′, G4′ and C6 bases of the CAGC sequence of DNA (Fig. 2), as well as forming similar hydrogen bonds with other sequences as well. In contrast, the hydroxyl group at the 4′ position on the ring does not appear to play nearly as large a role, while the methyl group at the 5′ ring position appears to make favorable hydrophobic contacts with the DNA backbone. The 14-OH also appears to play a minor role in binding as well, although in our models it presents somewhat unfavorable hydrophobic interactions with the DNA backbone.
Figure 2.
Stereo diagram of a HINT interaction map for the intercalation of doxorubicin with the CAGC base pair sequence of DNA, which displays, visually, the quality and magnitude of the various binding contacts involved in the interaction. The contour surfaces are color coded by interaction type at a constant map density value of ±75. Blue surfaces represent favorable polar interactions; red surfaces represent unfavorable polar interactions; green surfaces represent favorable hydrophobic interactions; magenta surfaces represent unfavorable hydrophobic interactions. The interatomic distance between the N3′ ammonium on the doxorubicin sugar ring and the carbonyl oxygen atoms of three surrounding base pairs of DNA is also indicated.
Upon selection of these key functional groups of doxorubicin, we were able to model five compounds versus a variety of DNA sequences that lacked one of the functional groups described above. All of these compounds were also compounds that Chaires et al. had analyzed and provided experimental binding data for. While there is somewhat significant deviation between the average calculated free energies (ΔG), which is often the case in computational studies, the differences in free energy (ΔΔG) caused by the loss of various functional groups (Table 2) showed relatively good agreement with the experimental data. This work is focused on these free energy differences because of the relevance to sequence specificity.
Table 2. Experimental free energy (ΔG) and the free energy difference (ΔΔG) between each compound and doxorubicin.
| Experimentala (kcal mol–1) | Calculatedb (kcal mol–1) | |||
|---|---|---|---|---|
| ΔG | ΔΔG | ΔG | ΔΔG | |
| Doxorubicin | –7.7 ± 0.3 | –9.6 ± 1.1 | ||
| Daunorubicin (loss of C14-OH) | –6.8 ± 0.3 | –0.9 ± 0.5 | –10.0 ± 0.9 | +0.4 ± 0.6 |
| Hydroxydoxorubicin (-NH3+ to -OH) | –7.0 ± 0.3 | –0.7 ± 0.5 | –5.9 ± 0.4 | –3.7 ± 1.1 |
| 9-Dehydroxydoxorubicin (loss of C9-OH) | –6.5 ± 0.3 | –1.2 ± 0.5 | –8.9 ± 1.0 | –0.7 ± 0.7 |
| Adriamycinone (loss of sugar ring) | –5.7 ± 0.3 | –2.0 ± 0.5 | –6.3 ± 0.6 | –3.2 ± 1.1 |
| Daunomycinone (loss of sugar ring and C14-OH) | –5.2 ± 0.3 | –2.5 ± 0.5 | –6.0 ± 0.5 | –3.6 ± 1.1 |
aExperimental data for the binding of doxorubicin analogs with calf thymus DNA was obtained from Chaires et al. (6).
bThe calculated data were obtained from molecular models of each analog intercalated into 32 DNA quartet sequences. A HINT interaction score was obtained and divided by –515 (units per kcal mol–1) and the average score from all 32 models per compound is reported. The ΔΔG for all compounds versus all sequences was also calculated and the average score from all 32 models per compound is reported.
We did not find a significant difference between the binding of doxorubicin and daunorubicin, which differ only by the presence of a hydroxyl group at position 14. In fact, our calculations indicate that the absence of this group has little effect (+0.4 ± 0.6 kcal mol–1). Also, the effect is opposite in sign to the experimental data that indicate that the energetic contribution of this group is approximately –0.9 ± 0.5 kcal mol–1. The difference in our calculated binding scores can be attributed to an adverse hydrophobic interaction between the oxygen atom of the C14 hydroxyl on doxorubicin with the carbon backbone of the DNA molecule, which can be visualized by the magenta colored surface in Figure 2. Daunorubicin, being identical to doxorubicin in every respect except that it lacks this group, therefore does not contain this adverse interaction, and we thus calculate an enhanced binding score for daunorubicin. It should be noted, however, that there were significant sequence-dependent variations in our calculations between doxorubicin and daunorubicin.
Importantly, our calculations detected more than one favorable conformation for this C14-OH arm of the molecule, so it is quite possible that the increased flexibility of this part of doxorubicin is a source of uncertainty in our free energy estimate, which is reflected in the relatively high standard deviation associated with the average calculated ΔΔG value. The crystal structure of this complex reveals that there may be solvent molecules in this vicinity that may be interacting with the C14-OH, thus adding additional entropy and uncertainty to the system (22).
To analyze this further, we can consider the ΔΔG between adriamycinone and daunomycinone (Table 2), which, like doxorubicin and daunorubicin, differ solely by the presence of the C14-OH. Both compounds also lack the entire daunosamine sugar as well, so this allows us to look more closely at the energetic contribution of the C14-OH without any effects of the daunosamine sugar. The ΔΔG for C14-OH as revealed by adriamycinone and daunomycinone is –0.3 ± 1.1 (calculated) and –0.5 ± 0.5 (experimental). This good agreement for the simpler des-daunosamine intercalators further reinforces our notion that there is conformational uncertainty in the C14-OH region and suggests that the sugar binding is a probable perturbing factor.
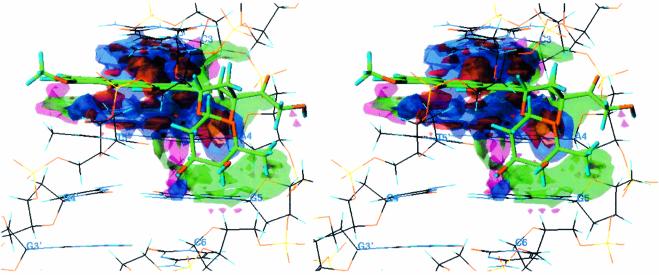
Our calculations also appear to overestimate the binding contribution of the N3′ ammonium by a fairly substantial amount. This is mainly attributable to the loss of three potent hydrogen bonds between the three hydrogen atoms of the amine cation and three carbonyl oxygen atoms of the DNA bases nearby. The neutral hydroxyl oxygen, with only a single hydrogen atom, does not have the same hydrogen bonding capability as this positively charged amine, and therefore we see a significantly reduced polar interaction for hydroxydoxorubicin (Fig. 3).
Figure 3.
Stereo diagram of a HINT interaction map for the intercalation of hydroxydoxorubicin with the CAGC base pair sequence of DNA, which displays, visually, the quality and magnitude of the various binding contacts involved in the interaction. The contour surfaces are color coded by interaction type at a constant map density value of ±75. Blue surfaces represent favorable polar interactions; red surfaces represent unfavorable polar interactions; green surfaces represent favorable hydrophobic interactions; magenta surfaces represent unfavorable hydrophobic interactions. The interatomic distance between the N3′ ammonium on the doxorubicin sugar ring and the carbonyl oxygen atoms of three surrounding base pairs of DNA is also indicated.
To analyze this discrepancy in more detail, ΔΔG was calculated for our compounds using the method of Andrews et al. (Table 3), which assigns partial free energy values to individual atoms or functional groups in a ligand molecule based solely on its structure and also incorporates entropy into the equation for free energy (24). In this way it is similar to HINT. Andrews et al. note through this algorithm that charged groups bind more strongly than polar groups, which in turn bind more strongly than non-polar groups (24). While it would be wise to exercise a certain degree of caution when comparing the Andrews et al. ΔΔG values to the Chaires et al. and HINT values, since they were obtained without consideration of the receptor and based solely on the structure of the intercalators themselves, we can nevertheless see a very significant free energy contribution associated with modification of the N3′ ammonium to a hydroxyl group. It should also be noted that this is consistent with a number of experimental observations. For example, a significant free energy difference (ΔΔG) of –4.4 kcal mol–1 is observed between the DHFR inhibitors methotrexate and the natural substrate folate. This difference is primarily attributed to the replacement of a hydroxyl (folate) with a protonated amine (methotrexate) (25).
Table 3. Free energy differences (ΔΔG) determined experimentally (Chaires et al.) and calculated using the HINT and Andrews et al. methods for our set of doxorubicin analogs.
| ΔΔG versus doxorubicin (kcal mol–1) | |||
|---|---|---|---|
| Chaires et al. | HINT | Andrews et al. | |
| Daunorubicin (loss of C14-OH) | –0.9 ± 0.5 | 0.4 ± 0.6 | –1.8 |
| Hydroxydoxorubicin (-NH3+ to -OH) | –0.7 ± 0.5 | –3.7 ± 1.1 | –9.7 |
| Hydroxydoxorubicin (-NH2 to -OH) | –0.7 ± 0.5 | –0.2 ± 0.3 | 0.6 |
| 9-Dehydroxydoxorubicin (loss of C9-OH) | –1.1 ± 0.5 | –0.7 ± 0.7 | –1.8 |
| Adriamycinone (loss of sugar ring) | –2.0 ± 0.5 | –3.2 ± 1.1 | –17.1 |
| Daunomycinone (loss of sugar ring and C14-OH) | –2.5 ± 0.5 | –3.6 ± 1.1 | –18.9 |
We also see a modest, even identical, free energy contribution in the Andrews et al. model for the C14-OH and C9-OH functional groups, which is more or less in agreement with experimental data and the HINT model. This should actually be expected since the changes, while different, are equivalent structurally (i.e. the same number of functional groups and degrees of freedom are lost in both molecules) in the Andrews et al. model.
With this knowledge that charged groups bind more strongly than polar groups, which in turn bind more strongly than non-polar groups, we were led to further questions regarding the exact protonation state of this N3′ ammonium of doxorubicin. With a pKa of 8.3 (26), we would expect the amine of doxorubicin to be ∼95% protonated with a +1 charge at neutral pH, according to the Henderson–Hasselbach equation. However, it is interesting to note that if the pH is increased to only 8.0, the protonation would be predicted to be only ∼50%. However, in our initial models we had assumed a 100% protonated amine, as is generally assumed (8,13). So, in order to reconsider the possibility that the amine is not 100% protonated, we recalculated free energy scores for doxorubicin containing an unprotonated -NH2 and a neutral charge (Table 3).
If we calculate the free energy difference (ΔΔG) between this unprotonated form of doxorubicin and hydroxydoxorubicin, we see a significantly reduced free energy difference. In fact, this new value is even lower then the experimental ΔΔG! Interestingly enough, we get a very similar free energy difference using the Andrews et al. method as well (Table 3). Considering that it is widely known that there can be variations in pH in solution, especially within biological systems, and considering the polyanionic, electron-rich nature of the DNA double helix, our computational analysis indicates that doxorubicin, in its intercalated state, may be considerably less than 100% protonated. In retrospect, the relatively small experimental ΔΔG attributed to the replacement of a charged ammonium functional group with a neutral hydroxyl group is itself evidence of a pKa shift between the free and bound forms of doxorubicin, and the ammonium must be largely in the amine form when bound.
In summary, we have demonstrated a relatively efficient and reasonably accurate method for analyzing the free energy contributions of various functional groups of doxorubicin. There appears to be a good correlation with experimental binding data, with a few explainable discrepancies. These results also indicate the reproducibility and accuracy of the computational studies. They suggest that the major source of uncertainty arises from conformational uncertainty in model building, while the lack of understanding of chemical effects and our ability to model them, e.g. the protonation of N3′, is a vexing cause of discrepancy between the calculated and experimental studies. With this test set of compounds calibrated to experimental data, we hope to further explore the sequence-dependent binding affinity of an expanded set of doxorubicin analogs. Thus, we are currently applying these methods to a wider array of analogs, and it is hoped that a rapid computational screen can ultimately be attained in the effort to search for additional sequence-specific and potent DNA binding agents.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge the helpful comments and discussions from Jason P. Rife in preparing this paper. The SYBYL software has been made available through a University Software Grant from Tripos Inc. The HINT program was developed in the laboratory of G.E.K. and has been licensed for distribution to for-profit corporations in which he has a financial interest. The algorithms in the HINT program have been fully disclosed and discussed in other publications (15,17–21).The HINT program may be obtained from Tripos Inc. as a SYBYL program module. This work was supported in part by the Massey Cancer Center of V.C.U. through an American Cancer Society Institutional Research Grant.
REFERENCES
- 1.Weiss R.B. (1992) The anthracyclines: will we ever find a better doxorubicin? Semin. Oncol., 19, 670–686. [PubMed] [Google Scholar]
- 2.Momparler R.L., Karon,M., Siegel,S.E. and Avila,F. (1976) Effect of adriamycin on DNA, RNA and protein synthesis in cell-free systems and intact cells. Cancer Res., 36, 2891–2895. [PubMed] [Google Scholar]
- 3.Tewey K.M., Rowe,T.C., Yang,L., Halligan,B.D. and Lui,L.F. (1984) Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science, 226, 466–468. [DOI] [PubMed] [Google Scholar]
- 4.Schneider E., Hsiang,Y. and Lui,L.F. (1990) DNA topoisomerases as anticancer drug targets. Adv. Pharmacol., 21, 149–183. [DOI] [PubMed] [Google Scholar]
- 5.Sander M. and Tsieh,T.-S. (1983) Double strand DNA cleavage by type II DNA topoisomerase from Drosophila melanogaster. J. Biol. Chem., 258, 8421–8428. [PubMed] [Google Scholar]
- 6.Chaires J.B., Priebe,W., Graves,D.E. and Burke,T.G. (1993) Dissection of the free energy of anthracycline antibiotic binding to DNA: electrostatic contributions. J. Am. Chem. Soc., 115, 5360–5364. [Google Scholar]
- 7.Chaires J.B. (1990) Daunomycin binding to DNA: from the macroscopic to the microscopic. In Pullman,B. and Jortner,J. (eds), Molecular Basis of Specificity in Nucleic Acid–Drug Interactions. Kluwer Academic, Dordrecht, The Netherlands, pp. 123–136. [Google Scholar]
- 8.Chen K.-X., Gresh,N. and Pullman,B.A. (1986) Theoretical investigation on the sequence selective binding of adriamycin to double-stranded polynucleotides. Nucleic Acids Res., 14, 2251–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trist H. and Phillips,D.R. (1989) In vitro transcription analysis of the role of flanking sequence on the DNA sequence specificity of adriamycin. Nucleic Acids Res., 17, 3673–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medhi C., Mitchell,J.B.O., Price,S.L. and Tabor,A.B. (1999) Electrostatic factors in DNA intercalation. Biopolymers, 52, 84–93. [DOI] [PubMed] [Google Scholar]
- 11.Fritzsche H. and Watler,A. (1990) Physico-chemical studies on anthracycline–DNA interactions: analysis of equilibrium, kinetic and structural data. In Kallenbach,N.R. (ed.), Chemistry and Physics of DNA–Ligand Interactions. Adenine Press, Schenectady, NY, pp. 1–35. [Google Scholar]
- 12.Pullman B. (1989) Molecular mechanisms of specificity in DNA–antitumour drug interactions. Adv. Drug Res., 18, 1–113. [Google Scholar]
- 13.Pullman B. (1991) Sequence specificity in the binding of anti-tumour anthracyclines to DNA: a success of theory. Anticancer Drug Des., 6, 95–105. [PubMed] [Google Scholar]
- 14.Gao Y.G., Priebe,W. and Wang,A.H.J. (1996) Substitutions at C2′ of daunosamine in the anticancer drug daunorubicin alter its DNA-binding sequence specificity. Eur. J. Biochem., 240, 331–335. [DOI] [PubMed] [Google Scholar]
- 15.Kellogg G.E., Scarsdale,J.N. and Fornari,F.A. (1998) Identification and hydropathic characterization of structural features affecting sequence specificity for doxorubicin intercalation into DNA double-stranded polynucleotides. Nucleic Acids Res., 26, 4721–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaires J.B., Satyanarayana,S., Suh,D., Fokt,I., Przewloka,T. and Priebe,W. (1996) Parsing the free energy of anthracycline antibiotic binding to DNA. Biochemistry, 35, 2047–2053. [DOI] [PubMed] [Google Scholar]
- 17.Burnett J.C., Kellogg,G.E. and Abraham,D.J. (2000) Computational methodology for estimating changes in free energies of biomolecular association upon mutation. The importance of bound water in dimer-tetramer assembly for β-37 mutant hemoglobins., Biochemistry, 39, 1622–1633. [DOI] [PubMed] [Google Scholar]
- 18.Cozzini P., Fornabaio,M., Marabotti,A., Abraham,D.J., Kellogg,G.E. and Mozzarelli,A. (2002) Simple, intuitive calculations of free energy of binding for protein–ligand complexes. 1. Models without explicit constrained water. J. Med. Chem., 45, 2469–2483. [DOI] [PubMed] [Google Scholar]
- 19.Kellogg G.E. and Abraham,D.J. (2000) Hydrophobicity: is log Po/w more than the sum of its parts? Eur. J. Med. Chem., 35, 651–661. [DOI] [PubMed] [Google Scholar]
- 20.Kellogg G.E., Scarsdale,J.N. and Cashman,D.J. (1999) Ligand docking and scoring in DNA oligonucleotides. Binding of doxorubicin and modeled analogs to optimize sequence specificity. Med. Chem. Res., 9, 592–603. [Google Scholar]
- 21.Abraham D.J., Kellogg,G.E., Holt,J.M. and Ackers,G.K. (1997) Hydropathic analysis of the non-covalent interactions between molecular subunits of structurally characterized hemoglobins. J. Mol. Biol., 272, 613–632. [DOI] [PubMed] [Google Scholar]
- 22.Frederick C.A., Williams,L.D., Ughetto,G., van der Marel,G.A., van Boom,J.H., Rich,A. and Wang,A.H.-J. (1990) Structural comparison of anticancer drug–DNA complexes: adriamycin and daunomycin. Biochemistry, 29, 2538–2549. [PubMed] [Google Scholar]
- 23.Hansch C. and Leo,A.J. (1979) Substituent Constants for Correlation Analysis in Chemistry and Biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 24.Andrews P.R., Craik,D.J. and Martin,J.L. (1984) Functional group contributions to drug–receptor interactions. J. Med. Chem., 27, 1648–1657. [DOI] [PubMed] [Google Scholar]
- 25.Cocco L., Temple,C., Montgomery,J.A., London,R.E. and Blakley,R.L. (1981) Protonation of methotrexate bound to the catalytic site of dihydrofolate reductase from Lactobacillus casei. Biochem. Biophys. Res. Commun., 100, 413–419. [DOI] [PubMed] [Google Scholar]
- 26.Speelmans G., Staffhorst,W.H.M., de Kruijff,B. and de Wolf,F.A. (1994) Transport studies of doxorubicin in model membranes indicate a difference in passive diffusion across and binding at the outer and inner leaflets of the plasma membrane. Biochemistry, 33, 13761–13768. [DOI] [PubMed] [Google Scholar]