Abstract
We report here the sequence of chromosome II from Trypanosoma brucei, the causative agent of African sleeping sickness. The 1.2-Mb pairs encode about 470 predicted genes organised in 17 directional clusters on either strand, the largest cluster of which has 92 genes lined up over a 284-kb region. An analysis of the GC skew reveals strand compositional asymmetries that coincide with the distribution of protein-coding genes, suggesting these asymmetries may be the result of transcription-coupled repair on coding versus non-coding strand. A 5-cM genetic map of the chromosome reveals recombinational ‘hot’ and ‘cold’ regions, the latter of which is predicted to include the putative centromere. One end of the chromosome consists of a 250-kb region almost exclusively composed of RHS (pseudo)genes that belong to a newly characterised multigene family containing a hot spot of insertion for retroelements. Interspersed with the RHS genes are a few copies of truncated RNA polymerase pseudogenes as well as expression site associated (pseudo)genes (ESAGs) 3 and 4, and 76 bp repeats. These features are reminiscent of a vestigial variant surface glycoprotein (VSG) gene expression site. The other end of the chromosome contains a 30-kb array of VSG genes, the majority of which are pseudogenes, suggesting that this region may be a site for modular de novo construction of VSG gene diversity during transposition/gene conversion events.
INTRODUCTION
The protozoan parasite Trypanosoma brucei is the causative agent of African sleeping sickness. More than a half a billion people live in areas inhabited by the tsetse fly that transmits the parasite, and hundreds of thousands are newly infected each year (1). Trypanosoma brucei exhibits unusual and complex mechanisms in the control of gene expression ranging from polycistronic transcription (2), trans-splicing of precursor RNAs (3,4) and mitochondrial RNA editing (5) to gene rearrangements during antigenic variation (6,7). Several of these phenomena have become the focus of intense research in this organism as well as in higher eukaryotes. In addition, the many elegant mechanisms used by trypanosomes and related parasites to evade the immune responses of their mammalian hosts have led to a better understanding of the diversity and complexity of host–parasite interactions. The sequences of the first two chromosomes (chromosomes I and II) of T.brucei are reported in this issue (8). Here we present an analysis of the 1.2-Mb DNA sequence of T.brucei chromosome II (chrII) fully annotated into one contig.
MATERIALS AND METHODS
Parasite DNA and chromosome sequencing
Trypanosomoa brucei TREU (Trypanosomiasis Research Edinburgh University) 927/4 (GPAL/KE/70/EATRO 1534) single VAT derivative GUTat 10.1 (9) (Tb927) was selected as the reference stock for the genome sequencing project. A bacterial artificial chromosome (BAC) library, RPCI93 (http://www.chori.org/bacpac/tbrucei93.htm), was the main substrate used for sequencing. Sheared DNA from selected BAC clones (1.6–2 kb) was cloned into a modified pUC18 vector via BstXI linkers. Sequences were assembled and gaps were closed using a combination of BAC walking, directed PCR or transposon insertion. The final assembly of the chromosome was verified by comparison with an XbaI optical restriction map. The restriction maps predicted from the sequence of all the internal BACs agreed with the optical map. Quality alignments could not be obtained in the subtelomeric regions. This can be explained by homologous chromosome pair polymorphisms in T.brucei that are not represented or are not resolved in the current optical map. We have used multiple combinations of unique primers from BACs RPCI93-3B10 (‘left’ end of the chrII) and RPCI93-36E18 (‘right’ end) and a known telomeric hexamer repeat to amplify and clone the telomeres from chrII. PCR products ranging from 5 to 12 kb in size have resisted repeated cloning attempts.
Annotation
A custom-modified version of Glimmer (10) was trained on 305 T.brucei genes and cDNAs taken from GenBank and then used to generated gene predictions on chrII. Following manual evaluation and model curation, genes on finished BACs were assigned systematic names based on the RPCI93 BAC from which they originated (e.g. 10C8.440, 10C8.445, etc.). Genes on the assembled chrII pseudomolecule were assigned systematic names according to a scheme agreed upon with the Sanger Institute (e.g. Tb927.2.3280, Tb927.2.3290, etc.) and reflecting organism (Tb), strain (927) and chromosome (2). Predicted proteins were searched against a non-redundant amino-acid database using BLASTP; other features were identified by specialised searches using the following programs and databases: InterPro (11), Pfam (12), Gene Ontology (GO) (13); transmembrane domains, TMHMM (14); signal peptides and signal anchors, SignalP-2.0 (15). The results of all analyses were reviewed using Manatee, a tool created at TIGR that interfaces with a relational database of all the information produced by the annotation software. Predicted gene products were manually assigned GO (13) terms. The annotation discussed in this report is also on the T.brucei Genome Annotation Database at TIGR (http://www.tigr.org/tdb/e2k1/tba1/tba1.shtml) and in GeneDB (http://www.genedb.org).
Genetic analysis
Mini- and microsatellite sequences were identified by analysing the chrII sequence with Tandem repeats finder (16) and a subset of these sequences were used to construct the genetic map. Primers were designed to unique sequences flanking three minisatellites and 16 microsatellites and these sequences are available on request. F1 progeny clones from a cross of T.brucei stocks Tb927 × Tb247 were used in this analysis (17). Further progeny clones were generated from cryopreserved uncloned populations that included products of mating as described (18). Genetically independent progeny were defined on the basis of either being derived from different tsetse flies or differing in genotype after screening with five unlinked mini- and microsatellite markers. Thirty-eight progeny were used to construct the map. Each clone was amplified by infection of mice, the trypanosomes purified from blood and DNA prepared. Each progeny clone was genotyped by PCR amplification of each locus and separation of the products by agarose gel electrophoresis, typically 3% Nusieve agarose gels.
RESULTS AND DISCUSSION
Gene content and structure of chrII
Sequencing was initiated using BACs that we mapped to chrII of T.brucei reference stock TREU927/4 GUTat10.1 (Tb927) by hybridisation to specific genetic markers (19). Subse quently, BAC end sequences and BAC fingerprint data allowed extension from three initial seed points and completion of the chromosome using a map-as-you-go approach (20). Ten BAC clones were sequenced and assembled into one contig representing 1 193 931 bp of non-redundant sequence terminating ∼5–20 kb from each of the telomeres.
Using a combination of gene prediction programs and database searches, the chromosome was manually annotated (Fig. 1). Four hundred and seventy-three putative coding sequences (CDSs) >200 bp were predicted on chrII (see Table S1 available as Supplementary Material at NAR Online). We have excluded from our discussion in this paper all hypothetical CDSs that are smaller than 450 bp and do not encode matches to previously characterised proteins or functional domains. The remaining 318 coding sequences occur at an average density of one gene per 3741 bp (Fig. 1 and Table 1). Of the 284 protein-encoding genes, 167 were designated as hypothetical. In addition, 34 pseudogenes, most of which resemble genes for variant surface glycoproteins (VSGs) and retrotransposon hot spot (RHS) genes, occur in the subtelomeric regions of the chromosome. Table 1 summarises the statistically salient features of chrII.
Figure 1.
(Opposite) Schematic representation of T.brucei chromosome II. Each predicted CDS >200 bp in length is represented by an arrow. The labels refer to the systematic name for each gene (see Table S1). The colours of the arrows represent the corresponding T.brucei GO assignment for each gene according to the Biological Process ontology (12). A set of high level GO categories were selected to represent the most informative high-level terms without overlapping paths in the GO hierarchy. Grey peaks reflect the GC skew scores calculated for a 2-kb window sliding in 1-kb increments. ‘Hypothetical proteins’ are proteins with no similarity to characterised proteins or with similarity to a hypothetical protein over less than 70% of the protein length. ‘Conserved hypothetical proteins’ are proteins with similarity to one or more hypothetical protein from an organism other than T.brucei over at least 70% of the protein length. ‘Trypanosoma brucei family hypothetical proteins’ are proteins with similarity with two or more T.brucei hypothetical proteins over at least 70% of the protein length. The predicted transmembrane domains are shown for proteins containing at least five domains. (Inset) Map of duplicated blocks on chrII. The copies of each putative duplicated block are classified by colour. Direction of arrowheads indicate the relative orientation of duplicated blocks. An enlarged version of this figure is included with the Supplementary Material (available at NAR Online).
Table 1. Chromosome II summary statistics.
| Chromosome II content | |
| Size (bp) | 1 193 931 |
| G+C content (%) | 44 |
| G+C content in protein coding region (%) | 49.8 |
| Gene density (bp per gene) | 3755 |
| Mean CDS length (bp) | 1831 |
| Number of predicted CDSs | 318 |
| Percent of protein coding region | 43.6 |
| Organisation features | |
| Number of directional gene clusters | 17 |
| Maximum length of gene cluster (bp) | 283 564 |
| Number of VSG clusters | 1 |
| Number of rRNA clusters | 3 |
| Structural RNA | |
| Number of rRNA genes | 24 |
| Number of tRNA genes | 0 |
| Proteome | |
| Number of predicted protein-coding genes | 284 |
| Number of pseudogenes | 34 |
| Hypothetical protein (total) | 167 |
| Hypothetical proteina | 109 |
| Conserved hypothetical proteinb | 5 |
| T.brucei family hypothetical proteinc | 49 |
| Conserved/T.brucei hypothetical proteind | 4 |
| Number of putative VSGs (total) | 8 |
| Number of VSG pseudogenes | 6 |
| Number of genes with InterPro match | 179 |
| Number of genes with Pfam match | 157 |
| Structural features | |
| Transmembrane domain(s)e | 80 |
| Signal peptide | 50 |
| Signal anchor | 26 |
| Non-secretory protein | 211 |
The analyses were performed on CDSs greater than 450 nucleotides. Using this criterion allows the exclusion of 155 hypothetical genes, the majority of which are not likely to code for a protein. It results, however, in the exclusion of genes for two small nuclear ribonucleoproteins, ribosomal protein L44 and a putative histone H4. Specialised searches used the following programs and databases: InterPro (11), Pfam (12), Gene Ontology (13); transmembrane domains, TMHMM (14); signal peptides and signal anchors SignalP-2.0 (15).
aHypothetical proteins are proteins with no similarity to characterised proteins or with similarity with a hypothetical protein over less than 70% of the protein length.
bConserved hypothetical proteins are proteins with similarity to one or more hypothetical protein from an organism other than T.brucei over at least 70% of the protein length.
cTrypanosoma brucei family hypothetical proteins are proteins with similarity to two or more T.brucei hypothetical proteins over at least 70% of the protein length.
dConserved/T.brucei hypothetical protein are proteins with similarity, over at least 70% of the protein length, to two or more T.brucei hypothetical proteins plus one or more matches with hypothetical proteins from other organism than T.brucei.
eNumber of predicted proteins with one or more transmembrane domains.
A remarkable feature of chrII is its gene organisation. All the genes are densely packed within a total of 17 directional clusters, the longest of which spans ∼284 kb and contains 92 protein-coding genes (Fig. 1). This unusual distribution of genes is reminiscent of bacterial operons and has been observed in Leishmania (21), albeit at a smaller scale; similar observations are reported for T.brucei chrI (8). Several lines of evidence from this and previous studies of trypanosomatid genes suggest, however, that the directional gene clusters are not regulated like conventional bacterial operons. First, unlike most prokaryotic and eukaryotic organisms, where gene expression is regulated primarily at the level of transcription, in trypanosomatids gene regulation is largely post-transcriptional (22). Expression data from a microarray containing 450 CDSs from chrII and probed with RNA from two developmental stages of the parasite (bloodstream forms circulating in the mammalian host versus procyclic form in the tsetse fly vector) reveal many instances of putative genes within the same directional cluster that exhibit a differential expression (data not shown). Second, the gene clusters do not appear to encode proteins that share a metabolic pathway as do genes of many bacterial operons. Instead, the gene products have apparently unrelated functions as illustrated by their assignments to different ‘biological processes’ within the GO classification system (Fig. 1). It is worth noting that no recognisable features of either eukaryotic or prokaryotic promoters were detected in the nucleotide sequences between the gene clusters where bidirectional transcription would presumably begin.
A statistical analysis of the nucleotide distribution of chrII reveals a remarkable correlation between the direction of the gene clusters and the sign of the GC-skew (Fig. 1). This (C–G)/(C+G) value, typically calculated in overlapping sliding windows along a sequence (23), has been used to detect strand bias or asymmetry in the base composition of almost all completely sequenced bacterial genomes (24–26). In most cases, a deviation switch at the origin and terminus of replication has strongly suggested a link with replication. On T.brucei chrII (and chrI, data not shown), the sign of the GC-skew correlates with the direction of the gene clusters, changing precisely and consistently at the boundaries of their convergence or divergence. This strongly suggests that the observed base composition asymmetry between the coding and non-coding strand is linked to transcription or transcription-coupled repair. Similar potential sources of strand asymmetry in other organisms were recently described (27–29). In order to rule out the possibility that polypyrimidine tracts found in Trypanosoma intergenic regions could be contributing to the GC-skew, we performed the same analysis on the concatenated coding and non-coding sequences and found no significant differences (data not shown). While consistent with nucleotide composition analyses of bacteria, our findings are in contrast with similar analyses of Leishmania major chromosome 1 (30). With so little known about nuclear DNA replication and RNA polymerase II transcription in trypanosmatids, the biological significance of the observed nucleotide bias remains to be determined.
Segmental duplications
We observe a high degree of gene redundancy in chrII with the presence of local gene duplications. These paralogous genes are present in tandem arrays. For example, the gene for the 65 kDa invariant surface glycoprotein is present as an array of six copies (10C8.435–10C8.460 in Fig. 1 inset, repeat class C); the genes for a nucleoside transporter and the iron/ascorbate oxidoreductase appear as a pair five times, ending with an extra copy of the nucleoside transporter gene (36E18.75–36E18.160, Fig. 1 inset, repeat class F); and a gene encoding a hypothetical protein is repeated eight times (30J2.95–30J2.105, 1F7.30–1F7.50, Fig. 1 inset, repeat class D). The nucleoside transporter-iron/ascorbate oxidoreductase units also contain intergene duplications but these segments have conserved very low nucleotide identity.
Another type of duplication observed in chrII involves segment duplications rather than single-gene copies. We have found three regions where 6–19 kb segmental copies are juxtaposed and have >95% identity (Fig. 1 inset, repeat classes A, B, E). Within these duplicated blocks, the homologous genes appear in the same order along the distinct segments. This type of duplicated blocks of genes was previously reported in other organisms such as Arabidopsis thaliana, Saccharomyces cerevisae and Caenorhabditis elegans (31–34). However, while in the case of A.thaliana and S.cerevisae most of the blocks are duplicated between chromosomes, in C.elegans they occur intra-chromosomally (35). Another interesting feature of most block duplications is that there is very little conservation between non-coding duplicated regions (32). In T.brucei chrII, we observe very high conservation of non-coding regions.
The duplicated ribosomal clusters (repeat class B1) and the 19-kb segments (repeat class E) contain inverted copies. In the case of the ribosomal RNA cluster, retroposon-like elements (Ingis) can be found at a certain distance on either side of a complete unit (Fig. 1) suggesting that these elements may have been involved in the duplication process. As for the 19-kb inverted segment, both units are an exact mirror image of each other, separated by 100 nucleotides of unique sequence. Only when the genome is completed will we be able to evaluate the existence of extensive duplication of blocks between chromosomes, and the extent to which mechanisms responsible for gene duplication are a mixture of ancient polyploidisation events versus unequal crossing over events.
Genetic map
Trypanosoma brucei parasites undergo genetic exchange when two stocks are co-transmitted through tsetse flies in the laboratory (18,35). The role of genetic exchange in field populations is controversial, however (36). Marker analysis of laboratory trypanosome crosses shows that the progeny are products of a diploid Mendelian genetic system and partial genetic maps have been constructed using anonymous amplified fragment length polymorphism (AFLP) markers (17).
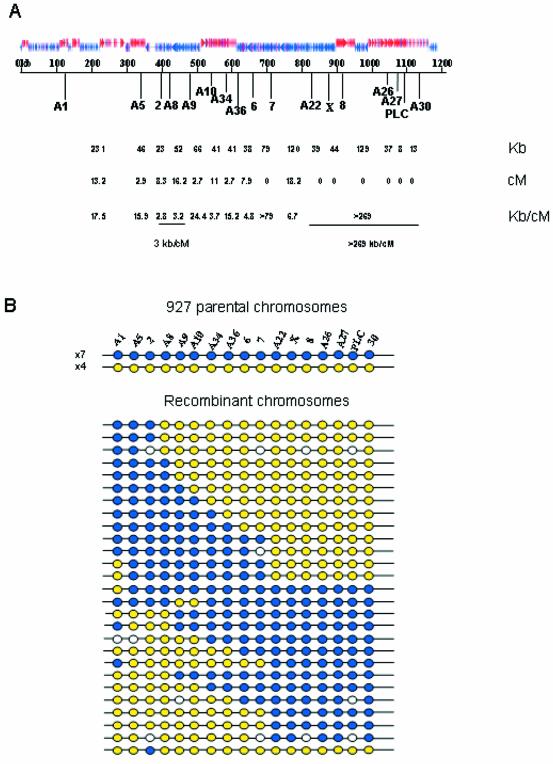
To identify polymorphic markers with which to construct a genetic map of chrII, we analysed the sequence for micro- and minisatellites. The former were defined as sequences containing >10 copies of a repeat motif of 2–5 nucleotides with >70% sequence identity and the latter were defined by the same criteria but with a repeat motif of >6 nucleotides. A total of 73 microsatellites and nine minisatellites were identified on the chromosome. Primers were designed to generate PCR products spanning a selection of these distributed across the chromosome and 17 were selected for map construction because they showed sufficient size variation, and were heterozygous for Tb927 and homozygous for stock Tb247. The inheritance of Tb927 alleles into a panel of 38 F1 progeny of a genetic cross between Tb927 × Tb247 enabled a genetic map to be constructed (Fig. 2). To do this, the parental haplotypes were determined on the basis of the most frequently inherited haplotypes and the positions of cross-over events were then identified in recombinant haplotypes. The genetic map of chrII is 83.1 cM in length, which gives an average density of approximately one marker per 4.9 cM. This map covers ∼1007 kb of the chromosome, giving an average physical size for a recombination unit of 12.1 kb/cM. This value is approximately half of that for chrI (8) but is within the range observed for single-celled eukaryotes (37). A genetic marker occurs approximately every 59 kb along chrII but comparison of the genetic and physical maps indicates that recombination events are unevenly distributed along its length (χ2 = 50.7, df = 11, P < 0.01; taking as our null hypothesis that there is an equal probability of crossovers in all regions of the chromosome). There is one recombination ‘hot spot’ (between markers 2 and A9) where crossovers occur readily (3.3 kb/cM). In contrast, between markers A22 and A30, in the ‘right hand’ half of the chromosome, resides a major recombination ‘cold spot’ where cross-overs are absent within a 269-kb region. In classical genetics a centromere may be defined by the absence of cross-over events in its vicinity; therefore, this cold spot may correspond to a candidate centromere.
Figure 2.
(A) The physical map of chromosome II with the directional gene clusters and position of polymorphic mini- and microsatellite markers indicated. The physical distances between markers are shown (kb) and the genetic distances (cM) were calculated from recombination frequencies. Genetic distances shown are uncorrected. The relationship of physical and genetic maps to show hot and cold spots of recombination is also indicated (kb/cM). (B) Diagrams of the haplotypes of stock Tb927 showing 11 parental haplotypes shown as blue or yellow, and 27 recombinant types. White circles are unscored markers.
Antigenic diversity by the recombination of pseudogenes
A distinctive feature near the ‘right’ end of chrII is the presence of a small subtelomeric array of putative VSGs, the majority of which are highly degenerate pseudo-VSGs disrupted by in-frame stop codons and frameshift mutations. The region spans ∼30 kb and contains seven putative VSGs, including six pseudo-VSGs (Fig. 1). African trypanosomes evade the immune response of their mammalian hosts by sequentially expressing different VSGs from telomere-linked expression sites. Although hundreds of transcriptionally silent VSGs are present in the genome, only one is usually expressed at a time in a given bloodstream parasite (38). The activation of a new VSG is often associated with one of three types of gene rearrangements. The best characterised rearrangement is the duplicative transposition of a silent, donor VSG from either an interior chromosomal location or a telomeric location to a telomeric-linked expression site, displacing the VSG already at that site. In many cases, this gene conversion is mediated on the 5′ side by homologous recombination between a few copies of a 70–76 bp repeat upstream of the donor gene and hundreds of copies of this same repeat in the expression site. On the 3′ side of the duplication, homologuous recombination often occurs between the segments of sequence similarities in the C-terminal coding regions or the 3′-untranslated regions.
We postulate that this region of chrII is a prototype of other similar storage areas in the genome that provide a repertoire of VSGs. Although previously speculated to exist (39), this is the first sequence of an array of VSGs that could potentially serve as gene conversion donors. A recent report by Robinson et al. (40) highlights the predominance of VSG conversion events in antigenic variation and Borst recently proposed an elegant VSG transposition model favouring a DNA synthesis- dependent, strand-annealing model over a break-induced, DNA-replication one (39). Unexpectedly, the VSG repertoire in this region consists mostly of pseudogenes. The six putative pseudo-VSGs contain 11 stop codons and five frameshift mutations evenly spread along the length of the sequences with a slight preference for the region encoding the C-terminal domain. The existence of expressed composite VSGs, derived from combinations of several donor genes or pseudogenes, has already been demonstrated (41–45). These recombinational processes have been proposed as a way for African trypanosomes to expand their repertoire of antigenic diversity by reassorting VSG sequences. The existence of a relatively large proportion of pseudo-VSGs shows there is no strong selection for the maintenance of intact VSGs in T.brucei. These data also predict that a modular de novo construction of VSG diversity during segmental conversion events may be more frequent than originally believed.
Retroposon hot spot (RHS) genes
A comparison of chrII and chrI reveals that about one-fifth of each chromosome (250 kb in chrII and 175 kb in chrI) is composed of a complex, repetitive and novel subtelomeric region that includes the ingi and RIME non-LTR retrotransposons (Fig. S1, see Supplementary Material). This ‘left’ subtelomeric region of chrII and its chrI counterpart each contain about half of their chromosome’s complement of RIME and ingi non-LTR retrotransposons (seven of 15 for chrII and eight of 13 for chrI). This region also contains genes and pseudogenes of two recently identified multigene families called LRRP1 (leucine rich repeat protein) and RHS (retrotransposon hot spot) (46,47). The chrII region has 4 LRRP1 and 29 RHS copies (including very degenerated RHS pseudogenes), whereas the chrI region has three LRRP1 and 15 RHS copies (Fig. S2) (8). The RHS multigene family is itself composed of six subfamilies called RHS1–RHS6 (47). Furthermore, all of the RHS4 repeat units in these regions contain a highly degenerated DNA-dependent RNA polymerase III pseudogene, and the LRRP1 repeat units contain ESAG4 (adenylate cyclase) pseudogenes (Fig. S2). The general organisation of this subtelomeric region in both chromosomes consists of large clusters of tandemly arranged RHS repeat units (chrII has two large RHS clusters composed of 13 and 10 units), separated by a single LRRP1 repeat unit (Fig. S2). In chrII, all 25 full-length RHS copies (with the exception of a RHS6 copy at the right extremity of the region) are on the same DNA strand, whereas all the LRRP1 copies are on the other strand (Fig. S2), indicating that the RHS clusters and LRRP1 repeat units are flanked by strand switches. Finally, all the ingi/RIME retroelements in these two regions are inserted inside the RHS (pseudo)genes, which contain a hot spot for retroelement insertions (47), demonstrating that the high prevalence of ingi/RIME previously observed in this region of chrIa (48) is associated with the presence of the RHS (pseudo)genes.
Interspersed with the RHS genes are six copies of truncated DNA-directed RNA polymerase pseudogenes, three expression site associated (pseudo)genes (ESAGs) 3 and one ESAG 4 (Fig. 1 and Table S1). Immediately upstream of one of the ESAG3 pseudogenes (3B10.55), there is a small cluster of 70–76 bp repeats. These data suggest that this subtelomeric region may be the vestige of a highly degenerate VSG expression site.
In summary, the 1.2-Mb chrII has three distinct regions. The 250-kb ‘left’ region (21% of the chromosome) is composed of a complex group of repeats, also found in chrI, whose biological significance is unclear. The 915-kb central region (76% of chrII) has 10 directional clusters of genes encoding many housekeeping and parasite-specific proteins and/or RNAs. No VSGs or pseudo-VSGs occur in these two regions. The remaining 30-kb ‘right’ region (3%) contains a small cluster of VSGs and pseudo-VSGs that could potentially serve as either or both a ‘salvage yard’ for retaining previously used VSGs or VSG segments and a repertoire for assembling new VSGs. Unlike at least some of the other megachromosomes of this organism, the chrII homolog sequenced here does not seem to possess telomere-linked VSG expression sites, so potential donor VSGs/partial VSGs on this chromosome need to undergo inter-chromosomal gene conversion to reach an expression site.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to acknowledge our colleagues at the Sanger Institute for useful discussions and close coordination of efforts, and Sarah McLellan for technical assistance. We are also grateful to the T.brucei research community worldwide for its continuous support of this project. In addition, we would like to thank the TIGR faculty, system administrators, sequencing facility, bioinformatics department and administrative staff. The work at the Institute for Genomic Research (TIGR) was supported by a grant from the National Institute for Allergy and Infectious Diseases, National Institutes of Health to N.E.S. (U01 AI43062). The genetic mapping studies carried out at University of Glasgow were supported by grants from The Wellcome Trust and The Sir Halley Stewart Trust.
REFERENCES
- 1.Barrett M.P. (1999) The fall and rise of sleeping sickness. Lancet, 353, 1113–1114. [DOI] [PubMed] [Google Scholar]
- 2.Johnson P.J., Kooter,J.M. and Borst,P. (1987) Inactivation of transcription by UV irradiation of T.brucei provides evidence for a multicistronic transcription unit including a VSG gene. Cell, 51, 273–281. [DOI] [PubMed] [Google Scholar]
- 3.Boothroyd J.C. and Cross,G.A. (1982) Transcripts coding for variant surface glycoproteins of Trypanosoma brucei have a short, identical exon at their 5′ end. Gene, 20, 281–289. [DOI] [PubMed] [Google Scholar]
- 4.Walder J.A., Eder,P.S., Engman,D.M., Brentano,S.T., Walder,R.Y., Knutzon,D.S., Dorfman,D.M. and Donelson,J.E. (1986) The 35-nucleotide spliced leader sequence is common to all trypanosome messenger RNA’s. Science, 233, 569–571. [DOI] [PubMed] [Google Scholar]
- 5.Stuart K. and Panigrahi,A.K. (2002) RNA editing: complexity and complications. Mol. Microbiol., 45, 591–596. [DOI] [PubMed] [Google Scholar]
- 6.Barry J.D. and McCulloch,R. (2001) Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol., 49, 1–70. [DOI] [PubMed] [Google Scholar]
- 7.Borst P. and Ulbert,S. (2001) Control of VSG gene expression sites. Mol. Biochem. Parasitol., 114, 17–27. [DOI] [PubMed] [Google Scholar]
- 8.Hall N., Berriman,M., Lennard,N.J., Harris,B.R., Hertz-Fowler,C., Bart-Delabesse,E.N., Gerrard,C.S., Atkin,R.J., Barron,A.J., Bowman,S. et al. (2003) The DNA sequence of chromosome I of an African trypanosome: gene content, chromosome organisation, recombination and polymorphism. Nucleic Acids Res., 31, 4864–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Deursen F.J., Shahi,S.K., Turner,C.M., Hartmann,C., Guerra-Giraldez,C., Matthews,K.R. and Clayton,C.E. (2001) Characterisation of the growth and differentiation in vivo and in vitro-of bloodstream-form Trypanosoma brucei strain TREU 927. Mol. Biochem. Parasitol., 112, 163–171. [DOI] [PubMed] [Google Scholar]
- 10.Delcher A.L., Harmon,D., Kasif,S., White,O. and Salzberg,S.L. (1999) Improved microbial gene identification with GLIMMER. Nucleic Acids Res., 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apweiler R., Attwood,T.K., Bairoch,A., Bateman,A., Birney,E., Biswas,M., Bucher,P., Cerutti,L., Corpet,F., Croning,M.D. et al. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res., 29, 37–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 16.Benson G. (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner C.M., Sternberg,J., Buchanan,N., Smith,E., Hide,G. and Tait,A. (1990) Evidence that the mechanism of gene exchange in Trypanosoma brucei involves meiosis and syngamy. Parasitology, 101, 377–386. [DOI] [PubMed] [Google Scholar]
- 18.Tait A., Masiga,D., Ouma,J., MacLeod,A., Sasse,J., Melville,S., Lindegard,G., McIntosh,A. and Turner,M. (2002) Genetic analysis of phenotype in Trypanosoma brucei: a classical approach to potentially complex traits. Philos. Trans. R. Soc. Lond. B Biol. Sci., 357, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Melville S.E., Leech,V., Gerrard,C.S., Tait,A. and Blackwell,J.M. (1998) The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol., 94, 155–173. [DOI] [PubMed] [Google Scholar]
- 20.Venter J.C., Smith,H.O. and Hood,L. (1996) A new strategy for genome sequencing. Nature, 381, 364–366. [DOI] [PubMed] [Google Scholar]
- 21.Myler P.J., Audleman,L., deVos,T., Hixson,G., Kiser,P., Lemley,C., Magness,C., Rickel,E., Sisk,E., Sunkin,S. et al. (1999) Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc. Natl Acad. Sci. USA, 96, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clayton C.E. (2002) Life without transcriptional control? From fly to man and back again. EMBO J., 21, 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobry J.R. (1996) Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol., 13, 660–665. [DOI] [PubMed] [Google Scholar]
- 24.Freeman J.M., Plasterer,T.N., Smith,T.F. and Mohr,S.C. (1998) Patterns of genome organization in bacteria. Science, 279, 1827a. [Google Scholar]
- 25.Mrazek J. and Karlin,S. (1998) Strand compositional asymmetry in bacterial and large viral genomes. Proc. Natl Acad. Sci. USA, 95, 3720–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLean M.J., Wolfe,K.H. and Devine,K.M. (1998) Base composition skews, replication orientation, and gene orientation in 12 prokaryote genomes. J. Mol. Evol., 47, 691–696. [DOI] [PubMed] [Google Scholar]
- 27.Beletskii A. and Bhagwat,A.S. (1998) Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol. Chem., 379, 549–551. [PubMed] [Google Scholar]
- 28.Francino M.P., Chao,L., Riley,M.A. and Ochman,H. (1996) Asymmetries generated by transcription-coupled repair in enterobacterial genes. Science, 272, 107–109. [DOI] [PubMed] [Google Scholar]
- 29.Frank A.C. and Lobry,J.R. (1999) Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene, 238, 65–77. [DOI] [PubMed] [Google Scholar]
- 30.McDonagh P.D., Myler,P.J. and Stuart,K. (2000) The unusual gene organization of Leishmania major chromosome 1 may reflect novel transcription processes. Nucleic Acids Res., 28, 2800–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langkjaer R.B., Cliften,P.F., Johnston,M. and Piskur,J. (2003) Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature, 421, 848–852. [DOI] [PubMed] [Google Scholar]
- 32.Grant D., Cregan,P. and Shoemaker,R.C. (2000) Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl Acad. Sci. USA, 97, 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe K.H. and Shields,D.C. (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature, 387, 708–713. [DOI] [PubMed] [Google Scholar]
- 34.Friedman R. and Hughes,A.L. (2003) The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol. Biol. Evol., 20, 154–161. [DOI] [PubMed] [Google Scholar]
- 35.Gibson W. and Stevens,J. (1999) Genetic exchange in the trypanosomatidae. Adv. Parasitol., 43, 1–46. [DOI] [PubMed] [Google Scholar]
- 36.MacLeod A., Tweedie,A., Welburn,S.C., Maudlin,I., Turner,C.M. and Tait,A. (2000) Minisatellite marker analysis of Trypanosoma brucei: reconciliation of clonal, panmictic, and epidemic population genetic structures. Proc. Natl Acad. Sci. USA, 97, 13442–13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mortimer R.K., Contopoulou,C.R. and King,J.S. (1992) Genetic and physical maps of Saccharomyces cerevisiae. Yeast, 8, 817–902. [DOI] [PubMed] [Google Scholar]
- 38.Van der Ploeg L.H., Valerio,D., De Lange,T., Bernards,A., Borst,P. and Grosveld,F.G. (1982) An analysis of cosmid clones of nuclear DNA from Trypanosoma brucei shows that the genes for variant surface glycoproteins are clustered in the genome. Nucleic Acids Res., 10, 5905–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borst P. (2002) In Craig,N.L. (ed.) Mobile DNA II. ASM Press, Washington, DC, pp. 953–971. [Google Scholar]
- 40.Robinson N.P., Burman,N., Melville,S.E. and Barry,J.D. (1999) Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol. Cell Biol., 19, 5839–5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pays E., Van Assel,S., Laurent,M., Darville,M., Vervoort,T., Van Meirvenne,N. and Steinert,M. (1983) Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell, 34, 371–381. [DOI] [PubMed] [Google Scholar]
- 42.Pays E., Delauw,M.F., Van Assel,S., Laurent,M., Vervoort,T., Van Meirvenne,N. and Steinert,M. (1983) Modifications of a Trypanosoma b. brucei antigen gene repertoire by different DNA recombinational mechanisms. Cell, 35, 721–731. [DOI] [PubMed] [Google Scholar]
- 43.Longacre S. and Eisen,H. (1986) Expression of whole and hybrid genes in Trypanosoma equiperdum antigenic variation. EMBO J., 5, 1057–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth C.W., Longacre,S., Raibaud,A., Baltz,T. and Eisen,H. (1986) The use of incomplete genes for the construction of a Trypanosoma equiperdum variant surface glycoprotein gene. EMBO J., 5, 1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thon G., Baltz,T. and Eisen,H. (1989) Antigenic diversity by the recombination of pseudogenes. Genes Dev., 3, 1247–1254. [DOI] [PubMed] [Google Scholar]
- 46.Berriman M., Hall,N., Sheader,K., Bringaud,F., Tiwari,B., Isobe,T., Bowman,S., Corton,C., Clark,L., Cross,G.A. et al. (2002) The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol., 122, 131–140. [DOI] [PubMed] [Google Scholar]
- 47.Bringaud F., Biteau,N., Melville,S.A., Hez,S., El-Sayed,N.M., Leech,V., Berriman,M., Hall,N., Donelson,J. and Baltz,T. (2002) A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryotic Cell, 1, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melville S.E., Gerrard,C.S. and Blackwell,J.M. (1999) Multiple causes of size variation in the diploid megabase chromosomes of African tyrpanosomes. Chromosome Res., 7, 191–203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.